Abstract
Smad proteins are critical intracellular mediators of signaling by growth and differentiation factors of the transforming growth factor β superfamily. We have isolated a member of the Smad family, Smad8, from a rat brain cDNA library and biochemically and functionally characterized its ability to transduce signals from serine kinase receptors. In Xenopus embryo, Smad8 is able to transcriptionally activate a subset of mesoderm target genes similar to those induced by the receptor serine kinase, activin receptor-like kinase (ALK)-2. Smad8 can be specifically phosphorylated by a constitutively active ALK-2 but not the related receptor serine kinase, ALK-4. In response to signaling from ALK-2, Smad8 associates with a common regulatory molecule, Smad4, and this association leads to a synergistic effect on gene transcription. Furthermore, Smad8 is able to rescue the expression of mesoderm genes blocked by truncated ALK-2 in the embryo. These results indicate that Smad8 can function as a downstream signaling mediator of ALK-2.
Keywords: activin receptor-like kinase, transforming growth factor β, Xnot, brachyury
Members of the transforming growth factor β (TGF-β) superfamily exert their biological effects by interacting with two types of transmembrane serine kinase receptors (1, 2). The ligands bind first to a type II receptor, and subsequently a type I serine kinase receptor is recruited to form a heteromeric complex on the cell surface (3, 4). In this complex, the serine and threonine residues in a glycine - and serine-rich region (GS domain) on the type I receptor are transphosphorylated by the kinase activity of the type II receptors, leading to activation of the type I receptor (5, 6). A mutation near the GS domain generates a constitutively active type I receptor that can signal in the absence of ligand and type II receptors (5, 6). Thus, the activated type I receptors are the transducers and responsible for the propagation of the signal to downstream targets. Thus far, seven type I receptors have been cloned and named ALK-1 through ALK-7, for activin receptor-like kinases (7–10).
Smads comprise a family of recently identified proteins that are key components of the TGF-β family signaling pathways that link activated membrane receptors/ligand complexes to transcriptional regulation of target genes (11). Biochemical studies have established two distinct TGF-β family pathways. ALK-3, acting through Smad1, mediates bone morphogenetic protein (BMP)-2/4 signals, whereas ALK-4 and ALK-5, acting through Smad2 or Smad3, mediate activin and TGF-β signals. In Xenopus embryo, BMP-4 induces ventral mesoderm and epidermal fates during mesoderm induction (12). ALK-3 and Smad1 have been shown to mediate these responses (13–16). In cell culture studies, BMP-2 and BMP-4 as well as activated ALK-3 were shown to be capable of phosphorylating Smad1 and promoting the translocation of Smad1 into the nucleus (17–19). Smad2, on the other hand, appears to mediate TGF-β responses in mammalian cells, as dominant negative Smad2 could block the TGF-β1 induced transcriptional activation of plasminogen activator inhibitor-1 (20). In addition, Smad2 has been shown to be a phosphorylation substrate for TGF-β receptors (20, 21). Smad3 appears to mediate TGF-β effects as well and may be an isoform of Smad2 (22, 23). In Xenopus, Smad2 can function in the activin pathway (16, 24) and ALK-4 and Smad2 can both activate the same set of transcriptional responses (A. B., unpublished data). The fact that both ALK-4 and ALK-5 can both act through Smad2 is not surprising because these receptors have almost identical kinase domains. If its type II receptor is present, the effects of TGF-β are indistinguishable in mesoderm induction from those of activin (25).
Characterization of other known type I receptors such as ALK-2 have languished mainly because of a lack of the downstream targets to correlate with ligand binding assays. For instance, ALK-2 was found to complex with TGF-β1, activin, or BMP-7/osteogenic protein-1, depending on the cell type and with which type II receptor it is coexpressed (7–10); however, whether these complexes are functional has not been established. To aid in characterizing the biological function of ALK-2, we report here the isolation and functional characterization of Smad8 and found that it can mediate effects of ALK-2 in vitro and in vivo.
MATERIALS AND METHODS
Cloning of Smad8.
Smad8 was cloned as a result of low stringency screening as described (23). The sequence of Smad8 was determined by sequencing both strands (Sequenase version 2, United States Biochemical). The full-length cDNA inserts of Smad8 were subcloned into pcDNA3 (Invitrogen) for expression in cell culture, and CS2+ vector for studies in Xenopus embryos.
Isolation of Xenopus ALK-2 cDNA.
Xenopus ALK-2 cDNA was isolated from screening a Xenopus neurula stage cDNA library with a full-length rat ALK-2 cDNA probe (10). The truncated ALK-2 receptor was generated by inserting a termination codon just past the transmembrane domain. The constitutively active form of the ALK-2 was generated by mutation of glu-203 into aspartic acid.
Animals, Reagents, and in Vitro Transcription.
Embryos were obtained from Xenopus laevis adult frogs (Nasco, Fort Atkinson, WI and Xenopus 1) by hormone-induced egg-laying and in vitro fertilization using standard protocols and staged according to Nieuwkoop and Faber. For in situ hybridization analysis, albino embryos were injected in one blastomere at the four-cell stage without any dorsal/ventral bias. All constructs for in vitro transcription were inserted into a CS2+ vector. Most of the 5′ and 3′ untranslated sequences of the constructs were minimized before cloning into the vector. RNA was synthesized in vitro from the linearized plasmids by SP6 polymerase.
