Abstract
Crystal structures and other biochemical data indicate that the N-terminal cap (NCap) region of the Abelson tyrosine kinase (c-Abl) is important for maintaining the downregulated conformation of the kinase domain. The exact contributions that NCap makes in stabilizing the various intramolecular interactions within c-Abl are less clear. While the NCap appears important for locking the SH3/SH2 domains to the back of the kinase domain, there may be other more subtle elements of regulation. Hydrogen exchange (HX) and mass spectrometry (MS) were used to determine if the NCap contributes to intramolecular interactions involving the Abl SH3 domain. Under physiological conditions, the Abl SH3 domain underwent partial unfolding and its unfolding half-life was slowed during binding to the SH2-kinase linker, providing a unique assay to test NCap-induced stabilization of the SH3 domain in various constructs. The results showed that NCap stabilizes the dynamics of the SH3 domain in certain constructs but does not increase the relative affinity of the SH3 domain for the native SH2-kinase linker. The stabilization effect was absent in constructs of just NCap + SH3 but was obvious when the SH2 domain and the SH2-kinase linker were present. These results suggest that interactions between NCap and the SH3 domain can contribute to c-Abl stabilization in constructs that contain at least the SH2 domain, an effect that may partially compensate for the absence of the negative regulatory C-terminal tail found in the related Src family of kinases.
The Abelson protein-tyrosine kinase (c-Abl) is a non-receptor tyrosine kinase ubiquitously expressed and highly conserved in metazoan evolution. c-Abl has many cellular functions including regulation of cell growth, survival, and differentiation, oxidative stress and DNA-damage responses, actin dynamics, and cell adhesion and migration (1, 2). Fusion of the gene encoding c-Abl on chromosome 9 with the breakpoint cluster region (Bcr) gene on chromosome 22 results in the formation of a fusion protein, Bcr-Abl, which has constitutive protein-tyrosine kinase activity (3, 4). The enhanced tyrosine kinase activity of the Bcr-Abl protein contributes to several disease states including chronic myelogenous leukemia (CML) and acute lymphocytic leukemia (ALL) (4). However regulation of both c-Abl and the Bcr-Abl fusion protein is still not well understood.
The tyrosine kinase core of c-Abl is composed of a number of regions including an N-terminal cap (NCap) region, an SH3 domain, an SH2 domain, and a kinase domain (see Fig. 1A). Multiple intramolecular interactions involving these regions occur in c-Abl and these interactions are crucial for maintaining the downregulated state (5). Although the c-Abl core is very similar to Src-family tyrosine kinases (SFKs) in structure (1, 6, 7), significant differences exist. In both c-Abl and SFKs, the SH3 domain binds the SH2-kinase linker, an interaction necessary to suppress kinase activity. However, intramolecular interactions involving the SH2 domain are different in c-Abl and SFKs. In downregulated SFKs, the SH2 domain binds to the C-terminal tail of the kinase when the tail is tyrosine-phosphorylated (8-11). In Abl, there is no equivalent C-terminal tail for interaction with the SH2 domain. In theory, there should be some compensation in c-Abl regulation to account for the loss of SH2 domain interactions, assuming that SFKs and Abl function in similar ways. We have previously shown (12) that the Abl SH2-kinase linker intramolecularly associates with the Abl SH3 domain in the absence of the kinase domain whereas in the SFK Hck, no such interaction could be detected (13). This observation suggests that the c-Abl SH3 domain may have a greater regulatory influence than its SH3 domain counterparts in SFKs.
Figure 1.
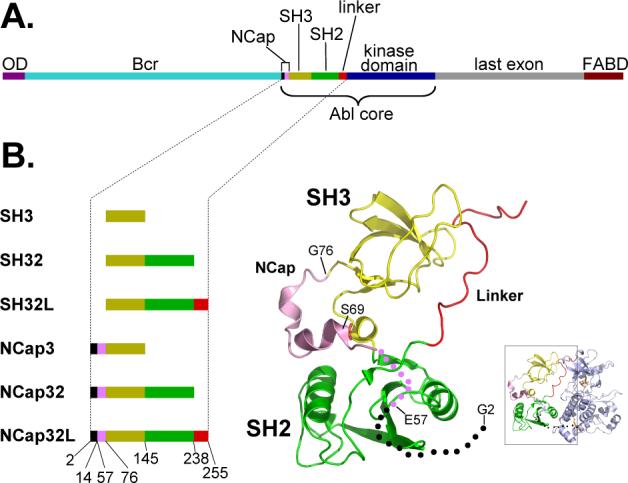
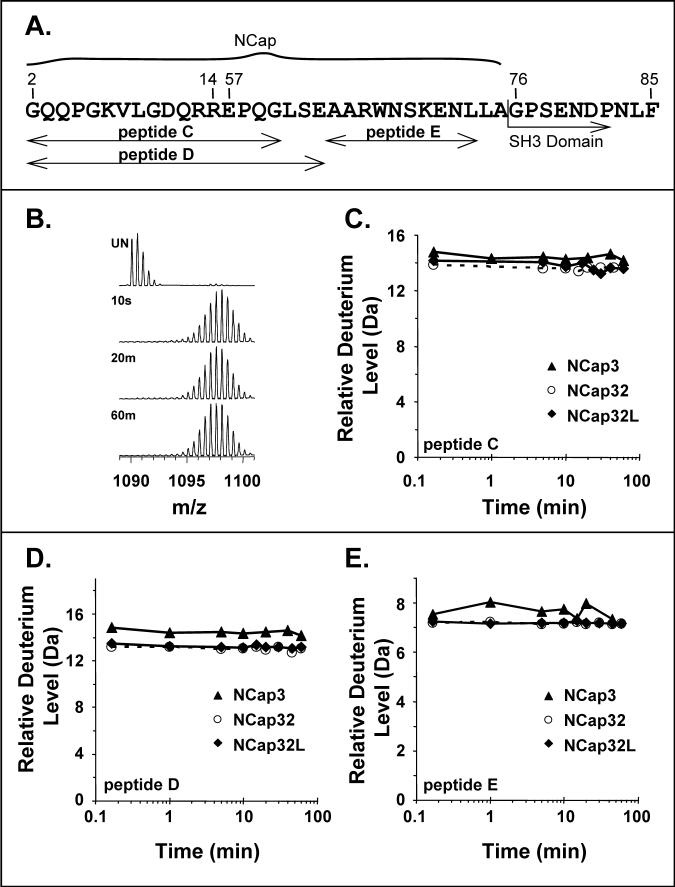
Schematic diagram of Bcr-Abl, Abl core, and other constructs used in this study. (A). The domains of Bcr-Abl are shown and the region referred to as Abl core is indicated. The color code is: dark purple = oligomerization domain (OD), light blue = breakpoint cluster region (Bcr), Black and magenta = NCap, dark yellow = SH3 domain, green = SH2 domain, red = SH2-kinase linker, blue = kinase domain, grey = last exon region, dark red = F-actin binding domain (FABD). (B). The domains of the constructs used in this study and their location in the crystal structure of c-Abl [inset, PDB code 2FO0, (16)]. Coloring is consistent with the schematic diagram in (A). The unstructured part of NCap is shown as dots with the same color as in the rest of the diagram. Numbers below the constructs indicate the residues at the boundaries of the various domains (see also Materials and Methods), as in (16).
