Abstract
An understanding of intermolecular interactions is essential for insight into how cells develop, operate, communicate and control their activities. Such interactions include several components: contributions from linear, angular, and torsional forces in covalent bonds, van der Waals forces, as well as electrostatics. Among the various components of molecular interactions, electrostatics are of special importance because of their long range and their influence on polar or charged molecules, including water, aqueous ions, and amino or nucleic acids, which are some of the primary components of living systems. Electrostatics, therefore, play important roles in determining the structure, motion and function of a wide range of biological molecules. This chapter presents a brief overview of electrostatic interactions in cellular systems with a particular focus on how computational tools can be used to investigate these types of interactions.
I. Introduction
An understanding of intermolecular interactions is essential for insight into how cells develop, operate, communicate and control their activities. Such interactions include several components: contributions from linear, angular, and torsional forces in covalent bonds, van der Waals forces, as well as electrostatics. Among the various components of molecular interactions, electrostatics are of special importance because of their long range and their influence on polar or charged molecules, including water, aqueous ions, and amino or nucleic acids, which are some of the primary components of living systems. Electrostatics, therefore, play important roles in determining the structure, motion and function of a wide range of biological molecules; for example, in ion-induced RNA folding (Cole et al., 1972; Römer and Hach, 1975; Stein and Crothers, 1976), low pH protein unfolding, electrostatic steering effects on biomolecule-small molecule binding, protein-biomembrane interactions (Wang et al., 2001a; Rauch et al., 2002; Wang et al., 2002), protein-nucleic acid binding (Bajaj et al., 1990; Record et al., 1991; Senear and Batey, 1991; Overman and Lohman, 1994), etc. This chapter presents a brief overview of electrostatic interactions in cellular systems (Sec. II) with a particular focus on how computational tools can be used to investigate these types of interactions (Secs. III and IV).
II. Electrostatics in cellular systems
Electrostatic interactions are ubiquitous for any system of charged or polar molecules, such as biomolecules in their aqueous environment. For example, proteins are made up of 20 types of amino acids, 11 of which are charged or polar in neutral solution. Nucleic acids contain long stretches of negative charges from the phosphate groups in nucleotides. Finally, sugars and related glycosaminoglycans can possess some of the highest charge densities of any biomolecules due to the presence of numerous negative functionalities, including carboxylate and sulfate groups. We will focus on a few specific examples of electrostatic interactions in cellular systems: biomolecule-ion, biomolecule-ligand, and biomolecule-biomolecule interactions. Each of these interactions will be discussed in more detail in the following sections.
II.A. Biomolecule-ion interactions
In cellular settings, biomolecules are immersed in solution along with water, ions, and numerous other small molecules and macromolecules. Ions influence biomolecular processes and interactions in several different ways, including long-range screening, site-specific ion binding, and preferential hydration effects. Long-range screening is a phenomenon in which the strength of electrostatic interactions within and between biomolecules is reduced by the presence of aqueous ions. This is a non-specific ion effect and is described well, at low salt charge and concentration, by Debye-Hückel theory (Debye and Hückel, 1923) and the related implicit solvent models described in Sec. III.B. In site-specific ion binding, ions interact with biomolecules by binding to specific sites in a manner similar to ligand binding (see Sec. II.B) (Draper et al., 2005). Preferential hydration or Hofmeister effects are species-specific competitions between ions and water for binding to non-specific sites on biomolecules (Hofmeister, 1888; Boström et al., 2006; Collins, 2006). This competition is between weak biomolecule-solvent and biomolecule-ion interactions and therefore observed only at very high salt concentrations (Eisenberg, 1976; Anderson and Record Jr., 2004). A similar effect involves competition between ionic species around charged biomolecules (Moore and Lohman, 1994; Reuter et al., 2005). Note that, although these effects can be important, preferential hydration and ion-ion competition are not routinely considered in simulations, mainly due to limitations in current computational methodology. While there is active work in improving the theoretical and computational treatment of these effects (Boström et al., 2003; Shimizu, 2004; Shimizu and Smith, 2004; Broering and Bommarius, 2005; Zhou, 2005; Boström et al., 2006), they are currently beyond the scope of this chapter.
Ion-induced RNA folding (Römer and Hach, 1975; Stein and Crothers, 1976; Cech and Bass, 1986; Dahm and Uhlenbeck, 1991; Misra and Draper, 2000, 2001; Draper et al., 2005) provides an excellent example of many of the ion-biomolecule interactions discussed above. RNA folding in the absence of salt is quite unfavorable due to a number of negative charges along its phosphodiester backbone. Bringing these negative charges together into a compact structure introduces a large energetic barrier to RNA folding. Positive ions promote folding by reducing the repulsion between these negative charges. However, some ions are more effective than others; for example, millimolar concentrations of Mg2+ can stabilize RNA tertiary structures that are only marginally stable in solution with a high concentration of monovalent cations, such as Na+ or K+ (Cole et al., 1972; Romer, 1975; Stein and Crothers, 1976). Accurately modeling the ion-RNA interactions is essential to explain this phenomenon. A major obstacle in modeling ion-RNA interactions is the presence of numerous different ion environments (Draper et al., 2005). Each environment is dominated by the different type of ion-biomolecule interactions described above and requires different approaches to evaluating the energies. For example, experimental results for Mg2+ effects on tRNAPhe folding can be modeled successfully while only considering long-range screening effects (Misra and Draper, 2000). However, the diffusive Mg2+ ion description provided by this model is not sufficient to describe folding of a 58-nt rRNA fragment. Instead, one Mg2+ ion must be explicitly included at a specific binding site (Misra and Draper, 2001). A comprehensive theoretical framework of ion-RNA interactions that accounts for the overall ion dependence of RNA folding is the aim of current RNA folding studies (Draper et al., 2005).
II.B. Biomolecule-ligand and -biomolecule interactions
Biomolecule-substrate recognition is central to nearly all biomolecular processes, including signal transduction, enzyme cooperativity, and metabolic regulation. The bimolecular binding process, from a kinetic perspective, can be reduced to two steps: diffusional association to form an initial encounter complex and non-diffusional rearrangement to form the fully bound complex. The diffusional association places an upper limit on the overall binding rate; so-called “perfect” enzymes operate at this diffusion-limited rate. Electrostatic forces have an important influence on biomolecular diffusional association: their long-range nature enables them to attract the substrate to its binding partner and orient the substrate properly for binding (Gabdoulline and Wade, 2002). It has been established that, for many biomolecular complexes, electrostatic interactions can significantly affect bimolecular association rates (Law et al., 2006). For example, by using Brownian dynamics (BD) simulations (Ermak and McCammon, 1978; Northrup et al., 1984) (Sec. IV.F) to calculate diffusional association rates, Gabdoulline and Wade demonstrated that, for fast-associating protein pairs, electrostatic interactions enhance association and are the dominant forces determining the rate of diffusional association (Radic et al., 1997; Gabdoulline and Wade, 2001). Using related methods, Sept et al. demonstrated the role of electrostatic interactions in determining the rates and polarity of actin polymerization (Sept et al., 1999; Sept and McCammon, 2001).
Electrostatic interactions also play an important role in determining thermodynamics of binding; i.e., binding affinity (Novotny and Sharp, 1992; Schreiber and Fersht, 1993, 1995; Zhu and Karlin, 1996; Chong et al., 1998; Sheinerman et al., 2000; Norel et al., 2001; Rauch et al., 2002). Substrate binding allows the formation of (potentially) favorable charge-charge interactions between the substrate and target, as well as stabilizing specific salt-bridges and hydrogen bonds (Schreiber and Fersht, 1993, 1995; Chong et al., 1998). However, at the same time, charges on the molecular binding surface must shed their bound water in order to allow close binding. This loss of water, or desolvation, is generally energetically unfavorable and offsets the favorable interactions formed upon binding. The binding affinities, from an electrostatic point of view, are determined by balance of these two energetic contributions (Xu et al., 1997; Lee and Tidor, 2001; Sheinerman and Honig, 2002; Russell et al., 2004; del Álamo and Mateu, 2005). Systematic studies of protein pairs, such as barnase and barstar (Schreiber and Fersht, 1993, 1995; Frisch et al., 1997; Dong et al., 2003), and fasciculin-2 (Radic et al., 1997), as well as protein kinase A and balanol (Wong et al., 2001), have shown that charged and polar residues at the protein-protein interfaces play important roles in binding energetics. Similarly, Sept et al. have demonstrated an important role for electrostatics in determining microtubule structure and stability (Sept et al., 2003). Finally, Wang et al (Wang et al., 2004) have demonstrated that nonspecific electrostatic interactions can provide a driving force for recruitment of proteins to intracellular membranes, an important step in signal transduction.
However, despite the role of electrostatics in protein-protein interactions, it is important to realize that the total interaction is also strongly influenced by shape complementarity at the protein-protein interface as well as nonpolar contributions to offset the penalties of desolvation (Janin and Chothia, 1990; Lo Conte et al., 1999; Ma et al., 2003; Vasker, 2004).
III. Models for biomolecular solvation and electrostatics
As described above, computer simulations can provide atomic-scale information on energetic and dynamic contributions to biomolecular structure and interactions. However, the capabilities of computer simulations are limited by the accuracy of the underlying models describing atomic interactions and also by the computational expense of adequately exploring all the relevant conformations of the biomolecule and surrounding water and ion. Therefore, most models of biomolecular solvation and electrostatics make a trade-off between these opposing considerations of atomic accuracy and computational expense.
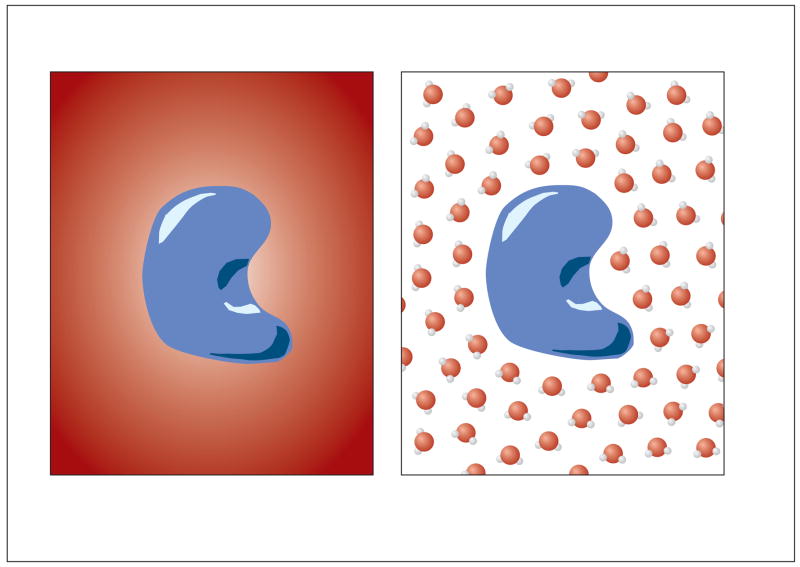
A variety of computational methods have been developed for studying electrostatic interactions in biomolecular systems. Popular methods for understanding electrostatic interactions in these systems can be loosely classified into two categories (see Fig. 1): explicit solvent methods (Burkert and Allinger, 1982; Jorgensen et al., 1983; Sagui and Darden, 1999; Ponder and Case, 2003; Horn et al., 2004), which treat the solvent in full atomic detail, and implicit solvent methods (Davis and McCammon, 1990b; Honig and Nicholls, 1995; Roux and Simonson, 1999; Roux, 2001; Baker, 2005b; Baker et al., 2006), which represent the solvent through its average effect on solute.
