Abstract
Mature blood cells develop from multipotent hematopoietic stem cells through a series of sequential intermediates in which the developmental potential for particular blood lineages is progressively extinguished. We previously reported the identification of one of these developmental intermediates, the common lymphoid progenitor (CLP), which can give rise to T cells, B cells, dendritic cells (DCs), and natural killer cells (NKs), but lacks myeloid and erythroid potential. Recently, several studies have suggested that the T-cell and DC potential of CLP is limited or absent, and/or that CLP contains significant myeloid potential. Here, we show that the originally identified CLP population can be divided into functionally distinct subsets based on the expression of the tyrosine kinase receptor, Flk2. The Flk2+ subset contains robust in vivo and in vitro T-cell, B-cell, DC, and NK potential, but lacks myeloid potential and, therefore, represents an oligopotent, lymphoid-restricted progenitor. This population of cells does not appear to be B cell–biased and robustly reconstitutes both B and T lineages in vivo, consistent with its being a physiologic progenitor of both of these subsets. Thus, Flk2 expression defines a homogeneous, readily obtainable subset of bone marrow CLP that is completely lymphoid-committed and can differentiate equivalently well into both B and T lineages.
Introduction
Hematopoietic stem cells (HSCs) have the unique ability to both self renew for the lifespan of an organism and to differentiate into more restricted progenitors.1 Through multiple rounds of proliferation and irreversible steps of lineage commitment, HSCs ultimately drive the production of all mature blood cells. In the current hierarchical model of hematopoiesis, the first lineage decision is made at the bifurcation between the myeloid/erythroid/megakaryocytic and the lymphoid lineages.2 This first step in lineage commitment is evidenced by the existence of 2 distinct progenitor populations, the common myeloid/erythroid progenitor (CMP)2 and common lymphoid progenitor (CLP). CLPs were originally defined as Lin−Sca-1intc-KitintIL-7Rα+ cells and as a population gave rise to T cells, B cells, natural killer (NK) cells, CD8+ and CD8− dendritic cells (DCs), Langerhans cells, and interferon-α producing DCs but not to macrophages, erythrocytes, or platelets.3–6
Both the lineage potential and physiologic relevance of CLPs have been questioned recently on the basis of results from several studies. Some studies have suggested limited or negligible T-cell production from CLPs, and have suggested that CLPs are primarily progenitors for B cells.7–9 In contrast, other studies have reported that CLP or their downstream progenitors retain myeloid potential, and are therefore not lymphoid committed (examples are Allman et al,7 Balciunaite et al,10 Perry et al,11 and Rumfelt et al,12 and reviewed in Ye and Graf,13 Pelayo et al,14 and Bhandoola et al15). If the CLP does not possess physiologically relevant levels of T-cell potential, or is not actually lymphoid committed, our current models of myeloid, T- and B-cell development will need to be reconsidered. Therefore, it will be an important advance in the field to unambiguously prove or disprove the existence of the CLPs, and to determine, whether CLPs are robust sources of all lymphoid lineages in vivo.
Expression of the cytokine receptor Flt3/Flk2 is restricted to early hematopoietic progenitors and dendritic cells.16,17 We and others have previously shown that short-term (ST)-HSCs and multipotent progenitors (MPPs) express Flk2 whereas long-term (LT)-HSCs do not.18,19 Surprisingly, when CLPs were reanalyzed following publication of Flk2 as a multipotent cell marker (recently confirmed by Forsberg et al20), we discovered that distinct subsets of CLPs could be identified on the basis of Flk2 expression,17 here labeled CLPF and CLPF− (following the convention we established in Forsberg et al20). We analyzed the developmental potential of CLPF and CLPF− and demonstrated that while the Flk2− subfraction of CLP contain only B-restricted progenitors, Flk2+ CLP possess robust T-cell, B-cell, NK, and DC potential, and completely lack myeloid potential. Furthermore, this oligopotency can be observed at the single cell level, definitively demonstrating that this population contains a common lymphoid-restricted progenitor. CLPF, when injected in vivo, robustly reconstitute both the B and T lineages. Therefore, CLPF represent the lymphoid branch of the hematopoietic tree, and Flk2 expression is a defining hallmark of these oligopotent progenitors.
Methods
Animals
All animal procedures were approved by the Institutional Animal Care and Use Committee and in compliance with the Public Health Service Policy for Humane Care and Use of Laboratory Animals. Balb/c, C57BL/Ka-Thy1.1CD45.2+ (BA), congenic C57BL/Ka-Thy1.1CD45.1+ (HZ), and C57BL/Ka rag2−/−γc−/− mice were bred and maintained at the Stanford Animal Facility according to the University's guidelines.
Cytokines and media
Sorted progenitor cells were cultured in Iscove-modified Dulbecco medium (IMDM) supplemented with 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA), L-Glutamine (5 mM; Invitrogen), 50 μM 2-mercaptoethanol, 1× nonessential amino acids, and penicillin G (100 U/mL)/streptomycin (100 μg/mL; Lonza Walkersville, Walkersville, MD). Recombinant murine IL-3, IL-7, IL-15, macrophage colony-stimulating factor (M-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), stem cell factor (SCF), and human Flt3L were purchased from R&D Systems (Minneapolis, MN) and reconstituted in sterile phosphate-buffered saline (PBS)/1% bovine serum albumin (BSA). Cell staining and sorting were done in PBS with 2% FBS.