Other elements of c-Abl are also believed to compensate for the lack of a C-terminal tail in Abl. In particular, the residues immediately N-terminal to the SH3 domain, termed the NCap, are required for c-Abl downregulation (14). Although mutagenesis experiments and crystal structures of the c-Abl core showed that the NCap is required for maintenance of the downregulated state (5, 15, 16), the mechanism is still poorly understood. The second crystal structure of the c-Abl kinase core (16) showed that the residues of the NCap interact with both SH3 and SH2 domains and appear to stabilize their interactions with the back of the kinase domain. In the c-Abl structures, the glycine residue at position 2 in the NCap was myristoylated and bound to a deep pocket in the C-lobe of the kinase domain, thus appearing to latch the SH2 and SH3 domains in the downregulatory position at the back of the kinase domain [see Ref (1)]. Ser69 in the NCap was also found to be phosphorylated and the crystal structure showed that Ser69 interacts with the SH3-SH2 linker (16). In contrast, the corresponding N-terminal region in SFKs (termed the SH4 domain) is crucial for membrane anchorage, but not for the regulation of kinase activity (17).
Using hydrogen exchange (HX) mass spectrometry (MS), we provide direct biophysical evidence that the NCap modulates the dynamics of the SH3 domain, a result that has direct implications for SH3 binding and downregulation of kinase activity. The effect was more pronounced in constructs containing the SH2 domain, pointing to a role for the NCap in locking SH3 onto SH2. In addition, no structure was detected in the NCap in solution which implies that NCap is highly mobile and presumably becomes conformationally restricted in the context of the Abl core which includes the kinase domain and the pocket for myristic acid insertion.
MATERIALS and METHODS
DNA Constructs and protein purification
The coding regions of the human c-Abl NCap3 (G2-R14, E57-S145), NCap32 (G2-R14, E57-K238), and NCap32L (G2-R14, E57-E255) were amplified by PCR and subcloned into the bacterial expression vector pET-41b (Novagen). NCap3, NCap32, and NCap32L proteins were overexpressed and purified as described previously (12). Abl core was purified from Sf9 insect cells upon co-expression with YopH, a protein tyrosine phosphatase. YopH prevents tyrosine phosphorylation of the Abl core (including at Y412 in the activation loop), promotes a downregulated conformation and permits high-yield purification from Sf9 cells (16). The Abl core construct was cloned into pVL1392 (BD Biosciences) and encompasses the residues 1−531 of human c-Abl 1b with residues 15−56 deleted. The construct contains a C-terminal cleavage site for the Tobacco etch virus (TEV) protease and a hexahistidine tag both introduced by PCR. Each construct was used to create high-titer recombinant baculovirus in Sf9 cells using Baculogold DNA and the manufacturer's protocol (BD Biosciences). For protein production, Sf9 cells (1 L) were grown in suspension in Grace's medium (Invitrogen) supplemented with 10% FBS and 50 μg/ml gentamycin. Sf9 cells were cultured to a density of 2 × 106 cells per mL and then co-infected with the Abl core and YopH baculoviruses at a multiplicity of infection of ten. Cells were grown for 48 hr, centrifuged, washed in PBS and stored at −80°C until further processing. Western blots with an anti-phosphotyrosine antibody revealed that the protein was not tyrosine phosphorylated. For purification, the Sf9 cell pellet was resuspended in buffer A (20 mM Tris/HCl, pH 8.3, 10% glycerol, 5 mM β-mercaptoethanol), lysed by sonication, and centrifuged at 16,000 rpm for 30 min. Recombinant Abl core was purified from the supernatant using a combination of ion-exchange and affinity chromatography as originally described by Schindler et al. for Hck (18). Upon purification, the protein was dialyzed against 20 mM Tris/HCl, pH 8.3, containing 100 mM NaCl, and 3mM DTT. The final protein was concentrated to 0.52 μg/μl as determined by SDS-PAGE and densitometry. The mass of the final Abl core was verified with electrospray mass spectrometry, which further indicated that the protein was stoichiometrically phosphorylated on Ser69 and that the glycine at position 2 was myristoylated (data not shown).
Deuterium exchange and MS analysis of deuterium incorporation
Proteins were incubated in 50 mM sodium phosphate buffer prepared in D2O (pD 8.3) at 22 °C (room temperature, verified for each sample preparation) for various lengths of time. The temperature of the room during deuteration did not fluctuate by more tha 1 °C over the course of the labeling timecouse. The reaction was quenched by adjusting the pH to 2.5 with 0.5 M HCl at each time point. Quenched samples were immediately frozen on dry ice and stored at −80 °C until analysis. Intact protein analysis of deuterium incorporation was as described previously (12). For peptic analysis, each 50 μl sample (100−150 pmol) was injected to a 50 mm×200 mm stainless steel column packed with pepsin immobilized on POROS-20AL beads (PerSeptive Biosystems) at 30−40 mg/mL (19). The resulting peptides were trapped on a C18 trap column. The total digestion time was 20 seconds. The trapped peptides were eluted from the C18 trap onto a Magic C-18 column (Michrom BioResources, Inc.) and directed into the mass spectrometer with a six minute gradient of 2−45% acetonitrile in H2O. The LC system used for all analyses was a 3-pump Shimadzu 10AD-VP. The injector, column and tubing were all cooled with an ice-bath (20). The pepsin column was located above the ice-bath at an approximate temperature of 18 °C. As for intact proteins, no correction was made for back-exchange. The average amount of back-exchange for peptides in this instrumental system was 18−22%, as measured with totally deuterated angiotensin and several other totally deuterated peptides. Intact protein analyses were performed with a Waters LCT premier mass spectrometer while peptide analyses were done with a Waters QTof Ultima instrument calibrated after each run with an infusion of myoglobin of glu-fibrinogen peptide. Peptic peptides were identified by a combination of exact mass analyses and MS/MS. For MS/MS, a peptic digest was prepared using the same online digestion conditions as for a deuterium exchange experiments but the peptides were collected from the six minute gradient rather than eluted into the mass spectometer. The collected peptides were reinjected in a separate experiment and separated with a 2 hour gradient of acetonitrile (2−45%). Each eluting peptide was systematically interrogated with data-dependent MS/MS and each MS/MS spectra was interpreted by hand to identify the peptide.
