Abstract
SYNOPSIS
Selenocysteine (Sec) is biosynthesized on its tRNA and incorporated into selenium-containing proteins (selenoproteins) as the 21st amino acid. Selenoprotein synthesis is dependent on Sec tRNA and the expression of this class of proteins can be modulated by altering Sec tRNA expression. The gene encoding Sec tRNA (Trsp) is a single copy gene and its targeted removal in liver demonstrated that selenoproteins are essential for proper function wherein their absence leads to necrosis and hepatocellular degeneration. In the present study, we found that the complete loss of selenoproteins in liver was compensated by an enhanced expression of several phase II response genes and their corresponding gene products. The replacement of selenoprotein synthesis in mice carrying mutant Trsp transgenes, wherein housekeeping, but not stress-related selenoproteins are expressed, led to normal expression of phase II response genes. Thus, this study provides evidence for a functional link between housekeeping selenoproteins and phase II enzymes.
Keywords: gene expression, liver, microarray, selenocysteine (Sec) tRNA, Trsp knockout, xenobiotic
INTRODUCTION
Several trace elements have important roles in human health and their over-abundance or reduced levels result in severe health problems. Selenium is one such essential micronutrient with antioxidant properties, whose deficiency has been associated with several disorders [1]. Selenium is incorporated into proteins (selenoproteins) as the amino acid Sec (selenocysteine) and its biological function is believed to be exerted in large part by these proteins [2]. To date, 25 selenoprotein genes have been identified in the human genome and 24 in the mouse genome [3]. The incorporation of Sec into proteins is a unique process in that it uses the stop codon, UGA, to decode this amino acid and involves a distinctive tRNA, designated tRNA[Ser]Sec. A number of other cis- and trans- acting factors are also required that form a complex with Sec-tRNA[Ser]Sec mediating the cotranslational incorporation of Sec into protein [2, 4, 5].
Higher vertebrates have two Sec tRNA[Ser]Sec isoforms that differ from each other by a single methyl group on the 2′-O-hydroxyribosyl moiety at position 34 [2]. This methyl group is designated Um34. Both isoforms also contain the base, 5′-methylcarboxylmethyluracil (mcm5U) at position 34. Since both isoforms contain mcm5U, but only one of them contains Um34, they are designated mcm5U (i.e., the isoform lacking Um34) and 5′-methylcarboxymethyl-2′-O-methyluridine (mcm5Um; i.e., the isoform containing Um34). Sec tRNA[Ser]Sec has three additional modified bases, pseudouridine at position 55, 1-methyladenosine at position 58 and N6-isopentenyladenosine (i6A) at position 37. The addition of Um34 is the last step in the maturation of Sec tRNA[Ser]Sec and this step is stringently dependent on the prior synthesis of all base modifications [6]. In addition, Um34 synthesis is influenced by selenium status and selenium deficiency leads to an enrichment of mcm5U as compared to mcm5Um, while selenium adequacy reverses this ratio [2]. The levels of the two isoforms modulate expression of different selenoproteins wherein some selenoproteins (e.g. GPx (glutathione peroxidase) 1 and 3 that function largely as stress-related proteins) are preferentially expressed in the presence of mcm5Um,, while others (e.g. TR (thioredoxin reductase) 1 and 3 that function as essential housekeeping proteins) are preferentially expressed in the presence of mcm5U [7, 8].
The gene encoding Sec tRNA[Ser]Sec (Trsp) is present in single copy and its expression is essential for the synthesis of all selenoproteins. Selenoproteins are the only known class of proteins in eukaryotes, whose expression is regulated by a single tRNA and manipulating the expression of Trsp in mice modulates selenoprotein synthesis. Since removal of Trsp is embryonic lethal [9, 10], the conditional knockout of Trsp [10] gave rise to several useful models for studying the role of selenium and selenoproteins in development and health [reviewed in 11]. In one of these models, we targeted the removal of Trsp in hepatocytes that demonstrated an essential role of selenoproteins in proper liver function [12]. Additionally, we rescued Trsp null mice with transgenic mice carrying a mutant Trsp transgene [7, 8]. In one mutant Trsp transgene, A37 was changed to G [7, 13] that resulted in loss of both i6A and Um34 [6]. We also produced a second transgenic mouse, where T34 was changed to A in the mutant transgene [8] and the resulting tRNA gene product also lacked Um34. Transgenic mice carrying mutant Trsp transgenes were used to replace selenoprotein synthesis in mice lacking Trsp in hepatocytes by matings between these two mouse lines as described elsewhere [8]. Introduction of either the A34 or G37 mutated transgene into the liver Trsp-knockout mice selectively replaced selenoproteins involved in housekeeping functions, but not those involved in stress-related functions [7, 8]. Furthermore, the number of gene copies of the mutant G37 transgene varied from 2 in one of the transgenic mouse lines we developed to 16 in the other transgenic mouse line.
In the present study, a comparative analysis of gene expression in the liver of the Trsp- knockout mice, designated ΔTrsp herein, and the A34 and G37 transgenic mice with gene expression of wild type mice was carried out using microarrays. These studies showed that the loss of selenoproteins in Trsp-knockout mice was associated with an enhanced expression of several phase II response genes and their corresponding enzymes. Phase II response genes are enzymes involved in detoxification as well as protection against oxidative stress. Interestingly, replacement of housekeeping selenoproteins in A34 or G37 transgenic mice resulted in the levels of phase II enzymes returning to normal. Taken together, the data suggest a functional association between housekeeping selenoproteins and phase II enzymes, wherein the loss of function of some housekeeping selenoproteins may be compensated by phase II enzymes in the liver of the knockout mouse.
EXPERIMENTAL PROCEDURES
Materials
NuPage polyacrylamide gels, polyvinylidene difluoride (PVDF) membranes, See-Blue Plus2 protein markers, Trizol and Superscript II reverse transcriptase were purchased from Invitrogen, SuperSignal West Dura extended duration substrate from Pierce and Cy3 and Cy5 Mono-reactive dyes from GE Healthcare. GSTA [GST (glutathione transferase) Alpha], GSTM (GST Mu) and EPHX1 (epoxide hydrolase 1) antibodies were obtained from Detroit R&D, Inc., HMOX1 (haem oxygenase) antibodies, anti-mouse and anti-rabbit HRP (horseradish peroxidase)-conjugated secondary antibodies from Santa Cruz Biotechnology, Inc., AOX1 antibodies from BD Biosciences and β-actin antibodies and anti-goat horseradish peroxidase-conjugated secondary antibodies from Abcam. Primers used for real-time PCR were procured from Sigma-Genosys. All other reagents were of the highest grade available and were obtained commercially.
Mouse lines and genotyping
The mice analyzed in this study were all males, 6-8 weeks of age in a B6/FVB genetic background and were fed a selenium sufficient diet. Each mouse line used in this study, preparation of the mutation carried in the A34 and G37 Trsp transgenes, and the manner in which these mouse lines were generated are described in detail elsewhere [7, 8], and their genotypes and designations are summarized in Table 1. The care of animals was in accordance with the National Institutes of Health institutional guidelines under the expert direction of Dr. Kyle Stump (NCI, National Institutes of Health, Bethesda, MD). DNA was extracted from mouse tail clippings and the genotype determined by PCR with the appropriate primers as described [7, 12].
Table 1.
Summary of mouse lines, their genotypes and designations
| Mice | Genotype | Designation f |
|---|---|---|
| Wild type a | Trsp+/+-AlbCre+/+ | Trsp |
| Trsp liver knockout b | Trspfl/fl-AlbCre+/+ | ΔTrsp |
| A34 transgenic (2 copies) c | Trspfl/fl-AlbCre+/+-A34t/t | A34 |
| G37 transgenic (2 copies) d | Trspfl/fl-AlbCre+/+-G37t/t | G37L |
| G37 transgenic (16 copies) e | Trspfl/fl-AlbCre+/+-G37t/t | G37H |
Wild type mice were homozygous for Trsp and albumin Cre.
Trsp liver knockout mice lacked Trsp in their liver.
A34 transgenic mice carrying 2 copies of the T34→A34 mutant transgene.
G37 transgenic mice carrying 2 copies of the A37→G37 mutant transgene.
G37 transgenic mice carrying 16 copies of the A37→G37 mutant transgene.
Designations used in the text to denote each mouse line.
Probe preparation
Total RNA from liver of wild type (Trsp), liver knockout (ΔTrsp), and transgenic (A34, G37L and G37H) mice was isolated using Trizol reagent according to the manufacturer’s protocol and labeled using the Fairplay® II Microarray Labeling kit (Stratagene). For indirect labeling of RNA, 15 μg of both control and experimental RNA were used to generate cDNA, using aminoallyl dNTP mix according to the manufacturer’s protocol. The resulting cDNA was purified using a MinElute column (Qiagen) and eluted from the column with 10 μL of MinElute elution buffer and dried by speed-vac for 15 min. Samples were next coupled to 5 μL of 2x Fairplay® coupling buffer and 5 μL of monofunctional dye and incubated at room temperature (22°C) in the dark for 30 min. Following incubation, the labeled cDNA was purified using a MinElute column and eluted with 10 μL of elution buffer.
Microarray hybridization
Mouse oligonucleotide glass arrays, containing 70mer oligonucleotides (printed on Corning epoxide slides), were procured from the NCI Microarray Facility, Frederick, MD. Each slide in these oligonucleotide arrays have 48 blocks containing 28 rows and 28 columns each, with 36960 oligonucleotide spots with a spacing of 155 μm.