Antibodies
The following monoclonal antibodies were produced in our laboratory: M1/70 (anti–Mac-1/CD11b), 8C5 (anti–Gr-1), 6B2 (anti-B220), KT-31 (anti-CD3), GK1.5 (anti-CD4), 53-7.3 (anti-CD5), 53-6.7 (anti-CD8), A7R34 (anti–IL-7Rα/CD127), 2B8 (anti-c-Kit/CD117), 3C11 (anti-c-Kit/CD117), E13-161-7 (anti–Sca-1), 19XE5 (anti-Thy-1.1), A20.1.7 (anti-CD45.1), AL1-4A2 (anti-CD45.2). Goat antirat IgG (phycoerythrin [PE] or Cy5-PE conjugated) was purchased from Invitrogen. Monoclonal antibodies to I-Ab, CD3, CD4, CD8, CD11b, CD11c, CD19, CD24, CD25, CD27, CD34, CD43, CD44, CD62L, CD93/AA4.1, Flk2, NK1.1, TCRβ, and isotype controls were purchased from BD Biosciences (San Jose, CA), eBioscience (San Diego, CA) or Biolegend (San Diego, CA). For visualization of biotinylated antibodies, streptavidin-conjugated PE, Cy5-PE, Cy7-PE, and Texas Red (Invitrogen, Carlsbad, CA) were used.
Isolation of hematopoietic progenitor cells from BM
Progenitor populations were isolated from bone marrow (BM) or thymus based on phenotype as previously described: hematopoietic stem cells (HSC; Lin−/CD127−/c-Kithi/Scahi/Thy1.1lo Flk2−); multipotent pogenitors (MPPF; Lin−/CD127−/c-Kithi/Scahi/Thy1.1−Flk2+); common lymphoid progenitors (CLP; Lin−/CD127+/c-Kitint/Scaint/Thy1.1−); thymic progenitors c-Kithi DN1 (Lin−/low/CD44hi/c-Kithi/CD25−); c-Kithi DN2 (Lin−/low/CD44hi/c-Kithi/CD25+); and pro B cells (B220+/CD43+/CD19+/IgM−). The lineage cocktail included antibodies directed against CD3, CD4, CD5, CD8, CD19, B220, Gr-1, Mac-1, NK1.1, and Ter-119. All populations were double sorted on a BD FACSVantage SE or BD FACSAria to high purity (> 99%; BD Biosciences). Lineage-specificity of progenitor populations was confirmed by flow cytometric analysis of spleen and thymus of mice that received transplants using a combination of donor- and host-specific anti-CD45 antibodies in combination with lineage specific antibodies, including CD19, TCRβ, NK1.1, Mac-1, Gr-1, and Ter-119.
For isolation of CLPF from progenitors with myeloid potential, high quality IL-7Rα stains are essential and require optimization. Anti-B220 should also be used in its own channel to distinguish CLP from early B lineage–committed progenitors. If these components are optimal, c-Kit and Sca-1 stains are confirmatory but not essential.
We have observed that CLPF are far more sensitive to serum and media differences, temperature fluctuations, and long delays after sorting than upstream MPP or downstream B progenitors (see Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Maintaining cells in ice-cold, serum-containing medium throughout preparation, sorting, and before transplantation or culturing is critical to preserving the oligopotency of CLPF.
Transplantation of congenic hematopoietic progenitor cells
Mice typically received 400 rad x-irradiation before intravenous injection of progenitors in 150 μL sterile PBS through the retro-orbital venous plexus. Rag2−/−γc−/− mice were used without conditioning. Intrathymic injections were performed as previously described.21
Colony formation assay
Cells were plated out in triplicates in 1 mL MethoCult M3231 methyl cellulose medium (StemCell Technologies, Vancouver, BC) containing SCF, Flt3L, IL-3, IL-6, IL-11, thrombopoietin (TPO), erythropoietin (Epo), and GM-CSF as previously described.2 Cultures were incubated for 12 days in a humidified chamber at 37°C, 5% CO2 and colonies counted on day 7 and day 12 using an inverted microscope.
Clonal and limiting dilution in vitro assays
For limiting dilution assays, CLPF were clone sorted directly onto 96-well plates plated with 5 × 103 to 2.5 × 104 of either S17 (B-cell assay) or OP9-DL1 (T-cell assay) stromal cell lines. Prior to plating, stroma cells were growth arrested by mitomycin C (Sigma-Aldrich, St Louis, MO) treatment (10 μg/mL for 3 hours). Cultures were supplemented with SCF, Flt3L, and IL-7. After 10 days, cultures were analyzed by flow cytometry to score for B cells (B220+CD19+) and/or T cells (Thy1.1+CD25+). Wells were scored positive when at least 20 B220+CD19+ and/or Thy1.1+CD25+ cells were detectable by flow cytometry. For clonal assays, cells were clone-sorted onto Terasaki wells plated with 200 cells/well OP9 stromal cells and supplemented with SCF, Flt3L, and IL-7. Single-cell deposit was confirmed right after sorting by microscopy, and only wells containing exactly one cell were further analyzed. After 3 days of culture, wells containing 4 or more cells were harvested and equal aliquots seeded into 96-well plates containing either B cell–inducing (OP9 stroma plus SCF, Flt3L, and IL-7) or T cell–inducing (OP9-DL1 stroma plus SCF, Flt3L, and IL-7). Clones that contained 8 or more cells were split into 4 wells, 1 with B cell–inducing, 1 with T cell–inducing, 1 with NK-inducing (OP9 stroma plus IL-2, IL-7, IL-15, SCF, and Flt3L) and 1 with DC/myeloid–inducing (AC6 stroma plus IL-3, GM-CSF, M-CSF, SCF, and Flt3L) conditions. All 3 stroma cell lines were left untreated in these experiments (no growth inhibition). After 8 days in culture, wells were analyzed by flow cytometry to identify B cells (CD19+B220+), T cells (Thy1.1+CD25+), NK cells (NK1.1+DX5+), DCs (I-Ab+CD11c+) and myeloid cells (Mac-1+Gr-1+). Only clones that produced 1 or more lineages were scored.