Figure 1.
A schematic comparison of implicit and explicit solvent models: (a) in the implicit solvent model, a low dielectric solute is surrounded by a continuum of high dielectric solvent; (b) in the explicit solvent model, solvent is represented by discrete water molecules.
III.A. Explicit solvent methods
Explicit solvent methods offer a very detailed description of biomolecular solvation. In explicit solvent methods, interactions between mobile ions, solvent, and solute atoms are typically described by molecular mechanics force fields (Wang et al., 2001b; Ponder and Case, 2003) which use classical approximations of quantum mechanical energies to describe the Coulombic (electrostatic), van der Waals, and covalent (bond, angle) interactions. Explicit solvent methods have the obvious advantage of offering the full details of solvent-solute and solvent-solvent interactions. These details can affect some aspects of biomolecular interactions. For example, the explicit representation of solvent structure can qualitatively change the detailed features of protein side chain interactions (Masunov and Lazaridis, 2003). Similarly, Yu et al have demonstrated the importance of including first shells of solvation to correctly describe the interaction of salt bridges in solution (Yu et al., 2004).
However, the explicit solvent methods are computationally expensive. In order to extract meaningful thermodynamic and kinetic parameters, all the numerous conformations of biomolecules, as well as the solvent and ions, must be explored. The extra degrees of freedom associated with the explicit solvent and ions dramatically increase the computational cost of explicit solvent methods and limit the temporal and spatial scales of biomolecular simulations.
III.B. Implicit solvent methods
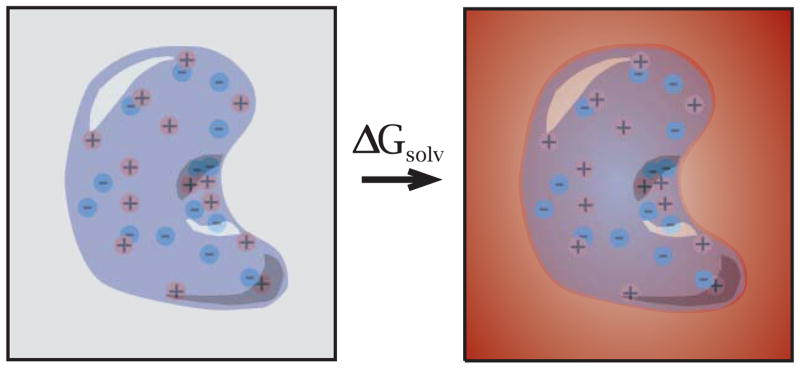
Implicit solvent methods have become popular, although lower-accuracy, alternatives to the computationally-expensive explicit solvent approaches (Davis and McCammon, 1990b; Honig and Nicholls, 1995; Gilson, 2000; Roux, 2001; Baker, 2005b; Baker et al., 2006). In implicit solvent methods, the molecules of interest are treated explicitly while the solvent is represented by its average effect on the solute (Roux and Simonson, 1999). Solute-solvent interactions are described by solvation energies; i.e., the free energy of transferring the solute from a vacuum to the solvent environment of interest (e.g., water at a certain ionic strength). This process is shown in more detail in Fig. 2, which consists of three steps: (1) solute charges are gradually reduced to zero in vacuum, (2) the uncharged solute is inserted into the solvent, and (3) solute charges are gradually increased back to their normal values in solvent. The free energy change in step (2) is called the nonpolar solvation energy. The sum of the energies associated with the step (1) and (3) is called the “charging” or polar solvation energy and represents the solvent’s effect on the solute charging process. In general, polar and nonpolar solvation terms act in opposing directions; nonpolar solvation favors compact structures with small areas and volumes, while polar solvation favors the maximum solvent exposure for all polar groups in the solute.
Figure 2.

A thermodynamic cycle illustrating the biomolecular solvation process. The steps are (1) un-charging the biomolecule in vacuum; (2) transferring the uncharged biomolecule from vacuum to solvent; (3) charging the biomolecule back to its normal value in solvent. The nonpolar solvation free energy is the free energy change in step (2). The polar solvation free energy is the sum of the free energy changes in steps (1) and (3).
III.B.1. Nonpolar solvation
One popular approximation for the nonpolar solvation free energy assumes a linear dependence between the nonpolar solvation energy and the solvent-accessible surface area (SASA), A, (Chothia, 1974; Eisenberg, 1976; Spolar et al., 1989; Sharp et al., 1991; Wesson and Eisenberg, 1992; Massova and Kollman, 2000; Swanson et al., 2004):
| (1) |
where γ is a “surface tension” which is typically chosen to reproduce the nonpolar solvation free energy of alkanes (Sharp et al., 1991; Simonson and Brunger, 1994; Sitkoff et al., 1994b) or model sidechain analogues (Eisenberg and McLachlan, 1986; Wesson and Eisenberg, 1992). The surface tension parameter may assume a single global value used for all atom types or different values may be assigned for each different type of atom. Although SASA methods have enjoyed surprising success, they are also subject to several caveats, including widely varying choices of surface tension parameter (Chothia, 1974; Eisenberg and McLachlan, 1986; Sharp et al., 1991; Sitkoff et al., 1994a; Elcock et al., 2001) as well as inaccurate descriptions of the detailed aspects of nonpolar solvation energy (Gallicchio and Levy, 2004), peptide conformations (Su and Gallicchio, 2004), and protein nonpolar solvation forces (Wagoner and Baker, 2004). Some of these problems have been fixed by new models which include the small but important attractive van der Waals interactions between solvent and solute (Gallicchio et al., 2000; Gallicchio et al., 2002; Gallicchio and Levy, 2004; Wagoner and Baker, 2006) as well as repulsive solvent-accessible volume terms (Wagoner and Baker, 2006).
III.B.2. Polar solvation
Implicit solvent methods have been used to study polar solvation and electrostatics for over 80 years, starting with work by Born on ion solvation (Born, 1920), Linderström-Lang (Linderström-Lang, 1924) and Tanford and Kirkwood (Tanford and Kirkwood, 1957) on protein titration, Manning on ion distributions surrounding nucleic acids (Manning, 1978), Flanagan et al (Flanagan et al., 1981) on the pH dependence of hemoglobin dimer assembly, and Warwicker and Watson (Warwicker and Watson, 1982) on the electrostatic potential of realistic protein geometries. Although they can be considerably different in their details and implementation, implicit solvent models generally treat the solvent as a high dielectric continuum, the aqueous ions as a diffuse cloud of charge, and the solute as a fixed array of point charges that are embedded in a lower dielectric continuum. Despite the limitations of these assumptions, implicit solvent models often give a good coarse-grained description of solvation energetics and have enjoyed widespread use over recent years.
Regardless of the particular type of implicit solvent model, the behavior of electrostatic interactions is generally determined by a few basic properties of the system, illustrated in Fig. 3: the charges, radii, and “dielectric constant” of the solute; the charges and radii of aqueous ionic species; and the radii and dielectric constant of the solvent. The relationship of these specific parameters to solvation energies and forces will be described in more detail in Secs. III.C and III.D.
Figure 3.
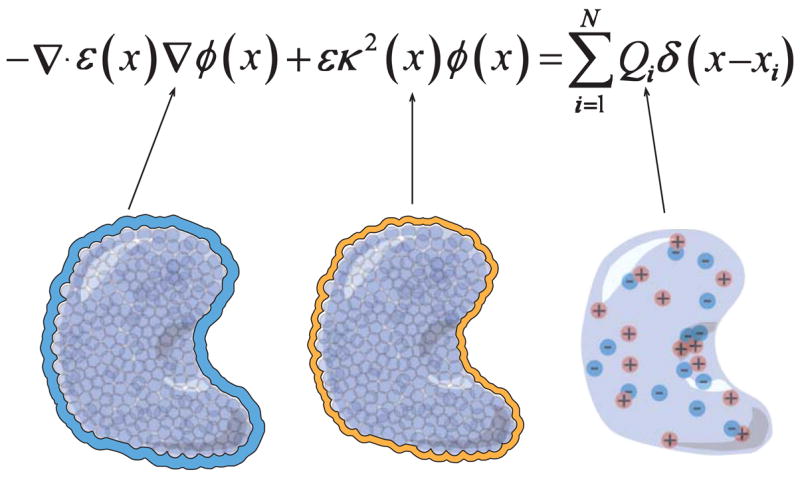
Description of the terms in the Poisson-Boltzmann equation: (a) the dielectric permittivity coefficient ε(x⃑) is much smaller inside the biomolecule than outside the biomolecule with a rapid change in value across the solvent-accessible biomolecular surface; (b) the ion-accessibility parameter κ2(x⃑) is proportional to the bulk ionic strength outside the ion-accessible biomolecular surface; (c) the biomolecular charge distribution is defined as the collection of point charges located at the center of each atom.
III.C. Poisson-Boltzmann methods
The Poisson-Boltzmann (PB) equation is a popular continuum description of electrostatics for biomolecular system. Although there are a number of ways to derive the PB equation based on statistical mechanics (Holm et al., 2001), the simplest derivation begins with Poisson’s equation (Jackson, 1975; Bockris and Reddy, 1998) (in SI units),
| (2) |
the basic equation for describing the electrostatic potential ϕ(x⃑) at point x⃑ generated by a charge distribution ρ(x⃑) in an environment with a dielectric permittivity coefficient ε(x⃑) (Jackson, 1975; Landau et al., 1982).
The coefficient ε(x⃑) is given by ε(x⃑) = ε0εr(x⃑) where ε0 = 8.8542×10−12 C2/(N · m2 is the electrostatic permittivity of a vacuum and εr(x⃑) is the dielectric coefficient or the relative electrostatic permittivity. The dielectric coefficient εr(x⃑) describes the local polarizability of the material; i.e., the generation of local dipole densities in response to the applied fields and changes in charge. The functional form of this coefficient depends on the shape of the biomolecule; εr(x⃑) assumes lower values of 2–20 in the biomolecular interior and higher values of approximately 80, the value for water at room temperature, in solvent-accessible regions. The distinction between biomolecular “interior” and “exterior” used to assign dielectric coefficients is imprecise; as a result, a variety of different definitions for the biomolecular surface and dielectric coefficient have been developed (Lee and Richards, 1971; Warwicker and Watson, 1982; Connolly, 1985; Im et al., 1998; Grant et al., 2001).