Slides were pre-hybridized for 1 hour at 42°C with 40 μL of pre-hybridization buffer (5x SSC, 1% BSA and 0.1% SDS). Pre-hybridization solution was removed by plunging the slides, first in deionized water and then in isopropanol, for 2 min each. The slides were air dried prior to hybridization. For hybridization, the Cy3 and Cy5 labeled cDNAs were combined and mixed with 1 μL COT-1 DNA, preheated at 100°C for 1 min to denature the targets and snap cooled on ice. This mixture was added to 20 μl of 2x F-hybridization buffer (50% formamide, 10x SSC, 0.2% SDS) and pre-warmed at 42°C. The total cDNA/hybridization solution mixture was loaded onto each prehybridized slide and covered with an M Series Lifterslip (Erie Scientific). The slides were placed in hybridization chambers and incubated overnight at 42°C. Humidity in each chamber was maintained by the addition of 20μl of 3x SSC solution. Post-hybridization washing included 5 min in 2x SSC + 0.1% SDS, 5 min in 1x SSC and 5 min in 0.2x SSC, after which the slides were dried by centrifugation (44 g for 5 min at 22°C).
Data processing and analysis
Microarray slides were scanned for each fluoroprobe at 10 μm using a Genepix® 4000B scanner and analyzed with GenePix Pro 3.0 software (Axon Instruments). Scanned images were exported as TIFF files to GenePix Pro 3.0 software for analysis. For data analysis, data files (in gpr format) and image (in jpeg format) were imported into the microarray database (mAdb) and analyzed by software tools provided by the National Cancer Institute, Center for Cancer Research in collaboration with the National Institutes of Health, Center for Information Technology, Bioinformatics and Molecular Analysis Section. Transcripts whose expression level varied at least two fold in ΔTrsp mice as compared to Trsp mice in more than 50% of the experiments with a P-value ≤ 0.05 were selected and the corresponding transcript levels were then analyzed in A34, G37L and G37H transgenic mice relative to Trsp mice. A hierarchical clustering analysis was performed on genes in the resultant analysis. Grouping of genes into different biological functions was performed using the David database (http://david.abcc.ncifcrf.gov) and/or the mAdb software.
Quantitative real-time PCR
Two-step quantitative real-time PCR (Q-PCR) was performed to validate relative expression of genes, using primer sequences outlined in Table 2. Two μg of total RNA from each sample was reverse transcribed to synthesize first strand cDNA using SuperScript II reverse transcriptase enzyme and random primers. The resulting cDNA was diluted, and in combination with 500 nM of each primer, iQ™ SYBR green supermix (Bio Rad Laboratories) and DNA Engine Opticon® 2 Real-Time PCR Detection System (MJ-Research), used for transcript quantification. The PCR reaction had an initial denaturation of 5 min at 95°C, followed by 40 cycles consisting of 20 sec at 94°C, 20 sec at 55°C and 30 sec at 72°C. The reactions were carried out in triplicate and the specificity of the primers was verified by melting curve analysis. RNA levels were normalized to β-glucuronidase (Gusb) and expression levels were compared to those of wild type mice.
Table 2.
Primers for assessing real-time PCRa
| Gene | Forward Sequence | Reverse Sequence |
|---|---|---|
| Aox1 | 5′-GAAGCTGGACAACGCTTACA-3′ | 5′-CCACATTTGATTGCCACTTC-3′ |
| Cd36 | 5′-GATTGTACCTGGGAGTTGGC-3′ | 5′-CATGAGAATGCCTCCAAACA-3′ |
| Ces1 | 5′-CAGAAGACAGCTGCATCCAT-3′ | 5′-TCCAATCAAGTCCAGGAACA-3′ |
| Ces2 | 5′-ATGTGAGGCTATGGATTCCC-3′ | 5′-TCCTCAGATGCCAACAACTC-3′ |
| Cyp2A5 | 5′-GAGATTGATCGGGTGATTGG-3′ | 5′-CGAAACTTGGTGTCCTTGGT-3′ |
| Ddc | 5′-CTGAATGGTGTGGAGTTTGC-3′ | 5′-TGAATCCTGAGTCCTGGTGA-3′ |
| Dmpk | 5′-CGTGTTCGCCTATGAGATGT-3′ | 5′-ACGAATGAGGTCCTGAGCTT-3′ |
| Ephx1 | 5′-GGGTCAAAGCCATCAGCCA-3′ | 5′-CCTCCAGAAGGACACCACTTT-3′ |
| Gsta1 | 5′-CGCAGACCAGAGCCATTCTC-3′ | 5′-TTGCCCAATCATTTCAGTCAGA-3′ |
| Gsta2 | 5′-CCCCTTTCCCTCTGCTGAAG-3′ | 5′-TGCAGCCACACTAAAACTTGA-3′ |
| Gsta4 | 5′-TTGAAATCGATGGGATGATG-3′ | 5′-ATCATCATCAGGTCCTGGGT-3′ |
| Gstm1 | 5′-CCAAACACACAGGTCAGTCC-3′ | 5′-CGTCACCCATGGTGTATCTC-3′ |
| Gstm2 | 5′-CCTATGACACTAGGTTACTGG-3′ | 5′-CACTGGCTTCGGTCATAGTCA-3′ |
| Gstm3 | 5′-TATGACACTGGGCTATTGGAAC-3′ | 5′-GGGCATCCCCCATGACA-3′ |
| Gstt3 | 5′-GGCAGAAGATGATGTTCCCT-3′ | 5′-TCAGCCACAGAAATATGGGA-3′ |
| Hmox1 | 5′-GCCACCAAGGAGGTACACAT-3′ | 5′-GCTTGTTGCGCTCTATCTCC-3′ |
| Htatip2 | 5′-GGCCAGGAGTCCTACTGTGT-3′ | 5′-GTTCAGCATCGCTCTAACCA-3′ |
| Ikbkg | 5′-CCTGGTAGCCAAACAGGAAT-3′ | 5′-CCTTCTTCTCCACCAGCTTC-3′ |
| Lgals1 | 5′-GCAACAACCTGTGCCTACAC-3′ | 5′-TGATGCACACCTCTGTGATG-3′ |
| Srxn1 | 5′-CCAGGGTGGCGACTACTACT-3′ | 5′-CAAGTCTGGTGTGGATGCTC-3′ |
| Ugdh | 5′-TGCTGTCCAATCCTGAGTTC-3′ | 5′-ACCCAGTGCTCATACACAGC-3′ |
| Ugt2b35 | 5′-AATGACCTTCTCGGTCATCC-3′ | 5′-CCACCATGTGTGCAATGTTA-3′ |
Primers designed for determining real-time PCR of each mRNA examined are shown in the table.
Western blotting
Protein extracts prepared from liver of Trsp, ΔTrsp, A34, G37L and G37H mice were electrophoresed on 10% polyacrylamide gels, transferred to PVDF membranes and immunoblotted with antibodies against GSTA (1:10,000 dilution), GSTM (1:10,000 dilution), EPHX1 (1:10,000 dilution), HMOX1 (1:500 dilution), AOX1 (1:250 dilution) and β-actin (1:1,000 dilution). Anti-goat HRP conjugated secondary antibody (1:40,000) was used for GSTA, GSTM, EPHX1 and β-actin, while anti-rabbit HRP-conjugated secondary antibody (1:25,000) was used for HMOX1 and anti-mouse HRP-conjugated secondary antibody (1:30,000) was used for AOX1. Following the attachment of the secondary antibody, membranes were washed with TBS (Tris-buffered saline, 20 mM Tris (pH 7.5), 150 mM NaCl) containing 0.1% Tween, incubated in SuperSignal West Dura Extended Duration Substrate and exposed to X-ray film.
