Abstract
Haloacetaldehydes can be employed for probing unpaired DNA structures involving cytosine and adenine residues. Using an enzyme that was structurally proven to flip its target cytosine out of the DNA helix, the HhaI DNA methyltransferase (M.HhaI), we demonstrate the suitability of the chloroacetaldehyde modification for mapping extrahelical (flipped-out) cytosine bases in protein–DNA complexes. The generality of this method was verified with two other DNA cytosine-5 methyltransferases, M.AluI and M.SssI, as well as with two restriction endonucleases, R.Ecl18kI and R.PspGI, which represent a novel class of base-flipping enzymes. Our results thus offer a simple and convenient laboratory tool for detection and mapping of flipped-out cytosines in protein–DNA complexes.
INTRODUCTION
Normally, the coding nucleobases in DNA are sheltered inside its double helical structure. However, the DNA helix can undergo conformational changes upon interactions with cellular proteins. A particularly remarkable example of a localized conformational distortion is so-called ‘base flipping’. It involves a complete rotation of a target nucleotide out of the DNA helix and into the catalytic site of an enzyme. Base flipping was first observed by X-ray crystallography for the bacterial DNA cytosine-5 methyltransferase HhaI (1), and subsequently for a few other DNA methyltransferases (MTases) (2,3) and various DNA repair enzymes (4). Numerous studies showed that base flipping is a fundamental mechanism in DNA modification and repair (5,6), and is also used by proteins responsible for the opening of the DNA or RNA helix during replication, transcription and recombination (7,8). However more recent findings, in which sequence-specific DNA recognition by restriction endonucleases have been shown to involve extrusion of both complementary bases in the centre of their target site (9,10), suggest that many other enzymes or DNA-binding proteins may employ this mechanism in their interactions with DNA.
The mechanism of base flipping has been intensively studied in several systems using a variety of methods, but the details of nucleotide rotation in many cases remain obscure. One of the most popular models for the base flipping studies is the HhaI MTase. A series of structures for binary and ternary M.HhaI reaction complexes at atomic resolution provided a structural framework for site-directed mutagenesis, biochemical analysis, scanning force microscopy, NMR spectroscopy and computational studies. Since many more proteins are expected to employ this mechanistic feature in their interactions with DNA, a fast method for initial screening would be very useful.
Although X-ray crystallography of protein–DNA complexes can provide the ultimate proof of base flipping, co-crystallization of proteins with their DNA substrates is often tedious or even impossible. Alternative methods to detect base flipping in aqueous solution are essential for extending studies of this phenomenon. One such method is fluorescence spectroscopy; however it requires modified bases to be introduced in the DNA. 2-aminopurine, a close structural analog of adenine, is one of the most widely used fluorescent probes (11–13). Several fluorescent analogs of cytosine, such as 2-pyrimidinone (14), pyrrolo-C (15), phenoxazine-C (16), or 4-amino-1H-benzo[g]quinazoline-2-one (17) had also been proposed. However these probes gained limited popularity for studies of enzyme-induced base-flipping due to substantial alterations of the base-pairing potential and/or steric bulk as compared with the natural cytosine base. On the other hand, fluorescence spectroscopy requires specialized equipment and may not always be sufficiently sensitive to serve as a routine laboratory tool. Previously it was found that KMnO4 is an efficient probe for detection of flipped out thymines in DNA-methyltransferase (18) and DNA-transposase complexes (19). However none of the above described approaches can directly detect flipping of cytosine, which is a natural target base for numerous DNA methyltransferases. Therefore, a cytosine-specific probe would clearly be a useful method for initial analysis of novel base-flipping systems.
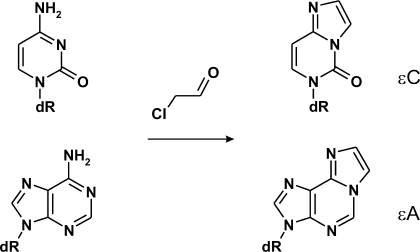
Here we aimed to assess the suitability of chemical methods that probe structural peculiarities of cytosine residues in DNA to detect flipped out bases in protein–DNA complexes. We considered those reactions that (i) proceed under mild conditions ensuring the integrity of protein–DNA complexes, and (ii) lead to a specific cleavage of the DNA strand and require no further enzymatic manipulations. Chloro- and bromoacetaldehyde are known to react with unpaired adenine and cytosine bases in DNA (20–22) yielding 1,N6-ethenoadenine or 3,N4-ethenocytosine derivatives, respectively (Figure 1). Such haloacetaldehyde-modified residues can be detected by piperidine-induced strand cleavage (22). Here we show that chloracetaldehyde (CAA) can be used to detect and map cytosine residues flipped out by the HhaI, AluI and SssI DNA cytosine-5 methyltransferases. Using this approach we also detect extrahelical cytosines in reaction complexes of two restriction endonucleases, R.Ecl18kI and R.PspGI, for which base flipping was observed by X-ray crystallography (9) or suggested from indirect observations (10,23).