In order to continue the derivation of the PB equation, we assume the charge distribution ρ(x⃑) includes two contributions: the solute charges ρf(x⃑) and the aqueous “mobile” ions ρm(x⃑). The solute charge distribution is generally described by a collection of N point charges located at each solute atom’s position x⃑i and scaled by that atom’s charge Qi ; i.e., the solute charge distribution is the summation of a set of delta functions . Neglecting explicit interactions between the aqueous ions (Holm et al., 2001), the mobile charges are modeled as a continuous “charge cloud” described by a Boltzmann distribution (McQuarrie, 1976). For m ion species with charges qj, bulk concentrations cj and steric potential Vj (x⃑) (a potential that prevents biomolecule-ion overlap), the mobile ion charge distribution is , where kB is Boltzmann’s constant and T is the absolute temperature. Combining both the solute and ion charge distributions with the Poisson equation, Eq. (2), gives the full PB equation:
| (3) |
A common simplification is that the exponential term exp can be approximated by the linear term in its Taylor series expansion − qjϕ((x⃑)/kBT for |qjϕ((x⃑)/kBT| ≪ 1. With this linearization and by assuming the steric occlusions are the same for all ion species (Vj = V for all j), Eq. (3) reduces to the linearized PB equation:
| (4) |
where κ2(x⃑), related to a modified inverse Debye-Hückel screening length (Debye and Hückel, 1923), is given by
| (5) |
where is the ionic strength and ec is the unit electric charge.
Once the PB equation is solved, the electrostatic potential is known for the entire system. Given this potential, the electrostatic free energy can be evaluated by a variety of integral formulations (Sharp and Honig, 1990; Gilson, 1995; Micu et al., 1997). The simplest, for the linearized PB equation, is
| (6) |
It is also possible to differentiate integral formulations of the electrostatic energy with respect to atomic position to obtain the electrostatic or polar solvation force on each atom (Gilson et al., 1993; Im et al., 1998).
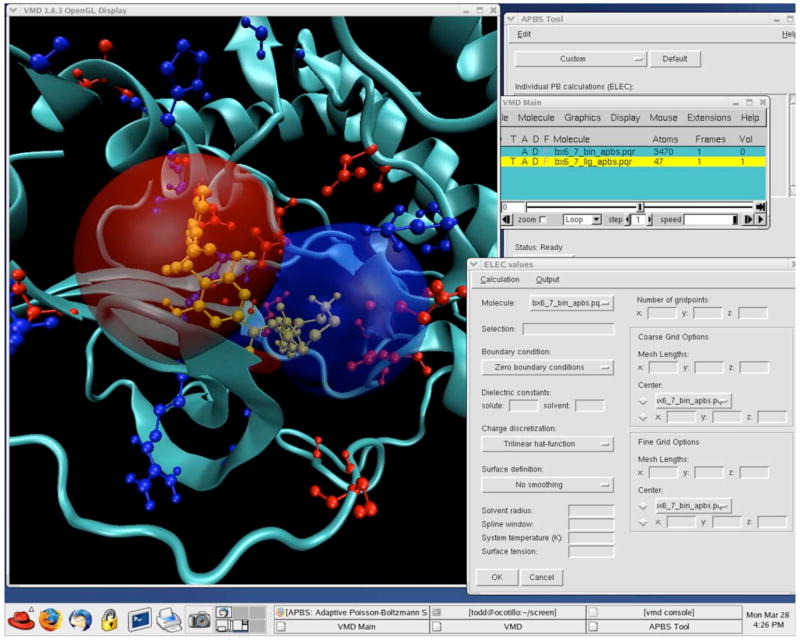
Analytical solutions of the PB equation are not available for biomolecules with realistic shapes and charge distributions. Numerical methods for solving PB equation were first introduced by Warwicker and Watson (Warwicker and Watson, 1982) to obtain the electrostatic potential at the active site of an enzyme. The most common numerical techniques for solving the PB equation are based on discretization of the domain of interest into small regions. Those methods include finite difference (Davis and McCammon, 1989; Nicholls and Honig, 1991; Holst and Saied, 1993; Holst and Saied, 1995; Baker et al., 2001), finite element (Cortis and Friesner, 1997a, 1997b; Baker et al., 2000; Holst et al., 2000; Baker et al., 2001; Dyshlovenko, 2002), and boundary element methods (Zauhar and Morgan, 1988; Juffer et al., 1991; Allison and Huber, 1995; Bordner and Huber, 2003; Boschitsch and Fenley, 2004), all of which continue to be developed to further improve the accuracy and efficiency of electrostatics calculations in the numerous biomolecular applications described below. The major software packages that can be used to solve the PB equation are listed in Table 1. Many of these packages are also used for visualization of the electrostatic potential around biomolecules. Such visualization can provide insight into biomolecular function and highlight regions of potential interest. Fig. 4 shows examples of the visualization of electrostatic potential calculated with APBS (Baker et al., 2001) and visualized with VMD (Humphrey et al., 1996).
Table 1.
Major PB equation solver
| Software package | URL |
|---|---|
| APBS (Baker et al., 2001) | http://apbs.sf.net/ |
| Delphi (Rocchia et al., 2002) | http://trantor.bioc.columbia.edu/delphi/ |
| MEAD (Bashford, 1997) | http://www.scripps.edu/mb/bashford/ |
| ZAP (Grant et al., 2001) | http://www.eyesopen.com/products/toolkits/zap.html |
| UHBD (Madura et al., 1995) | http://mccammon.ucsd.edu/uhbd.html |
| Jaguar (Cortis and Friesner, 1997a, 1997b) | http://www.schrodinger.com/ |
| CHARMM (MacKerell et al., 1998) | http://yuri.harvard.edu |
| Amber (Luo et al., 2002) | http://amber.scripps.edu |
Figure 4.
Examples of the visualization of the balanol electrostatic potential in the binding site of protein Kinase A as calculated by APBS (Baker et al., 2001) and visualized with VMD (Humphrey et al., 1996).
III.D. Simpler models
In addition to the PB methods, simpler approximate models have also been constructed for continuum electrostatics, including distance-dependent dielectric functions (Leach, 2001; MacKerell and Nilsson, 2001), analytic continuum methods (Schaeler and Karplus, 1996), and generalized Born models (Still et al., 1990; Osapay et al., 1996; Dominy and Brooks III, 1999; Bashford and Case, 2000; Onufriev et al., 2002). Among these simpler methods, generalized Born (GB) is currently the most popular. The GB model was introduced by Still et al. in 1990 and subsequently refined by several other researchers (Osapay et al., 1996; Dominy and Brooks III, 1999; Bashford and Case, 2000; Onufriev et al., 2002). The model shares the same continuum representation of the solvent as the Poisson or PB theories. However, the GB model is based on the analytical solvation energy obtained from the solution of the Poisson equation for a simple sphere (Born, 1920). The biomolecular electrostatic solvation free energy is approximated by a modified form of the analytical solvation energy for a sphere (Still et al., 1990):
| (7) |
where the self terms as , are the “effective Born radii” and the cross terms as , are the effective interaction distances. The most common form of (Still et al., 1990) is
| (8) |
where Ri are the effective radii of the atoms and rij are the distance between atom i and j. Efficiently and accurately calculating effective radii Ri is essential for GB methods. “Perfect” GB radii, which reproduce the atom i’s self energy obtained by solving Poisson equation for the biomolecule-solvent system with only atom i charged, have demonstrated the ability to accurately follow the results of more detailed models such as PB (Onufriev et al., 2002). However using such “perfect” radii does not directly provide any computational advantage over solving the Poisson equation. In the absence of perfect radii for every biomolecular conformation, GB methods fail to capture some aspects of molecular structure included in more detailed models, such as the PB equation. Nonetheless, GB methods have become increasingly popular because of their computational efficiency.
III.E. Limitations of implicit solvent methods
Although implicit solvent methods offer simpler descriptions of the system and greater computational efficiency, it is important to recall that these reductions of complexity and effort are obtained at the cost of substantial simplification of the description of the solvent. In particular, implicit solvent methods are only capable of describing non-specific interactions between solvent and solute. In general, explicit solvent methods should be used wherever the detailed interactions between solvent and solute are important, such as solvent finite size effects in ion channels (Nonner et al., 2001), strong solvent-solute interactions (Bhattacharrya et al., 2003), strong solvent coordination of ionic species (Figueirido et al., 1994; Yu et al., 2004), and saturation of solvent polarization near a membrane (Lin J.-H. et al., 2002). Similarly, as mentioned earlier in the context of RNA-ion interactions, implicit descriptions of mobile ions can also become questionable in some cases, such as high ion valency or strong solvent coordination, specific ion-solute interactions, and high local ion densities (Holm et al., 2001), where the ions interact with each other or with the solute directly.
IV. Applications
In the previous section, we discussed the basic concepts behind the computational tools that can be used to simulate the electrostatic interactions in cellular systems. In this section, we will illustrate the use of these methods, especially PB methods, to deal with the various biomolecular problems.
IV.A. Solvation free energy
As mentioned in Sec. III.B, the solvation free energy is the free energy of transferring a solute from a uniform dielectric continuum (a constant dielectric) to an inhomogeneous medium (a low dielectric solute surrounded by a high dielectric solvent), which is often divided into two terms: a nonpolar term and polar term. The nonpolar term is usually estimated using either SASA or the improved methods discussed in Sec. III.B.1. For the polar term, as shown in Fig. 5, two PB calculations are usually performed: (1) calculating biomolecular electrostatic free energy in a homogeneous medium with a constant dielectric equal to the solute dielectric coefficient and (2) calculating the biomolecular electrostatic free energy in the inhomogeneous medium of interest; e.g., a protein in aqueous medium. The polar contribution to the solvation free energy is then given by:
| (9) |
Figure 5.
Schematic of a polar solvation free energy calculation; in the initial state, the dielectric coefficient is a constant throughout the entire system and equal to the solute dielectric coefficient; in the final state, the dielectric coefficient is inhomogeneous and smaller in the solute than in the bulk solvent.
Additionally, solving the PB equation twice helps to cancel the numerical artifacts which arise from the discretization used in finite difference and finite element methods, i.e. it reduces the grid size dependence. Although, in most cases, polar solvation free energy alone is not sufficient to explain the biological phenomenon, it is the foundation for the other, more complex, electrostatic calculations described below.
IV.B. Electrostatic free energy
The total electrostatic free energy can be easily obtained from the polar solvation free energy by adding the electrostatic free energy of the biomolecule in a homogeneous medium with a constant dielectric equal to the solute dielectric coefficient using Coulomb’s law:
| (10) |
where
| (11) |
where rij is the distance between charge Qi and Qj and εp is the dielectric coefficient of the solute. The resulting electrostatic free energies are the basis for nearly all applications of continuum electrostatics methods to biomolecular systems.
As a specific example, such electrostatic free energy calculations have been used to study the electrostatic sequestration of phosphatidylinositol 4,5-bisphosphate (PIP2) by membrane-adsorbed basic peptides (Wang et al., 2004). PIP2 is a very important lipid in the cytoplasmic leaflet of the plasma membrane (De Camilli et al., 1996; Toker, 1998; Raucher et al., 2000; Martin, 2001; Payrastre et al., 2001; Cantley, 2002; Irvine, 2002; McLaughlin et al., 2002; Yin and Janmey, 2003) with a net charge of −4e on the lipid head group. By calculating the electrostatic free energy of laterally sequestering a PIP2 lipid from a region of “bulk” membrane to a region in the vicinity of a membrane-absorbed basic peptide, Wang et al demonstrated that nonspecific electrostatic interactions provide a driving force for the lateral sequestration of PIP2 by membrane-adsorbed basic peptides (Wang et al., 2001a; Rauch et al., 2002; Wang et al., 2002; Wang et al., 2004). Such lateral sequestration of PIP2 is thought to contribute to the regulation of PIP2 function by controlling its accessibility to other proteins (Laux et al., 2000; McLaughlin et al., 2002).