Results
CLP can be divided into Flk2+ and Flk2− subfractions
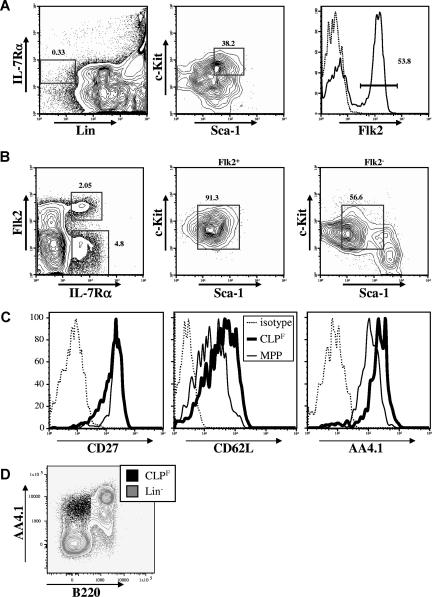
We originally described the CLP as Lin−IL-7Rα+c-KitloSca-1lo and showed more recently that this population expressed variable levels of Flk2.17 In 6-week-old Bl/6 mice, depending on the strictness of lineage marker exclusion, roughly 60% of the CLPs express high levels of Flk2, whereas the remaining 40% were negative (Figure 1A). We previously showed that most of the dendritic-cell potential is retained within the Flk2+ fraction.4,5 Nevertheless, the full lymphoid developmental potential of these 2 CLP fractions was never formally investigated. After altering the sequence of the gating strategy by first gating on Lin− live cells and then gating on the IL-7Rα+ Flk2−/+ fractions, we found that the Flk2+ cells were a homogenous population with respect to Sca-1 and c-Kit expression and matched the original Sca-1 and c-Kit expression of conventionally isolated CLPs (Figure 1B). In contrast, the Flk2− fraction was more heterogeneous and these cells only partially overlapped with the original Sca-1/c-Kit gate (Figure 1B). We also tested the expression of several other surface molecules that have been used to describe early lymphoid or multipotent progenitors including CD24/HSA, CD27, CD34, CD43, CD44, CD62L/L-selectin, and CD93/AA4.1, and found that all Flk2+ CLPs expressed homogeneous levels of these markers (Figure 1C and data not shown). Interestingly, we found that Flk2+ CLP expressed lower levels of CD93 (also known as AA4.1) than downstream B220+ populations (Figure 1D), calling into question results from other studies that identified putative CLP based upon high expression of AA4.1 and low or absent expression of Sca1.7–9,12,22–25 We also tested Balb/c mice and found a population with nearly identical phenotype, suggesting that Flk2+ and Flk2− fractions of CLPs is not unique to the Bl/6 background (see Figure S2).
Figure 1.
CLPs can be divided into Flk2+ and Flk2− fractions. (A) Strategy originally used to isolate CLPs by first gating on Lin−IL-7Rα+ cells (left panel) and subsequently on c-KitintSca-1int cells (middle panel) reveals heterogeneous expression of Flk2 (right panel). (B) Changing the gating strategy by first gating on lineage negative cells (not shown) reveals 2 distinct IL-7Rα+ populations (left panel). The Flk2+ fraction shows homogeneous expression of c-Kit and Sca-1 (middle panel) whereas the Flk2− fraction shows a more heterogeneous expression pattern (right panel). Numbers on plots are percentages of gated cells. (C) Surface expression of CD27, CD62L and CD93/AA4.1 on CLPF (bold lines) in comparison to MPP (solid thin lines), or isotype controls (dashed lines). (D) FACS plot overlay of CLPF (black dot plot) and lineage negative (excluding B220, gray contour plot) populations in C57Bl/6 BM. AA4.1hi cells are predominantly B220+.
Only Flk2+ CLP can produce B, T, DC, and NK cells in vivo
Next, we determined the full lineage potential of these 2 CLP subpopulations in vivo. Both populations were isolated by 2 rounds of fluorescence-activated cell sorting (FACS) to achieve high purity (> 99%) from the bone marrow of Bl/6 Ly5.2 mice, and 103 cells were transplanted intravenously into sublethally irradiated (400 rad) congenic Bl/6 Ly5.1 mice. Three weeks after transplantation, a time when B-cell lineage production is robust and T-cell export from the thymus into the secondary lymphoid organs has just begun, we analyzed donor cell contributions to the spleen and thymus of recipient mice. In the spleen, the overall magnitude of reconstitution from CLPF was much higher than from the Flk2− fraction (4% vs 0.02%; Figure 2A). In addition, we found that all progeny derived from the Flk2− fraction were B-lineage cells as determined by expression of CD19+, whereas no T, NK or myeloid cells (Mac-1+Gr-1+) were detected (Figure 2B-D). In contrast, CLPF produced TCRβ+ T cells and Nk1.1+ NK cells as well as large numbers of CD19+ B cells. Similar results were obtained when bone marrow and blood were analyzed (data not shown). Within the thymus CLPF, but not the Flk2− fraction, gave rise to donor-derived T-lineage cells, contributing nearly 70% of thymocytes at 3 weeks after transplantation (Figure 2E). Even when both subpopulations were injected directly into the thymus, only CLPF gave rise to T-lineage progeny (Figure 2F).