Animal Pole Explant Assay.
Two-cell stage embryos were injected in both blastomeres and allowed to develop until animal pole explants were cut at blastula (stage 9). RNase protection was performed with 10 animal caps per sample with a Direct Protect Lysate RPA kit (Ambion, Austin, TX). The 32P-labeled RNA probes in the assay were generated by in vitro transcription with the linearized DNA template and purified from a denaturing polyacrylamide gel before use. The protection products were separated on 6% denaturing polyacrylamide gel and detected by autoradiography. The relative amount of RNA in each sample was normalized by monitoring the levels of EF-1α RNA.
Whole-Mount in Situ Hybridization.
The whole-mount in situ hybridization protocol developed by Harland was used with minor modifications. Digoxigenin-labeled RNA probes were generated by in vitro transcription. Hybridization was detected with an alkaline-phosphatase-coupled anti-digoxigenin antibody and visualized using nitroblue tetrazolium/5-bromo-4-chloro-3-indoyl phosphate p-toluidine salt as a substrate (Boehringer Mannheim). Stained embryos were fixed overnight in MEMFA, washed in ethanol, and photographed directly.
Generation of Epitope-Tagged Smad2, Smad4, and Smad8.
To generate the N-terminal myc epitope-tagged Smad8, an oligonucleotide containing the myc tag sequence (EEQKLISEEDLL) was used in PCR with a downstream primer and the Smad8 cDNA as template. The Flag-tagged Smad4 (26) was generated by PCR with a upstream primer encoding the Flag tag (DYKDDDDK) and a downstream primer. A consensus Kozak translation initiation sequence (27) was introduced into the beginning of the tag sequences and the first ATG of both Smad8 and Smad4 were removed. The PCR products were cloned into the N-terminal regions of the Smad8 and Smad4, respectively, to generate the in-frame myc- or Flag-tagged constructs. The tagged Smad8 and Smad4 cDNA were then cloned into pcDNA3 for expression studies. The hemagglutinin (HA)-tagged Smad2 was generated by cloning the coding region of Smad2 (24) into a pRC/CMV vector (Invitrogen) with a HA tag sequence in the 5′ end. The first ATG of Smad2 was also removed during the cloning so the transcription would start from the Kozak sequence in the front of the HA tag.
Cell Culture, Transfection, Immunoprecipitation, Immunoblotting, and Phosphate Labeling.
The Chinese hamster ovary cells (CHO-K1) and human embryonic kidney 293 cells were cultured in DMEM containing 10% fetal bovine serum. Transfection was performed by the lipofectamine method (GIBCO/BRL) for CHO cells and calcium phosphate method for the 293 cells. Forty-eight hours after transfection, cells were lysed with 1× lysis buffer that contains 150 mM NaCl, 20 mM Tris (pH 7.5), 1 mM EDTA and 1% Nonidet P-40 plus protease inhibitors. For the coimmunoprecipitation of Smad8 with Smad4, the cell lysate was incubated with 9E10 mouse anti-myc antibody (2 μg/ml) in the presence of a secondary rabbit anti-mouse antibody and protein-A Sepharose. After incubation at 4°C for 4 h, the Sepharose beads were washed with the 1× lysis buffer and separated by 8% SDS/PAGE. The protein was then transferred to nitrocellulose membrane, blotted with a M2 mouse anti-Flag antibody (Kodak) followed by incubation with a secondary horse radish peroxidase-conjugated antibody (Bio-Rad), and detected by Chemiluminescence Reagent Plus (NEN). For Western blot analysis, the cell lysate was separated on 8% SDS/PAGE and detected by either 9E10 anti-myc or M2 anti-Flag antibodies. For in vivo phosphorylation experiment, transfected cells were labeled for 3 h with [32P]phosphate (0.5 mCi/ml; 1 Ci = 37 GBq) in the phosphate-free medium with 0.5% fetal bovine serum. The cell lysate was immunoprecipitated with 9E10 anti-myc antibody (to detect phosphorylation of the myc-tagged Smad8) or 12CA5 anti-HA antibody (to detect phosphorylation of the HA-tagged Smad2). The immune precipitatewas washed 4 times with the 1× lysis buffer and subjected to electrophoresis on SDS/PAGE followed by autoradiography.
RESULTS
Cloning of Smad8, a Novel Member of the Smad Family.
We carried out a low stringency screen of a rat brain cDNA library with a probe made from the full-length cDNA of rat Smad1 (23). We isolated Smad1 and three other related molecules. Two of these were the rat homologues of Smad3 (23) and Smad5 (Y. C., unpublished data). The third clone represented a previously uncharacterized Smad that we named Smad8. Very recently, the sequence of another human Smad was reported that appears to be the human homolog of Smad8, though no functional characterization was described (28). The comparison between Smad8 and other known Smads is shown in a phylogenetic tree (Fig. 1A). The cDNA sequence of Smad8 encoded a putative protein with a length of 434 amino acids and predicted molecular weight of 49 kDa. Smad8 shares the common structural motifs as other family members. These include the highly homologous N- and C-terminal regions (MH1 and MH2) and a highly variable linker domain in the middle. Smad8 possesses a SSXS motif at the end of the C terminus, identical to those of Smad1, 2, 3, and 5.