IV.C. Folding free energies
Biomolecular native (folded) structure is very important for proper performance of their biological functions. However, accurately determining the mechanism by which electrostatic interactions affect the stability of bimolecular native structure is still a challenging experimental and computational question. The electrostatic contribution to the biomolecular folding stability is usually defined as the difference in electrostatic free energy between folded and unfolded states:
| (12) |
If , from the electrostatic point of view, the folded structure is more stable than the unfolded structure. If reduces in response to a mutation, i.e., if , this mutation makes folded protein more stable. This method has been widely used to study electrostatic contribution to protein folding stabilities through mutations that involve charged or polar residues. For example, Bacillus caldolyticus cold shock protein (Bc-Csp) is a thermophilic protein, which differs from Bacillus subtilis cold shock protein B (Bs-CspB), its mesophilic homolog, in 11 of its 66 residues (Mueller et al., 2000; Delbruck et al., 2001). Through mutational studies which reduced the sequence differences between these two protein molecules, both experiment (Mueller et al., 2000; Pace, 2000; Perl et al., 2000; Delbruck et al., 2001; Perl, 2001) and PB calculations (Zhou and Dong, 2003) demonstrated that the difference in stability of these two proteins arises mostly from the interactions among three residues: Arg 3, Glu 46, and Leu 66 in Bc-Csp, as compared with Glu 3, Ala 46, and Glu 66 in Bs-CspB. The removal of the repulsion between Glu 3 and Glu 66 and creation of a favorable salt bridge between Arg 3 and Glu 46 are the main reasons that Bc-Csp is more stable than Bs-CspB at higher temperatures. Moreover, the excellent agreement between PB calculations and experimental data (the correlation coefficient is 0.98) implies that electrostatic interactions dominate the thermostability of thermophilic proteins (Zhou and Dong, 2003).
IV.D. Binding free energies
The binding of biomolecules is fundamental to cellular activity. The simplest type of binding energy calculations are performed on the biomolecular complex assuming a rigid conformation; i.e., without any conformational changes upon binding, which is clearly not realistic, but often provides useful initial estimates for relative biomolecular binding affinities. Fig. 6 illustrates the procedure to calculate the polar contribution to the binding free energy which is given by
Figure 6.
Thermodynamic cycle illustrating the standard procedure for calculating the electrostatic contribution to the binding free energy of a complex with rigid-body. The steps are (1) transfer the isolated molecule from a inhomogeneous dielectric into a homogeneous dielectric, the free energy change is ; (2) form the complex from isolated molecules in a homogeneous dielectric, the free energy change is , (3) transfer the complex from the homogeneous dielectric into the inhomogeneous dielectric, the free energy change is .
| (13) |
where is the polar solvation free energy change upon binding with the values calculated according to Eq. (9) above. The quantity is the Coulombic free energy change upon binding with components calculated according to Eq. (10) above.
For the more general situation in which biomolecules experience conformational changes during the binding process, MM/PBSA and MM/GBSA methods (Kollman et al., 2000; Swanson et al., 2004) are commonly used to calculate the binding free energy. The nature of this method can be best understood through its acronym: MM stands for the molecular mechanics force fields used to calculate the intramolecular and direct intermolecular contributions to binding free energies; PB and GB refer to the implicit solvent methods used to calculate the electrostatic contributions, and SA stands for solvent accessible surface area (SASA) methods used to calculate the nonpolar contributions to binding free energies.
Binding free energy calculations using continuum solvation models have been successfully performed on many different biomolecular complexes (Eisenberg and McLachlan, 1986; Murray et al., 1997; Misra et al., 1998; Sept et al., 1999; Massova and Kollman, 2000; Wong et al., 2001; Dong et al., 2003; Sept et al., 2003; Wang et al., 2004; Green and Tidor, 2005). As specific examples, binding free energy calculations have been performed to investigate the roles of charged residues at the interface of the barnase (and extracellular ribonuclease) and barstar (a protein inhibitor), which have been a popular test case for both computational (Gabdoulline and Wade, 1997, 1998, 2001; Lee and Tidor, 2001; Sheinerman and Honig, 2002; Dong et al., 2003; Wang and Wade, 2003; Spaar and Helms, 2005; Spaar et al., 2006) and experimental studies of protein-protein interactions (Schreiber and Fersht, 1993, 1995; Frisch et al., 1997). In particular, PB calculations (Dong et al., 2003) successfully reproduced the experimental result (Schreiber and Fersht, 1993, 1995; Frisch et al., 1997) that cross-interface salt-bridges and hydrogen bonds dominate the binding affinities of barnase and barstar (Dong et al., 2003).
IV.E. pKa calculations
The presence of ionizable sites, which can exchange protons with their environment, produces pH-dependent phenomena in proteins and has a significant influence on the protein’s function. The correct prediction of protein titration states is important for the analysis of enzyme mechanisms, protein stability, and molecular recognition. As mentioned earlier, efforts have been underway for more than 80 years (Linderström-Lang, 1924; Antosiewicz et al., 1996b; Bastyns et al., 1996; Luo et al., 1998; Nielsen and Vriend, 2001; Fitch et al., 2002; Georgescu et al., 2002; Li et al., 2002; Alexov, 2003; Nielsen and McCammon, 2003; Li et al., 2004; Jensen et al., 2005; Krieger et al., in press) to correctly predict protein titration states and understand the determinants of pKa s for amino acids in protein environments (see chapter by Whitten, et al. in this volume).
The free energy change, ΔG, for protonation of a single ionizable site at a given pH may be written as (Linderström-Lang, 1924; Tanford and Kirkwood, 1957)
| (14) |
where is the equilibrium constant for dissociation of proton H+ and it’s conjugate site A−kB is Boltzmann’s constant; and T is the absolute temperature. A widely-used assumption in pKa predictions is that any pKa differences of an ionizable site when located in a protein versus in a model compound are solely determined by the difference in the electrostatic free energy required to protonate that site in the protein versus the model compound. Thus, the pKa of the single ionizable site in protein is given by
| (15) |
where ΔΔG = ΔGprotein − ΔGmod el and pK0 is the pKa of the isolated ionizable site in the model compound. In general, proteins have multiple ionizable sites and the protonation energetics of these different sites are coupled, as discussed below. Single site pKa predictions have successfully reproduced measured pKa s for different residues in several different proteins (Dong and Zhou, 2002; Dong et al., 2003) and therefore have some predictive power. However, a more complete treatment of ionizable residues in proteins considers the coupling between all the ionizable sites. There are a number of techniques for treating such coupling (Tanford and Roxby, 1972; Beroza et al., 1991; Antosiewicz et al., 1996a; Antosiewicz et al., 1996b; Bashford, 2004) and thereby determining the complete titration state of the protein. Unfortunately, such methods are complex and are beyond the scope of the current discussion.
IV.F. Biomolecular association rates
Brownian dynamics (BD) calculations are popular methods to simulate the diffusional motion between two solute particles and thereby estimate the rate of diffusion-controlled rate of binding between two molecules (Ermak and McCammon, 1978; Northrup et al., 1984). Given the importance of electrostatic interactions in biomolecular association, BD simulations are usually combined with continuum electrostatic calculations to provide the most accurate estimates of diffusion-limited encounter rates (Allison and McCammon, 1985; Davis and McCammon, 1990a; Ilin et al., 1995; Madura et al., 1995; Sept et al., 1999; Gabdoulline and Wade, 2001, 2002). Such calculations have been used in numerous diffusional encounter rate calculations, including simulations of small molecule interactions with enzymes (Allison and McCammon, 1985; Madura and McCammon, 1989; Davis et al., 1991; Sines et al., 1992; Luty et al., 1993; Tan et al., 1993; Elcock et al., 1996; Radic et al., 1997; Tara et al., 1998), simulations of protein-protein encounter (Gabdoulline and Wade, 1997; Elcock et al., 1999; Sept et al., 1999; Elcock et al., 2001; Gabdoulline and Wade, 2001; Spaar et al., 2006), as well as functional assessment of differences in protein electrostatics (Livesay et al., 2003).
V. Conclusion and future directions
Computer simulation is becoming an increasingly routine way to help with drug discovery or other applications requiring a detailed understanding of molecular interactions. A correct understanding of the energetic interactions within and between biomolecules is essential for such simulations. Among the various contributions to these energies, electrostatic interactions are of special importance because of their long range and strength. In this chapter, we have covered some of the computational methods that are currently available to model the electrostatic interactions biomolecular systems, ranging from highly detailed explicit solvent methods to simpler PB and GB methods. There are several reviews available on all of these methods which provide a more in-depth discussion of the different solvation approaches. The reviews of Ponder and Case (Ponder and Case, 2003) as well as the texts of Becker et al (Becker et al., 2001), Leach (Leach, 2001), and Schlick (Schlick, 2002) provide excellent background on explicit solvent methods. There also are several reviews available for implicit solvent methods see (Honig and Nicholls, 1995; Roux and Simonson, 1999; Bashford and Case, 2000; Simonson, 2003; Baker, 2005a), including a particularly thorough treatment by Lamm (Lamm, 2003), a discussion of current PB limitations by Baker (Baker, 2005b) and an up-to-date discussion by Feig and Brooks (Feig and Brooks III, 2004) of current challenges for GB methods. For additional background and more in-depth discussion of the principles and limitations of continuum electrostatics, interested readers should see the general volume by Jackson (Jackson, 1975) and Landau et al. (Landau et al., 1982), the electrochemistry text of Bockris et al. (Bockris and Reddy, 1998), the colloid theory treatise by Verwey and Overbeek (Verwey and Overbeek, 1999), or the excellent collection of condensed matter electrostatics articles assembled by Holm et al. (Holm et al., 2001)
Acknowledgments
The authors would like to thank Baker group members for their reading of this manuscript. This work was supported by National Institutes of Health grant R01 GM069702.
Contributor Information
Feng Dong, Dept. of Biochemistry and Molecular Biophysics, Center for Computational Biology, Washington University in St. Louis. 700 S. Euclid Ave., Campus Box 8036, St. Louis, MO 63110. E-mail: fdong@ccb.wustl.edu.
Brett Olsen, Dept. of Biochemistry and Molecular Biophysics, Center for Computational Biology, Washington University in St. Louis. 700 S. Euclid Ave., Campus Box 8036, St. Louis, MO 63110. E-mail: bnolsen@artsci.wustl.edu.
Nathan A. Baker, Dept. of Biochemistry and Molecular Biophysics, Center for Computational Biology, Washington University in St. Louis. 700 S. Euclid Ave., Campus Box 8036, St. Louis, MO 63110. Tel: 314-362-2040, Fax: 314-362-0234, E-mail: baker@ccb.wustl.edu. Corresponding author..