Figure 1.
Chloroacetaldehyde reacts with unpaired cytosine and adenine nucleobases in DNA producing 3,N4-ethenocytosine (εC) and 1,N6-ethenoadenine (εA), respectively.
MATERIALS AND METHODS
SAM-free M.HhaI was prepared as described earlier (24). Protein concentration was estimated by absorption at λ = 280 nm. S-adenosyl-L-homocysteine (SAH) was purchased from Sigma.
M.AluI (50 units/μl) and M.SssI (4 units/μl) were purchased from New England Biolabs (USA). M.HpaII was obtained from Fermentas Life Sciences, Lithuania (1500 units/μl) or New England Biolabs (4 units/μl).
Restriction endonucleases R.Ecl18kI, R.PspGI and R.BcnI were purified and their concentrations were determined as described (10,25).
The following duplex oligodeoxyribonucleotides, 5′-33P-labeled at the upper strands, were used for analysis (target bases on the upper strand are underlined, recognition sequences for respective enzymes are boldface; M stands for 5-methylcytosine):
25-mer cognate duplex for M.HhaI and M.SssI, GCGC/GMGC
5′-TACAGTATCAGGCGCTGACCCACAA
3′- TGTCATAGTCCGMGACTGGGTGTTG
25-mer mismatch duplex for M.HhaI, GCGC/GMCC
5′-TACAGTATCAGGCGCTGACCCACAA
3′- TGTCATAGTCCCMGACTGGGTGTTG
42-mer duplex for M.AluI and M.HpaII, AGCT/AGCT
5′-TAATAGACTGCACGACGCGCCAGGCCGGCGAGCTTT
3′-ATTATCTGACGTGCTGCGCGGTCCGGCCGCTCGAAA
ACGAT-3′
TGCTAT-5′
31-mer duplex for R.Ecl18kI, R.PspGI and R.BcnI, CCCGG/CCGGG
5′-TGACCCACGCTCGCCCGGCGACACATTACGT
3′-ACTGGGTGCGAGCGGGCCGCTGTGTAATGCA
Oligodeoxyribonucleotide strands (HPLC purified) were obtained from IDT DNA (USA) or Metabion (Germany). Oligonucleotides were 5′-labeled using an Oligonucleotide 5′-labeling kit (Fermentas Life Sciences, Lithuania) and [γ-33P]ATP (Hartmann Analytic, Germany). DNA duplexes were prepared by annealing appropriate oligonucleotide strands as described (11).
M.HhaI-binding reactions contained 10–500 nM labeled 25-mer duplex, 12–500 nM M.HhaI, 100–200 μM SAH, if any, and 5% glycerol in 10–25 μl of binding buffer (50 mM MOPS, 50 mM MES pH 7.0, 1 mM Na2EDTA, 15 mM NaCl, 0.2 mg/ml bovine serum albumin). M.AluI-binding reactions contained 100–150 nM labeled 42-mer duplex, 1.5–10 units/µl M.AluI and 5% glycerol in M.HhaI-binding buffer (pH 7.5). M.SssI-binding reactions contained 20–180 nM labeled 25-mer or 42-mer duplex, 0.06–0.8 units/µl M.SssI and 5–10% glycerol in M.HhaI-binding buffer (pH 7.5). M.HpaII-binding reactions contained 50–150 nM labeled 42-mer duplex, 0.2–43 units/µl M.HpaII and 5% glycerol in M.HhaI-binding buffer (pH 7.5). R.Ecl18kI-binding reactions contained 10–900 nM labeled 31-mer duplex, 30–2000 nM R.Ecl18kI, 10 mM CaCl2 and 5% glycerol in 10–25 µl of binding buffer (33 mM Tris-Acetate pH 7.9 (25°C), 66 mM K-Acetate, 0.2 mg/ml bovine serum albumin). R.PspGI-binding reactions contained 100–900 nM labeled 31-mer duplex, 200–2000 nM R.PspGI and 5% glycerol in R.Ecl18kI-binding buffer. R.BcnI-binding reactions contained 100 nM labeled 31-mer duplex, 2000 nM R.BcnI and 5% glycerol in R.Ecl18kI-binding buffer. Typically, reactions were incubated for 20 min at 20°C or 37°C, and 1–3 μl samples were then loaded onto an 8% polyacrylamide gel (37.5:1 crosslink ratio). Electrophoresis was performed in 45 mM Tris-borate (pH 8.3), 1 mM Na2EDTA for 1–2 h at 10 V/cm (for M.HhaI, M.AluI, M.SssI and M.HpaII) or 40 mM Tris-Acetate (pH 8.3), 10 mM Ca-Acetate for 2–3 h at 5 V/cm (for R.Ecl18kI, R.PspGI and R.BcnI).