RESULTS
Gene expression profile in liver of ΔTrsp mice
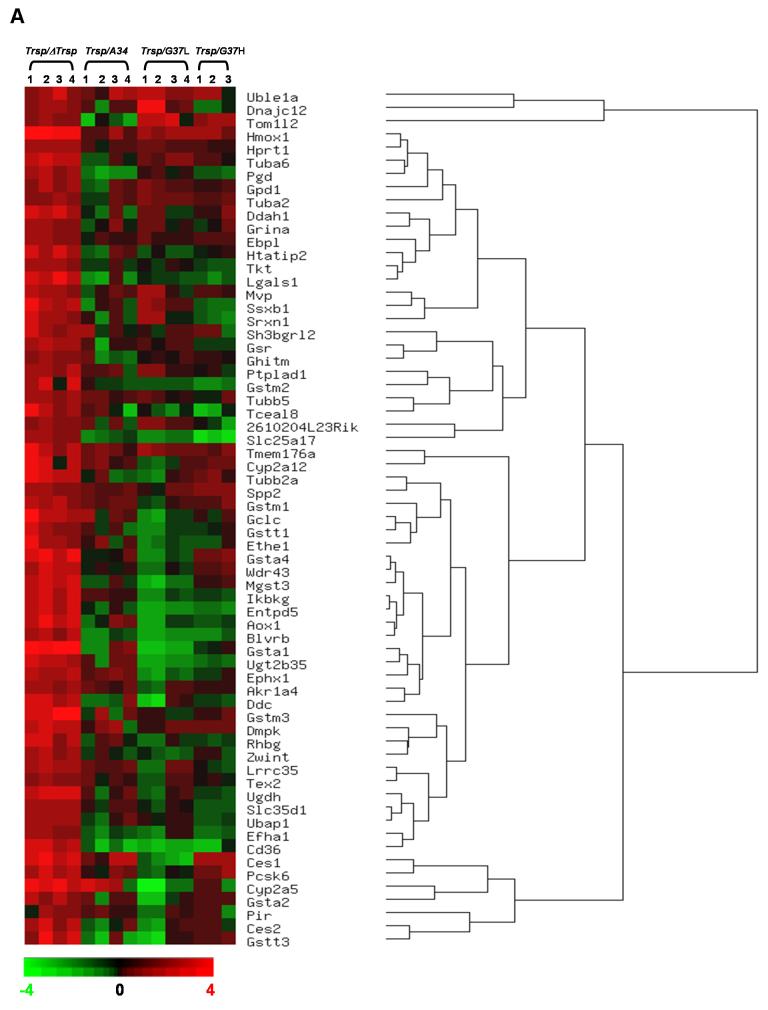
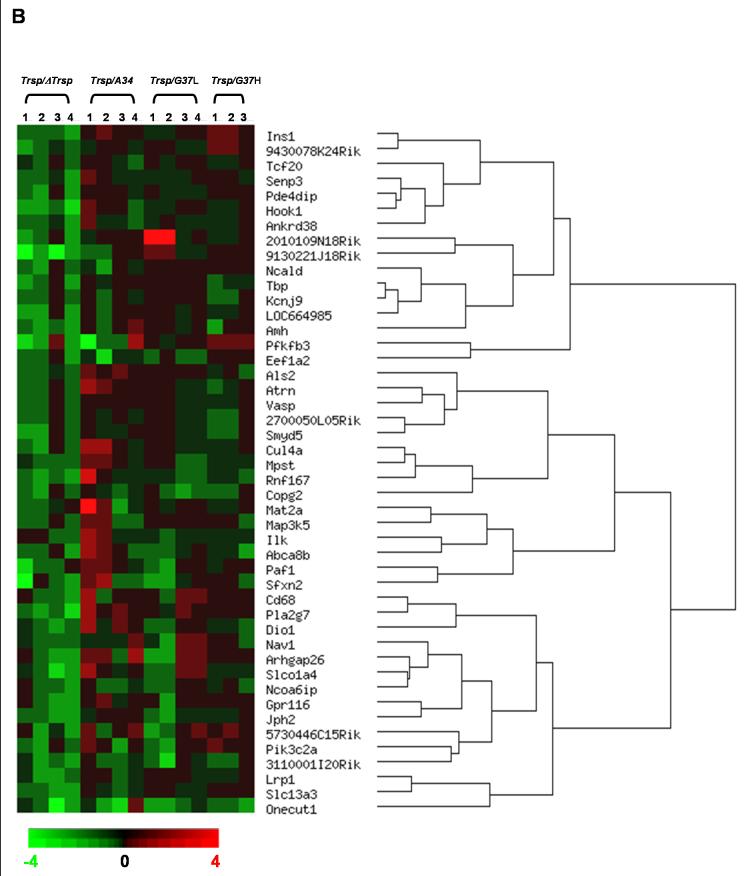
The overall gene expression profile associated with the conditional knockout of Trsp in mouse liver (ΔTrsp) and in liver of mice following the selective replacement of selenoproteins with mutated Trsp transgenes was examined (Figure 1). An analysis of gene expression in livers from Trsp, ΔTrsp, A34, G37L and G37H mice showed that the loss of Trsp was associated with altered levels of some mRNAs, reflected through changes in gene expression and/or mRNA stability. Initially, gene expression in ΔTrsp mice was compared to those in Trsp mice and genes displaying a greater than two-fold change in the microarray analysis with a P-value ≤ 0.05 were selected. These genes were then segregated as upregulated (Table 3) or downregulated (Table 4) and ordered by their pattern of gene expression by hierarchical clustering (Figure 1). Transcripts upregulated in ΔTrsp mice are shown in Figure 1A and those downregulated in Figure 1B, along with the relative expression of these transcripts in A34, G37L and G37H transgenic mice. Following filtering, genes upregulated in ΔTrsp mice were grouped under 6 major hierarchical clusters, while those downregulated were grouped into 5 major hierarchical clusters.
Figure 1. Hierarchical clustering analysis of gene alterations following Trsp removal in liver.
Hierarchical dendrogram representing the expression profiles of significantly altered genes, following data filtering as described in Results. (A) upregulated or (B) downregulated in knockout mice. The genes are ordered by clustering tightness, with a distance measure of 1 - Pearson correlation coefficient and a P-value threshold of 0.05. Each column represents data from one experimental set, and rows indicate individual genes. Increases and decreases in transcript expression levels are represented by shades of red and green, respectively.
Table 3.
Transcripts in elevated levels within liver of knockout mice relative to wild type mice a
| ΔTrsp vs Trsp | A34 vs Trsp | G37L vs Trsp | G37H vs Trsp | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| UniGene | Description | Folds | P value | Folds | P value | Folds | P value | Folds | P value | Function/Gene Ontology |
| Defense stress and detoxification b | ||||||||||
| Mm.28191 | carboxylesterase 2 (Ces2) | 3.8 | 0.02 | -0.7 | 0.07 | -0.5 | 0.17 | 0.5 | 0.53 | Ester hydrolase activity |
| Mm.389848 | Cytochrome P450, family 2, subfamily a, polypeptide 5 (Cyp2a5) | 8.2 | 0.01 | 3.0 | 0.30 | -5.6 | 0.03 | 0.2 | 0.51 | Degradation of environmental toxins and mutagens |
| Mm.32550 | DnaJ (Hsp40) homolog, subfamily C, member 12 (Dnajc12) | 2.2 | 0.00 | 0.4 | 0.51 | 5.5 | 0.11 | -1.5 | 0.00 | Heat shock protein binding, protein folding |
| Mm.218639 | sulfiredoxin 1 homolog (S. cerevisiae) (Srxn1) | 3.3 | 0.03 | -0.2 | 0.19 | 0.9 | 0.96 | -1.6 | 0.00 | Antioxidant activity, response to oxidative stress |
| Metabolism b | ||||||||||
| Mm.30085 | aldo-keto reductase family 1, member A4 (aldehyde reductase) (Akr1a4) | 2.4 | 0.00 | 1.3 | 0.03 | -0.1 | 0.18 | 1.1 | 0.01 | Glycerolipid metabolism |
| Mm.26787 | aldehyde oxidase 1 (Aox1) | 6.5 | 0.01 | 0.7 | 0.73 | -1.8 | 0.00 | -1.1 | 0.00 | Xenobiotic metabolism |
| Mm.24021 | Biliverdin reductase B (Blvrb) | 3.4 | 0.01 | -1.5 | 0.00 | -2.1 | 0.00 | -1.7 | 0.00 | Porphyrin and chlorophyll metabolism |
| Mm.22720 | carboxylesterase 1 (Ces1) | 8.0 | 0.01 | 2.8 | 0.09 | -2.4 | 0.00 | 3.0 | 0.00 | Alkaloid biosynthesis |
| Mm.12906 | dopa decarboxylase (Ddc) | 5.1 | 0.00 | -0.5 | 0.15 | -2.0 | 0.15 | -1.1 | 0.00 | Amino acid and derivative metabolic process |
| Mm.22758 | emopamil binding protein-like (Ebpl) | 2.2 | 0.00 | 0.6 | 0.48 | 1.2 | 0.09 | 1.3 | 0.00 | Sterol metabolism |
| Mm.10211 | ectonucleoside triphosphate diphosphohydrolase 5, transcript variant 1 (Entpd5) | 5.4 | 0.02 | -0.7 | 0.03 | -2.3 | 0.00 | -1.6 | 0.00 | Purine metabolism; pyrimidine metabolism |
| Mm.9075 | epoxide hydrolase 1, microsomal (Ephx1) | 3.3 | 0.00 | 1.5 | 0.02 | -1.8 | 0.00 | -0.3 | 0.14 | Xenobiotic metabolism |
| Mm.252391 | glycerol-3-phosphate dehydrogenase 1 (soluble) (Gpd1) | 3.1 | 0.02 | -0.1 | 0.24 | 1.4 | 0.02 | 1.2 | 0.08 | Carbohydrate metabolism |
| Mm.283573 | Glutathione reductase 1 (Gsr) | 3.2 | 0.00 | 0.2 | 0.42 | 1.2 | 0.02 | -1.1 | 0.00 | Glutathione metabolism |
| Mm.197422 | Glutathione S-transferase, alpha 1 (Ya) (Gsta1) | 23.4 | 0.00 | -0.2 | 0.33 | -3.6 | 0.00 | -0.3 | 0.14 | Glutathione metabolism; xenobiotic metabolism |
| Mm.422778 | Glutathione S-transferase, alpha 2 (Yc2) (Gsta2) | 2.9 | 0.001 | 0.45 | 0.55 | -2.3 | 0.06 | 1.2 | 0.00 | Glutathione metabolism; xenobiotic metabolism |
| Mm.2662 | Glutathione S-transferase, alpha 4 (Gsta4) | 8.6 | 0.00 | 0.6 | 0.51 | -1.5 | 0.00 | 1.8 | 0.05 | Glutathione metabolism; xenobiotic metabolism |
| Mm.37199 | Glutathione S-transferase, mu 1 (Gstm1) | 3.2 | 0.01 | 1.4 | 0.00 | 0.0 | 0.20 | 1.5 | 0.10 | Glutathione metabolism; xenobiotic metabolism |
| Mm.37199 | Glutathione S-transferase, mu 2 (Gstm2) | 4.8 | 0.01 | -0.6 | 0.03 | -1.2 | 0.00 | -1.8 | 0.00 | Glutathione metabolism; xenobiotic metabolism |
| Mm.37199 | Glutathione S-transferase, mu 3 (Gstm3) | 12.0 | 0.01 | 0.4 | 0.54 | 0.0 | 0.16 | 1.2 | 0.21 | Glutathione metabolism; xenobiotic metabolism |
| Mm.5731 | Glutathione S-transferase, theta 3 (Gstt3) | 5.1 | 0.04 | -1.8 | 0.07 | -2.4 | 0.15 | 1.3 | 0.01 | Glutathione metabolism |
| Mm.276389 | heme oxygenase (decycling) 1 (Hmox1) | 17.3 | 0.00 | 1.6 | 0.08 | 2.4 | 0.00 | 2.2 | 0.02 | Porphyrin and chlorophyll metabolism; xenobiotic metabolism |
| Mm.299381 | hypoxanthine guanine phosphoribosyl transferase 1 (Hprt1) | 2.8 | 0.00 | 0.6 | 0.54 | 1.3 | 0.00 | 1.1 | 0.12 | Purine metabolism |