Figure 2.
Only CLPF have multilymphoid potential in vivo. (A-D) FACS analysis of spleens 3 weeks after intravenous transplantation of 103 CLPF or CLPF− into Ly5 congenic, sublethally (400 rad) irradiated hosts and control mice that received sham transplants. (A) Host cells are Ly5.1+ whereas donor-derived cells are Ly5.2+. (B,C) FACS plots showing T cell (TCRαβ+), B cell (CD19+), or NK cell (Nk1.1+) cell reconstitution of the spleen after pregating on either Ly5.1+ host cells (left panels) or Ly5.2+ donor-derived cells (middle and right panels). Both Flk2− and Flk2+ CLPs gave rise to B cells but donor-derived T cells and NK cells were only detectable in CLPF reconstituted mice. (D) FACS plots showing absence of myeloid cells (Mac-1+Gr-1+) from donor-derived Flk2− fraction cells or CLPF. (E) Thymi from irradiated (400 rads) host Ly5.2+ animals 3 weeks after intravenous (i.v.) transplantation of 103 CLPF or Flk2− fraction cells in comparison to animals that received sham transplants (PBS) were analyzed for donor derived Ly5.1+ progeny. 3 weeks after transplantation CLPF derived thymocytes were responsible for roughly two-thirds of the thymus cellularity in mice that received transplants whereas the Flk2− fraction yielded no thymic cells. (F) Thymic reconstitution after intrathymic injection of 103 Ly5.1+ CLPF or Flk2− fraction cells into irradiated (400 rads) Ly5.2+ recipient mice. Numbers on plots are percentages of gated cells.
Robust T-cell output of Flk2+ CLP
While all groups have recognized the ability of CLP to produce B cells, some have questioned whether CLP have enough T-cell potential to be considered a potential major source of these cells. Because T-cell potential segregates with Flk2 expression within the original CLP population, we performed a time course experiment to determine the kinetics and magnitude of T- and B-cell production from CLPF and the Flk2− fraction (Figure 3A). As our initial results predicted, the Flk2− fraction gave rise only to B cells in peripheral tissues at all time points. While few T cells were detected in the spleen at 3 weeks or earlier from Flk2+ CLP, a much larger number were observed at 4 weeks and beyond, suggesting a maturation period of approximately 3 weeks within the thymus before donor T-cell emigration to secondary lymphoid tissues (Figure 3A left). Underscoring the robustness of the T-cell potential of CLP, CLPF-derived splenic T cells outnumbered splenic B cells by week 4. In the blood, CLPF derived B- and T-cell contributions ranged from 0.5% to 2% at 3 weeks after transplantation and were roughly parallel over the time course, strongly supporting the idea of CLPF being a major contributor to both B and T lineages (Figure 3A right).
Figure 3.
Kinetics of CLPF-driven T cell reconstitution. (A) Kinetic analyses of B-cell (squares) and T-cell (diamonds) reconstitution of spleen (left panel) and blood (right panel) of sublethally irradiated mice that received transplants of 103 CLPF (black) or Flk2− fraction cells (white). Results shown are mean values of 3 to 5 animals analyzed per time point. For blood analysis, the same animals were bled periodically for 10 weeks. (B) FACS plots illustrating the CD4/CD8 profile of host (top row) and CLPF derived donor cells (bottom row) as well as reconstitution with thymic B cells (B220+), γδ T cells, and NK cells showing the lymphoid potential of CLPFs. (C) FACS plots illustrating the T-lineage development of intravenously injected CLPF in the thymus, from 1 to 5 weeks. Detailed analysis of CD4/CD8 development (top row), and within the CD4/CD8 double negative fraction, CD44 and CD25 development (middle row) or CD25 and c-Kit development (bottom row), after previously gating on lineage negative cells. Shown are representative FACS plots from 3 mice that received transplants per time point. (D) Thymic reconstitution 6 days after intrathymic (i.t.) injection transfer of 103 CLPF into congenic unirradiated mice. Events shown are gated on lineage negative donor-derived cells. FACS analysis reveals generation of c-Kithi DN1 (CD25−c-Kit+) and c-Kithi DN2(CD25+c-Kit+) directly derived from CLPF. Numbers on plots are percentages of gated cells.
Within the thymus, donor chimerism exceeded 50% by 3 weeks (Figure 2E), with the vast majority (99%) of donor-derived cells belonging to the α/β T-cell lineage (as determined by expression of either or both CD4 and CD8), with limited TCRγδ, NK, and B220+ cells (Figure 3B). T-cell development of CLPF-derived cells proceeded sequentially from the CD4−CD8− (DN) stage to the CD4+CD8+ (DP) and CD4+CD8− or CD8+CD4− (SP) stages, and more mature SP cells were predominant by 3 weeks post injection (Figure 3C top row). The very earliest thymocyte stages (DN1 and DN2) express CD44 and c-Kit, and these were not observed, even at the 1 week time point. Instead, CLPF-derived cells at the DN3 and DN4 stages (CD44−CD25+ and CD44−CD25−, respectively) predominated at 1 week, suggesting that CLPF either directly seeded the thymus, or gave rise to thymus seeding cells rapidly after injection, and that these cells had differentiated beyond the DN2 stage by 1 week (Figure 3C bottom panels). We tested whether CLP could give rise to the earliest thymocyte stages (DN1 and DN2), by directly injecting CLPF intrathymically into nonirradiated recipients, and determining the differentiation status of the donor cells 4 days after injection (Figure 3D). We found that intrathymically injected CLPF differentiate into DN1 and DN2 at this time point, consistent with the intravenous injection time course, indicating that CLP can develop down the T lineage through the classic T-cell development pathway.