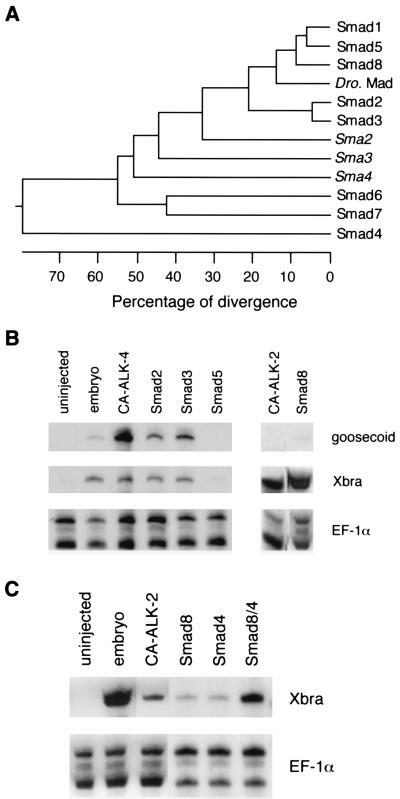
Figure 1.
Smad8 activates Xbra in animal pole explants. (A) Phylogenetic tree of the Smad proteins. The numbers below the tree indicate the percent divergence among the amino acid sequences. The sequences used here include rat Smad1, Smad3, and Smad8, mouse Smad2 and Smad5, human Smad4, Smad6, and Smad7, Drosophila Mad and three Caenorhabditis elegans Mads, sma-2, sma-3 and sma-4. (B) Smad8 induces Xbra in animal pole explants. Xenopus embryos at two-cell stage were injected with 1–2 ng RNA of Smad2, Smad3, Smad5, Smad8 and 10–20 pg of CA ALK-2 or CA ALK-4. The embryos were dissected at stage 9, and the explants were cultured until stage 12. The animal pole explants were analyzed by RNase protection with probes of Xbra, gsc, and EF-1α. The ubiquitously expressed EF-1α was used as a loading control to monitor the total RNA levels among samples. Embryos at stage 12 and the animal pole explants from the uninjected embryos were used as controls. (C) Smad8 synergizes with Smad4 in Xbra activation. Xenopus embryos at two-cell stage were injected with 50 pg of CA ALK2 RNA or 0.2 ng RNA of Smad8 alone, Smad4 alone, or Smad8 together with Smad4. The embryos were dissected at stage 9 and the explants were cultured until stage 12. The animal pole explants were analyzed by RNase protection with probes of Xbra and EF-1α.
Smad8 Is Able to Induce Brachyury (Xbra) in Animal Pole Explants.
Because specification of cell fate in early development of Xenopus is known to involve serine kinase receptors and their ligands, we first tested the potential signaling activity of Smad8 by injecting in vitro-transcribed RNA into the animal pole of Xenopus embryos and analyzed the ectodermal explants for the transcriptional activation of mesoderm-specific genes, a pan-mesodermal marker brachyury (29), and goosecoid (gsc), a marker specifically expressed in the dorsal marginal zone (30). We compared the activity of Smad8 with Smad2, Smad3, and Smad5 as well as two constitutively active (CA) forms of serine kinase receptors, ALK-2 and ALK-4. The CA forms of the Xenopus ALK-2 and ALK-4 were generated by mutations in the GS domain analogous to those reported for other mammalian type I receptors (5, 6). As shown in Fig. 1B, Smad8 and CA ALK-2 were able to activate high levels of Xbra but not gsc. On the other hand, CA ALK-4, Smad2, and Smad3 activated both gsc and Xbra, consistent with the recent findings that Smad2 may function in the ALK-4 pathway in Xenopus embryos (A. B., unpublished). The ability of Smad3 to activate gsc suggested that Smad3 may have a mesoderm-inducing activity similar to Smad2. Smad5 failed to activate either of these markers in this assay, which is consistent with the observation that Smad5 is not involved in dorsal mesoderm induction (31). Thus, the activity of Smad8 in the animal pole explants was clearly different from that of the other Smad molecules and appeared to be similar to the activity of ALK-2.
According to structural criteria, Smad8 belongs to a subgroup in the Smad family that contains a SSXS motif in the C-terminus. Other members of this subgroup, Smad1, Smad2, and Smad3, have been shown to mediate effects of specific TGF-β superfamily members. Smad4 belongs to another subgroup that is thought to be a shared partner for SSXS-containing Smads and necessary for mediating the activity of specific receptors (32, 33). We explored whether Smad4 would functionally interact with Smad8. We first tested if Smad8 and Smad4 acted synergistically to induce Xbra when expressed at low level in Xenopus embryos. Because injection of high levels (1–2 ng) of Smad8 RNA alone was capable of inducing Xbra, we injected approximately 10-fold less Smad8 and Smad4 RNA separately and together into the animal poles of the two-cell stage embryos and analyzed the excised explants for the activation of Xbra. As shown in Fig. 1C, Smad8 and Smad4 at these levels barely activate transcription of Xbra in the animal pole explants when injected alone. Coinjection of Smad8 and Smad4, however, activated Xbra RNA levels that were comparable to the explants from the embryos injected with the activated form of ALK-2. This result showed that Smad8 could synergize with Smad4 and thus Smad8 behaves like its homologous Smad members of the SSXS-containing subgroup in functionally interacting with the common regulatory partner, Smad4.
Constitutively Active ALK-2 Stimulates Heteromerization Between Smad8 and Smad4.