| Mm.218286 | microsomal glutathione S-transferase 3 (Mgst3) | 5.8 | 0.00 | -0.5 | 0.05 | -2.7 | 0.00 | 1.1 | 0.04 | Glutathione metabolism; xenobiotic metabolism |
| Mm.344831 | UDP-glucose dehydrogenase (Ugdh) | 6.4 | 0.002 | 0.5 | 0.56 | -0.1 | 0.16 | -1.2 | 0.00 | Nucleotide sugar metabolism; starch and sucrose metabolism |
| Mm.312095 | UDP glucuronosyltransferase 2 family, polypeptide B35 (Ugt2b35) | 5.0 | 0.01 | 0.6 | 0.61 | -2.4 | 0.00 | -1.4 | 0.00 | Sugar metabolism |
| Intracellular communication/signal transduction b | ||||||||||
| Mm.18628 | CD36 antigen (Cd36) | 5.6 | 0.00 | -2.6 | 0.00 | -3.2 | 0.00 | -3.8 | 0.00 | Adipocytokine signaling pathway; PPAR signaling pathway |
| Mm.6529 | Dystrophia myotonica-protein kinase (Dmpk) | 5.0 | 0.00 | 1.0 | 0.97 | 1.3 | 0.06 | 1.5 | 0.00 | Protein amino acid phosphorylation; regulation of small GTPase mediated signal transduction |
| Mm.12967 | inhibitor of kappaB kinase gamma, transcript variant 2 (Ikbkg) | 5.1 | 0.02 | 1.2 | 0.01 | -1.6 | 0.00 | -1.1 | 0.00 | Activation of NF-kappaB-inducing kinase |
| Mm.294007 | PREDICTED: proprotein convertase subtilisin/kexin type 6, transcript variant 4 (Pcsk6) | 3.7 | 0.00 | 0.9 | 0.94 | -0.7 | 0.03 | 1.8 | 0.04 | Determination of left/right symmetry; transmembrane receptor protein tyrosine kinase signaling pathway |
| Mm.308180 | protein tyrosine phosphatase-like A domain containing 1 (Ptplad1) | 3.0 | 0.00 | 0.7 | 0.64 | 1.6 | 0.09 | 0.3 | 0.41 | I-kappaB kinase/NF-kappaB cascade; JNK cascade; Rho protein signal transduction |
| Cell cycle/Growth and differentiation b | ||||||||||
| Mm.20801 | HIV-1 tat interactive protein 2, homolog (human) (Htatip2) | 4.2 | 0.01 | -0.1 | 0.22 | -0.7 | 0.03 | 0.4 | 0.40 | Regulation of angiogenesis and apoptosis; cell differentiation |
| Mm.43831 | lectin, galactose binding, soluble 1 (Lgals1) | 6.0 | 0.01 | -1.4 | 0.03 | -1.1 | 0.00 | -1.5 | 0.00 | Myoblast differentiation; sugar binding |
| Mm.62876 | ZW10 interactor (Zwint) | 3.0 | 0.01 | -0.5 | 0.04 | -1.2 | 0.00 | 0.3 | 0.43 | cell cycle; cell division |
| Mm.290692 | transketolase (Tkt) | 2.6 | 0.00 | -0.5 | 0.03 | 0.5 | 0.40 | -1.2 | 0.00 | Metal ion binding; regulation of growth |
| Cellular transport and transport mechanism b | ||||||||||
| Mm.103777 | Rhesus blood group-associated B glycoprotein (Rhbg) | 4.8 | 0.02 | 0.1 | 0.24 | -0.8 | 0.03 | -1.1 | 0.00 | Ammonium transporter activity |
| Mm.222536 | solute carrier family 25 (mitochondrial carrier, peroxisomal membrane protein), member 17 (Slc25a17) | 2.4 | 0.00 | -0.9 | 0.02 | -1.9 | 0.00 | -5.8 | 0.00 | mitochondrial transport; transporter activity |
| Mm.281800 | Solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1 (Slc35d1) | 2.8 | 0.00 | 0.6 | 0.53 | 0.5 | 0.37 | -1.2 | 0.00 | Nucleotide-sugar transporter |
| Mm.218875 | target of myb1-like 2 (chicken), transcript variant 1 (Tom1l2) | 2.1 | 0.00 | -2.1 | 0.06 | 5.4 | 0.00 | 2.5 | 0.01 | Golgi vesicle-mediated transport; intracellular protein transport |
| Transcription/Translation/Protein modification b | ||||||||||
| Mm.298030 | synovial sarcoma, X member B, breakpoint 1 (Ssxb1) | 5.1 | 0.02 | -0.2 | 0.20 | 1.7 | 0.05 | -1.4 | 0.00 | Regulation of transcription |
| Mm.34483 | leucine rich repeat containing 35 (Lrrc35) | 3.2 | 0.00 | 1.4 | 0.03 | -0.1 | 0.25 | -0.4 | 0.12 | Metal ion binding; protein modification |
| Miscellaneous (mixed functions) b | ||||||||||
| Mm.234247 | Dimethylarginine dimethylaminohydrolase 1 (Ddah1) | 4.5 | 0.01 | -0.6 | 0.08 | 0.2 | 0.33 | 1.4 | 0.28 | nitric oxide biosynthesis; protein amino acid nitrosylation |
| Mm.26834 | EF hand domain family A1 (Efha1) | 2.4 | 0.00 | -1.3 | 0.00 | -0.4 | 0.15 | -1.3 | 0.00 | Calcium ion binding |
| Mm.182912 | growth hormone inducible transmembrane protein (Ghitm) | 2.2 | 0.00 | -0.8 | 0.07 | 1.1 | 0.09 | 1.1 | 0.10 | Interacts with FtsH |
| Mm.41665 | Glutamate receptor, ionotropic, N-methyl D-asparate-associated protein 1 (glutamate binding) (Grina) | 2.3 | 0.00 | 0.1 | 0.29 | 0.7 | 0.69 | 0.5 | 0.54 | Receptor activity |
| Mm.228797 | major vault protein (Mvp) | 3.2 | 0.01 | 0.6 | 0.57 | 1.5 | 0.62 | 1.1 | 0.00 | Calcium ion binding; ribonucleoprotein complex |
| Mm.252080 | PREDICTED: phosphogluconate dehydrogenase, transcript variant 1 (Pgd) | 3.0 | 0.00 | -2.0 | 0.00 | 0.5 | 0.45 | -1.2 | 0.00 | pentose-phosphate shunt, oxidative branch |
| Mm.293463 | pirin (Pir) | 2.2 | 0.05 | 1.2 | 0.01 | -0.1 | 0.18 | 0.2 | 0.46 | metal ion binding |
| Mm.173058 | secreted phosphoprotein 2 (Spp2) | 2.3 | 0.00 | 1.3 | 0.01 | 0.8 | 0.74 | 1.7 | 0.00 | Bone remodeling |
| Unknown b | ||||||||||
| Mm.22109 | RIKEN cDNA 2610204L23 gene (2610204L23Rik) | 2.3 | 0.00 | 0.2 | 0.34 | 1.6 | 0.01 | -0.8 | 0.14 | Unknown |
| Mm.100125 | SH3 domain binding glutamic acid-rich protein like 2 (Sh3bgrl2) | 3.5 | 0.03 | 0.8 | 0.85 | 0.6 | 0.55 | 0.5 | 0.62 | Unknown |
| Mm.102407 | testis expressed gene 2 (Tex2) | 2.4 | 0.00 | 0.7 | 0.61 | 0.0 | 0.20 | 0.3 | 0.39 | Unknown |
| Mm.289795 | ubiquitin-associated protein 1 (Ubap1) | 2.6 | 0.00 | 0.5 | 0.44 | 0.6 | 0.43 | -1.2 | 0.00 | Unknown |
| Mm.257762 | PREDICTED: WD repeat domain 43, transcript variant 9 (Wdr43) | 4.1 | 0.00 | 0.6 | 0.43 | -0.8 | 0.03 | 1.3 | 0.04 | Unknown |
Genes elevated ≥ 2.0 fold in ΔTrsp mice compared to Trsp mice with a P-value ≤ 0.05 were assessed and shown in the table. The corresponding transcript levels were also analyzed in A34, G37L and G37H replacement mice relative to Trsp mice. Gene Unigene Accession Number, gene description, fold change with P value, and gene function(s) are shown. Four control and four experimental animals were used for comparing ΔTrsp, A34 and G37L with Trsp, while three control and three experimental animals were used for comparing G37H with Trsp.