Modification with CAA (Fluka, Germany) was performed by adding CAA to a final concentration of 0.001–0.5 M for M.HhaI, M.AluI, M.SssI and M.HpaII or 0.1–1.2 M for R.Ecl18kI, R.PspGI and R.BcnI-binding reactions. The samples were incubated for 15–105 min at room temperature or 37°C. DNA was then precipitated with ethanol, redissolved in 1 M piperidine, heated at 90°C for 30 min, and lyophilized. Lyophilized material was resuspended in gel-loading solution (STOP solution, Fermentas Life Sciences) and applied to a 15–20% polyacrylamide gel (19:1 crosslink ratio) with 7 M urea. Electrophoresis was performed in 90 mM Tris-borate (pH 8.3), 2 mM Na2EDTA at 60 W constant power for 1–2 h. Gels were dried on Whatman 3 MM paper and radioactive bands were autoradiographed to an imaging plate (Fujifilm, Japan) followed by scanning with a FLA-5100 phosphoimager. DNA bands were quantitated using MultiGauge software (Fujifilm).
Purine-specific (G+A) tracks were generated by traditional Maxam-Gilbert sequencing reactions (26).
RESULTS
Reactivity of cytosines in M.HhaI-DNA complexes
A 25-mer duplex DNA containing a hemimethylated target sequence for M.HhaI (GCGC/GMGC), and 5′-labeled on the upper strand was used for screening. Premethylation of the cytosine on the bottom strand served to ensure docking of the enzyme in a single orientation targeting the upper strand (27). M.HhaI-DNA complexes were formed by adding a slight access of M.HhaI to ensure complete binding, and were then treated with CAA for 15–105 min at room temperature or 37°C. DNA strand cleavage at CAA-modified sites was achieved by heating with 1 M piperidine and followed by denaturing gel electrophoresis. In parallel, the integrity of the M.HhaI-DNA complexes was verified by analyzing reaction aliquots using gel mobility shift assay under native conditions. A series of reaction conditions were tested to achieve maximum reactivity of the target cytosine and at least 50% survival of the initial complex.
Examination of the M.HhaI-GCGC/GMGC complex with 50–200 mM CAA showed an enhanced reactivity of the target cytosine residue. Addition of M.HhaI led to a selective and significant enhancement of the reactivity of the target base, whereas a control reaction containing no enzyme showed background levels of reactivity (Figure 2). The presence of the enzyme had no effect on the CAA reactivity of any other cytosine residue on either strand. Importantly, addition of the Q237G mutant, which was indirectly shown to have abolished base flipping activity but retained binding affinity (28), showed no enhanced reactivity with CAA, serving as a control reaction (Figure 2).
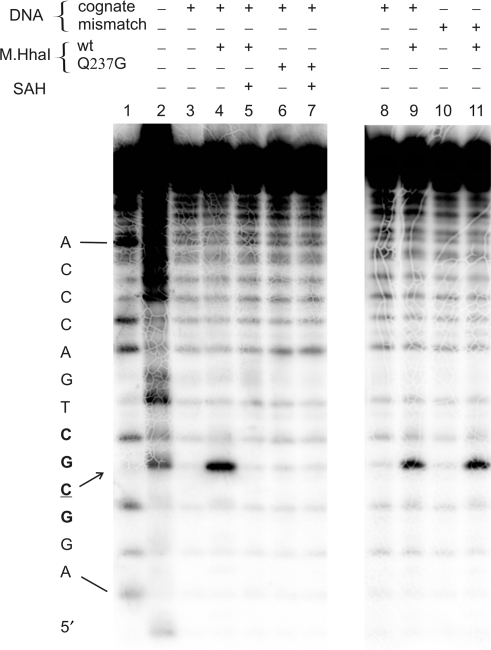
Figure 2.