Data processing
The deuterium uptake curves were plotted using HX-Express (21). The peak width was measured at full-width half maximum (FWHM) with HX-Express for each time point and plotted against the deuterium labeling time (22). Linear regression was performed on each side of the peak in each peak-width plot. The intersection point of the two linear equations was determined and used as the half life of unfolding [as described in (12)]. When more than one measurement was made for each construct, the t1/2 value reported corresponds to the average of the replicates. The 95% confidence interval was also calculated when more than 4 independent experiments were made.
RESULTS
Overview of recombinant Abl protein constructs and HX MS
The NCap region is critical for maintaining c-Abl in its downregulated state. Deleting this region from c-Abl resulted in high kinase activity (14) whereas removal of the N-terminal residues upstream of the SH3 domain in SFKs does not alter kinase activity (17). A number of constructs were created to determine the mechanism by which the NCap facilitates intramolecular interactions in c-Abl (Figure 1B). The NCap that was attached to the N-terminal end of the SH3 domain was the modified, shorter version that was used in crystal structure determinations (5, 16). Deletion of the residues between amino acids 15 and 56 was not shown to have any effect on the regulation of Abl (15). All constructs (with the exception of the full-length Abl core) were expressed in E. coli (see Material and Methods); hence, for all constructs except for the Abl core, Ser69 was not phosphorylated and the protein was not myristoylated at Gly2. The Abl core was made in Sf9 as in previous x-ray crystallography and functional studies (16) and was phosphorylated on Ser69 and myristoylated at Gly2, as confirmed by mass spectrometry (data not shown). Each protein was purified and then separately incubated in D2O buffer under identical conditions. The deuterium exchange-in reaction proceeded for predetermined amounts of time and was quenched by lowering the pH to 2.5 and the temperature to 0 °C. Each quenched reaction was analyzed by HPLC MS to determine the extent of labeling at each D2O incubation time.
In hydrogen/deuterium exchange, the isotope exchange rate is determined by several factors including protein dynamics (23). We previously used HX MS to show that the Abl SH3 domain undergoes partial cooperative unfolding that is sensitive to ligand binding (12). By monitoring the appearance of the unfolding event during the deuterium labeling time-course, the half-life (t1/2) of unfolding can be determined. Analysis of HX MS in intact proteins also provided information about global differences in deuterium uptake in the various constructs. To obtain conformation and dynamics information about specific regions, pepsin digestion and peptide analysis were performed and the deuterium levels in each short peptic fragment were analyzed [as described (23)]. Supplementary Figure 1s shows the peptic peptides that were identified from the NCap32L construct. The peptides cover 100% of NCap32L sequence and many of them are overlapping. In summary, by monitoring both the unfolding dynamics that present themselves in the appearance of the mass spectra and the actual deuterium levels at each exchange time point, we were able to characterize a number of features of the NCap regulatory mechanism, as detailed in the following sections.
NCap is mostly unstructured
We first determined whether the NCap region had any significant protection from exchange in solution. Protection from exchange is indicative of structure and/or intramolecular interactions (24). In the crystal structures, there were some regions that appeared to have helical content with the NCap pinned to the body of the SH3/SH2 domains (see also Figure 1B). The deuterium uptake in several peptic peptides that span the NCap region (Figure 2A) was determined in recombinant proteins that contained the SH3 domain (NCap3), the SH3 and SH2 domains (NCap32) and the SH3, SH2 and linker (NCap32L). The deuterium levels in the NCap peptic peptides were similar in the NCap3, NCap32 and NCap32L constructs. There was a large amount of deuteration after the shortest time point used in this study (10 seconds) and the deuterium level remained constant for the entire time course. A representative example of the mass spectra showing this is found in Figure 2B for peptide “C” (the peptides are termed C, D, and E in Figure 2A, to match the data for each peptide in panels C, D, E of Figure 2). After determining the amount of deuterium that had been incorporated at each exchange point and preparing deuterium exchange-in curves, it was obvious that all peptides from the NCap region were rapidly deuterated and maintained the same deuterium levels throughout the time course. (Figure 2C, D, E). In addition, by monitoring the deuterium levels in the NCap peptides in various constructs containing the NCap, it became clear that addition of the SH2 domain or addition of both SH2 and the natural linker to NCap3 generally did not influence the deuterium incorporation in NCap peptides C, D & E. In peptide D, there was a small but measurable change to the deuterium levels when SH2 or SH2-linker was attached. These results may imply that SH2 can influence exchange within the linker, and hence the structure, at certain residues. Because such an effect was not seen in peptide C, which is just 3 residues shorter than peptide D at the C-terminal end, the backbone amide hydrogens that must be altered upon SH2 inclusion lie within residues LSE, the extra residues in peptide D. The location of the 61LSE63 sequence is shown in Figure 1B and may directly contact the SH2 domain. These data therefore imply slight protection of residues 61−63 in NCap but also indicate that all other residues in the NCap besides these are not influenced by the presence of SH2 or SH2-linker.
Figure 2.
Analysis of the NCap. (A). Sequence in NCap region and the peptic peptides that were followed from the NCap. Sequence numbering is as in Figure 1B. Three peptides were followed and the naming of these peptides (peptide C, peptide D, peptide E) corresponds to data in panels C, D, E, respectively of this figure. (B). Representative electrospray mass spectra of deuterium-labeled peptide C (m/z=1090.4, +2) in NCap3. The spectra for this peptide from both NCap32 and NCap32L are similar to that of NCap3. (C-E). Relative deuterium uptake with time for the peptides C, D and E shown as in (A). These results have not been adjusted for back exchange. The maximum of the y-axis in each case is the maximum amount of deuterium that could be incorporated into each peptide.
The major conclusion from HX MS analysis of the NCap deuterium incorporation, notwithstanding slight protection when SH2 is present, is that the NCap is highly dynamic, mostly unstructured, or both. Each peptide was rapidly deuterated and remained so for the duration of the time course, results consistent with no structure and high solvent mobility (23-25). We speculate that in the absence of the myristic acid binding pocket in the large lobe of the kinase domain, which is obviously absent even in the largest of the constructs tested here (NCap32L), the NCap region is not pinned to the SH3 and SH2 domains as shown in the crystal structure. One would predict that this would mimic what would happen to this part of c-Abl were the myristic acid pocket blocked or the myristic acid absent.