VI. Reference
- Alexov E. Role of the protein side-chain fluctuations on the strength of pair-wise electrostatic interactions: comparing experimental with computed pKas. Proteins. 2003;50:94–103. doi: 10.1002/prot.10265. [DOI] [PubMed] [Google Scholar]
- Allison SA, Huber GA. Modeling the electrophoresis of rigid polyions: application of lysozyme. Biophys J. 1995;68:2261–2270. doi: 10.1016/S0006-3495(95)80408-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison SA, McCammon JA. Dynamics of substrate binding to copper zinc superoxide dismutase. J Phys Chem. 1985;89:1072–1074. [Google Scholar]
- Anderson CF, Record MT., Jr Gibbs–Duhem-based relationships among derivatives expressing the concentration dependences of selected chemical potentials for a multicomponent system. Biophys Chem. 2004;112:165–175. doi: 10.1016/j.bpc.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Antosiewicz J, Briggs JM, Elcock AH, Gilson MK, McCammon JA. Computing ionization states of proteins with a detailed charge model. J Comput Chem. 1996a;17:1633–1644. [Google Scholar]
- Antosiewicz J, McCammon AJ, Gilson MK. The determinants of pKas in proteins. Biochemistry. 1996b;35:7819–7833. doi: 10.1021/bi9601565. [DOI] [PubMed] [Google Scholar]
- Bajaj NP, McLean MJ, Waring MJ, Smekal E. Sequence-selective, pH-dependent binding to DNA of benzophenanthridine alkaloids. J Mol Recognit. 1990;3:48–54. doi: 10.1002/jmr.300030106. [DOI] [PubMed] [Google Scholar]
- Baker N, Holst M, Wang F. Adaptive multilevel finite element solution of the poisson-boltzmann equation ii. Refinement at solvent-accessible surfaces in biomolecular systems. J Comput Chem. 2000;21:1343–1352. [Google Scholar]
- Baker NA. Biomolecular applications of Poisson-Boltzmann methods. In: Lipkowitz KB, Larter R, Cundari TR, editors. Reviews in Computational Chemistry. Wiley-VCH, John Wiley & Sons, Inc; Hoboken, NJ: 2005a. pp. 349–379. [Google Scholar]
- Baker NA. Improving implicit solvent simulations: a Poisson-centric view. Curr Opin Struct Biol. 2005b;15:137–143. doi: 10.1016/j.sbi.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Baker NA, Bashford D, Case D. Implicit solvent electrostatics in biomolecular simulation. In: Leimkuhler B, Chipot C, Elber R, et al., editors. New Algorithms for Macromolecular Simulation. Springer-Verlag; Berlin: 2006. pp. 263–295. [Google Scholar]
- Baker NA, Sept D, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashford D. Macroscopic electrostatic models for protonation states in proteins. Frontiers Biosci. 2004;9:1082–1099. doi: 10.2741/1187. [DOI] [PubMed] [Google Scholar]
- Bashford D, Case DA. Generalized Born models of macromolecular solvation effects. Annu Rev Phys Chem. 2000;51:129–152. doi: 10.1146/annurev.physchem.51.1.129. [DOI] [PubMed] [Google Scholar]
- Bastyns K, Froeyen M, Diaz JF, Volckaert G, Engelborghs Y. Experimental and theoretical study of electrostatic effects on the isoelectric pH and pKa of the catalytic residue His-102 of the recombinant ribonuclease from Bacillus amyloliquefaciens (barnase) Proteins. 1996;24:370–378. doi: 10.1002/(SICI)1097-0134(199603)24:3<370::AID-PROT10>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Becker O, MacKerell AD, Jr, Roux B, Watanabe M. Computational Biochemistry and Biophysics. Marcel Dekker; New York: 2001. [Google Scholar]
- Beroza P, Fredkin DR, Okamura MY, Feher G. Protonation of interacting residues in a protein by Monte Carlo method: Application to lysozyme and the photosynthetic reaction center of Rhodobacter sphaeroides. Proc Natl Acad Sci U S A. 1991;88:5804–5808. doi: 10.1073/pnas.88.13.5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharrya SM, Wang Z-G, Zewail AH. Dynamics of water near a protein surface. J Phys Chem B. 2003;107:13218–13228. [Google Scholar]
- Bockris JO, Reddy KN. Modern Electrochemistry: Ionics. Plenum Press; New York: 1998. [Google Scholar]
- Bordner AJ, Huber GA. Boundary element solution of linear Poisson-Boltzmann equation and a multipole method for the rapid calculation of forces on macromolecules in solution. J Comput Chem. 2003;24:353–367. doi: 10.1002/jcc.10195. [DOI] [PubMed] [Google Scholar]
- Born M. Volumen und hydratationswarme der ionen. Z Phys. 1920;1:45–48. [Google Scholar]
- Boschitsch AH, Fenley MO. Hybrid boundary element and finite difference method for solving the nonlinear Poisson-Boltzmann equation. J Comput Chem. 2004;25:935–955. doi: 10.1002/jcc.20000. [DOI] [PubMed] [Google Scholar]
- Boström M, Deniz V, Ninham BW. Ion specific surface forces between membrane surfaces. J Phys Chem B. 2006;110:9645–9649. doi: 10.1021/jp0606560. [DOI] [PubMed] [Google Scholar]
- Boström M, Williams DRM, Stewart PR, Ninham BW. Hofmeister effects in membrane biology: The role of ionic dispersion potentials. Phys Rev E. 2003;68:041902–041907. doi: 10.1103/PhysRevE.68.041902. [DOI] [PubMed] [Google Scholar]
- Broering JM, Bommarius AS. Evaluation of Hofmeister Effects on the Kinetic Stability of Proteins. J Phys Chem B. 2005;109:20612–20619. doi: 10.1021/jp053618+. [DOI] [PubMed] [Google Scholar]
- Burkert U, Allinger NL. Molecular Mechanics. American Chemical Society; Washington, D.C: 1982. [Google Scholar]
- Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- Cech TR, Bass BL. Biological catalysis by RNA. Annu Rev Biochem. 1986;55:599–630. doi: 10.1146/annurev.bi.55.070186.003123. [DOI] [PubMed] [Google Scholar]
- Chong LT, Dempster SE, Hendsch ZS, Lee LP, Tidor B. Computation of electrostatic complements to proteins: A case of charge stabilized binding. Protein Sci. 1998;7:206–210. doi: 10.1002/pro.5560070122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C. Hydrophobic bonding and accessible surface area in proteins. Nature. 1974;248:338–339. doi: 10.1038/248338a0. [DOI] [PubMed] [Google Scholar]
- Cole PE, Yang SK, Crothers DM. Conformational changes of transfer ribonucleic acid. Equilibrium phase diagrams. Biochemistry. 1972;11:4358–4368. doi: 10.1021/bi00773a024. [DOI] [PubMed] [Google Scholar]
- Collins KD. Ion hydration: implications for cellular function, polyelectrolytes, and protein crystallization. Biophys Chem. 2006;119:271–281. doi: 10.1016/j.bpc.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Connolly ML. Computation of molecular volume. J Am Chem Soc. 1985;107:1118–1124. [Google Scholar]
- Cortis CM, Friesner RA. An automatic three-dimensional finite element mesh generation system for Poisson-Boltzmann equation. J Comput Chem. 1997a;18:1570–1590. [Google Scholar]
- Cortis CM, Friesner RA. Numerical solution of the Poisson-Boltzmann equation using tetrahedral finite-element meshes. J Comput Chem. 1997b;18:1591–1608. [Google Scholar]
- Dahm SC, Uhlenbeck OC. Role of divalent metal ions in the hammerhead RNA cleavage reaction. Biochemistry. 1991;30:9464–9469. doi: 10.1021/bi00103a011. [DOI] [PubMed] [Google Scholar]
- Davis ME, Madura JD, Sines J, Luty BA, Allison SA, McCammon JA. Diffusion-controlled enzymatic reactions. Methods Enzymol. 1991;202:473–497. doi: 10.1016/0076-6879(91)02024-4. [DOI] [PubMed] [Google Scholar]
- Davis ME, McCammon JA. Solving the finite difference linearized Poisson-Boltzmann equation: a comparison of relaxation and conjugate gradient methods. J Comput Chem. 1989;10:386–391. [Google Scholar]
- Davis ME, McCammon JA. Calculating electrostatic forces from grid-calculated potentials. J Comput Chem. 1990a;11:401–409. [Google Scholar]
- Davis ME, McCammon JA. Electrostatics in biomolecular structure and dynamics. Chem Rev. 1990b;90:509–521. [Google Scholar]
- De Camilli P, Emr SD, McPherson PS, Novick P. Phosphoinositides as regulators in membrane traffic. Science. 1996;271:1533–1539. doi: 10.1126/science.271.5255.1533. [DOI] [PubMed] [Google Scholar]
- Debye P, Hückel E. Zur Theorie der Elektrolyte. I Gefrierpunktserniedrigung und verwandte Erscheinungen. Physikalische Zeitschrift. 1923;24:185–206. [Google Scholar]
- del Álamo M, Mateu MG. Electrostatic repulsion, compensatory mutations, and long-range non-additive effects at the dimerization interface of the HIV capsid protein. J Mol Biol. 2005;345:893–906. doi: 10.1016/j.jmb.2004.10.086. [DOI] [PubMed] [Google Scholar]
- Delbruck H, Mueller U, Perl D, Schmid FX, Heinemann U. Crystal structures of mutant forms of the Bacillus caldolyticus cold shock protein differing in thermal stability. J Mol Biol. 2001;313:359–369. doi: 10.1006/jmbi.2001.5051. [DOI] [PubMed] [Google Scholar]
- Dominy BN, Brooks CL., III Development of a Generalized Born model parameterization for proteins and nucleic acids. J Phys Chem B. 1999;103:3765–3773. [Google Scholar]
- Dong F, Vijayakumar M, Zhou H-X. Comparison of calculation and experiment implicates significant electrostatic contributions to the binding stability of barnase and barstar. Biophys J. 2003;85:49–60. doi: 10.1016/S0006-3495(03)74453-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F, Zhou H-X. Electrostatic contributions to T4 lysozyme stability: solvent-exposed charges versus semi-buried salt bridges. Biophys J. 2002;83:1341–1347. doi: 10.1016/S0006-3495(02)73904-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper DE, Grilley D, Soto AM. Ions and RNA folding. Annu Rev Biophys Biomol Struct. 2005;34:221–243. doi: 10.1146/annurev.biophys.34.040204.144511. [DOI] [PubMed] [Google Scholar]
- Dyshlovenko PE. Adaptive numerical method for Poisson-Boltzmann equation and its application. Comp Phys Commun. 2002;147:335–338. [Google Scholar]
- Eisenberg D, McLachlan AD. Solvation energy in protein folding and binding. Nature. 1986;319:199–203. doi: 10.1038/319199a0. [DOI] [PubMed] [Google Scholar]
- Eisenberg H. Biological Macromolecules and Polyelectrolytes in Solution, Chapter 2. Clarendon; Oxford: 1976. [Google Scholar]
- Elcock AH, Gabdoulline RR, Wade RC, McCammon JA. Computer simulation of protein-protein association kinetics: Acetylcholinesterase-fasciculin. J Mol Biol. 1999;291:149–162. doi: 10.1006/jmbi.1999.2919. [DOI] [PubMed] [Google Scholar]