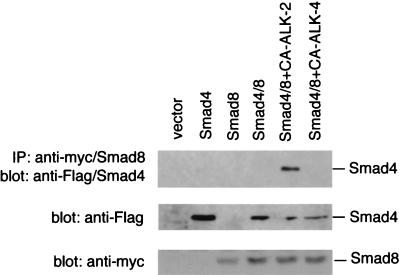
The synergistic effects of Smad8 and Smad4 on gene induction in animal pole explant assays led us to examine whether Smad8 and Smad4 could form a complex and if this association depended on signaling from ALK-2. The ability of Smad8 to associate with Smad4 was tested by using a N-terminal myc-tagged Smad8 and a N-terminal Flag-tagged Smad4. These tagged Smads were cotransfected with a CA ALK-2 or CA ALK-4 into CHO cells and used in immunoprecipitation with an anti-myc/Smad8 antibody. The immunoprecipitate was separated on SDS/PAGE and blotted with an anti-Flag/Smad4 antibody. As shown in Fig. 2, immunoprecipitation of Smad8 with an anti-myc antibody could bring down the Flag-tagged Smad4 only in the presence of CA-ALK-2, indicating that only activation of ALK-2 but not ALK-4 could induce the heteromerization of Smad8 with Smad4. The absence of Smad4/Smad8 association in other samples is not caused by the decreased expression of Smad8 or Smad4 proteins in the transfection cells, because Western blot analysis with the anti-Flag or anti-myc antibody with the cell lysate showed comparable protein expression levels among different groups. Thus, activation of the ALK-2 pathway promotes the specific association between Smad8 and Smad4. The ability of Smad8 to associate with Smad4 upon receptor activation is similar to two other SSXS motif-containing Smads, Smad1 and Smad2, that have been shown to associate directly with Smad4 in response to BMP-4 and TGF-β, respectively (32). Furthermore, this finding also suggests that Smad8 plays a role in the signaling pathway of ALK-2.
Figure 2.
CA ALK-2 induces association of Smad8 with Smad4. CHO cells were transiently transfected with control vector (pcDNA3), a Flag-tagged Smad4 (Flag/Smad4) alone, a myc-tagged Smad8 (myc/Smad8) alone, myc/Smad8 and Flag/Smad4, or myc/Smad8 and Flag/Smad4 with CA ALK-2 or CA ALK-4. The transfected cells were lysed, and myc/Smad8 was purified by immunoprecipitation with an anti-myc antibody. The immunoprecipitate was separated on SDS/PAGE, and Flag/Smad4 that complexed with Smad8 was detected by an anti-Flag antibody. The crude cell lysate (1/30th equivalent of the amount used in immunoprecipitation) from above transfection also were separated on SDS/PAGE and detected with anti-Flag or anti-myc antibody to determine the protein expression level of Flag/Smad4 and myc/Smad8.
Constitutively Active ALK-2 Phosphorylates Smad8.
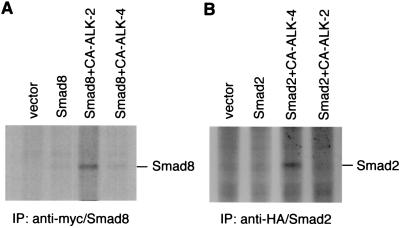
We next tested whether signaling from the serine kinase receptor ALK-2 is correlated to the phosphorylation of Smad8. We expressed the myc-tagged Smad8 into the CHO cells alone or together with either CA ALK-2 or ALK-4 and labeled the cells with [32P]phosphate. The Smad8 protein was purified by immunoprecipitation with an anti-myc antibody and separated by SDS/PAGE. The phosphorylation products were detected by autoradiography. When transfected alone into these cells, Smad8 did not reveal any detectable phosphorylation (Fig. 3A). However, when cotransfected with CA ALK-2, Smad8 was phosphorylated. Cotransfection of Smad8 with CA ALK-4, by contrast, did not result in the same level of phosphorylation. Because the wild-type ALK-2 did not lead to any detectable phosphorylation (data not shown), the ALK-2-mediated phosphorylation of Smad8 appears to require the activation of the receptor.
Figure 3.
CA ALK-2 specifically phosphorylates Smad8. (A) CA ALK-2 but not CA ALK-4 phosphorylates Smad8. CHO cells were transiently transfected with control vector (pcDNA3), myc/Smad8, or myc/Smad8 with either CA ALK-2 or CA ALK-4. The transfected cells were labeled with [32P]phosphorus for 3 h before harvesting. The cell lysate was immunoprecipitated with an anti-myc antibody, separated on SDS/PAGE, and detected by autoradiography. The migration of myc/Smad8 on the gel was determined by anti-myc Western blot analysis with the lysate from 32P-unlabeled cells that expressed myc/Smad8 (data not shown). (B) CA ALK-4 but not CA ALK-2 phosphorylates Smad2. 293 cells were transiently transfected with control vector (pcDNA3), a HA-tagged Smad2 (HA/Smad2), and HA/Smad2 with either CA ALK-4 or CA ALK-2. The transfected cells were labeled with [32P]phosphorus, and the cell lysate was immunoprecipitated with an anti-HA antibody followed by separation on SDS/PAGE. The phosphorylation products were detected by autoradiography.