NCap effect on SH3 dynamics
We have previously used HX MS to examine the dynamics of the Abl SH3 domain in isolation and bound to ligands (12). Briefly, some SH3 domains exhibit partial cooperative unfolding and such unfolding is a domain dependent phenomenon (26). In the Abl SH3 domain, unfolding occurs with a half-life of about 4.5−5.0 minutes. By monitoring the appearance of and changes to the isotopic pattern (which reveals the unfolding event) in various constructs over time, intramolecular interactions involving SH3 can be ascertained. Ligand binding to SH3 substantially slows the unfolding rate and the relative strength of binding can be determined by the magnitude of the slowing (12). Protein dynamics that are altered in general by global stabilization of the structure in an allosteric fashion will also change the deuterium incorporation and peak appearance and can also be monitored.
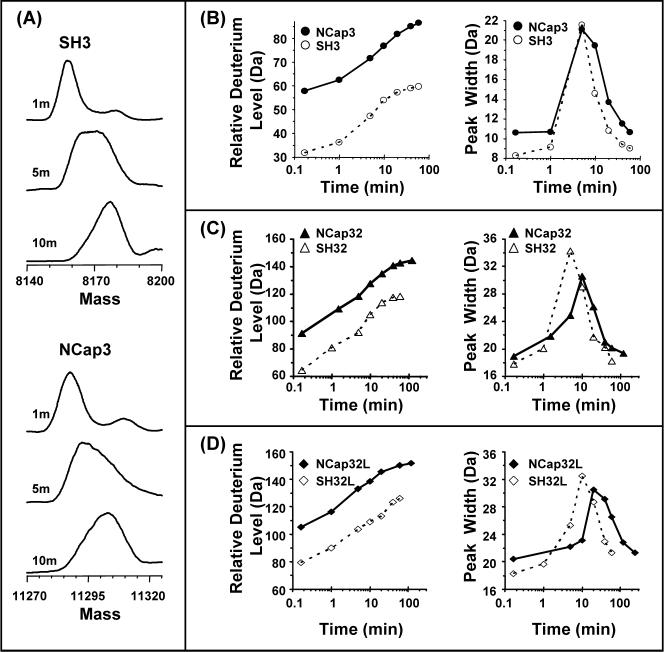
To determine the effects, if any, of the NCap region on SH3 domain dynamics, a construct of Abl that included just NCap and the SH3 domain (NCap3) (see also Fig 1B) was investigated. In this investigation, results from HX MS of NCap3 were compared to HX MS results from the isolated SH3 domain. The spectra for SH3 alone during deuteration showed that there was characteristic broadening of the isotope distribution (12), gradually changing from a binomial isotopic distribution to a bimodal distribution characteristic of an EX1 unfolding event (Figure 3A). The amount of deuterium incorporated into the protein after each exchange time point was determined. NCap3 was more deuterated than SH3 (Figure 3B, left) because it contained more residues. While apparent in the raw spectra, the change in peak width with time that is indicative of EX1 kinetics and partial unfolding is most obvious in a peak-width plot where an increase in width correlates with the unfolding event (22). The centroid of the peak in a peak-width plot is the approximate unfolding half-life. The width of each isotopic distribution at each deuterium labeling time point in Abl SH3 and Abl NCap3 was measured (Figure 3B, right). The Abl SH3 alone showed peak broadening characteristic of EX1 unfolding with half-life of 4.61 minutes, as shown previously (12). The unfolding half-life of NCap3 (5.13 min) was very similar to the unfolding half-life of Abl SH3 without the NCap. These results indicate that in the NCap3 construct, NCap does not interact with the SH3 domain to slow the SH3 unfolding rate.
Figure 3.
Intact protein HX MS analyses. (A). Transformed electrospray mass spectra of deuterium labeled Abl SH3 (top) and NCap3 (bottom). The peak broadening characteristic of EX1 kinetics is visible after 5 min for both SH3 and NCap3. Relative deuterium uptake and peak width change during the time-course of deuterium labeling for (B) Abl SH3 and NCap3; (C) Abl SH32 and NCap32; (D) Abl SH32L and NCap32L. These results have not been adjusted for back exchange. Mass spectral peak width was measured at FWHM for each time point (see Materials and Methods).
An alternative to analysis of whole, undigested proteins by HX MS is analysis of peptic peptides. While this alternative is less important for small constructs, it provides confirmatory results for larger proteins where measurement of peak width for the intact protein can be compromised by salt adducts. We have used analysis of exchange into peptides as a confirmatory test in smaller constructs and as the sole test in larger constructs (e.g. NCap32L). The consistency of results from intact protein analysis or peptide analysis validates the use of either as a good measure of unfolding (see also below).
On-line pepsin digestion HX MS analyses (see Materials and Methods) of both NCap3 and Abl SH3 were carried out concurrently under the same experimental conditions. Note that the peptides are produced after incubating NCap3 and Abl SH3 in D2O and stopping the labeling (23). The peptide fragment (ion of m/z = 809.9, +2 charge state, representing residues 119CEAQTKNGQGWVPSN133, see also Supplementary Fig 1s and Supplementary Table 1s) in the Abl SH3 domain directly relates to the SH3 domain unfolding (26). We define this peptide as the “reporter peptide”. The amount of deuterium that was exchanged-in after each time point was determined for the reporter peptide for both NCap3 and Abl SH3. A summary of peak-width plot data showed that the average half-life of unfolding for the reporter peptide in NCap3 was 5.15 min (Figure 4, top panel) while the average half-life of unfolding for the same peptide fragment in Abl SH3 was 4.71 min (Figure 4, top panel). These results show, as did the data for the intact proteins described above, that no significant changes in protein dynamics occur in the SH3 domain when the NCap region is covalently attached to the SH3 domain. NCap does not appear to associate with the SH3 domain in any way. To affect the dynamics of SH3 domain, NCap may require other mechanisms.
Figure 4.