- Elcock AH, Potter MJ, Matthews DA, Knighton DR, McCammon JA. Electrostatic channeling in the bifunctional enzyme dihydrofolate reductase-thymidylate synthase. J Mol Biol. 1996;262:370–374. doi: 10.1006/jmbi.1996.0520. [DOI] [PubMed] [Google Scholar]
- Elcock AH, Sept D, McCammon JA. Computer simulation of protein-protein interactions. J Phys Chem B. 2001;105:1504–1518. [Google Scholar]
- Ermak DL, McCammon JA. Brownian dynamics with hydrodynamic interactions. J Chem Phys. 1978;69:1352–1360. [Google Scholar]
- Feig M, Brooks CL., III Recent advances in the development and application of implicit solvent models in biomolecule simulations. Curr Opin Struct Biol. 2004;14:217–224. doi: 10.1016/j.sbi.2004.03.009. [DOI] [PubMed] [Google Scholar]
- Figueirido F, Delbuono GS, Levy RM. Molecular mechanics and electrostatic effects. Biophys Chem. 1994;51:235–241. doi: 10.1016/0301-4622(94)00044-1. [DOI] [PubMed] [Google Scholar]
- Fitch CA, Karp DA, Lee KK, Stites WE, Lattman EE, Garcia-Moreno EB. Experimental pKa values of buried residues: analysis with continuum methods and role of water penetration. Biophys J. 2002;82:3289–3304. doi: 10.1016/s0006-3495(02)75670-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan MA, Ackers GK, Matthew JB, Hanania GIH, Gurd FRN. Electrostatic contributions to energetics of dimer-tetramer assembly in human hemoglobin: pH dependence and effect of specifically bound chloride ions. Biochemistry. 1981;20:7439–7449. doi: 10.1021/bi00529a018. [DOI] [PubMed] [Google Scholar]
- Frisch C, Schreiber G, Johnson CM, Fersht AR. Thermodynamics of the interaction of barnase and barstar: changes in free energy versus changes in enthalpy on mutation. J Mol Biol. 1997;267:696–706. doi: 10.1006/jmbi.1997.0892. [DOI] [PubMed] [Google Scholar]
- Gabdoulline RR, Wade RC. Simulation of the diffusional association of barnase and barstar. Biophys J. 1997;72:1917–1929. doi: 10.1016/S0006-3495(97)78838-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabdoulline RR, Wade RC. Brownian dynamics simulation of protein-protein diffusional encounter. Methods Enzymol. 1998;14:329–341. doi: 10.1006/meth.1998.0588. [DOI] [PubMed] [Google Scholar]
- Gabdoulline RR, Wade RC. Protein-protein association: investigation of factors influencing association rates by Brownian dynamics simulations. J Mol Biol. 2001;306:1139–1155. doi: 10.1006/jmbi.2000.4404. [DOI] [PubMed] [Google Scholar]
- Gabdoulline RR, Wade RC. Biomolecular diffusional association. Curr Opin Struct Biol. 2002;12:204–213. doi: 10.1016/s0959-440x(02)00311-1. [DOI] [PubMed] [Google Scholar]
- Gallicchio E, Kubo MM, Levy RM. Enthalpy-entropy and cavity decomposition of alkane hydration free energies: Numerical results and implications for theories of hydrophobic solvation. J Phys Chem B. 2000;104:6271–6285. [Google Scholar]
- Gallicchio E, Levy RM. AGBNP: An analytic implicit solvent model suitable for molecular dynamics simulations and high-resolution modeling. J Comput Chem. 2004;25:479–499. doi: 10.1002/jcc.10400. [DOI] [PubMed] [Google Scholar]
- Gallicchio E, Zhang LY, Levy RM. The SGB/NP hydration free energy model based on the surface generalized born solvent reaction field and novel nonpolar hydration free energy estimators. J Comput Chem. 2002;21:86–104. doi: 10.1002/jcc.10045. [DOI] [PubMed] [Google Scholar]
- Georgescu RE, Alexov EGRGM. Combining conformational flexibility and continuum electrostatics for calculating pKas in proteins. Biophys J. 2002;83:1731–1748. doi: 10.1016/S0006-3495(02)73940-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson M. Introduction to continuum electrostatics. In: Beard DA, editor. Biophysics textbook online. Biophysical society; Bethesda, MD: 2000. [Google Scholar]
- Gilson M, Davis ME, Luty BA, McCammon JA. Computation of electrostatic forces on solvated molecules using the Poisson-Boltzmann equation. J Phys Chem. 1993;97:3591–3600. doi: 10.1063/1.1924448. [DOI] [PubMed] [Google Scholar]
- Gilson MK. Theory of electrostatic interactions in macromolecules. Curr Opin Struct Biol. 1995;5:216–223. doi: 10.1016/0959-440x(95)80079-4. [DOI] [PubMed] [Google Scholar]
- Grant JA, Pickup BT, Nicholls A. A smooth permittivity function for Poisson-Boltzmann solvation methods. J Comput Chem. 2001;22:608–640. [Google Scholar]
- Green DF, Tidor B. Design of improved protein inhibitors of HIV-1 cell entry: Optimization of electrostatic interactions at the binding interface. Proteins. 2005;60:644–657. doi: 10.1002/prot.20540. [DOI] [PubMed] [Google Scholar]
- Hofmeister F. Zur lehre von der wirkung der salze. zweite mittheilung. Arch Exp Pathol Pharmakol. 1888;24:247–260. [Google Scholar]
- Holm C, Kekicheff P, Podgornik R. Electrostatic effects in soft matter and biophysics. Kluwer academic publishers; Boston, MA: 2001. [Google Scholar]
- Holst M, Baker N, Wang F. Adaptive multilevel finite element solution of the Poisson-Boltzmann equation i. Algorithms and examples. J Comput Chem. 2000;21:1319–1342. [Google Scholar]
- Holst M, Saied F. Multigrid solution of the Poisson-Boltzmann equation. J Comput Chem. 1993;14:105–113. [Google Scholar]
- Holst MJ, Saied F. Numerical solution of nonlinear Poisson-Boltzmann equation: developing more robust and efficient methods. J Comput Chem. 1995;16:337–364. [Google Scholar]
- Honig BH, Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- Horn HW, Swope WC, Pitera JW, Madura JD, Dick TJ, Hura GL, Head-Gordon T. Development of an improved four-site water model for biomolecular simulations: TIP4P-Ew. Journal of Chemical Physics. 2004;120:9665–9678. doi: 10.1063/1.1683075. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD -- visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Ilin A, Bagheri B, Scott LR, Briggs JM, McCammon JA. Parallelization of Poisson-Boltzmann and Brownian Dynamics calculations. American Chemical Society Symposium Series. 1995;592:170–185. [Google Scholar]
- Im W, Beglov D, Roux B. Continuum solvation model:electrostatic forces from numerical solutions to the Poisson-Boltzmann equation. Comp Phys Commun. 1998;11:59–75. [Google Scholar]
- Irvine RF. Nuclear lipid signaling. SciSTKE. 2002;150:1–12. doi: 10.1126/stke.2002.150.re13. [DOI] [PubMed] [Google Scholar]
- Jackson JD. Classical Electrodynamics. Wiley; New York: 1975. [Google Scholar]
- Janin J, Chothia C. The structure of protein-protein recognition sites. J Biol Chem. 1990;265:16027–16030. [PubMed] [Google Scholar]
- Jensen JH, Li H, Robertson AD, Molina PA. Prediction and rationalization of protein pKa values using QM and QM/MM methods. J Phys Chem A. 2005;109:6634–6643. doi: 10.1021/jp051922x. [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- Juffer AH, Botta EFF, van Keulen BAM, van der Ploeg A, Berendsen HJC. The electric potential of a macromolecule in solvent: a fundamental approach. J Comput Phys. 1991;97:144–171. [Google Scholar]
- Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham ITE. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- Krieger E, Nielsen JE, Spronk CAEM, Vriend G. Fast empirical pKa prediction by Ewald summation. J Mol Graph Model. doi: 10.1016/j.jmgm.2006.02.009. in press. [DOI] [PubMed] [Google Scholar]
- Lamm G. The Poisson-Boltzmann Equation. In: Lipkowitz KB, Larter R, Cundari TR, editors. Reviews in Computational Chemistry. John Wiley and Sons, Inc; Hoboken, N.J: 2003. pp. 147–366. [Google Scholar]
- Landau LD, Lifshitz EM, Pitaevskii LP. Electrodynamics of Continuous Media. Butterworth-Heinenann; Boston, MA: 1982. [Google Scholar]
- Laux T, Fukami K, Thelen M, Golub T, Frey D, Caroni P. GAP43, MARCKS, CAP23 modulate PI(4,5)P2 at plasmalemmal rafts, and regulate cell cortex actin dynamics through a common mechanism. J Cell Biol. 2000;149:1455–1472. doi: 10.1083/jcb.149.7.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law MJ, Linde ME, Chambers EJ, Oubridge C, Katsamba PS, Nilsson L, Haworth IS, Laird-Offringa IA. The role of positively charged amino acids and electrostatic interactions in the complex of U1A protein and U1 hairpin II RNA. Nucleic Acids Res. 2006;34:275–285. doi: 10.1093/nar/gkj436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach AR. Molecular Modeling: Principles and Applications. Prentice Hall; Harlow, England: 2001. [Google Scholar]
- Lee B, Richards FM. The interpretation of protein structures: Estimation of static accessibility. J Mol Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Lee LP, Tidor B. Optimization of binding electrostatics: Charge complementarity in the barnase-barstar protein complex. Protein Sci. 2001;10:362–377. doi: 10.1110/ps.40001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Hains AW, Everts JE, Robertson AD, Jensen JH. The prediction of protein pKa’s using QM/MM: The pKa of lysine 55 in turkey ovomucoid third domain. J Phys Chem B. 2002;106:3486–3494. [Google Scholar]
- Li H, Robertson AD, Jensen JH. The determinants of carboxyl pKa values in turkey ovomucoid third domain. Proteins. 2004;55:689–704. doi: 10.1002/prot.20032. [DOI] [PubMed] [Google Scholar]
- Lin J-H, Baker NA, McCammon JA. Bridging the implicit and explicit solvent approaches for membrance electrostatics. Biophys J. 2002;83:1374–1379. doi: 10.1016/S0006-3495(02)73908-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linderström-Lang K. On the ionisation of proteins. Comptes-rend Lab Carlaberg. 1924;15:1–29. [Google Scholar]
- Livesay DR, Jambeck P, Rojnuckarin A, Subramaniam S. Conservation of electrostatic properties within enzyme families and superfamilies. Biochemistry. 2003;42:3464–3473. doi: 10.1021/bi026918f. [DOI] [PubMed] [Google Scholar]
- Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- Luo R, Head MS, Moult J, Gilson MK. pKa shifts in small molecules and HIV protease: electrostatics and conformation. J Am Chem Soc. 1998;120:6138–6146. [Google Scholar]
- Luty BA, Elamrani S, McCammon JA. Simulation of the bimolecular reaction between superoxide and superoxide dismutase - synthesis of the encounter and reaction steps. J Am Chem Soc. 1993;115:11874–11877. [Google Scholar]