CAA reactivity of cytosines in cognate (GCGC/GMGC) and mismatch (GCGC/GMCC) 25-mer DNA duplexes upon interaction with M.HhaI (or its Q237G mutant) and cofactor product SAH. Autoradiograph of degradation products of the 25-mer duplexes 5′-labeled on the target (upper) strand after separation by electrophoresis on a 15% sequencing gel. Lane 1, G + A nucleotide cleavage marker; lanes 2–7, reactions with 200 mM CAA for 45 min at 20°C, containing 300 nM protein, 280 nM DNA and 0.1 mM SAH as follows: lane 2, upper GCGC strand alone (treated at 37°C); lane 3, GCGC/GMGC; lane 4, GCGC/GMGC + M.HhaI, lane 5, GCGC/GMGC + M.HhaI + SAH; lane 6, GCGC/GMGC + M.HhaI(Q237G); lane 7, GCGC/GMGC +M.HhaI (Q237G) + SAH; lanes 8–11, CAA reactions containing 30 nM protein and 10 nM DNA as follows: lane 8, GCGC/GMGC; lane 9, GCGC/GMGC + M.HhaI; lane 10, GCGC/GMCC; lane 11, GCGC/GMCC + M.HhaI.
Notably, there was no detectable CAA modification of the flipped out target cytosines in the ternary M.HhaI complex with DNA and cofactor product SAH (SAM was not used as it causes enzymatic turnovers) (Figure 2). The most likely explanation of this observation is that the ternary complex is more compact as compared to the binary M.HhaI-DNA complexes. Cofactor SAH stimulates closure of the mobile catalytic loop in the enzyme, tightly locking the flipped out base in the catalytic site (1,29,30). This leads to a much lower accessibility of the target base to exogenous compounds such as acrylamide (31).
In addition, we analyzed the CAA reactivity of a mismatched target cytosine paired with C on the complementary strand of the DNA duplex. The weaker C:C base pair may exist in unpaired, partially flipped out conformations and is thus likely to become accessible to chemical modification. However, no CAA modification of the mismatched target cytosine was detected under the assay conditions in the absence of enzyme. This observation indicates that a cytosine residue is modified with CAA only when stabilized in an extrahelical conformation by the enzyme (Figure 2). On the other hand, cytosines in single-stranded DNA (absence of complementary strand) are readily detectable under these conditions (Figure 2).
Reactivity of cytosines in complexes with other DNA cytosine-5 methyltransferases
Having successfully demonstrated chemical mapping of extrahelical cytosines in a well proven system, we went on to examine the generality of this approach by analyzing a series of DNA cytosine-5 MTases. To date, only two DNA cytosine-5 MTases, M.HhaI (1) and M.HaeIII (2), were crystallographically proven to flip out their target bases. However, due to a high sequence and structural similarity of their numerous bacterial and eukaryotic homologs, they are also expected to operate using similar mechanisms (5). We chose commercially available recombinant bacterial DNA cytosine-5 methyltransferases M.AluI (recognition target AGCT), M.HpaII (CCGG) and M.SssI (CG) for our studies (32). First, commercial enzyme preparations were checked for the presence of bound endogenous SAM (33), which would interfere with our assay due to enzymatic turnovers (not shown). The CAA modification analysis was carried out on a 42-mer DNA duplex that contained unmethylated target sites for M.AluI and M.HpaII or on the GCGC/GMGC 25-mer, which contained the hemimethylated M.SssI/M.HhaI site. The chemical modification reactions involved initial treatment of the DNA duplex with CAA in the presence of saturating amounts of the MTases and the cofactor SAH, if any, followed by hydrolysis with piperidine to generate DNA strand breaks at modified sites. Reaction conditions were again optimized to achieve maximal cytosine modification with CAA in a control ssDNA, while maintaining the integrity of the MTase-DNA complexes (examined in a gel mobility shift assay, not shown).
Analysis with 50–300 mM CAA showed an enhanced reactivity of their respective target cytosine residues in the presence of saturating amounts of M.AluI and M.SssI (Figure 3). Best results were achieved with 50 mM CAA at 37°C for M.AluI and 100 mM CAA at 20°C for M.SssI. As observed for M.HhaI, addition of SAH abolished the CAA reaction in both cases (data not shown).
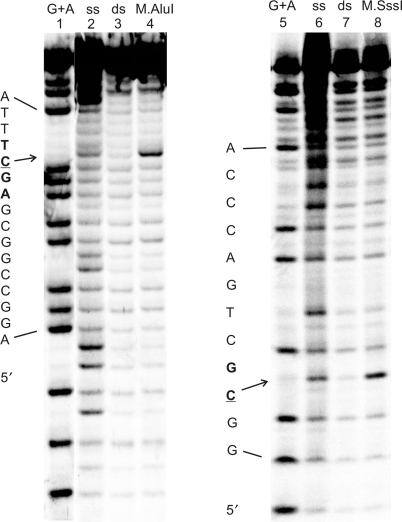
Figure 3.