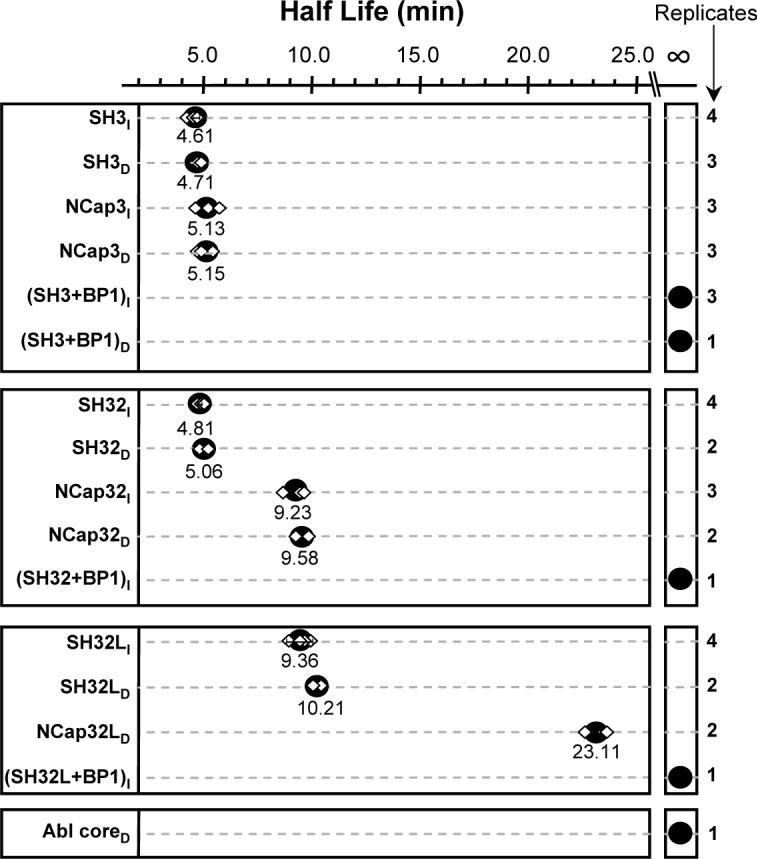
Summary of the Abl SH3 domain unfolding half-life in various constructs. The top box summarizes the data for SH3 with and without NCap, the second box from the top summarizes the data for SH32 with and without NCap, and the third box from the top summarizes data for SH32L with and without NCap. The subscript I represents Intact analyses such as the data in Figure 3 and the subscript D represents on-line pepsin Digestion analyses such as the data in Figure 5. The unfolding half-life determinations in on-line digestion experiments are based on HX MS data of the reporter peptide fragment (m/z 809.9, +2) following the same method as the intact half-life analyses (see Materials and Methods). Each independent measurement of unfolding half-life is shown as a diamond in the plot and the number of measurements that were obtained for each construct is shown on the right-hand side of the figure. Replicate measurements were independent determinations from independent protein purifications and labeling experiments. When 4 measurements were made, the 95% confidence interval is shown surrounding the average half-life (large black dot). For 3 or less replicates, no confidence interval is shown and the large black dot is the average of the determinations. The average half-life value is indicated under each black dot. The various constructs are described in Figure 1B. BP1 binding to SH3 is used as a positive control for tight binding, as described previously (12).
NCap alters SH3 dynamics when SH2 is present
We used the same methodology to investigate if NCap affected SH3 unfolding in the SH32 construct by attaching NCap to the SH32 construct (construct termed NCap32, see Fig 1B). The mass spectra during the deuterium labeling time course for intact SH32 alone (no NCap, data not shown) indicated that this construct had peak width properties similar to those of the intact SH3 alone (Figure 3C, right). The half-life for SH3 domain unfolding in both cases was still around 5 minutes. However, intact protein analyses showed the SH3 unfolding half-life within the NCap32 construct averaged 9.23 min, double the unfolding half-life of SH32 (Figure 3C and Figure 4, middle panel). Raw HX MS data (Figure 5A) for the SH3 reporter peptide from the SH32 and NCap32 constructs showed subtle changes in the relative deuterium uptake in this region (Figure 5B, triangles). The peak width plot (from which the summary t1/2 results in Figure 4 were obtained) demonstrates an obvious shift in the dynamics within the reporter peptide in SH32 when NCap is attached. The effect is obvious both in the peak width plot (Figure 5B) and when comparing the spectra (compare the isotope distribution at the 10 minute time point for SH32 versus NCap32). The unfolding half-life of the reporter peptide in NCap32 was about 9.58 min, double the unfolding half-life of the same peptide fragment in SH32 (5.06 min), see Figure 4. These results, as well as the intact protein data, show that the NCap alters dynamics in the SH3 domain and makes the partial unfolding half-life of the SH3 domains twice as long compared to SH3 in SH32 without the NCap. Note that this effect only occurred in the SH32 construct when NCap was attached and not in the SH3 construct when NCap was attached (see above). Taken together, these data suggest that in the SH32 construct, NCap alters the dynamics of the SH3 domain only when the SH2 domain is present.
Figure 5.
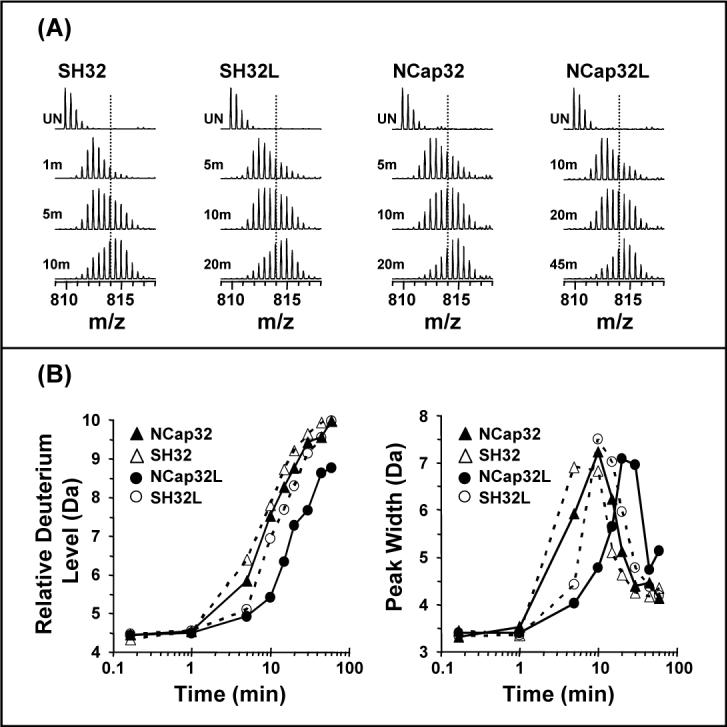
Analyses of the reporter peptide from HX labeling of various constructs. (A). Electrospray mass spectra of deuterium labeled reporter peptide in Abl SH32, SH32L, NCap32, and NCap32L. The relative deuterium uptake (B) and peak width change (C) during the time-course of deuterium labeling is shown for the same four constructs. Back exchange adjustment and peak width measurement were as reported in Figure 3.