In a parallel experiment, we studied whether CA ALK-4 is capable of phosphorylating Smad2. Functional studies of Smad2 in Xenopus embryos have suggested that Smad2 acts downstream of ALK-4 (16). A HA-tagged Smad2 was transfected into 293 cells with either CA ALK-4 or CA ALK-2. After labeling with [32P]phosphate, the transfected cells were lysed and Smad2 was purified by immunoprecipitation with an anti-HA antibody. The immunoprecipitate was separated on SDS/PAGE and exposed to an x-ray film. As shown in Fig. 3B, expression of Smad2 alone or together with CA ALK-2 did not reveal appreciable phosphorylation of Smad2. However, cotransfection of Smad2 with CA ALK-4 was able to induce phosphorylation of Smad2. Thus, activation of ALK-4 specifically phosphorylates Smad2, and activation of ALK-2 specifically phosphorylates Smad8, clearly indicating that these two receptors are activating two distinct pathways through two distinct Smad molecules.
Truncated ALK-2 Blocks a Subset of Mesoderm Genes in Early Xenopus Embryo.
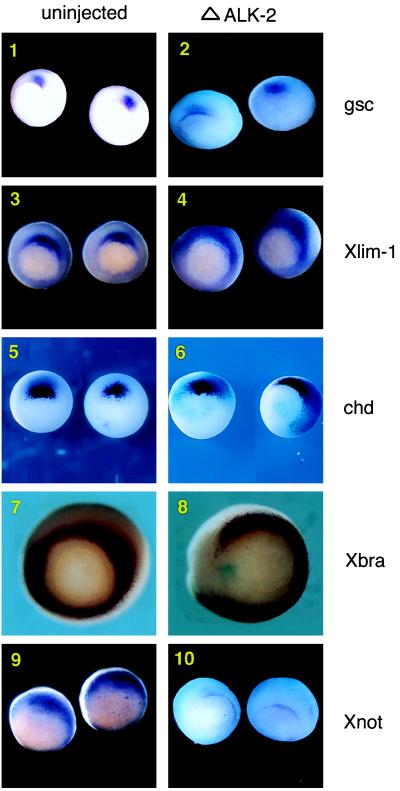
To further examine the relationship between ALK-2 and Smad8, we characterized the signaling of ALK-2 in Xenopus embryos by constructing a C-terminal truncated form of ALK-2 (ΔALK-2) and utilized this construct to block the signaling mediated by ALK-2 in the embryo. It has been shown that the truncated type I receptor is able to behave as a dominant-negative form in specifically blocking the downstream signaling of the receptor (13). We examined embryos that were injected with ΔALK-2 with in situ hybridization with a set of mesoderm markers that reflect the patterning of mesoderm at the late blastula stage. gsc, Xlim-1, and chordin (chd) represent organizer-specific genes that are expressed in the dorsal marginal zone (DMZ) of the embryos at the beginning of gastrulation (Fig. 4 1, 3, and 5) (30, 34). Xbra is a pan mesoderm marker expressed radially in the marginal zone (Fig. 4-7) (29). Xnot is expressed in the prospective midline cells (Fig. 4-9) (35). Albino embryos at the two-cell stage were injected with ΔALK-2 (along with lacZ transcripts) and analyzed at stage 10.5 by whole-mount in situ hybridization for the transcriptional activation of the markers mentioned above. The lacZ transcripts was served as a marker to indicate the distribution of the injected mRNA. When ΔALK-2 was injected in the DMZ, the expression of gsc, Xlim-1, and chd were unaffected (Fig. 4 2, 4, and 6). However, the expression of Xbra in these embryos was lost around the site of injection and thus no longer radial (Fig. 4-8). Xnot expression was also inhibited in gastrula stage embryos (Fig. 4-10). These results indicate that ALK-2 activity is not required for the specification of organizer-specific genes but is involved in the activation of a subset of mesoderm patterning genes.
Figure 4.
Truncated ALK-2 blocks a subset of mesoderm genes in Xenopus embryo. Albino embryos were injected with ΔALK-2 RNA and a lacZ transcript in the marginal zone at the four-cell stage without dorsal/ventral bias. The lacZ transcript was used in all injections to indicate the distribution of the injected truncated receptor. Embryos were cultured until the 10.5 stage and used in whole-mount in situ hybridization with probes of gsc, Xlim-1, chd, Xbra, and Xnot. Representative embryos are shown here (vegetal view with dorsal side up). (1) gsc expression in uninjected embryos. (2) gsc expression in ΔALK-2 injected embryos. (3) Xlim-1 expression in uninjected embryos. (4) Xlim-1 expression in ΔALK-2 injected embryos. (5) chd expression in uninjected embryos. (6) chd expression in ΔALK-2 injected embryos. (7) Xbra expression in uninjected embryos. (8) Xbra expression in ΔALK-2 injected embryos. (9) Xnot expression in uninjected embryos. (10) Xnot expression in ΔALK-2 injected embryos.
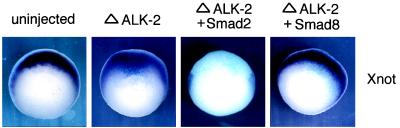
Smad8 Can Rescue Xnot Expression in ΔALK-2-Injected Embryos.