Genes are grouped into classes according to function.
Table 4.
Transcripts in reduced levels within liver of knockout mice relative to wild type mice a
| ΔTrsp vs Trsp | A34 vs Trsp | G37L vs Trsp | G37H vs Trsp | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| UniGene | Description | Folds | P value | Folds | P value | Folds | P value | Folds | P value | Function/Gene Ontology |
| Cellular transport and transport mechanism b | ||||||||||
| Mm.381860 | coatomer protein complex, subunit gamma 2 (Copg2) | -2.3 | 0.00 | -0.8 | 0.05 | -2.5 | 0.00 | -1.7 | 0.00 | Protein transport |
| Mm.261168 | Potassium inwardly-rectifying channel, subfamily J, member 9 (Kcnj9) | -2.3 | 0.00 | -0.5 | 0.08 | 1.1 | 0.01 | -1.7 | 0.00 | Ion transport; voltage-gated ion channel activity |
| Mm.283370 | Neurocalcin delta (Ncald) | -2.3 | 0.00 | -1.5 | 0.00 | 1.5 | 0.00 | 1.2 | 0.00 | Vesicle-mediated transport |
| Mm.296837 | sideroflexin 2 (Sfxn2) | -3.8 | 0.01 | 0.1 | 0.49 | -1.4 | 0.05 | 0.0 | 0.43 | Ion transport |
| Mm.250738 | solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 (Slc13a3) | -2.4 | 0.00 | -0.8 | 0.04 | -1.2 | 0.00 | 1.0 | 0.00 | Dicarboxylic acid transport; ion transport; symporter activity; |
| Mm.255586 | solute carrier organic anion transporter family, member 1a4 (Slco1a4) | -2.5 | 0.00 | 1.0 | 0.98 | -0.3 | 0.20 | -1.2 | 0.00 | Ion transport; organic anion transporter activity |
| Intracellular communication/Signal transduction b | ||||||||||
| Mm.376094 | Anti-Mullerian hormone (Amh) | -2.3 | 0.00 | -0.0 | 0.41 | -0.1 | 0.24 | -1.0 | 0.32 | Transforming growth factor beta receptor binding |
| Mm.271854 | Low density lipoprotein receptor-related protein 1 (Lrp1) | -2.3 | 0.00 | -0.1 | 0.23 | 0.0 | 0.18 | -1.3 | 0.00 | Negative regulation of Wnt receptor signaling pathway |
| Mm.6595 | mitogen activated protein kinase kinase kinase 5 (Map3k5) | -2.3 | 0.00 | -0.2 | 0.35 | 1.2 | 0.00 | 0.1 | 0.43 | Activation of MAPK activity |
| Mm.3810 | phosphatidylinositol 3-kinase, C2 domain containing, alpha polypeptide (Pik3c2a) | -2.2 | 0.00 | 0.4 | 0.58 | -0.7 | 0.23 | 1.6 | 0.00 | Intracellular signaling cascade |
| Metabolism b | ||||||||||
| Mm.46269 | insulin I (Ins1) | -2.3 | 0.00 | 1.7 | 0.02 | -1.1 | 0.00 | 2.1 | 0.01 | Carbohydrate metabolism |
| Mm.34459 | junctophilin 2 (Jph2) | -2.7 | 0.00 | 0.3 | 0.34 | -1.8 | 0.00 | -1.2 | 0.00 | Carbohydrate metabolism |
| Mm.19669 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (Pfkfb3) | -2.3 | 0.05 | -2.3 | 0.19 | 0.7 | 0.60 | 2.1 | 0.01 | Fructose and mannose metabolism |
| Mm.9277 | phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) (Pla2g7) | -2.8 | 0.00 | 2.2 | 0.14 | 0.7 | 0.60 | 1.1 | 0.03 | Lipid catabolism |
| Cellular structural organization b | ||||||||||
| Mm.49994 | hook homolog 1 (Drosophila) (Hook1) | -3.1 | 0.00 | 0.4 | 0.59 | 0.0 | 0.19 | -1.1 | 0.00 | Cytoskeleton organization and biogenesis; spermatid development |
| Mm.129840 | phosphodiesterase 4D interacting protein transcript variant 1 (Pde4dip), | -2.4 | 0.00 | 0.5 | 0.56 | -1.2 | 0.00 | 1.0 | 0.21 | Association with spindle pole body microtubules |
| Mm.9684 | vasodilator-stimulated phosphoprotein (Vasp) | -2.0 | 0.00 | 1.2 | 0.01 | -1.2 | 0.00 | -1.1 | 0.00 | Actin cytoskeleton organization and biogenesis |
| Transcription/Translation b | ||||||||||
| Mm.174044 | RIKEN cDNA 2700050L05 gene, transcript variant 2 (2700050L05Rik) | -2.0 | 0.00 | -1.2 | 0.00 | -1.4 | 0.00 | -1.8 | 0.00 | Transcription regulation |
| Mm.2645 | eukaryotic translation elongation factor 1 alpha 2 (Eef1a2) | -2.4 | 0.00 | -2.4 | 0.02 | -1.8 | 0.00 | 1.3 | 0.03 | Protein biosynthesis; translational elongation |
| Mm.303355 | One cut domain, family member 1 (Onecut1) | -4.0 | 0.04 | -1.8 | 0.09 | -2.1 | 0.00 | -2.5 | 0.00 | Regulation of transcription |
| Mm.7916 | Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) (Paf1) | -2.7 | 0.00 | 0.7 | 0.77 | -0.9 | 0.16 | 0.0 | 0.31 | Regulation of transcription |
| Mm.244820 | TATA box binding protein (Tbp) | -2.6 | 0.00 | 0.4 | 0.45 | 1.2 | 0.00 | -1.4 | 0.00 | Regulation of transcription |
| Miscellaneous (Mixed functions) b | ||||||||||
| Mm.28796 | RIKEN cDNA 5730446C15 gene, (5730446C15Rik) | -2.1 | 0.00 | 0.7 | 0.74 | -0.5 | 0.24 | 1.7 | 0.00 | Peptidase activity |
| Mm.71924 | ankyrin repeat domain 38 (Ankrd38) | -2.3 | 0.00 | -0.7 | 0.01 | -1.3 | 0.00 | -1.1 | 0.00 | Protein-protein interaction |
| Mm.119936 | attractin (Atrn) | -2.0 | 0.00 | 2.2 | 0.01 | -1.1 | 0.00 | -1.7 | 0.00 | Inflammatory response; protein binding; sugar binding |
| Mm.212861 | cullin 4A (Cul4a) | -2.2 | 0.00 | 1.7 | 0.62 | -1.4 | 0.00 | -1.1 | 0.00 | Cell cycle; induction of apoptosis by intracellular signals; ubiquitin cycle |
| Mm.171323 | nuclear receptor coactivator 6 interacting protein (Ncoa6ip) | -2.4 | 0.00 | -0.8 | 0.03 | -0.4 | 0.17 | -0.6 | 0.14 | S-adenosylmethionine-dependent methyltransferase activity; protein binding |
| Mm.261818 | Ring finger protein 167 (Rnf167) | -2.8 | 0.00 | 1.2 | 0.91 | -1.5 | 0.00 | -1.5 | 0.00 | Metal ion binding; protein binding; peptidase activity |
| Mm.284592 | SUMO/sentrin specific peptidase 3 (Senp3) | -2.2 | 0.00 | 0.2 | 0.35 | -0.6 | 0.04 | 0.3 | 0.43 | Peptidase activity; protein metabolism; ubiquitin-specific protease activity |
| Unknown b | ||||||||||
| Mm.416885 | RIKEN cDNA 2010109N18 gene | -2.9 | 0.00 | 0.3 | 0.36 | 3.5 | 0.27 | -1.4 | 0.00 | Unknown |
| Mm.138091 | PREDICTED: RIKEN cDNA 3110001I20 gene, transcript variant 1 (3110001I20Rik) | -2.3 | 0.00 | -0.9 | 0.05 | -1.7 | 0.10 | -0.9 | 0.13 | Unknown |
| Mm.395958 | RIKEN cDNA 9130221J18 gene | -4.9 | 0.00 | -0.3 | 0.14 | 0.3 | 0.52 | 1.4 | 0.01 | Unknown |
| Mm.395042 | RIKEN cDNA 9430078K24 gene | -2.1 | 0.00 | -0.3 | 0.13 | -1.2 | 0.00 | 1.8 | 0.00 | Unknown |
| Mm.425114 | PREDICTED: similar to gonadotropin inducible ovarian transcription factor 1, transcript variant 3 (LOC664985) | -2.9 | 0.00 | 0.2 | 0.35 | 0.1 | 0.24 | -1.2 | 0.00 | Unknown |
| Mm.219946 | SET and MYND domain containing 5 (Smyd5) | -2.7 | 0.00 | 0.0 | 0.21 | -1.1 | 0.00 | -2.0 | 0.00 | Unknown |
Genes depleted ≥ 2.0 fold in ΔTrsp mice as compared to Trsp mice with a P-value ≤ 0.05 were assessed and shown in the table. The corresponding transcript levels were also analyzed in A34, G37L and G37H replacement mice relative to Trsp mice. Gene Unigene Accession Number, gene description, fold change with P value, and gene function(s) are shown. Four control and four experimental animals were used for comparing ΔTrsp, A34 and G37L with Trsp, while three control and three experimental animals were used for comparing G37H with Trsp.
Genes are grouped into classes according to function.