CAA reactivity of cytosines in the 42-mer AGCT/AGCT duplex upon interaction with M.AluI (left panel) and in the 25-mer GCGC/GMGC duplex upon interaction with M.SssI (right panel). Lanes 1 and 5, G + A nucleotide cleavage marker; lanes 2–4, reactions with 50 mM CAA for 50 min at 37°C, containing 10 units/µl M.AluI and 105 nM DNA as follows: lane 2, upper AGCT strand alone; lane 3, AGCT/AGCT; lane 4, AGCT/AGCT + M.AluI; lanes 6–8, reactions with 100 mM CAA for 50 min at 20°C, containing 0.4 units/µl M.SssI and 20 nM DNA as follows: lane 6, upper GCGC strand alone (treated at 37°C); lane 7, GCGC/GMGC; lane 8, GCGC/GMGC + M.SssI.
Similar experiments with the HpaII methyltransferase using 50–500 mM CAA gave no detectable modification at the target cytosine (data not shown). Consistent with previous reports (34,35), no discrete band corresponding to a specific binary complex between M.HpaII and the 42-mer DNA duplex was detectable at accessible enzyme concentrations (up to 1.5 µM) in control gel-shift experiments (data not shown). Therefore, assessment of the M.HpaII-DNA complex survival under CAA reaction conditions was not possible.
Reactivity of cytosines in other base flipping systems
To further assess the generality of the new assay, we extended our studies to a recently discovered class of base-flipping enzymes—restriction endonucleases. Three endonucleases were tested.
Restriction endonuclease Ecl18kI (R.Ecl18kI) recognizes the CCNGG sequence in DNA and cleaves it before the outer C. The enzyme has recently been proven using X-ray crystallography (9) to flip out both nucleotides (N) of the central base-pair upon binding the substrate. Therefore we examined the susceptibility of the central cytosine (underlined) in the sequence CCCGG to the CAA reagent. The restriction endonuclease PspGI (R.PspGI), which recognizes the CC(A/T)GG sequence in DNA, is homologous to the R.Ecl18kI (36). Using 2-aminopurine fluorescence analysis, R.PspGI has been shown to unstack nucleotides of the central base-pair upon binding to DNA (10). We used a non-cognate DNA substrate for R.PspGI, CC(C/G)GG, since it has recently been suggested that R.PspGI can interact with CC(C/G)GG sites in B-DNA and flip out the central cytosine in vivo (23). In contrast, the BcnI restriction endonuclease (R.BcnI), which recognizes and cuts the CC(C/G)GG target site, exerts no base flipping upon binding its substrate DNA (25). This system was therefore expected to provide a negative control.
CAA reaction conditions were optimized to achieve maximal cytosine modification, at the same time maintaining the R.BcnI-DNA, R.Ecl18kI-DNA or R.PspGI-DNA complexes during the modification reaction. Similarly as with M.HhaI, the chemical modification reactions involved initial treatment with CAA followed by hydrolysis with piperidine to generate DNA strand breaks at modified sites. Fragmentation at the ‘target’ cytosine sites in R.Ecl18kI-DNA was observed, indicating an extrahelical conformation of those cytosines in complexes with R.Ecl18kI (Figure 4) in solution. Notably, the modification was observed only in a relatively high CAA concentration range of 0.2–1.2 M. However gel binding experiments confirmed that the substrate DNA was bound with high affinity under these conditions. Cytosines at positions other than the ‘target’ site were not modified by CAA. The central cytosine in R.PspGI-DNA complexes was also modified by CAA, indicating an extrahelical position for this nucleotide. In both cases, the strand cleavage was CAA-dependent and occurred at positions different from those observed during nucleolytic cleavage in the presence of Mg2+ ions (data not shown).