To address whether Smad8 can function downstream of ALK-2, we tested if Smad8 could rescue Xnot expression in embryos injected with ΔALK-2. As shown in Fig. 4, injection of ΔALK-2 mRNA in dorsal blastomeres can block Xnot expression as detected by whole-mount in situ hybridization in mid-gastrula stage embryos. However, when Smad8 mRNA was coinjected with ΔALK-2 mRNA into the DMZ of the embryos, Xnot expression could be detected (Fig. 5). Therefore, injection of Smad8 was able to restore the transcriptional activation of Xnot blocked by ΔALK-2. We also tested another Smad family member, Smad2, in this assay. In contrast to Smad8, Smad2 was unable to rescue Xnot expression and appeared to augment the inhibition of Xnot expression. Thus, this experiment demonstrates that Smad8, but not Smad2, acts downstream of ALK-2 and is sufficient to activate Xnot.
Figure 5.
Smad8 rescues the block of Xnot by truncated ALK-2. Albino embryos at the four-cell stage were injected in the marginal zone with in vitro transcribed RNA of ΔALK-2 alone or ΔALK-2 with either Smad2 or Smad8. The embryos were cultured until stage 10.5 and used in whole-mount in situ hybridization with a Xnot probe. The lacZ transcripts was used to indicate the distribution of the injected RNA. Representative embryos are shown (vegetal view with dorsal side up).
DISCUSSION
The TGF-β family members initiate a signaling cascade by the formation of a ligand-induced heteromeric complex on the cell surface that includes two types of serine kinase receptors, known as type I and II (3). Upon formation of this complex, the type II receptor phosphorylates the type I receptor on a cytoplasmic region, known as the GS domain (4). This phosphorylation is thought to trigger a conformation change that activates the type I receptor and thereby allows the type I receptor to transduce the signal into the cell. Studies with the Smad family of transcriptional regulators have provided insight concerning the events that link activation of the type I receptors on the cell surface to specific activation of gene transcription in the nucleus. In this report, we present the isolation and characterization of Smad8 and establish its position in the signaling pathways by functionally characterizing its transcriptional activity in Xenopus embryos. We show that Smad8 and ALK-2 activate the same transcriptional targets. We also demonstrate that Smad8 associates with Smad4, and this association is dependent on specific signaling from ALK-2. In addition, Smad8 is specifically phosphorylated by CA ALK-2. Overexpression of Smad8 is also sufficient to activate its targets in the absence of ALK-2 signaling. Thus, Smad8 mediates the signaling by receptor kinase ALK-2.
Characterization of ALK-2 and Smad8 Signaling Pathway.
The serine kinase receptor ALK-2 was originally isolated as a result of a search for members homologous to TGF-β and activin receptors (7–10). However, the functional characterization of ALK-2 has been elusive for two reasons. First, the downstream target genes whose transcriptional activation is mediated by ALK-2 signaling had not been discovered, thus impairing functional analysis of ALK-2 signaling. Our work here provides two targets that are transcriptionally regulated by ALK-2 signaling. Second, the physiological ligand of ALK-2 had not been defined. We have generated a CA mutant form of ALK-2 that provided a tool for the characterization of ALK-2 signaling in the absence of its ligand and type II receptor.
The search for the ligand of ALK-2 in the past relied exclusively on cross-linking and binding assays. Because type I receptors of the TGF-β family do not bind ligand directly or bind with low affinity, these assays are only suggestive. In addition, the binding ability of the type I receptor depends on the nature of the type II receptors present on the cell surface. When coexpressed with the type II receptors of TGF-β and activin, ALK-2 was shown to be able to form a heteromeric complex with TGF-β and activin, respectively (7–9). Expression of ALK-2 alone has also been reported to bind BMP-7/osteogenic protein-1 (36). The isolation of a downstream mediator of ALK-2, Smad8, as well as the characterization of the target genes of ALK-2, will undoubtedly assist in the further characterization of this pathway. The present finding makes it possible to test the capability of potential ligands to activate the transcriptional targets of ALK-2 and to regulate the phosphorylation of Smad8.
Smad8 Cooperates with Smad4.
The members of the Smad family can be categorized into three subgroups. Smad1, Smad2, Smad3, Smad5, and now Smad8 belong to a subgroup containing the SSXS motif at the C-terminal end. The SSXS motifs of Smad1 and Smad2 have been shown to be the direct targets for phosphorylation by their cognate type I receptors. In addition, these SSXS-containing Smads appear to be pathway-specific, as these Smads are phosphorylated by specific serine kinase type I receptors (17, 19, 20). Furthermore, the activities of these Smads can be correlated with the activity of different serine kinase type I receptors. Smad2/Smad3 can function downstream of ALK-4 and ALK-5 (16, 20–23) and Smad1/Smad5 can function downstream of ALK-3 (16–18, 31). Results presented here suggest that Smad8 can function downstream of ALK-2. Because Smad8 shares considerable structural similarity to Smad1 and Smad5, further biochemical and functional studies will be needed to determine if Smad8 can also transduce the signals for BMP type I receptors ALK-3 and ALK-6.
All members of the SSXS-containing Smad subgroup associate with a second class of Smads represented by Smad4. Smad4 is the most structurally divergent member in the Smad family and does not contain the SSXS motif at its C-terminus. Smad4 also does not appear to specifically mediate any signaling but is capable of cooperating with Smad1, Smad2, and Smad8 to activate their specific genes when overexpressed in the animal pole explants of Xenopus embryos (ref. 33 and this manuscript). Smad4 is required for the SSXS-containing subgroup to mediate the specific signaling from the serine kinase receptors, though its exact role in this process is poorly understood. Lastly, the recently described Smad7 forms the third class of Smads. Smad7 differs from the pathway-specific subgroup because it lacks the SSXS motif. It also differs from Smad4 as it has been shown to bind TGF-β type I receptor ALK-5 directly and act as an antagonist of TGF-β signaling (41).