Genes elevated or repressed in ΔTrsp mice in comparison to Trsp mice were grouped according to their functions, while the transcript levels of corresponding genes in A34, G37L and G37H transgenic mice were analyzed relative to Trsp and represented in Tables 3 and 4. Genes elevated in ΔTrsp (Table 3) were mainly involved in detoxification, stress response, xenobiotic metabolism, intracellular communication, cellular transport and cell growth and differentiation. Some genes that were significantly upregulated in ΔTrsp mice are involved in detoxification and xenobiotic metabolism and include epoxide hydrolase 1 (Ephx1); carboxylesterase 1 and 2 (Ces1, Ces2); cytochrome P450, family 2, subfamily a, polypeptide 5 (Cyp2a5) members of glutathione S-transferase family (Gst); haem oxygenase 1 (Hmox1) and aldehyde oxidase 1 (Aox1). Interestingly, the levels of expression of these genes in A34, G37L and G37H mice were similar to Trsp.
Genes that were repressed in ΔTrsp mice and compared to Trsp mice were grouped in a similar manner as those manifesting enhanced expression and their relative transcript levels measured in transgenic mice (Table 4). While most of the genes downregulated in ΔTrsp mice had diverse or unknown functions, some of them could be grouped as being involved in transcription, intracellular communication and cellular transport.
Quantitative real time PCR validation of elevated genes
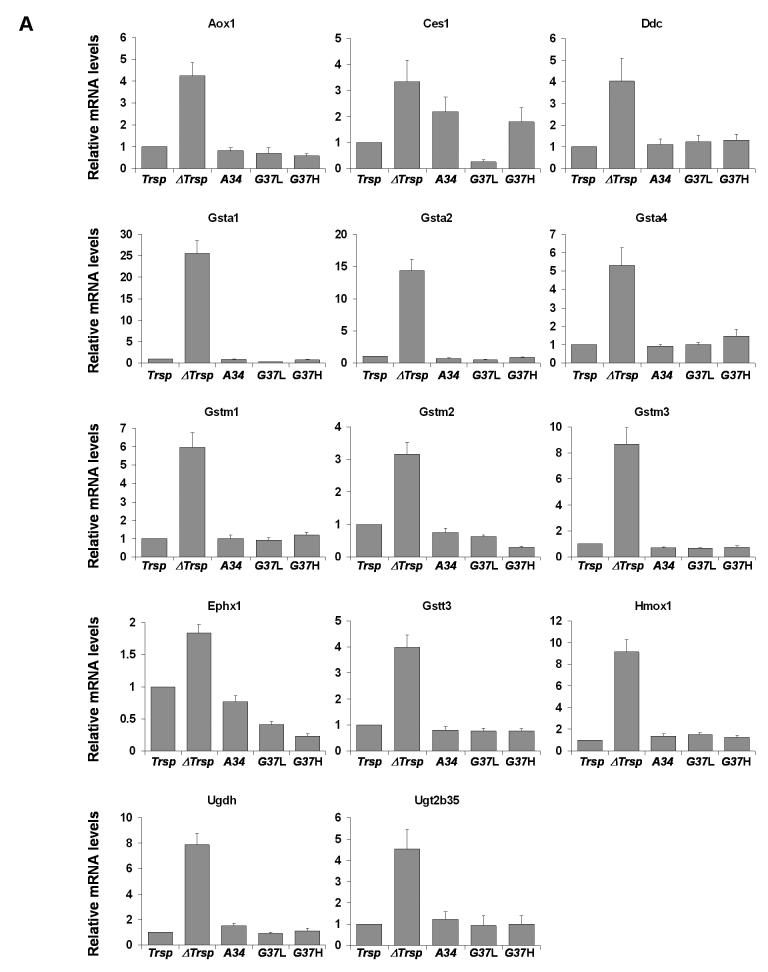
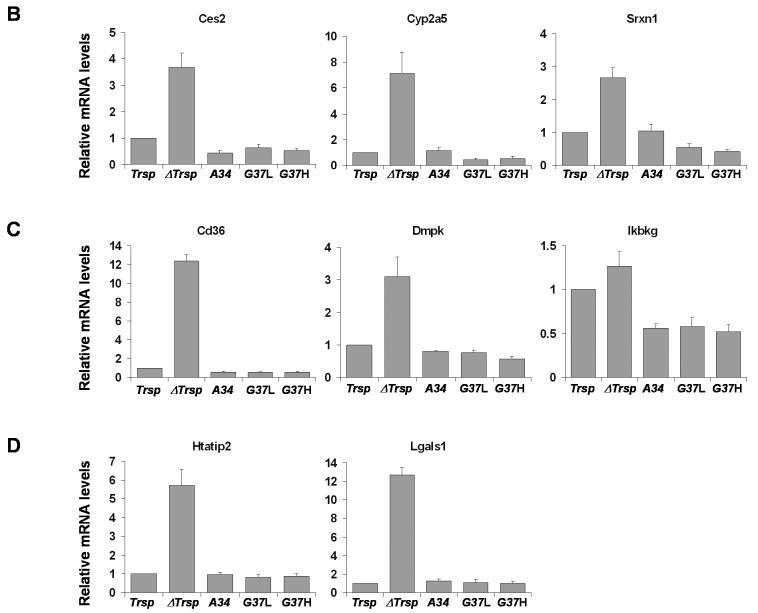
The expression levels of 22 genes elevated in ΔTrsp mice were verified by Q-PCR (Figure 2) and were in excellent agreement with the microarray analysis. Expression of the corresponding transcripts in transgenic mice was similar to that in Trsp mice. The genes analyzed by Q-PCR were grouped according to their function (Table 3) as (A) metabolism, (B) defense stress and detoxification, (C) intracellular communication/signal transduction, and (D) cell cycle/growth and differentiation (Figure 2).
Figure 2. Quantitative PCR of upregulated genes following Trsp removal in liver.
The relative expression of genes upregulated in ΔTrsp mice (Table 3) was examined by Q-PCR, normalized to the expression of Gusb as described in Experimental Procedures. The normalized value for each mRNA in liver of ΔTrsp, A34, G37L and G37H mice was then compared to Trsp mice and plotted along with error bars. Upregulated genes analyzed are grouped according to their function (see Table 3): (A) metabolism; (B) defense stress and detoxification; (C) intracellular communication/signal transduction; and (D) cell cycle/growth and differentiation. Results represent 3-4 independent experiments, each carried out in triplicate.
Expression of phase II enzymes
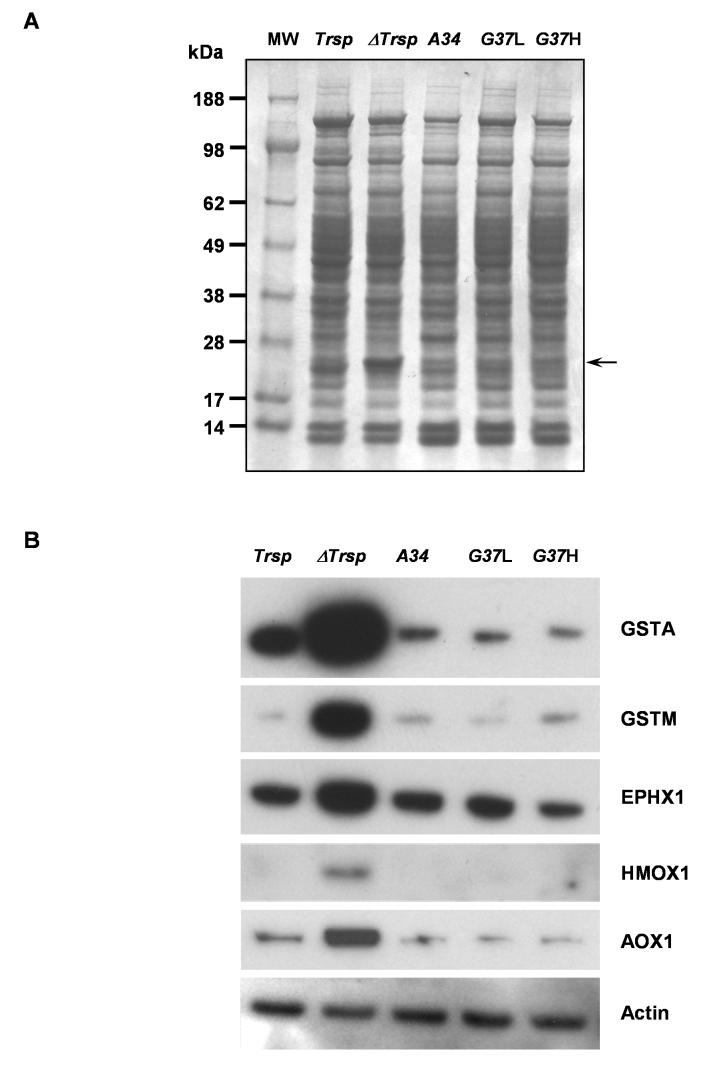
Protein expression profiles from the five mouse lines appeared similar in liver samples as observed on Coomassie Blue stained gels with the exception of a prominently enriched band of approximately 25 kDa in ΔTrsp mice (Figure 3A, indicated by the arrow). This observation was also noted in an earlier study and the elevated band was sequenced and identified as glutathione S-transferase (GST) [12]. As expected, the mRNA levels of the Gst isoforms were also increased (Figure 2). To verify that the induced mRNA levels also gave rise to a consequential increase in the corresponding protein levels, we analyzed the amounts of several phase II enzymes by western blotting (Figure 3B). Indeed, a marked increase in two of the GST isoforms, GSTA and GSTM, were observed in ΔTrsp mice as compared to Trsp. Furthermore, several other Phase II proteins, EPHX1, HMOX1 and AOX1, were increased in ΔTrsp mice compared to Trsp. The increase in amounts of these proteins in ΔTrsp mice paralleled their induced mRNA levels. Most interestingly, the protein levels of these enzymes in the transgenic mice were virtually the same as in Trsp mice providing strong evidence that the link between enhanced Phase II protein expression and loss of selenoprotein expression is due to the absence of one or more housekeeping selenoproteins. These observations are further considered in the Discussion. β-actin was examined by western blotting as a control protein and its level was unaffected in the five mouse lines (see lower panel in Figure 3B).