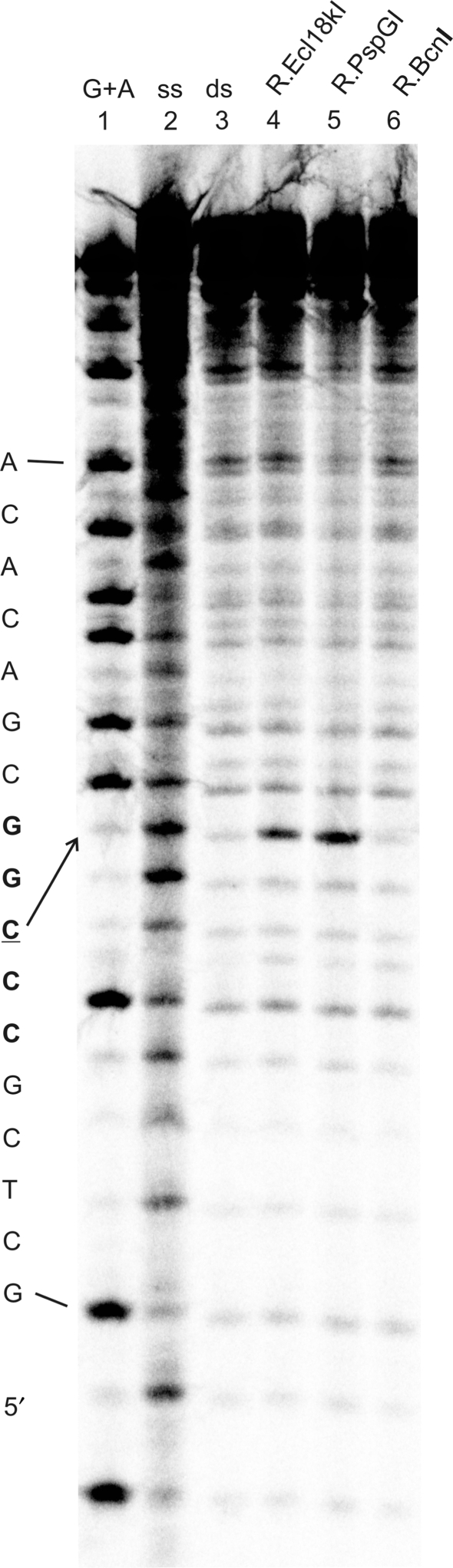
Figure 4.
CAA reactivity of cytosines in the 31-mer CCCGG/CCGGG duplex (100 nM) upon interaction with restriction endonucleases R.Ecl18kI, R.PspGI and R.BcnI (2000 nM). Complexes were treated with 400 mM CAA for 40 min at 37°C followed by analysis as described in Figure 2. Lane 1, purine nucleotide cleavage marker; lane 2, upper CCCGG strand alone (treated with 200 mM CAA for 10 min at 37°C); lane 3, CCCGG/CCGGG; lane 4, CCCGG/CCGGG + R.Ecl18kI; lane 5, CCCGG/CCGGG + R.PspGI; lane 6, CCCGG/CCGGG + R.BcnI.
The control reactions involving R.BcnI showed no CAA-dependent modification and strand cleavage at the expected cytosine residue under similar conditions. Gel binding experiments confirmed that, under the conditions of the chemical reaction, the protein–DNA complex remained intact. Since R.BcnI interacts with all its target base pairs without much disturbing the double helical DNA structure (25), our observation confirms that the CAA modification reaction is specific for unpaired extrahelical cytosines.
DISCUSSION
Our results demonstrate the applicability of the CAA modification reaction for probing single extrahelical cytosines in enzyme–DNA complexes. We did not observe significant adenine reactivity in single-strand DNA controls (Figures 2–4) using our assay conditions (CAA modification and piperidine cleavage). Most likely, unpaired adenines are modified with CAA (Figure 1), but the modified residues are not well converted to strand-cleavage products during the piperidine treatment. In the majority of published procedures (22,37,38), an intermediate reaction with formic acid or hydrazine is performed following the CAA modification and prior to piperidine cleavage. This leads to the appearance of G+A or C tracks, respectively, along with enhanced bands at the CAA-modified residues. Using our two-step procedure, the CAA-modified cytosines were clearly revealed without an overlay with nucleotide-specific bands. This simple and sensitive assay can be easily performed in any laboratory and thus can serve as convenient chemical tool to study enzyme-induced flipping of cytosine residues in duplex DNA.
The sensitivity of this assay is potentially very high when radioactive (32P, 33P or 35S) or fluorophore end-labeling of the target strand is used. However, the reaction depends on the formation and stability of a specific enzyme–protein complex in the assay, which means that the affinity of such interaction (KD value) puts a lower limit to accessible concentrations for each particular system. The use of hemimethylated target sites for DNA methyltransferases (Figure 2) might appear favorable due to enhanced binding and unique binding orientation of enzyme (27), however, ordinary unmethylated DNA duplexes gave good results with M.HhaI, M.SssI (data not shown) and M.AluI (Figure 3).