Linker affinity for SH3 in the presence of NCap
We next investigated whether the presence of the NCap attenuates the binding of SH3 to the SH2-kinase linker, thereby playing a role in SH3-mediated downregulation of kinase activit To test this idea, a version of SH32L in which the NCap was covalently attached (termed NCap32L; see Fig 1B) was expressed, purified and probed with HX MS. Exchange into SH32L was used as a reference to determine the effect of NCap. Our previous investigations (12) of SH32L showed that there is weak affinity of the SH3 domain for the linker in Abl such that the SH3 unfolding half-life is approximately double when the linker is present. In the current analyses of intact SH32L, summarized in Figure 3D and Figure 4, the SH3 unfolding half-life was about 9.36 min, or double the unfolding half-life in intact Abl SH32 (4.81 min). The results for the reporter peptide from Abl SH32 and SH32L digestion experiments were similar at 5.06 min and 10.21 min, respectively. These protein and peptide results show that the SH3 dynamics are as twice as slow when the SH2-kinase linker was covalently attached to the SH32 construct and are consistent with the previous analyses of the intact SH32 vs. SH32L constructs (12). However, when the NCap was covalently attached to SH32L, there was less deuterium uptake at each data point in the reporter peptide of NCap32L than that in SH32L (Figure 5B, circles) and the peak-width plot (Figure 5B, circles) showed that SH3 unfolding in NCap32L was much slower (23.11 min) than that in SH32L (10.21 min). Again, the raw mass spectra clearly show the time-shift in the deuterium uptake: compare Figure 5A NCap32L at 20 min to NCap32 at 10 min). [Note that due to the increasing size of the NCap32L construct and its propensity to form salt adducts during electrospray, it was not possible to analyze the intact NCap32L protein for peak-width changes. As we have demonstrated above, there is a high degree of consistency between reporter peptide data and intact protein analyses. Only reporter peptide data is presented for the NCap32L protein and we are confident that such analyses are valid.] To summarize, SH3 dynamics in SH32 are twice as slow in the presence of NCap while SH3 dynamics are four times slower in the NCap32L construct. These results indicate that NCap does not disrupt or enhance the interaction between the SH3 domain and the SH2-kinase linker. Rather, the NCap and linker appear to independently slow SH3 domain dynamics. That is, the linker already slowed dynamics in SH3 by two-fold (compare SH32 with SH32L, Figure 4). Addition of the NCap to the SH32L construct slows dynamics by two-fold more.
HX in the regulatory domains
Although the dynamics of the Abl SH3 domain are altered by the presence of the NCap, the deuteration of most parts of the NCap32L construct does not seem to be affected by the presence of the NCap. As shown in Supporting Information Figures 2s and 3s, the deuterium incorporation remains essentially the same in most other parts of the regulatory apparatus in the presence of the NCap. NCap does not affect exchange into the SH2-kinase linker either (Supporting Information Figure 2s, panel o) as exchange in this region is already very fast, again implying that the linker is very dynamic in solution and mostly unstructured. The only significant effect NCap seems to elicit on the regulatory apparatus in terms of hydrogen exchange is a change to the dynamics of the SH3 domain. Changes are obvious in the reporter peptides and a few other smaller peptides that are related to SH3 partial unfolding. Changes to SH3 domain dynamics could be transmitted to the rest of the protein, namely the kinase domain, and play a role in kinase activity by altering the way in which the SH3 domain interacts with the SH2-kinase linker. At the same time, the overall structure of the regulatory domains could remain unaffected by the NCap.
Abl core
We next investigated HX MS within the entire Abl core, which contains the NCap just as in the smaller constructs (see Figure 1B), together with the SH3, SH2, linker and kinase domain. The Abl core was expressed in Sf9 insect cells (see Material and Methods) and the purified protein was labeled with deuterium under identical experimental conditions to those used for the smaller constructs. Note that this protein is stoichiometrically myristoylated on its N-terminus, and was expressed in the presence of the YopH phosphatase, to promote isolation of the downregulated conformation as per Nagar et al.(16). Labeled sample was digested using an on-line pepsin column under quench conditions. For the purposes of this paper, the only data analyzed from the Abl core protein was that of the SH3 domain reporter peptide. Additional HX MS data from the Abl core will be reported elsewhere. While the reporter peptide in NCap32L showed peak broadening characteristic of EX1 unfolding with a half-life 23.11 min, the mass spectra of the reporter peptide in Abl core showed that the unfolding was totally abolished (Figure 6A). The spectra of the reporter peptide in Abl core were very similar to those of the reporter peptide from Abl SH3 incubated with the high-affinity SH3 peptide control BP1 (12) which is used as the positive control for Abl SH3 binding (Figure 6A, right). Deuterium uptake was much less in Abl core reporter peptide than in NCap32L reporter peptide (Figure 6B) and intermediate to that of reporter peptide in SH3 bound to BP1. The peak-width plot for Abl core was nearly flat, again similar to the reporter peptide from SH3 bound to BP1 (Figure 6C). These results suggest that the SH3 domain has strong intramolecular interactions with the other parts of the Abl core (linker and kinase domain) and that these interactions result in domain stabilization similar to that observed when Abl SH3 interacts with a tight-binding peptide. A similar phenomenon has been observed in HX MS analyses of downregulated, near-full length Src-family kinases Hck and Lck (J.R. Engen, T.E. Smithgall, unpublished results).
Figure 6.