Figure 3. Western blot analysis of phase II enzymes.
Protein extracts were prepared from liver of Trsp, ΔTrsp, A34, G37L and G37H mice and electrophoresed on 10% polyacrylamide gels. (A) Coomassie Blue stained gel. The elevated band corresponding to GST in ΔTrsp mice is indicated by an arrow on the right side of the gel and molecular weight (MW) markers were run in lane 1 and their sizes indicated on the left. (B) The proteins were transferred to PVDF membranes and the membranes probed with antibodies specific for the indicated phase II enzymes and β-actin as described in Experimental Procedures. β-Actin served as a loading control.
DISCUSSION
Biochemical and in silico studies have identified 25 selenoprotein genes in humans and 24 in mice [3, 14]. The functions of many of these selenoproteins have not been identified and most that have been characterized serve as oxidoreductases associated with various metabolic pathways, e.g., free radical scavenging, maintenance of intracellular redox status, and repair of oxidized methionine residues [15, 16].
Our previous studies have shown that selective knockout of Trsp in mouse hepatocytes resulted in the virtual absence of selenoproteins in liver, and a pronounced reduction in selenium levels even though the low molecular weight selenocompounds were little affected [12]. These data demonstrated that selenoproteins are essential for proper liver function and their absence causes severe necrosis and hepatocellular degeneration, accompanied by necrosis of peritoneal and retroperitoneal fat [12]. Subsequently, we replaced the selenoprotein population in this knockout mouse with either one of two mutant transgenes that produce tRNA gene products lacking i6A and Um34, or mcm5U and Um34, respectively, demonstrating that, while most of the selenoproteins were absent or diminished in the knockout mice, some were selectively replaced in the transgenic mice [8]. These replaced selenoproteins were housekeeping selenoproteins which are essential for liver function [8]. To assess the consequences of selenoprotein loss in ΔTrsp mice and their subsequent partial replacement with mutant transgenes, we examined gene expression in Trsp, ΔTrsp, A34, G37L and G37H transgenic mice by microarray. These analyses showed an elevated expression of several members of the phase II enzyme family in ΔTrsp mice. This change was validated through Q-PCR and western blotting of the corresponding proteins. Several major phase II response genes that were upregulated in ΔTrsp mice included Gsta1, Gsta2, Gsta4, Gstm1, Gstm2, Gstm3, Cyp2a5, Ephx1, Hmox1 and Aox1.
Phase II enzymes conjugate xenobiotics or Phase I products to small donor molecules, such as glutathione, making them water soluble and easily excretable from the body, thus assisting in chemoprotection and detoxification [17]. They can be induced in animals by (a) chemical compounds which can react with a sulfhydryl group; (b) regulation of common promoter elements (e.g., antioxidant responsive element or ARE); and (c) reactions leading to catalysis of electrophiles and reactive oxygen species (ROS) [reviewed in 17]. Induction of phase II enzymes in tissues has been shown to protect against carcinogens [18]. GST isozymes conjugate electrophilic compounds to glutathione, thus preventing their interaction with DNA [19], whereas Ephx1 is a bifunctional protein, that metabolizes polycyclic aromatic hydrocarbons [20] and mediates sodium-dependent uptake of bile acids [21]. HMOX1 is a cytoprotective enzyme, which degrades haem to biliverdin, which is further reduced to bilirubin [22] with both biliverdin and bilirubin acting as antioxidants [23]. CYP2A5 metabolizes toxic xenobiotic compounds, such as nitrosamines and aflatoxins [24, 25], takes part in the degradation of bilirubin [26] and is induced during hepatic pathogenesis [27]. AOX1 is a molybdenum containing flavoprotein which plays an important role in ethanol-induced hepatic lipoperoxidation [28]. The expression of AOX1 may determine the susceptibility of liver cells to some pharmacologic agents and the levels of ROS produced under certain pathophysiological conditions [29].
The effect of dietary selenium on hepatic chemoprotective enzymes or xenobiotic enzymes in rodents have been extensively studied over the last few decades and results indicated a role of this element in the regulation of several phase II enzymes, including GST [30, 31], epoxide hydrolase [32] and haem oxygenase 1 [33]. Deficiency of selenium has been associated with an increase in these enzymes in rodents [30-33]. Our results indicate a similar elevation in the levels of phase II enzymes in Trsp knockout mice, suggesting this phenomenon to be a result of loss of selenoproteins rather than a reduction in dietary selenium. Interestingly, the levels of phase II response genes were normal when housekeeping selenoproteins were replaced in transgenic mice providing strong evidence of their upregulation being a consequence of deficiency in housekeeping selenoproteins. In contrast, reduced expression of stress-related selenoproteins, such as GPx1 or SELR, had no role in upregulation of phase II enzymes. An earlier study reported that inhibition of thioredoxin reductase (TR) by aurothioglucose leads to induction of hepatic HMOX1 activity [34]. These investigators postulated that the lack of TR, or a TR related reaction, induces hepatic haem oxygenase 1. GPx and GST are both responsible for detoxifications of xenobiotic electrophiles by the addition of reduced glutathione (GSH) and possess similar enzyme folds in the GSH-binding site. Earlier studies have demonstrated that the alpha-class GST isoenzymes also exhibit selenium-independent glutathione peroxidase activity in rodents [35, 36], and these isozymes are very effective at reducing hydroperoxides, thus providing protection against membrane lipid peroxidation [37]. In the liver of ΔTrsp mice, the elevated levels of GST might functionally compensate for GPx and/or another selenoprotein or selenoproteins that might also be involved in detoxification.
The present study shows that an interplay exists between the loss of one or more housekeeping selenoproteins and enrichment in members of the phase II response protein class. The fact that several members of the phase II protein class manifesting a wide variety of functions are upregulated suggests that several members of the housekeeping selenoprotein class are likely involved in this interplay. Thus, our data provide strong evidence of a functional link between housekeeping selenoproteins and phase II enzymes.
Acknowledgements
This work was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health (D. L. H), and by National Institutes of Health grants (V. N. G.).