- Ma B, Elkayam T, Wolfson H, Nussinov R. Protein-protein interactions: structurally conserved residues distinguish between binding sites and exposed protein surfaces. Proc Natl Acad Sci U S A. 2003;100:5772–5777. doi: 10.1073/pnas.1030237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell ADJ, Nilsson L. Nucleic Acid Simulation. In: Becker OM, MacKerell ADJ, Roux B, Watanabe M, editors. Computational biochemistry and biophysics. Marcel Dekker; New York: 2001. pp. 441–463. [Google Scholar]
- Madura JD, Briggs JM, Wade RC, Davis ME, Luty BA, Antosiewicz IJ, Gilson MK, Bagheri N, Scott LR, McCammon JA. Electrostatics and diffusion of molecules in solution-simulations with the University of Houston Brownian Dynamics program. Comp Phys Commun. 1995;91:57–95. [Google Scholar]
- Madura JD, McCammon JA. Brownian dynamics simulation of diffusional encounters between triose phosphate isomerase and d-glyceraldehyde phosphate. J Phys Chem. 1989;93:7285–7587. [Google Scholar]
- Manning GS. The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Q Rev Biophys. 1978;11:179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- Martin TF. PI(4,5)P2 regulation of surface membrane traffic. Curr Opin Cell Biol. 2001;13:493–499. doi: 10.1016/s0955-0674(00)00241-6. [DOI] [PubMed] [Google Scholar]
- Massova I, Kollman PA. Combined molecular mechanical and continuum solvent approach (MM-PBSA/GBSA) to predict ligand binding. Perspect Drug Discov Des. 2000;18:113–135. [Google Scholar]
- Masunov A, Lazaridis T. J Am Chem Soc. 2003;125:1722–1730. doi: 10.1021/ja025521w. [DOI] [PubMed] [Google Scholar]
- McLaughlin S, Wang J, Gambhir A, Murray D. PIP2 and proteins: interactions, organization and information flow. Annu Rev Biophys Biomol Struct. 2002;31:151–175. doi: 10.1146/annurev.biophys.31.082901.134259. [DOI] [PubMed] [Google Scholar]
- McQuarrie DA. Statistical Mechanics. HarperCollins; 1976. [Google Scholar]
- Micu AM, Bagheri B, Ilin AV, Scott LR, Pettitt BM. Numerical considerations in the computation of the electrostatic free energy of interaction within the poisson-boltzmann theory. J Comput Phys. 1997;136:263–271. [Google Scholar]
- Misra VK, Draper DE. Mg(2+) binding to tRNA revisited: the nonlinear Poisson-Boltzmann model. J Mol Biol. 2000;299:1135–1147. doi: 10.1006/jmbi.2000.3769. [DOI] [PubMed] [Google Scholar]
- Misra VK, Draper DE. A thermodynamic framework for Mg2+ binding to RNA. Proc Natl Acad Sci U S A. 2001;98:12456–12461. doi: 10.1073/pnas.221234598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra VK, Hecht JL, Yang A-S, Honig B. Electrostatic contributions to the binding free energy of λcI repressor to DNA. Biophys J. 1998;75:2262–2273. doi: 10.1016/S0006-3495(98)77671-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KJM, Lohman TM. Kinetic mechanism of adenine nucleotide binding to and hydrolysis by the Escherichia coli Rep monomer. 2 Application of a kinetic competition approach. Biochemistry. 1994;33:14565–78. doi: 10.1021/bi00252a024. [DOI] [PubMed] [Google Scholar]
- Mueller U, Perl D, Schmid FX, Heinemann U. Thermal stability and atomic-resolution crystal structure of the Bacillus caldolyticus cold shock protein. J Mol Biol. 2000;297:975–988. doi: 10.1006/jmbi.2000.3602. [DOI] [PubMed] [Google Scholar]
- Murray D, Ben-Tal N, Honig B, McLaughlin S. Electrostatic interaction of myristoylated proteins with membranes: simple physics, complicated biology. Structure. 1997;5:985–989. doi: 10.1016/s0969-2126(97)00251-7. [DOI] [PubMed] [Google Scholar]
- Nicholls A, Honig B. A rapid finit difference algorithm, utilizing successive over-relaxation to solve the Poisson-Boltamann equation. J Comput Chem. 1991;12:435–445. [Google Scholar]
- Nielsen JE, Vriend G. Optimizing the hydrogen-bond network in poisson-boltzmann equation-based pk(a) calculations. Proteins. 2001;43:403–412. doi: 10.1002/prot.1053. [DOI] [PubMed] [Google Scholar]
- Nielsen JE, McCammon JA. On the evaluation and optimization of protein x-ray structures for pKa calculations. Protein Sci. 2003;12:313–326. doi: 10.1110/ps.0229903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonner W, Gillespe D, Henderson D, Eisenberg D. Ion accumulation in biologycal calcium channel: effects of solvent and confining pressure. J Phys Chem B. 2001;105:6427–6436. [Google Scholar]
- Norel R, Sheinerman F, Petrey D, Honig B. Electrostatic contributions to protein-protein interactions: Fast energetic filters for docking and their physical basis. Protein Sci. 2001;10:2147–2161. doi: 10.1110/ps.12901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northrup SH, Allison SA, McCammon JA. Brownian dynamics simulation of diffusion-influenced biomolecular reactions. J Chem Phys. 1984;80:1517–1524. [Google Scholar]
- Novotny J, Sharp K. Electrostatic fields in antibodies and antibody/antigen complexes. Prog Biophys Mol Biol. 1992;58:203–224. doi: 10.1016/0079-6107(92)90006-r. [DOI] [PubMed] [Google Scholar]
- Onufriev A, Case DA, Bashford D. Effective born radii in the Generalized Born approximation: the importance of being perfect. J Comput Chem. 2002;23:1297–1304. doi: 10.1002/jcc.10126. [DOI] [PubMed] [Google Scholar]
- Osapay K, Young WS, Bashford D, Brooks CL, III, Case DA. Dielectric continuum models for hydration effects on peptide conformation transitions. J Phys Chem. 1996;100:2698–2705. [Google Scholar]
- Overman LB, Lohman TM. Lingkage of pH, aion and cation effectx in protein-nucleic acid equilibria. Escherichia coli SSB protein-single strand nucleic acid interactions. J Mol Biol. 1994;236:165–178. doi: 10.1006/jmbi.1994.1126. [DOI] [PubMed] [Google Scholar]
- Pace CN. Single surface stabilizer. Nat Struct Biol. 2000;7:345–346. doi: 10.1038/75100. [DOI] [PubMed] [Google Scholar]
- Payrastre B, Missy K, Giuriato S, Bodin S, Plantavid M, Gratacap M. Phosphoinositides: key players in cell signalling, in time and space. Cell Signal. 2001;13:377–387. doi: 10.1016/s0898-6568(01)00158-9. [DOI] [PubMed] [Google Scholar]
- Perl D, Mueller U, Heinemann U, Schmid FX. Two exposed amino acid residues confer thermostability on a cold shock protein. Nat Struct Biol. 2000;7:380–383. doi: 10.1038/75151. [DOI] [PubMed] [Google Scholar]
- Perl D, Schmid FX. Electrostatic stabilization of a thermophilic cold shock protein. J Mol Biol. 2001;213:343–357. doi: 10.1006/jmbi.2001.5050. [DOI] [PubMed] [Google Scholar]
- Ponder JW, Case DA. Force fields for protein simulations. Adv Protein Chem. 2003;66:27–85. doi: 10.1016/s0065-3233(03)66002-x. [DOI] [PubMed] [Google Scholar]
- Radic Z, Kirchhoff PD, Quinn DM, McCammon JA, Taylor P. Electrostatic influence on the kinetics of ligand binding to acetylcholinesterase. J Biol Chem. 1997;272:23265–23277. doi: 10.1074/jbc.272.37.23265. [DOI] [PubMed] [Google Scholar]
- Rauch ME, Ferguson CG, Prestwich GD, Cafiso D. Myristoylated alanine-rich C kinase Substrate (MARCKS) sequesters spin-labeled phosphatidylinositol-4,5-bisphosphate in lipid bilayers. J Biol Chem. 2002;277:14068–14076. doi: 10.1074/jbc.M109572200. [DOI] [PubMed] [Google Scholar]
- Raucher D, Stauffer T, Chen W, Shen K, Guo S, York JD, Sheetz MP, Meyer T. Phosphatidylinositol 4,5-bisphosphate functions as a second messenger that regulates cytoskeleton-plasma membrane adhesion. Cell. 2000;100:221–228. doi: 10.1016/s0092-8674(00)81560-3. [DOI] [PubMed] [Google Scholar]
- Record MT, Ha J-H, Fisher MA. Analysis of equilibrium and kinetic measurements to determine thermodynamic origins of stability and specificity and mechanism of formation of site-specific complexes between proteins and helical DNA. Methods Enzymol. 1991;208:291–343. doi: 10.1016/0076-6879(91)08018-d. [DOI] [PubMed] [Google Scholar]
- Reuter H, Pott C, Goldhaber JI, Henderson SA, Philipson KD, Schwinger RH. Na(+)--Ca2+ exchange in the regulation of cardiac excitation-contraction coupling. Cardiovasc Res. 2005;67:198–207. doi: 10.1016/j.cardiores.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Römer R, Hach R. tRNA conformation and magnesium binding. A study of a yeast phenylalanine-specific tRNA by a fluorescent indicator and differential melting curves. Eur J Biochem. 1975;55:271–284. doi: 10.1111/j.1432-1033.1975.tb02160.x. [DOI] [PubMed] [Google Scholar]
- Romer R, Hach R. tRNA conformation and magnesium binding. A study of yeast phenylalanine-specific tRNA by fluorecent indicator and differential melting curves. Eur J Biochem. 1975;55:271–284. doi: 10.1111/j.1432-1033.1975.tb02160.x. [DOI] [PubMed] [Google Scholar]
- Roux B. Implicit solvent models. In: Becker OM, Mackerell AD Jr, Roux B, Watanabe M, editors. Computational Biochemistry and biophysics. Marcel Dekker; New York: 2001. pp. 133–152. [Google Scholar]
- Roux B, Simonson T. Implicit solvent models. Biophys Chem. 1999;78:1–20. doi: 10.1016/s0301-4622(98)00226-9. [DOI] [PubMed] [Google Scholar]
- Russell RB, Alber F, Aloy P, Davis FP, Korkin D, Pichaud M, Topf M, Sali A. A structural perspective on protein-protein interactions. Curr Opin Struct Biol. 2004;14:313–324. doi: 10.1016/j.sbi.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Sagui C, Darden TA. Molecular dynamics simulation of biomolecules: long-range electrostatic effects. Annu Rev Biophys Biomol Struct. 1999;28:155–179. doi: 10.1146/annurev.biophys.28.1.155. [DOI] [PubMed] [Google Scholar]
- Schaeler M, Karplus M. A comprehensive analytical treatment of continuum electrostatics. J Phys Chem. 1996;100:1578–1599. [Google Scholar]
- Schlick T. Molecular Modeling and Simulation: An Interdisciplinary Guide. Springer-Verlag; New York: 2002. [Google Scholar]
- Schreiber G, Fersht AR. Energetics of protein-protein interactions: analysis of the barnase-barstar interface by single mutations and double mutant cycles. J Mol Biol. 1995;248:478–486. doi: 10.1016/s0022-2836(95)80064-6. [DOI] [PubMed] [Google Scholar]
- Schreiber G, Fersht AR. Interaction of barnase with its polypeptide inhibitor barstar studied by protein engineering. Biochemistry. 1993;32:5145–5150. doi: 10.1021/bi00070a025. [DOI] [PubMed] [Google Scholar]