Flk2+ CLP do not produce myeloid cells in vitro or in vivo
Several groups have reported that CLP, or progenitors presumed downstream of CLP, have significant myeloid potential. These studies contradict the original description of CLP, which showed that CLP lack myeloid potential. Because the CLPF subset has robust B- and T-lineage potential and is a more homogeneous population than has been previously identified, it enabled us to address this issue in a more convincing manner than previous studies. We therefore examined myeloid readout in intravenous transplants of CLPF. We did observe a small number of Mac1+ cells that others have proposed represent myeloid cells (Figure 2D). However we found that nearly all these cells were CD11c+, and thus were of the DC lineage, consistent with the lymphoid restriction of CLP (Figure 4A). We did not observe any Mac1+Gr-1+ cells derived from CLPF (Figure 2D). To exclude low levels of early myeloid cell production, we analyzed mice 2, 4 and 7 days after transplantation of 104 cells and stained bone marrow, spleen, and blood for Mac-1+Gr-1+ cells but found no donor-derived granulocytes at any time point (data not shown). At 4 weeks after transplantation, we also observed Mac1+ B-1b cells (Figure 4B). In addition to the in vivo transplantations, we used colony formation assays in methylcellulose, which are very sensitive assays for detection of myeloid or erythroid potential. We used various numbers of CLPF cells and compared them to the more primi-tive c-KithiLin−Sca-1hiFlk2+ (MPPF) cells. Using a cocktail of 8 different cytokines to enable simultaneous readout of erythroid, myeloid, and megakaryocytic potential, we detected approximately 1 colony per every 103 seeded CLPF cells (Figure 4C), which were 99% pure after double sorting (data not shown). In stark contrast, 100 MPPF cells had a plating efficiency of above 80%, giving rise to mostly mixed colonies. Therefore, CLPF do not give rise to myeloid cells at frequencies above the calculated impurity levels in our sorted preparations.
Figure 4.
CLPF produce Mac-1+ B1b cells and Mac-1+CD11c+ dendritic cells, but not myeloid cells. (A) Congenic sublethally irradiated mice received transplants of 103 CLPF cells. Three weeks after the transfer spleens were analyzed for donor-derived cells. FACS plots shown are previously gated on recipient (left) or donor-derived cells (right) and are B220−CD19−Gr-1−. Percentage of Mac-1+CD11c+ dendritic cells are shown. (B) Analysis of Mac-1+ B-1b cell differentiation from CLPF in the spleen 4 weeks after transplantation. Cells are pregated on CD5−, and the percentage of Mac-1+IgD− B-1b cells are shown. Numbers on plots are percentages of gated cells. (C) Methylcellulose myeloid differentiation assay. c-Kit+Lin−Sca-1+Flk2+ (MPPF) cells, CLPF, and Flk2− fraction cells (F− frac.) from bone marrow were double sorted and plated out at 100 cells (MPPF) and 500 to 1000 cells (CLPF and F− frac.) in triplicates in methyl cellulose medium containing SCF, Flt3L, GM-CSF, Epo, Tpo, IL-3, IL-6, and IL-11. The average number of colonies yielded and their lineage content as colony forming units (CFU) from 2 combined experiments are shown. G, granulocyte; E, erythrocyte; M, macrophage; B, B cell; and Σ, sum.
Determination of progenitor frequencies in vitro and in vivo by limiting dilution
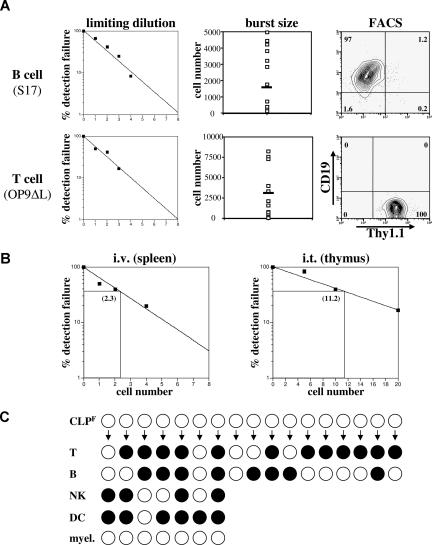
It is clear from the data above that only the Flk2+ fraction of CLP has any significant combined B- and T-cell potential. We therefore sought to determine the clonal frequency of T- and B-cell progenitor activity within this population by in vitro and in vivo limiting dilution assays. When limiting dilutions of Flk2+ CLP were cultured on S17 stroma in the presence of SCF, Flt3L, and IL-7, the frequency of CLPF that had B-cell progenitor potential was approximately 1 of 3 (Figure 5A top left). When we counted the total number of progeny from a single CLPF after 10 days we calculated an average burst size of approximately 1900 per CLPF, demonstrating the large proliferative capacity of these cells when differentiating down the B lineage (Figure 5A top middle). To determine the clonal T-cell potential of CLPF, we cultured CLPF in SCF, Flt3L, and IL-7 on OP9 stromal cells expressing delta-like 1 (OP9-DL1), a ligand for Notch.26 We found that on average 1 of 2 CLPF cells gave a T-cell readout (Figure 5A bottom left). Aliquots of some of the positive wells were further cultured for an additional 2 weeks on OP9-DL1 stroma and positive staining for CD4 and CD8 confirmed their T-cell lineage commitment (data not shown). We also measured the proliferative potential of single CLPF cells that gave a positive readout and found expansion capacities of up to 8000 T cells within 10 days with an average of approximately 3500 cells generated per seeded CLPF (Figure 5A bottom middle).