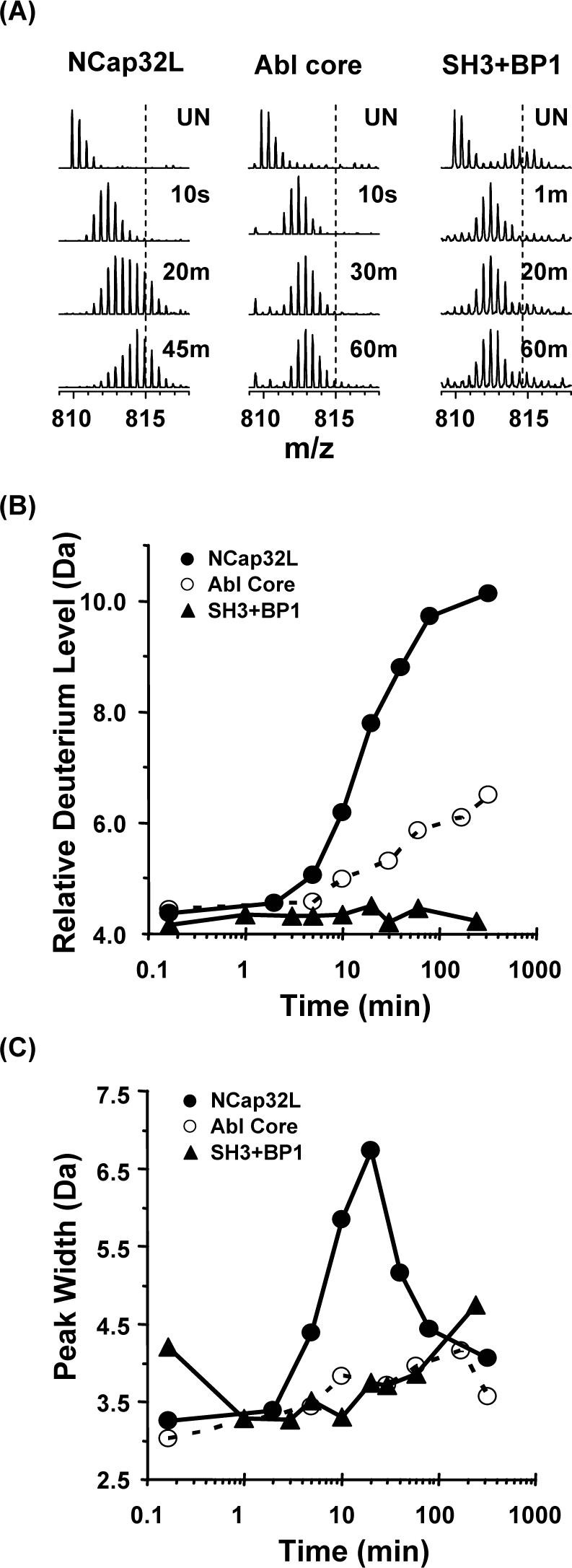
Analyses of deuterium incorporation in the reporter peptide in NCap32L versus Abl core. (A). Electrospray mass spectra of deuterium labeled reporter peptide in NCap32L, Abl core, and SH3 bound to BP1 [as a positive control for the effects of tight SH3 binding, see (12)]. (B) Relative deuterium uptake and (C) peak width change with time for the reporter peptide in the three constructs. Back exchange adjustment and peak width measurement were as reported in Figure 3.
DISCUSSION and CONCLUSIONS
c-Abl is thought to be maintained in a inactive conformation by a number of intramolecular interactions. Mutagenesis studies have shown that the interactions between the SH3 domain and the SH2-kinase linker were very important for the inactive conformation (27). Previous HX MS analyses showed that the SH3 domain associates with the SH2-kinase linker in the absence of the kinase domain (12) whereas such an interaction does not occur in the Src-family kinase Hck (13). The NCap region, directly N-terminal to the SH3 domain, is important for maintaining the downregulated state in biological assays (14, 15). We hypothesized that NCap interactions strengthen downregulatory interactions in the inactive state and used HX MS to probe protein unfolding and dynamics in different constructs to determine whether NCap stabilizes the unfolding of the SH3 domain in the absence of the kinase domain.
The hydrogen exchange data showed that the NCap region was fully exchanged within 10 sec. with only very minor protection from exchange in the presence of the SH2 domain (Fig. 2C, D, and E). These results indicate that the NCap is unstructured and dynamic in solution in the absence of the kinase domain. In the first crystal structure, much of the NCap region was disordered and could not be modeled into the electron density (5). Only after deleting a portion of the residues between 15−56 and adding the myristoyl group in trans, a few residues of NCap could be modeled in the second crystal structure (16) and also revealed a phosphorylated serine residue (Ser-69) which interacts with the SH3-SH2 linker. These crystallography results support the idea that the NCap is a highly dynamic part of the c-Abl structure.
Our data (summarized in Figure 4) indicate that covalent attachment of the NCap to SH3 does not affect the conformation or protein dynamics in the SH3 domain alone. However, when the NCap is covalently attached to SH32, which also includes the SH2 domain, there are significant changes in both the relative deuterium uptake of some specific regions (for example, the SH3 domain reporter peptide) and the unfolding half-life of the SH3 domain. These results imply that NCap stabilizes the SH3 domain through allosteric mechanisms involving the SH2 domain. In other words, NCap stabilization of the SH3 domain requires the SH2 domain. When NCap was covalently attached to SH32L, which includes the SH2-kinase linker, there were also significant differences in the relative deuterium uptake in the reporter peptide in the SH3 domain. The unfolding half-life of the reporter peptide was doubled in NCap32L versus SH32L. This further indicates that the presence of the NCap stabilizes the SH3 domain. The extent to which the NCap slowed SH3 domain unfolding suggests that it has an equivalent effect on SH3 as does linker binding to SH3 in the absence of the kinase domain [recall that comparison of SH32 and SH32L showed that the SH3 unfolding in SH32L was twice as slow as in SH32, see also Figure 4]. Through these interactions, the SH3 domain dynamics may be coupled to the SH2 domain and thus SH3 unfolding is slowed both in NCap32 or NCap32L.
In general, hydrogen exchange was not vastly altered by the presence of the NCap as in most regions, the deuterium incorporation was nearly the same with or without the NCap (see Supporting Information Figure 2s). The dynamics of the SH3 domain were the main thing affected. These results further indicate that the NCap is a highly mobile structural element and that the overall conformation of the regulatory domains is unaffected by the NCap.
HX MS showed that when SH3 was alone and unliganded, the unfolding half-life was around 5 minutes (Figure 4). When a weak ligand bound to SH3, such as in the constructs SH32L or NCap32L, the unfolding half-life was shifted to longer times. Stronger stabilizing forces, including those encountered upon binding to the linker or from the presence of the NCap, made SH3 dynamics much slower. At the extreme, binding of the high-affinity peptide ligand BP1 to SH3, SH32, and SH32L in trans (Figure 4) resulted in a half-life of SH3 unfolding that was so slow that we could not measure it in an 8 hour experiment. Similarly, SH3 unfolding in the Abl core was not measurable even after eight hours, indicating that SH3 strongly interacts with the linker and may be strongly stabilized by both the NCap and the presence of the kinase domain. It seems, therefore, that the kinase domain plays a critical role in the ability of the NCap to downregulate activity through SH3 binding to the linker and the backside of the kinase domain. However, the kinase domain does not contribute all of the downregulatory influence as the presence of NCap in the SH32L construct contributed significantly to the decreased dynamics of the SH3 domain in the absence of the kinase domain. In Bcr-Abl, the N-terminal 46 residues of the Abl core (which include the NCap region) are replaced by Bcr (1). NCap's stabilizing influence on the SH3 domain is also likely to be disrupted by fusion to Bcr sequences. Future analyses of constructs containing the Bcr fusion junction will begin to address this question. In addition, identification of small molecules that restore or mimic the stabilizing influence of the NCap on SH3:linker interaction may provide new leads for Bcr-Abl inhibitors.
ACKNOWLEDGEMENTS
Dr. S. Brier is acknowledged for his assistance in preparing some of the NCap constructs. This work is contribution number 921 from the Barnett Institute.