ALK-2 and Smad8 Signaling Pathway in the Embryo.
The role of the ALK-2 and Smad8-mediated signaling in early embryogenesis is not addressed here, though this study does provide some insights on the significance of this pathway. The inability of the dominant negative ALK-2 to block organizer-specific gene expression suggests that this pathway is not involved in the induction of the classical Spemann’s organizer. The identification of the targets regulated by ALK-2, however, does provide clues that the ALK-2/Smad8 pathway may be involved in patterning and subdividing mesoderm during gastrulation. Both Xnot and Xbra are expressed in prospective notochord cells and have been shown to be involved in the formation of the notochord (29, 35, 37, 38). The Xnot homologue in Zebrafish has been shown to be essential for notochord development (39). The brachyury mutant in mouse, T mutants, lack posterior notochord (40). The role of ALK-2/Smad8 pathway in early vertebrate development and notochord formation, as well as its functional interaction with other serine kinase receptor pathways, e.g., ALK-3/Smad1 and ALK-4/Smad2 in the embryo remain to be explored.
Acknowledgments
We are grateful to Dr. Chris Kintner for allowing A.B. to use the instruments in his laboratory and for invaluable discussions. We thank K. Cho for gsc probe, E. M. DeRobertis for chd probe, G. Gaudriault for the HA-tagged pRC/CMV vector, R. Harland for the mouse Smad2 clone, D. Kimelman for Xnot probe, M. Schutte for the human Smad4 (DPC4) clone, and J. Smith for Xbra probe. Y.C. is an American Cancer Society postdoctoral fellow. W.V. is a Foundation for Medical Research Senior Investigator. A.B. is supported by National Institutes of Health National Research Service Award Fellowship HD07969. This work was supported by National Institutes of Health Grant HD13527 and the Foundation for Medical Research, Inc.
ABBREVIATIONS
- ALK
activin receptor-like kinase
- TGF-β
transforming growth factor β
- GS domain
glycine- and serine-rich region
- HA
hemagglutinin
- BMP
bone morphogenetic protein
- CA
constitutively active
- Xbra
brachyury
- gsc
goosecoid
- chd
chordin
- DMZ
dorsal marginal zone
Footnotes
References
- 1.Mathews L S, Vale W W. Cell. 1991;65:973–982. doi: 10.1016/0092-8674(91)90549-e. [DOI] [PubMed] [Google Scholar]
- 2.Lin H Y, Wang X F, Ng-Eaton E, Weinberg R A, Lodish H F. Cell. 1992;68:775–785. doi: 10.1016/0092-8674(92)90152-3. [DOI] [PubMed] [Google Scholar]
- 3.Attisano L, Wrana J L, Lopez C F, Massague J. Biochim Biophys Acta. 1994;1222:71–80. doi: 10.1016/0167-4889(94)90026-4. [DOI] [PubMed] [Google Scholar]
- 4.Wrana J L, Attisano L, Carcamo J, Zentella A, Doody J, Laiho M, Wang X F, Massague J. Genes Dev. 1996;10:1880–1889. [Google Scholar]
- 5.Wieser R, Wrana J L, Massague J. EMBO J. 1995;4:2199–2208. doi: 10.1002/j.1460-2075.1995.tb07214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Attisano L, Wrana J L, Montalvo E, Massague J. Mol Cell Biol. 1996;16:1066–1073. doi: 10.1128/mcb.16.3.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.ten Dijke P, Yamashita H, Ichijo H, Franzen P, Laiho M, Miyazono K, Heldin C H. Science. 1994;264:101–104. doi: 10.1126/science.8140412. [DOI] [PubMed] [Google Scholar]
- 8.Ebner R, Chen R H, Lawler S, Zioncheck T, Derynck R. Science. 1993;262:900–902. doi: 10.1126/science.8235612. [DOI] [PubMed] [Google Scholar]
- 9.Attisano L, Carcamo J, Ventura F, Weis F M, Massague J, Wrana J L. Mol Cell Biol. 1994;14:3810–3821. doi: 10.1128/mcb.14.6.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsuchida K, Mathews L S, Vale W W. Proc Natl Acad Sci USA. 1993;90:11242–11246. doi: 10.1073/pnas.90.23.11242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Massague J, Hata A, Liu F. Trends Cell Biol. 1997;7:187–192. doi: 10.1016/S0962-8924(97)01036-2. [DOI] [PubMed] [Google Scholar]
- 12.Smith J C. Curr Opin Cell Biol. 1995;7:856–861. doi: 10.1016/0955-0674(95)80070-0. [DOI] [PubMed] [Google Scholar]