Figure 5.
CLPF clonally give rise to B and T cells. (A) B-cell (top panel) and T-cell (bottom panel) progenitor frequency of CLPF was assessed by culturing them on either S17 (for B cells) and delta-like 1 expressing OP9 (for T cells) stroma cells in the presence of IL-7, SCF, and Flt3L for 10 days in 96-well plates at densities between 0 and 8 cells per well. The left panel shows the percentage of failure of detection of either CD19+ or Thy1.1+ progeny by FACS. The middle panel shows the number of progeny cells obtained from a single CLPF in these assays with the black bar marking the average burst size. Right panel shows a representative CD19/Thy1.1 FACS plot of a B- and T-cell culture. Horizontal bars in the middle panels represent means. Numbers on right plots are percentages of gated cells. (B) Limiting dilution analysis of donor cells after transfer of CLPF into sublethally irradiated hosts. For B cells, CLPF were intravenously injected and spleens analyzed at 4 weeks. For T cells, CLPF were intrathymically injected and thymi analyzed at 4 days after injection. Three to 5 mice per cell dose were analyzed and numbers shown within the plots represent the calculated limiting number. (C) Results of a clonal assay to simultaneously detect all 4 lymphoid lineages from a single cell (for experimental design and technical details, see Figure S3). Positive clones are symbolized by black circles and a negative result by white circles. Of the 17 clones analyzed, each derived from a single CLPF cell, 8 of 17 gave rise to B cells and 12 of 17 to T cells when cultured under appropriate conditions. Of these positive clones a total of 35% showed simultaneous development of T and B cells, demonstrating the bipotential of CLPF. Furthermore, 7 clones of the 17 underwent 3 or more cell divisions and therefore could also be analyzed for NK, DC, and myeloid potential. Whereas 4 of 7 showed NK cell and 6 of 7 DC potential, none of the clones generated any myeloid cells (0/7). Of the 7 clones analyzed for all 4 lymphoid lineages 2 (28%) simultaneously readout all 4 lymphoid lineages, proving that a single CLPF can possess the full lymphoid potential.
We next determined if this high progenitor potential and enormous burst size accurately reflected in vivo repopulating frequencies. We transplanted between 1 and 4 CLPF cells intravenously into sublethally irradiated congenic mice in a limiting dilution experiment (Figure 5B left). Based on these data, we determined the progenitor frequency among CLPF to be roughly 1 of 2 cells, thus confirming the in vitro results (Figure 5B). When very low numbers of CLPF were transplanted, donor cells were largely limited to the B lineage in the spleen and bone marrow. Occasionally, small numbers of NK cells were detectable in the spleen, but T cells were not. To more sensitively detect T-cell potential in vivo, we transplanted CLPF by intrathymic injection and analyzed thymi 3 weeks later by FACS. Using this assay we determined the T-cell progenitor frequency among CLPF to be 1 of 11 cells (Figure 5B right).
Although the high frequency of T- and B-cell readout from CLPF suggests that at least some CLPF are bipotent, we sought to show this directly at the clonal level. Using automated cell deposition, single CLPF cells were cultured in Terasaki wells on top of OP9 cells in the presence of SCF, Flt3L, and IL-7 (experimental layout in Figure S3A). After 3 days of culture, wells containing 4 or more cells were split into separate wells containing either B cell– or T cell–promoting differentiation conditions. Clones that contained 8 or more cells were split into 4 wells containing, B-cell, T-cell, NK-cell, or DC/myeloid conditions. After 8 additional days of culture, wells were analyzed by flow cytometry using a panel of antibodies to identify committed cells including B, T, NK, DC, and myeloid cells (see Figure S3B for representative stains). Under these conditions, 35% of single CLPF-derived colonies gave rise to both B and T cells (Figure 5C), thus directly proving that a substantial fraction of CLPF contains combined clonal B- and T-cell potential. When we analyzed clones that were additionally cultured under NK cell– and DC-inducing conditions we found that 28% (2/7) showed a combined readout of all 4 lymphoid lineages stemming from a single CLPF. Under the DC promoting conditions, which also supports myeloid cell generation (compare MPP control sample in Figure S3B), no CD11c− myeloid cells were detectable. Although the technical limitations of these experiments may cause lineage restriction decisions to be made during the first 3 days of culture and thus prevent 100% of CLPF from differentiating into all 4 lineages, it clearly demonstrates that at least a portion of single CLPF cells exist that can differentiate into all 4 lymphoid lineages, but not the myeloid lineage. To be precise, these experiments do not prove that every single CLPF cell possesses the full lymphoid potential but it demonstrates that true clonogenic CLPs exist within this population and at least that all cells are restricted to one of the 4 lymphoid lineages.
Discussion
The identification of the CLP, one of the key intermediates in lymphoid differentiation, has been a point of contention within the recent literature, with much debate as to (1) whether the T-cell potential of this population is physiologically relevant, and (2) whether or not this population is truly lymphoid committed. In this study, we address both of these issues, first by identifying key CLP markers that enable unprecedented purification of this population, and second, by performing a series of in vivo and clonal in vitro assays of lineage potential using these highly purified cells.