Another key factor for the detection of extrahelical cytosines is their accessibility to endogenous reagents in a protein–DNA complex. DNA methyltransferases flip out their target bases into the active site to carry out the methylation reaction. Unfortunately, no binary co-crystal structure with DNA is yet available for a DNA cytosine-5 methyltransferase. But it is well documented for M.HhaI (30,31) that the target base is much more readily accessible in a binary complex as compared to the tight ternary complex with SAH. However, the dramatic inhibitory effect of SAH on the target cytosine reactivity (Figure 2) is in apparent discord with the results of permanganate probing of M.HhaI–DNA complexes containing thymine at the target base position (18). A slight enhancement of the target thymine reactivity observed upon addition of SAH or other cofactor analogues indicates that there is little change in accessibility of the flipped out thymine base upon addition of SAH. Biochemical studies and co-crystalization trials of M.HhaI with T/G mismatched substrates (29,39) indirectly suggest that thymine, as opposed to cytosine, cannot bind in the catalytic site, which precludes the formation a tight ternary complex (18).
A different mode of base flipping is observed in restriction endonucleases, some of which extrude a nucleotide pair in order to shift the register of sequence-specific recognition with the remaining bases of a target site. Recent crystal structures of the R.Ecl18kI–DNA complex reveal that both nucleotides of the central A:T base pair of the CC(A/T)GG target sequence are completely flipped out from the DNA helix and bound in two symmetrical pockets of the homodimeric protein (9). Although the flipped out thymine base is sandwiched between Trp and Arg residues in the pocket, its Watson–Crick edge is exposed to solvent. If extrusion of the central cytosine from the CC(C/G)GG duplex proceeds in a similar manner, it would be well accessible to CAA modification in the complex. The structure of the R.PspGI enzyme is not yet known, but modeling studies (23) suggest significant similarities to R.Ecl18kI. Solution 2-aminopurine fluorescence studies confirmed the presence of extrahelical bases in complexes involving both R.Ecl18kI and R.PspGI (10). The results of the present study are thus in accord with these findings and predict that the environment (solvent accessibility) of the flipped out cytosine is similar with both restriction endonucleases (Figure 4).
Clearly, the overall accessibility and local conformational peculiarities of a flipped out cytosine are likely to determine the reaction conditions (CAA concentration, exposure time and reaction temperature) required to achieve its sufficient reactivity. On the other hand, one should bear in mind that CAA is a highly reactive electrophile with respect to numerous nucleophilic centers present in proteins. In most cases, we observed dose-dependent inactivation of the DNA methyltransferases under the assay conditions (data not shown). Therefore, an upper limit of useful CAA concentrations (as well as exposure time and reaction temperature) is determined by the chemical durability of a protein–DNA complex concerned. Taken together, it can be concluded that reaction conditions may require some optimization in different systems to achieve a desired signal-to-noise ratio of the assay (see Table 1).
Table 1.
Reaction conditions for chloroacetaldehyde probing of extrahelical cytosine residues in enzyme-DNA complexes
| Enzyme | CAA concentration range (mM) | Reaction temperature (°C) | Incubation time (min) |
|---|---|---|---|
| M.HhaI | 25–200 | 20–37 | 15–105 |
| M.AluI | 50–200 | 20–37 | 50 |
| M.SssI | 100–300 | 20 | 50 |
| R.Ecl18kI, R.PspGI, R.BcnI | 200–1200 | 37 | 20–60 |
Besides chemical piperidine treatment, other methods can be employed for the mapping of haloacetaldehyde-modified bases. Since CAA modification is expected to block base-pairing with guanine, enzymatic strand cleavage with single-stranded specific nuclease S1 under mild conditions can be used (40). Alternatively, polymerase extension-based approaches (41) should in principle be useful even with unlabeled CAA-modified templates, extending this assay to ex vivo or in vivo studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors thank Virginijus Siksnys for discussions and comments on the manuscript. This work was supported by the Lithuanian State and Study Foundation (grant T-17/2007 to D.D.). The Open Access publication charges were waived by OUP.
Conflict of interest statement. None declared.