- 13.Graff J M, Thies R S, Song J J, Celeste A J, Melton D A. Cell. 1994;79:169–179. doi: 10.1016/0092-8674(94)90409-x. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki A, Thies R S, Yamaji N, Song J J, Wozney J M, Murakami K, Ueno N. Proc Natl Acad Sci USA. 1994;91:10255–10259. doi: 10.1073/pnas.91.22.10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maeno M, Ong R C, Suzuki A, Ueno N, Kung H F. Proc Natl Acad Sci USA. 1994;91:10260–10264. doi: 10.1073/pnas.91.22.10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graff J M, Bansal A, Melton D A. Cell. 1996;85:479–487. doi: 10.1016/s0092-8674(00)81249-0. [DOI] [PubMed] [Google Scholar]
- 17.Hoodless P A, Haerry T, Abdollah S, Stapleton M, O’Connor M B, Attisano L, Wrana J L. Cell. 1996;85:489–500. doi: 10.1016/s0092-8674(00)81250-7. [DOI] [PubMed] [Google Scholar]
- 18.Liu F, Hata A, Baker J C, Doody J, Carcamo J, Harland R M, Massague J. Nature (London) 1996;381:620–623. doi: 10.1038/381620a0. [DOI] [PubMed] [Google Scholar]
- 19.Kretzschmar M, Liu F, Hata A, Doody J, Massague J. Genes Dev. 1997;11:984–995. doi: 10.1101/gad.11.8.984. [DOI] [PubMed] [Google Scholar]
- 20.Macias-Silva M, Abdollah S, Hoodless P A, Pirone R, Attisano L, Wrana J L. Cell. 1996;87:1215–1224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- 21.Eppert K, Scherer S W, Ozcelik H, Pirone R, Hoodless P, Kim H, Tsui L C, Bapat B, Gallinger S, Andrulis I L, Thomsen G H, Wrana J L, Attisano L. Cell. 1996;86:543–552. doi: 10.1016/s0092-8674(00)80128-2. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Feng X, We R, Derynck R. Nature (London) 1996;383:168–172. doi: 10.1038/383168a0. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y, Lebrun J J, Vale W. Proc Natl Acad Sci USA. 1996;93:12992–12997. doi: 10.1073/pnas.93.23.12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baker J C, Harland R M. Genes Dev. 1996;10:1880–1889. doi: 10.1101/gad.10.15.1880. [DOI] [PubMed] [Google Scholar]
- 25.Bhushan A, Lin H Y, Lodish H F, Kintner C R. Mol Cell Biol. 1994;14:4280–4285. doi: 10.1128/mcb.14.6.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hahn S A, Schutte M, Hoque A T, Moskaluk C A, da Costa L T, Rozenblum E, Weinstein C L, Fischer A, Yeo C J, Hruban R H, Kern S E. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 27.Kozak M. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 28.Watanabe T, Kawai A, Shimizu F, Shinomiya H, Nishino N, Taniguchi Y, Hirano H, Fujiwara T, Kanemoto N, Okuno S, Kyushiki H, Kuga Y, Shimada Y, Nagata M, Takaichi A, Horie M, Saito A, Maekawa H, Takahashi E. Genomics. 1997;42:446–451. [Google Scholar]
- 29.Smith J C, Price B M, Green J B, Weigel D, Herrmann B G. Cell. 1991;67:79–87. doi: 10.1016/0092-8674(91)90573-h. [DOI] [PubMed] [Google Scholar]
- 30.Cho K W, Blumberg B, Steinbeisser H, De R E. Cell. 1991;67:1111–1120. doi: 10.1016/0092-8674(91)90288-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki A, Chang C, Yingling J M, Wang X F, Hemmati-Brivanlou A. Dev Biol. 1997;84:402–405. doi: 10.1006/dbio.1997.8548. [DOI] [PubMed] [Google Scholar]
- 32.Lagna G, Hata A, Hemmati-Brivanlou A, Massague J. Nature (London) 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Musci T, Derynck R. Curr Biol. 1997;7:270–276. doi: 10.1016/s0960-9822(06)00123-0. [DOI] [PubMed] [Google Scholar]
- 34.Sasai Y, Lu B, Steinbeisser H, Geissert D, Gont L K, De R E. Cell. 1994;79:779–790. doi: 10.1016/0092-8674(94)90068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Dassow G, Schmidt J E, Kimelman D. Genes Dev. 1993;7:355–366. doi: 10.1101/gad.7.3.355. [DOI] [PubMed] [Google Scholar]
- 36.ten Dijke P, Yamashita H, Sampath T K, Reddi A H, Estevez M, Riddle D L, Ichijo H, Heldin C H, Miyazono K. J Biol Chem. 1994;269:16985–16988. [PubMed] [Google Scholar]
- 37.Gont L K, Fainsod A, Kim S H, De R E. Dev Biol. 1996;174:174–178. doi: 10.1006/dbio.1996.0061. [DOI] [PubMed] [Google Scholar]
- 38.Conlon F L, Sedgwick S G, Weston K M, Smith J C. Development (Cambridge, UK) 1996;122:2427–2435. doi: 10.1242/dev.122.8.2427. [DOI] [PubMed] [Google Scholar]
- 39.Talbot W S, Trevarrow B, Halpern M E, Melby A E, Farr G, Postlethwait J H, Jowett T, Kimmel C B, Kimelman D. Nature (London) 1995;378:150–7. doi: 10.1038/378150a0. [DOI] [PubMed] [Google Scholar]
- 40.Wilkinson D G, Bhatt S, Herrmann B G. Nature (London) 1990;343:657–659. doi: 10.1038/343657a0. [DOI] [PubMed] [Google Scholar]
- 41.Hayashi H, Abdollah S, Qiu Y, Cai J, Xu Y B W, Grinnell B G, Richardson M A, Topper J N, Gimbrone M A J, Wrana J L, Falb D. Cell. 1997;89:1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]