We demonstrate that the originally defined CLP population can be separated into 2 major subsets on the basis of Flk2 expression, and that while the Flk2+ subset (CLPF) contains lymphoid oligopotent cells, the Flk2− subset is mainly composed of weak B progenitors that lack T-cell potential. When transplanted in vivo, CLPF have all the hallmarks of a lymphoid committed progenitor that gives rise equally well to both B and T lineages. Even a single intravenously transplanted CLPF can give rise to B cells in the spleen, and as few as 10 CLPF intrathymically injected give rise to T cells in the thymus. These numbers illustrate a remarkably high efficiency of CLPF to engraft and differentiate into both B and T lineages in vivo.
While in vivo assays are the gold standard for determining the lineage potential of progenitor populations, in vitro assays on single cells must be performed to determine clonal lineage potentials. When plated as single cells in conditions that enabled differentiation into multiple lymphoid and myeloid lineages, single CLPF gave rise to T, B, NK, and DC lineages, but did not give rise to any macrophages or neutrophils. Approximately 50% of single CLPF clones gave rise to both B- and T-lineage cells in a clonal assay, and while this does not definitively prove that every single phenotypic CLPF contains the full lymphoid potential, it is likely that the frequency of oligopotent cells is much higher than this assay reveals. Nevertheless, this assay indicates that the CLPF population contains a significant proportion of cells that are lymphoid committed and lymphoid oligopotent at the single cell level.
One of the disputed properties of the CLP is whether its T-lineage potential is physiologically relevant. Because Flk2 expression enriched for cells with T potential within the previously defined CLP population, it was important to revisit this issue. When 1000 CLPF were transplanted intravenously, these cells contributed more than 50% of recipient's thymocytes and produced around 2 to 8 × 107 thymocytes at 3 weeks after transplantation. At 4 weeks after transplantation, after CLPF-derived thymic T cells have matured and exited the thymus, the contribution of CLPF-derived T cells to the spleen was numerically similar to the B lineage contribution. Moreover, the clonal frequency and burst size of CLPF that gave rise to B and T cells in vitro were similar. By these measures, the T and B potential of CLPF, both in vitro and in vivo, are robust and of similar magnitude.
Nevertheless, these data do not address whether CLPF are the major source or even a requisite step of T- or B-cell development. While the large proliferative capacity and differentiation potential suggest that CLPF are a likely candidate for this role, this question will require more detailed analysis of thymus seeding, timing of development, and progenitor progeny relationships between CLP and other putative progenitors in both the B and T lineages. Other studies have attempted to address this issue in the past through limiting dilution analysis of CLP and other candidate populations, although the CLP that were used in those studies were isolated using markers that we show here were probably inadequate. Our isolation and characterization of CLPF strongly suggest that the markers used to purify CLP in many previous studies, specifically those that used high expression of the marker AA4.1, would lead to isolation of a population of cells highly enriched for committed B progenitors (Figure 1D). Thus, the identification and characterization of a homogeneous CLPF population provides an explanation for previous conflicting studies regarding the lineage potential of this progenitor.
While the CLP was originally described as being lymphoid committed, this has been subject to variable results from other groups. We addressed this issue in vitro and in vivo, using CLPF, and found that CLPF was lymphoid committed. It should be noted that most of the studies that have identified myeloid potential within the CLP have identified cells as myeloid based solely on expression of Mac-1, which is insufficient to unequivocally identify myeloid cells. CLPF do give rise to Mac-1+ cells, however, these are all either CD11c+ dendritic cells, or B-1b cells that coexpress Mac1. Moreover, it is well known that NK cells express low levels of Mac1.27
CLPF represents a homogeneous, lymphoid-restricted oligopotent progenitor that gives rise equally well to both B and T lineages. Flk2 expression has heretofore been an underappreciated hallmark of these cells. Its use enriches for T potential by depleting B-committed cells from the previously purified populations. This has enabled the clarification of the lineage potential of these cells, and as a result should aid the field in moving forward from conflicting data, largely due to the prior difficulty of isolating pure CLP. With CLPF at the branch point between all lymphoid lineages, this will also be an ideal population of cells to study the processes of lineage commitment.
Supplementary Material
Acknowledgments
The authors thank Christina Richter and Stephanie Smith-Berdan for antibody preparations and Libuse Jerabek for excellent laboratory management.
Support to T.S. was provided though a Ruth L. Kirschstein National Research Service Award (1 F32 AI 58521). D.B. was supported by a fellowship from the Cancer Research Institute and the National Institute of Health (T32AI0729022 and 5K01DK078318), and M.A.I. was supported by a fellowship from the California Institute for Regenerative Medicine (T1-00001). This work was supported by National Institutes of Health grant AI47458 to I.L.W.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: H.K. and I.L.W. designed the experiments; H.K., T.S., D.B., and M.A.I. performed the experiments; H.K., D.B., M.A.I., and T.S. collected and analyzed data; all authors interpreted data; H.K. wrote the paper and all authors edited it.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Holger Karsunky, Cellerant Therapeutics, Inc., 1561 Industrial Road, San Carlos, CA 94070; e-mail: hkarsunky@cellerant.com.