Abbreviations used
- AOX
aldehyde oxidase
- EPHX
epoxide hydrolase
- GPx
glutathione peroxidase
- GST
glutathione transferase
- GSTA
GST Alpha
- GSTM
GST Mu
- HMOX
haem oxygenase
- HRP
horseradish peroxidase
- i6A
N6-isopentenyladenosine
- mcm5U
5′-methylcarboxylmethyluracil
- mcm5Um
5′-methylcarboxymethyl-2′-O-methyluridine
- Q-PCR
quantitative real-time PCR
- ROS
reactive oxygen species
- RT
reverse transcriptase
- Sec
selenocysteine
- TR
thioredoxin reductase
- Um34
2′-O-methylribose at position 34 in selenocysteine tRNA
REFERENCES
- 1.Hatfield DL, Berry MJ, Gladyshev VN, editors. Selenium: Its Molecular Biology and Role in Human Health. Springer Science Business Media LLC; New York, NY: 2006. [Google Scholar]
- 2.Hatfield DL, Gladyshev VN. How selenium has altered our understanding of the genetic code. Mol. Cell Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 4.Berry MJ. Insights into the hierarchy of selenium incorporation. Nat. Genet. 2005;37:1162–1163. doi: 10.1038/ng1105-1162. [DOI] [PubMed] [Google Scholar]
- 5.Driscoll DM, Copeland PR. Mechanism and regulation of selenoprotein synthesis. Annu. Rev. Nutr. 2003;23:17–40. doi: 10.1146/annurev.nutr.23.011702.073318. [DOI] [PubMed] [Google Scholar]
- 6.Kim LK, Matsufuji T, Matsufuji S, Carlson BA, Kim SS, Hatfield DL, Lee BJ. Methylation of the ribosyl moiety at position 34 of selenocysteine tRNA[Ser]Sec is governed by both primary and tertiary structure. RNA. 2000;6:1306–1315. doi: 10.1017/s1355838200000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlson BA, Xu XM, Gladyshev VN, Hatfield DL. Selective rescue of selenoprotein expression in mice lacking a highly specialized methyl group in selenocysteine tRNA. J. Biol. Chem. 2005;280:5542–5548. doi: 10.1074/jbc.M411725200. [DOI] [PubMed] [Google Scholar]
- 8.Carlson BA, Moustafa ME, Sengupta A, Schweizer U, Shrimali R, Rao M, Zhong N, Wang S, Feigenbaum L, Lee BJ, Gladyshev VN, Hatfield DL. Selective restoration of the selenoprotein population in a mouse hepatocyte selenoproteinless background with different mutant selenocysteine tRNAs lacking Um34. J. Biol. Chem. 2007;282:32591–32602. doi: 10.1074/jbc.M707036200. [DOI] [PubMed] [Google Scholar]
- 9.Bosl MR, Takaku K, Oshima M, Nishimura S, Taketo MM. Early embryonic lethality caused by targeted disruption of the mouse selenocysteine tRNA gene (Trsp) Proc. Natl. Acad. Sci. U.S.A. 1997;94:5531–5534. doi: 10.1073/pnas.94.11.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumaraswamy E, Carlson BA, Morgan F, Miyoshi K, Robinson GW, Su D, Wang S, Southon E, Tessarollo L, Lee BJ, Gladyshev VN, Hennighausen L, Hatfield DL. Selective removal of the selenocysteine tRNA [Ser]Sec gene (Trsp) in mouse mammary epithelium. Mol. Cell Biol. 2003;23:1477–1488. doi: 10.1128/MCB.23.5.1477-1488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatfield DL, Carlson BA, Xu XM, Mix H, Gladyshev VN. Selenocysteine incorporation machinery and the role of selenoproteins in development and health. Prog. Nucleic Acid Res. Mol. Biol. 2006;81:97–142. doi: 10.1016/S0079-6603(06)81003-2. [DOI] [PubMed] [Google Scholar]
- 12.Carlson BA, Novoselov SV, Kumaraswamy E, Lee BJ, Anver MR, Gladyshev VN, Hatfield DL. Specific excision of the selenocysteine tRNA[Ser]Sec (Trsp) gene in mouse liver demonstrates an essential role of selenoproteins in liver function. J. Biol. Chem. 2004;279:8011–8017. doi: 10.1074/jbc.M310470200. [DOI] [PubMed] [Google Scholar]
- 13.Moustafa ME, Carlson BA, El-Saadani MA, Kryukov GV, Sun QA, Harney JW, Hill KE, Combs GF, Feigenbaum L, Mansur DB, Burk RF, Berry MJ, Diamond AM, Lee BJ, Gladyshev VN, Hatfield DL. Selective inhibition of selenocysteine tRNA maturation and selenoprotein synthesis in transgenic mice expressing isopentenyladenosine-deficient selenocysteine tRNA. Mol. Cell Biol. 2001;21:3840–3852. doi: 10.1128/MCB.21.11.3840-3852.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Driscoll DM, Chavatte L. Finding needles in a haystack. In silico identification of eukaryotic selenoprotein genes. EMBO Rep. 2004;5:140–141. doi: 10.1038/sj.embor.7400080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gromer S, Eubel JK, Lee B,L, Jacob J. Human selenoproteins at a glance. Cell Mol. Life Sci. 2005;62:2414–2437. doi: 10.1007/s00018-005-5143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rederstorff M, Krol A, Lescure A. Understanding the importance of selenium and selenoproteins in muscle function. Cell Mol. Life Sci. 2006;2006;63:52–59. doi: 10.1007/s00018-005-5313-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Talalay P. Chemoprotection against cancer by induction of phase 2 enzymes. Biofactors. 2000;12:5–11. doi: 10.1002/biof.5520120102. [DOI] [PubMed] [Google Scholar]
- 18.Kwak MK, Egner PA, Dolan PM, Ramos-Gomez M, Groopman JD, Itoh K, Yamamoto M, Kensler TW. Role of phase 2 enzyme induction in chemoprotection by dithiolethiones. Mutat. Res. 2001;480-481:305–315. doi: 10.1016/s0027-5107(01)00190-7. [DOI] [PubMed] [Google Scholar]
- 19.Hayes JD, Pulford DJ. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit. Rev. Biochem. Mol. Biol. 1995;30:445–600. doi: 10.3109/10409239509083491. [DOI] [PubMed] [Google Scholar]
- 20.Fretland AJ, Omiecinski CJ. Epoxide hydrolases: biochemistry and molecular biology. Chem. Biol. Interact. 2000;129:41–59. doi: 10.1016/s0009-2797(00)00197-6. [DOI] [PubMed] [Google Scholar]
- 21.von DP, Amoui M, Alves C, Levy D. Na(+)-dependent bile acid transport by hepatocytes is mediated by a protein similar to microsomal epoxide hydrolase. Am. J. Physiol. 1993;264:G528–G534. doi: 10.1152/ajpgi.1993.264.3.G528. [DOI] [PubMed] [Google Scholar]
- 22.Otterbein LE, Choi AM. Heme oxygenase: colors of defense against cellular stress. Am. J. Physiol. Lung Cell. Mol. Physiol. 2000;279:L1029–L1037. doi: 10.1152/ajplung.2000.279.6.L1029. [DOI] [PubMed] [Google Scholar]
- 23.Wu TW, Carey D, Wu J, Sugiyama H. The cytoprotective effects of bilirubin and biliverdin on rat hepatocytes and human erythrocytes and the impact of albumin. Biochem. Cell Biol. 1991;69:828–834. doi: 10.1139/o91-123. [DOI] [PubMed] [Google Scholar]
- 24.Camus AM, Geneste O, Honkakoski P, Bereziat JC, Henderson CJ, Wolf CR, Bartsch H, Lang MA. High variability of nitrosamine metabolism among individuals: role of cytochromes P450 2A6 and 2E1 in the dealkylation of N-nitrosodimethylamine and N-nitrosodiethylamine in mice and humans. Mol. Carcinog. 1993;7:268–275. doi: 10.1002/mc.2940070410. [DOI] [PubMed] [Google Scholar]
- 25.Pelkonen P, Lang MA, Negishi M, Wild CP, Juvonen RO. Interaction of aflatoxin B1 with cytochrome P450 2A5 and its mutants: correlation with metabolic activation and toxicity. Chem. Res. Toxicol. 1997;10:85–90. doi: 10.1021/tx960078m. [DOI] [PubMed] [Google Scholar]
- 26.Abu-Bakar A, Moore MR, Lang MA. Evidence for induced microsomal bilirubin degradation by cytochrome P450 2A5. Biochem. Pharmacol. 2005;70:1527–1535. doi: 10.1016/j.bcp.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 27.Camus-Randon AM, Raffalli F, Bereziat JC, McGregor D, Konstandi M, Lang MA. Liver injury and expression of cytochromes P450: evidence that regulation of CYP2A5 is different from that of other major xenobiotic metabolizing CYP enzymes. Toxicol. Appl. Pharmacol. 1996;138:140–148. doi: 10.1006/taap.1996.0107. [DOI] [PubMed] [Google Scholar]
- 28.Shaw S, Jayatilleke E. The role of aldehyde oxidase in ethanol-induced hepatic lipid peroxidation in the rat. Biochem. J. 1990;268:579–583. doi: 10.1042/bj2680579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neumeier M, Weigert J, Schaffler A, Weiss TS, Schmidl C, Buttner R, Bollheimer C, Aslanidis C, Scholmerich J, Buechler C. Aldehyde oxidase 1 is highly abundant in hepatic steatosis and is downregulated by adiponectin and fenofibric acid in hepatocytes in vitro. Biochem. Biophys. Res. Commun. 2006;350:731–735. doi: 10.1016/j.bbrc.2006.09.101. [DOI] [PubMed] [Google Scholar]
- 30.Christensen MJ, Nelson BL, Wray CD. Regulation of glutathione S-transferase gene expression and activity by dietary selenium. Biochem. Biophys. Res. Commun. 1994;202:271–277. doi: 10.1006/bbrc.1994.1923. [DOI] [PubMed] [Google Scholar]
- 31.Liu JZ, Zhang BZ, Milner JA. Dietary selenite modifies glutathione metabolism and 7,12-dimethylbenz(a)anthracene conjugation in rats. J. Nutr. 1994;124:172–180. doi: 10.1093/jn/124.2.172. [DOI] [PubMed] [Google Scholar]
- 32.Reddy CC, Thomas CE, Scholz RW, Massaro EJ. Effects of inadequate vitamin E and/or selenium nutrition on enzymes associated with xenobiotic metabolism. Biochem. Biophys. Res. Commun. 1982;107:75–81. doi: 10.1016/0006-291x(82)91671-0. [DOI] [PubMed] [Google Scholar]
- 33.Mostert V, Hill KE, Ferris CD, Burk RF. Selective induction of liver parenchymal cell heme oxygenase-1 in selenium-deficient rats. Biol. Chem. 2003;384:681–687. doi: 10.1515/BC.2003.076. [DOI] [PubMed] [Google Scholar]
- 34.Mostert V, Hill KE, Burk RF. Loss of activity of the selenoenzyme thioredoxin reductase causes induction of hepatic heme oxygenase-1. FEBS Lett. 2003;541:85–88. doi: 10.1016/s0014-5793(03)00309-0. [DOI] [PubMed] [Google Scholar]
- 35.Hurst R, Bao Y, Jemth P, Mannervik B, Williamson G. Phopholipid hydroperoxide glutathione peroxidase activity of human glutathione transferases. Biochem. J. 1998;332:97–100. doi: 10.1042/bj3320097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Y, Sharma R, Zimniak P, Awasthi YC. Role of alpha class glutathione S-transfe-rase as antioxidant enzymes in rodent tissues. Toxicol. Appl. Pharmacol. 2002;182:105–115. doi: 10.1006/taap.2002.9450. [DOI] [PubMed] [Google Scholar]
- 37.Prabhu KS, Reddy PV, Gumpricht E, Hildenbrandt GR, Scholtz RW, Sordillo LM, Reddy CC. Microsomal glutathione S-transferase A1-1 with glutathione peroxidase activity from sheep liver: molecular cloning, expression and characterization. Biochem. J. 2001;360:345–354. doi: 10.1042/0264-6021:3600345. [DOI] [PMC free article] [PubMed] [Google Scholar]