ABBREVIATIONS
- HX
hydrogen exchange
- MS
mass spectrometry
- SFK
Src-family kinase
Footnotes
We are pleased to acknowledge generous financial support from the NIH: GM070590 (to J.R.E.) and CA101828 (to T.E.S.)
SUPPORTING INFORMATION AVAILABLE A Supporting Table (Table 1s) of the peptic fragments of Abl NCap32L, a Figure (Figure 1s) indicating the pepsin digestion map of Abl NCap32L and additional deuterium incorporation data (Figure 2s, panels a-o; Figure 3s) for peptides in Abl NCap32L is provided. This material is available free of charge via the Internet at http://pubs.acs.org.
Supplementary Material
REFERENCES
- 1.Hantschel O, Superti-Furga G. Regulation of the c-Abl and Bcr-Abl tyrosine kinases. Nat Rev Mol Cell Biol. 2004;5:33–44. doi: 10.1038/nrm1280. [DOI] [PubMed] [Google Scholar]
- 2.Pendergast AM. The Abl family kinases: mechanisms of regulation and signaling. Adv Cancer Res. 2002;85:51–100. doi: 10.1016/s0065-230x(02)85003-5. [DOI] [PubMed] [Google Scholar]
- 3.de Klein A, van Kessel AG, Grosveld G, Bartram CR, Hagemeijer A, Bootsma D, Spurr NK, Heisterkamp N, Groffen J, Stephenson JR. A cellular oncogene is translocated to the Philadelphia chromosome in chronic myelocytic leukaemia. Nature. 1982;300:765–767. doi: 10.1038/300765a0. [DOI] [PubMed] [Google Scholar]
- 4.Heisterkamp N, Stephenson JR, Groffen J, Hansen PF, de Klein A, Bartram CR, Grosveld G. Localization of the c-ab1 oncogene adjacent to a translocation break point in chronic myelocytic leukaemia. Nature. 1983;306:239–242. doi: 10.1038/306239a0. [DOI] [PubMed] [Google Scholar]
- 5.Nagar B, Hantschel O, Young MA, Scheffzek K, Veach D, Bornmann W, Clarkson B, Superti-Furga G, Kuriyan J. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell. 2003;112:859–871. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 6.Courtneidge SA. Cancer: Escape from inhibition. Nature. 2003;422:827–828. doi: 10.1038/422827a. [DOI] [PubMed] [Google Scholar]
- 7.Harrison SC. Variation on an Src-like theme. Cell. 2003;112:737–740. doi: 10.1016/s0092-8674(03)00196-x. [DOI] [PubMed] [Google Scholar]
- 8.Brown MT, Cooper JA. Regulation, substrates and functions of src. Biochim Biophys Acta. 1996;1287:121–149. doi: 10.1016/0304-419x(96)00003-0. [DOI] [PubMed] [Google Scholar]
- 9.Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 10.Xu W, Doshi A, Lei M, Eck MJ, Harrison SC. Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Mol Cell. 1999;3:629–638. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- 11.Xu W, Harrison SC, Eck MJ. Three-dimensional structure of the tyrosine kinase c-Src. Nature. 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 12.Chen S, Brier S, Smithgall TE, Engen JR. The Abl SH2-kinase linker naturally adopts a conformation competent for SH3 domain binding. Protein Sci. 2007;16:572–581. doi: 10.1110/ps.062631007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hochrein JM, Lerner EC, Schiavone AP, Smithgall TE, Engen JR. An examination of dynamics crosstalk between SH2 and SH3 domains by hydrogen/deuterium exchange and mass spectrometry. Protein Sci. 2006;15:65–73. doi: 10.1110/ps.051782206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pluk H, Dorey K, Superti-Furga G. Autoinhibition of c-Abl. Cell. 2002;108:247–259. doi: 10.1016/s0092-8674(02)00623-2. [DOI] [PubMed] [Google Scholar]
- 15.Hantschel O, Nagar B, Guettler S, Kretzschmar J, Dorey K, Kuriyan J, Superti-Furga G. A Myristoyl/Phosphotyrosine Switch Regulates c-Abl. Cell. 2003;112:845–857. doi: 10.1016/s0092-8674(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 16.Nagar B, Hantschel O, Seeliger M, Davies JM, Weis WI, Superti-Furga G, Kuriyan J. Organization of the SH3-SH2 unit in active and inactive forms of the c-Abl tyrosine kinase. Mol Cell. 2006;21:787–798. doi: 10.1016/j.molcel.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 17.Koegl M, Courtneidge SA, Superti-Furga G. Structural requirements for the efficient regulation of the Src protein tyrosine kinase by Csk. Oncogene. 1995;11:2317–2329. [PubMed] [Google Scholar]
- 18.Schindler T, Sicheri F, Pico A, Gazit A, Levitzki A, Kuriyan J. Crystal structure of Hck in complex with a Src family-selective tyrosine kinase inhibitor. Mol Cell. 1999;3:639–648. doi: 10.1016/s1097-2765(00)80357-3. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Pan H, Smith DL. Hydrogen exchange-mass spectrometry: optimization of digestion conditions. Mol Cell Proteomics. 2002;1:132–138. doi: 10.1074/mcp.m100009-mcp200. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weis DD, Engen JR, Kass IJ. Semi-automated data processing of hydrogen exchange mass spectra using HX-Express. J Am Soc Mass Spectrom. 2006;17:1700–1703. doi: 10.1016/j.jasms.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 22.Weis DD, Wales TE, Engen JR, Hotchko M, Ten Eyck LF. Identification and characterization of EX1 kinetics in H/D exchange mass spectrometry by peak width analysis. J Am Soc Mass Spectrom. 2006;17:1498–1509. doi: 10.1016/j.jasms.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 23.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 24.Smith DL, Deng Y, Zhang Z. Probing the non-covalent structure of proteins by amide hydrogen exchange and mass spectrometry. J Mass Spectrom. 1997;32:135–146. doi: 10.1002/(SICI)1096-9888(199702)32:2<135::AID-JMS486>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 25.Hoofnagle AN, Resing KA, Ahn NG. Protein analysis by hydrogen exchange mass spectrometry. Annu. Rev. Biophys. Biomol. Struct. 2003;32:1–25. doi: 10.1146/annurev.biophys.32.110601.142417. [DOI] [PubMed] [Google Scholar]
- 26.Wales TE, Engen JR. Partial unfolding of diverse SH3 domains on a wide timescale. J Mol Biol. 2006;357:1592–1604. doi: 10.1016/j.jmb.2006.01.075. [DOI] [PubMed] [Google Scholar]
- 27.Barila D, Superti-Furga G. An intramolecular SH3-domain interaction regulates c-Abl activity. Nat Genet. 1998;18:280–282. doi: 10.1038/ng0398-280. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.