References
- 1.Kondo M, Wagers AJ, Manz MG, et al. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu Rev Immunol. 2003;21:759–806. doi: 10.1146/annurev.immunol.21.120601.141007. [DOI] [PubMed] [Google Scholar]
- 2.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 3.Traver D, Akashi K, Manz M, et al. Development of CD8alpha-positive dendritic cells from a common myeloid progenitor. Science. 2000;290:2152–2154. doi: 10.1126/science.290.5499.2152. [DOI] [PubMed] [Google Scholar]
- 4.Mende I, Karsunky H, Weissman IL, Engleman EG, Merad M. Flk2+ myeloid progenitors are the main source of Langerhans cells. Blood. 2006;107:1383–1390. doi: 10.1182/blood-2005-05-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karsunky H, Merad M, Mende I, Manz MG, Engleman EG, Weissman IL. Developmental origin of interferon-alpha-producing dendritic cells from hematopoietic precursors. Exp Hematol. 2005;33:173–181. doi: 10.1016/j.exphem.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 6.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 7.Allman D, Sambandam A, Kim S, et al. Thymopoiesis independent of common lymphoid progenitors. Nat Immunol. 2003;4:168–174. doi: 10.1038/ni878. [DOI] [PubMed] [Google Scholar]
- 8.Harman BC, Miller JP, Nikbakht N, Gerstein R, Allman D. Mouse plasmacytoid dendritic cells derive exclusively from estrogen-resistant myeloid progenitors. Blood. 2006;108:878–885. doi: 10.1182/blood-2005-11-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwarz BA, Bhandoola A. Circulating hematopoietic progenitors with T lineage potential. Nat Immunol. 2004;5:953–960. doi: 10.1038/ni1101. [DOI] [PubMed] [Google Scholar]
- 10.Balciunaite G, Ceredig R, Massa S, Rolink AG. A B220+ CD117+ CD19- hematopoietic progenitor with potent lymphoid and myeloid developmental potential. Eur J Immunol. 2005;35:2019–2030. doi: 10.1002/eji.200526318. [DOI] [PubMed] [Google Scholar]
- 11.Perry SS, Welner RS, Kouro T, Kincade PW, Sun XH. Primitive lymphoid progenitors in bone marrow with T lineage reconstituting potential. J Immunol. 2006;177:2880–2887. doi: 10.4049/jimmunol.177.5.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rumfelt LL, Zhou Y, Rowley BM, Shinton SA, Hardy RR. Lineage specification and plasticity in CD19- early B cell precursors. J Exp Med. 2006;203:675–687. doi: 10.1084/jem.20052444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye M, Graf T. Early decisions in lymphoid development. Curr Opin Immunol. 2007;19:123–128. doi: 10.1016/j.coi.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 14.Pelayo R, Welner R, Perry SS, et al. Lymphoid progenitors and primary routes to becoming cells of the immune system. Curr Opin Immunol. 2005;17:100–107. doi: 10.1016/j.coi.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 15.Bhandoola A, von Boehmer H, Petrie HT, Zuniga-Pflucker JC. Commitment and developmental potential of extrathymic and intrathymic T cell precursors: plenty to choose from. Immunity. 2007;26:678–689. doi: 10.1016/j.immuni.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 16.Lyman SD, Jacobsen SE. c-kit ligand and Flt3 ligand: stem/progenitor cell factors with overlapping yet distinct activities. Blood. 1998;91:1101–1134. [PubMed] [Google Scholar]
- 17.Karsunky H, Merad M, Cozzio A, Weissman IL, Manz MG. Flt3 ligand regulates dendritic cell development from Flt3+ lymphoid and myeloid-committed progenitors to Flt3+ dendritic cells in vivo. J Exp Med. 2003;198:305–313. doi: 10.1084/jem.20030323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adolfsson J, Borge OJ, Bryder D, et al. Upregulation of Flt3 expression within the bone marrow Lin(-)Sca1(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity. Immunity. 2001;15:659–669. doi: 10.1016/s1074-7613(01)00220-5. [DOI] [PubMed] [Google Scholar]
- 19.Christensen JL, Weissman IL. Flk-2 is a marker in hematopoietic stem cell differentiation: a simple method to isolate long-term stem cells. Proc Natl Acad Sci U S A. 2001;98:14541–14546. doi: 10.1073/pnas.261562798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forsberg EC, Serwold T, Kogan S, Weissman IL, Passegue E. New evidence supporting megakaryocyte-erythrocyte potential of flk2/flt3+ multipotent hematopoietic progenitors. Cell. 2006;126:415–426. doi: 10.1016/j.cell.2006.06.037. [DOI] [PubMed] [Google Scholar]
- 21.Jerabek L, Weissman IL. Intrathymic injection for analysis of T-cell progenitor activity. In: Klug CA, Jordan CT, editors. Hematopoietic stem cell protocols. Totowa, NJ: Humana Press; 2002. pp. 161–165. [DOI] [PubMed] [Google Scholar]
- 22.Izon D, Rudd K, DeMuth W, et al. A common pathway for dendritic cell and early B cell development. J Immunol. 2001;167:1387–1392. doi: 10.4049/jimmunol.167.3.1387. [DOI] [PubMed] [Google Scholar]
- 23.Borghesi L, Hsu LY, Miller JP, et al. B lineage-specific regulation of V(D)J recombinase activity is established in common lymphoid progenitors. J Exp Med. 2004;199:491–502. doi: 10.1084/jem.20031800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borghesi L, Aites J, Nelson S, Lefterov P, James P, Gerstein R. E47 is required for V(D)J recombinase activity in common lymphoid progenitors. J Exp Med. 2005;202:1669–1677. doi: 10.1084/jem.20051190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller JP, Izon D, DeMuth W, Gerstein R, Bhandoola A, Allman D. The earliest step in B lineage differentiation from common lymphoid progenitors is critically dependent upon interleukin 7. J Exp Med. 2002;196:705–711. doi: 10.1084/jem.20020784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmitt TM, Zuniga-Pflucker JC. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17:749–756. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 27.Huntington ND, Vosshenrich CA, Di Santo JP. Developmental pathways that generate natural-killer-cell diversity in mice and humans. Nat Rev Immunol. 2007;7:703–714. doi: 10.1038/nri2154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.