- Senear DF, Batey R. Comparison of operator-specific and nonspecific interactions of lambda cI repressor: [KCL] and pH effects. Biochemistry. 1991;30:6677–6688. doi: 10.1021/bi00241a007. [DOI] [PubMed] [Google Scholar]
- Sept D, Baker NA, McCammon JA. The physical basis of microtubule structure and stability. Protein Sci. 2003;12:2257–2261. doi: 10.1110/ps.03187503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sept D, Elcock AH, McCammon JA. Computer simulations of actin polymerization can explain the barbed-pointed end asymmetry. J Mol Biol. 1999;294:1181–1189. doi: 10.1006/jmbi.1999.3332. [DOI] [PubMed] [Google Scholar]
- Sept D, McCammon JA. Thermodynamics and kinetics of actin filament nucleation. Biophys J. 2001;81:667–674. doi: 10.1016/S0006-3495(01)75731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp KA, Honig BH. Electrostatic interactions in macromolecules: theory and applications. Annu Rev Biophys Biophys Chem. 1990;19:301–332. doi: 10.1146/annurev.bb.19.060190.001505. [DOI] [PubMed] [Google Scholar]
- Sharp KA, Nicholls A, Fine RF, Honig B. Reconciling the magnitude of the microscopic and macroscopic hydrophobic effects. Science. 1991;252:106–109. doi: 10.1126/science.2011744. [DOI] [PubMed] [Google Scholar]
- Sheinerman FB, Honig B. On the role of electrostatic interactions in the design of protein-protein interfaces. J Mol Biol. 2002;318:161–177. doi: 10.1016/S0022-2836(02)00030-X. [DOI] [PubMed] [Google Scholar]
- Sheinerman FB, Norel R, Honig B. Electrostatic aspects of protein-protein interactions. Curr Opin Struct Biol. 2000;10:153–159. doi: 10.1016/s0959-440x(00)00065-8. [DOI] [PubMed] [Google Scholar]
- Shimizu S. Estimating hydration changes upon biomolecular reactions from osmotic stress, high pressure, and preferential hydration experiments. Proc Natl Acad Sci U S A. 2004;101:1195–1199. doi: 10.1073/pnas.0305836101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Smith D. Preferential hydration and the exclusion of cosolvents from protein surfaces. J Chem Phys. 2004;121:1148–1154. doi: 10.1063/1.1759615. [DOI] [PubMed] [Google Scholar]
- Simonson T. Electrostatics and dynamics of proteins. Rep Prog Phys. 2003;66:737–787. [Google Scholar]
- Simonson T, Brunger AT. Solvation free energies estimated from macroscopic continuum theory: an accuracy assessment. J Phys Chem. 1994;98:4683–4694. [Google Scholar]
- Sines JJ, McCammon JA, Allison SA. Kinetic effects of multiple charge modifications in enzyme-substrate reactions - Brownian Dynamics simulations of Cu, Zn superoxide dismutase. J Comput Chem. 1992;13:66–69. [Google Scholar]
- Sitkoff D, Sharp KA, Honig B. Correlating solvation free energies and surface tensions of hydrocarbon solutes. Biophys Chem. 1994a;51:397–409. doi: 10.1016/0301-4622(94)00062-x. [DOI] [PubMed] [Google Scholar]
- Sitkoff D, Sharp KA, Honig BH. Accurate calculation of hydration free energies using macroscopic solvent models. J Phys Chem. 1994b;98:1978–1988. [Google Scholar]
- Spaar A, Dammer C, Gabdoulline RR, Wade RC, Helms V. Diffusional encounter of barnase and barstar. Biophys J. 2006;90:1913–1924. doi: 10.1529/biophysj.105.075507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaar A, Helms V. Free energy landscape of protein-protein encounter resulting from brownian dynamics simulations of barnase:Barstar. J Chem Theory Comput. 2005;1:723–736. doi: 10.1021/ct050036n. [DOI] [PubMed] [Google Scholar]
- Spolar RS, Ha JH, Record MTJ. Hydrophobic effect in protein folding and other noncovalent processes involving proteins. Proc Natl Acad Sci U S A. 1989;86:8382–8385. doi: 10.1073/pnas.86.21.8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A, Crothers DM. Conformational changes of transfer RNA. The role of magnesium(II) Biochemistry. 1976;15:160–167. doi: 10.1021/bi00646a025. [DOI] [PubMed] [Google Scholar]
- Still WC, Tempczyk A, Hawley RC, Hendrickson T. Semianalytical treatment of solvation for molecular mechanics and dynamics. J Am Chem Soc. 1990;112:6127–6129. [Google Scholar]
- Su Y, Gallicchio E. The non-polar solvent potential of mean force for the dimerization of alanine dipeptide: the role of solute-solvent van der Waals interactions. Biophys Chem. 2004;109:251–260. doi: 10.1016/j.bpc.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Swanson JMJ, Henchman RH, McCammon JA. Revisiting free energy calculations: A theoretical connection to MM/PBSA and direct calculation of the association free energy. Biophys J. 2004;86:67–74. doi: 10.1016/S0006-3495(04)74084-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan RC, Truong TN, McCammon JA, Sussman JL. Acetylcholinesterase - electrostatic steering increases the rate of ligand binding. Biochemistry. 1993;32:401–403. doi: 10.1021/bi00053a003. [DOI] [PubMed] [Google Scholar]
- Tanford C, Kirkwood JG. Theory of protein titration curves. I General equations for impenetrable spheres. J Am Chem Soc. 1957;79:5333–5339. [Google Scholar]
- Tanford C, Roxby R. Interpretation of protein titration curves. Biochemistry. 1972;11:2192–2198. doi: 10.1021/bi00761a029. [DOI] [PubMed] [Google Scholar]
- Tara S, Elcock AH, Kirchhoff PD, Briggs JM, Radic Z, Taylor P, McCammon JA. Rapid binding of a cationic active site inhibitor to wild type and mutant mouse acetylcholinesterase: Brownian dynamics simulation including diffusion in the active site gorge. Biopolymers. 1998;46:465–474. doi: 10.1002/(SICI)1097-0282(199812)46:7<465::AID-BIP4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Toker A. The synthesis and cellular roles of phosphatidylinositol 4,5-bisphosphate. Curr Opin Cell Biol. 1998;10:254–261. doi: 10.1016/s0955-0674(98)80148-8. [DOI] [PubMed] [Google Scholar]
- Vasker IA. Protein-protein interfaces are special. Structure. 2004;12:910–912. doi: 10.1016/j.str.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Verwey EJW, Overbeek JTG. Theory of Stability of Lyophobic Colloids. Dover Publications, Inc; Mineola, New York: 1999. [Google Scholar]
- Wagoner J, Baker NA. Solvation forces on biomolecular structures: a comparison of explicit solvent and Poisson-Boltzmann models. J Comput Chem. 2004;25:1623–1629. doi: 10.1002/jcc.20089. [DOI] [PubMed] [Google Scholar]
- Wagoner JA, Baker NA. Assessing implicit models for nonpolar mean solvation forces: the importance of dispersion and volume terms. Proc Natl Acad Sci U S A. 2006;103:8331–8336. doi: 10.1073/pnas.0600118103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Arbuzova A, Hangyas-Mihalyne G, McLaughlin S. The effector domain of myristoylated alanine-rich C kinase substrate (MARCKS) binds strongly to phosphatidylinositol 4,5-bisphosphate (PIP2) J Biol Chem. 2001a;276:5012–5019. doi: 10.1074/jbc.M008355200. [DOI] [PubMed] [Google Scholar]
- Wang J, Gambhir A, Hangyas-Mihalyne G, Murray D, Golebiewska U, McLaughlin S. Lateral sequestration of phosphatidylinositol 4,5-bisphosphate by the basic effector domain of myristoylated alanine-rich C kinase substrate is due to nonspecific electrostatic interactions. J Biol Chem. 2002;277:34401–34412. doi: 10.1074/jbc.M203954200. [DOI] [PubMed] [Google Scholar]
- Wang J, Gambhir A, McLaughlin S, Murray D. A computational model for the electrostatic sequestration of PI(4,5)P2 by membrane-adsorbed basic peptides. Biophys J. 2004;86:1969–1986. doi: 10.1016/S0006-3495(04)74260-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wade RC. Implicit solvent models for flexible protein-protein docking by molecular dynamics simulation. Proteins. 2003;50:158–169. doi: 10.1002/prot.10248. [DOI] [PubMed] [Google Scholar]
- Wang W, Donini O, Reyes CM, Kollman PA. Biomolecular simulations: recent development in force fields, simulationa of enzyme catalysis, protein-ligand, protein-protein, and protein-nucleic acid noncovalent interactions. Annu Rev Biophys Biomol Struct. 2001b;30:211–243. doi: 10.1146/annurev.biophys.30.1.211. [DOI] [PubMed] [Google Scholar]
- Warwicker J, Watson HC. Calculation of the electric potential in the active site cleft due to alphs-helix dipoles. J Mol Biol. 1982;157:671–679. doi: 10.1016/0022-2836(82)90505-8. [DOI] [PubMed] [Google Scholar]
- Wesson L, Eisenberg D. Atomic solvation parameters applied to molecular dynamics of proteins in solution. Protein Sci. 1992;1:227–235. doi: 10.1002/pro.5560010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CF, Hünenberger PH, Akamine P, Narayana N, Diller T, McCammon JA, Taylor S, Xuong N-H. Computational analysis of PKA-balanol interactions. J Med Chem. 2001;44:1530–1539. doi: 10.1021/jm000443d. [DOI] [PubMed] [Google Scholar]
- Xu D, Lin SL, Nussinov R. Protein binding versus protein folding: The role of hydrophilic bridges in protein associations. J Mol Biol. 1997;265:68–84. doi: 10.1006/jmbi.1996.0712. [DOI] [PubMed] [Google Scholar]
- Yin HL, Janmey PA. Phosphoinositide regulation of the actin cytoskeleton. Annu Rev Physiol. 2003;65:761–789. doi: 10.1146/annurev.physiol.65.092101.142517. [DOI] [PubMed] [Google Scholar]
- Yu Z, Jacobson MP, Josovitz J, Rapp CS, Friesner RA. First-shell solvation of ion pairs: Correction of systematic errors in implicit solvent models. J Phys Chem B. 2004;108:6643–6654. [Google Scholar]
- Zauhar RJ, Morgan RJ. The rigorous computation of the molecular electric potential. J Comput Chem. 1988;9:171–187. [Google Scholar]
- Zhou HX. Interactions of macromolecules with salt ions: an electrostatic theory for the Hofmeister effect. Proteins. 2005;61:69–78. doi: 10.1002/prot.20500. [DOI] [PubMed] [Google Scholar]
- Zhou HX, Dong F. Electrostatic contributions to the stability of a thermophilic cold shock protein. Biophys J. 2003;84:2216–2222. doi: 10.1016/S0006-3495(03)75027-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu ZY, Karlin S. Clusters of charged residues in protein three-dimensional structures. Proc Natl Acad Sci U S A. 1996;93:8350–8355. doi: 10.1073/pnas.93.16.8350. [DOI] [PMC free article] [PubMed] [Google Scholar]