REFERENCES
- 1.Klimasauskas S, Kumar S, Roberts RJ, Cheng X. Hhal methyltransferase flips its target base out of the DNA helix. Cell. 1994;76:357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 2.Reinisch KM, Chen L, Verdine GL, Lipscomb WN. The crystal structure of HaeIII methyltransferase covalently complexed to DNA: an extrahelical cytosine and rearranged base pairing. Cell. 1995;82:143–153. doi: 10.1016/0092-8674(95)90060-8. [DOI] [PubMed] [Google Scholar]
- 3.Goedecke K, Pignot M, Goody RS, Scheidig AJ, Weinhold E. Structure of the N6-adenine DNA methyltransferase M.TaqI in complex with DNA and a cofactor analog. Nat. Struct. Biol. 2001;8:121–125. doi: 10.1038/84104. [DOI] [PubMed] [Google Scholar]
- 4.Roberts RJ, Cheng X. Base flipping. Annu. Rev. Biochem. 1998;67:181–198. doi: 10.1146/annurev.biochem.67.1.181. [DOI] [PubMed] [Google Scholar]
- 5.Cheng X, Roberts RJ. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 2001;29:3784–3795. doi: 10.1093/nar/29.18.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stivers JT, Jiang JS. A mechanistic perspective on the chemistry of DNA repair glycosylases. Chem. Rev. 2003;103:2729–2759. doi: 10.1021/cr010219b. [DOI] [PubMed] [Google Scholar]
- 7.Lim HM, Lee HJ, Roy S, Adhya S. A “master” in base unpairing during isomerization of a promoter upon RNA polymerase binding. Proc. Natl Acad. Sci. USA. 2001;98:14849–14852. doi: 10.1073/pnas.261517398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005;309:2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- 9.Bochtler M, Szczepanowski RH, Tamulaitis G, Grazulis S, Czapinska H, Manakova E, Siksnys V. Nucleotide flips determine the specificity of the Ecl18kl restriction endonuclease. EMBO J. 2006;25:2219–2229. doi: 10.1038/sj.emboj.7601096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamulaitis G, Zaremba M, Szczepanowski RH, Bochtler M, Siksnys V. Nucleotide flipping by restriction enzymes analyzed by 2-aminopurine steady-state fluorescence. Nucleic Acids Res. 2007;35:4792–4799. doi: 10.1093/nar/gkm513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holz B, Klimasauskas S, Serva S, Weinhold E. 2-Amino purine as a fluorescence probe for DNA base flipping by methyltransferases. Nucleic Acids Res. 1998;26:1076–1083. doi: 10.1093/nar/26.4.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allan BW, Beechem JM, Lindstrom WM, Reich N. Direct real time observation of base flipping by the EcoRI DNA methyltransferase. J. Biol. Chem. 1998;273:2368–2373. doi: 10.1074/jbc.273.4.2368. [DOI] [PubMed] [Google Scholar]
- 13.Neely RK, Daujotyte D, Grazulis S, Magennis SW, Dryden DT, Klimasauskas S, Jones AC. Time-resolved fluorescence of 2-aminopurine as a probe of base flipping in M.HhaI-DNA complexes. Nucleic Acids Res. 2005;33:6953–6960. doi: 10.1093/nar/gki995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Y, Ts’o P.OP. Solid-phase synthesis of oligo-2-pyrimidinone-2′-deoxyribonucleotides and oligo-2-pyrimidinone-2′-deoxyriboside methylphosphonates. Nucleic Acids Res. 1996;24:2652–2659. doi: 10.1093/nar/24.14.2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tinsley RA, Walter NG. Pyrrolo-C as a fluorescent probe for monitoring RNA secondary structure formation. RNA. 2006;12:522–529. doi: 10.1261/rna.2165806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin K-Y, Jones RJ, Matteucci M. Tricyclic 2'-deoxycytidine analogs: syntheses and incorporation into oligodeoxynucleotides which have enhanced binding to complementary RNA. J. Am. Chem. Soc. 1995;117:3873–3874. [Google Scholar]
- 17.Godde F, Toulme J.-J, Moreau S. 4-amino-1H-benzo[g]quinazoline-2-one: a fluorescent analog of cytosine to probe protonation sites in triplex forming oligonucleotides. Nucleic Acids Res. 2000;28:2977–2985. doi: 10.1093/nar/28.15.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serva S, Weinhold E, Roberts RJ, Klimasauskas S. Chemical display of thymine residues flipped out by DNA methyltransferases. Nucleic Acids Res. 1998;26:3473–3479. doi: 10.1093/nar/26.15.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bischerour J, Chalmers R. Base-flipping dynamics in a DNA hairpin processing reaction. Nucleic Acids Res. 2007;35:2584–2595. doi: 10.1093/nar/gkm186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kusmierek JT, Singer B. Chloroacetaldehyde-treated ribo- and deoxyribopolynucleotides. 1. Reaction products. Biochemistry. 1982;21:5717–5722. doi: 10.1021/bi00265a050. [DOI] [PubMed] [Google Scholar]
- 21.Kusmierek JT, Singer B. Chloroacetaldehyde-treated ribo- and deoxyribopolynucleotides. 2. Errors in transcription by different polymerases resulting from ethenocytosine and its hydrated intermediate. Biochemistry. 1982;21:5723–5728. doi: 10.1021/bi00265a051. [DOI] [PubMed] [Google Scholar]
- 22.Kohwi-Shigematsu K, Kohwi Y. Detection of non-B-DNA structures at supercoiled DNA and chromatin with haloacetaldehyde and diethyl pyrocarbonate. In: Lilley D.MJ, Dahlberg JE, editors. Methods Enzymol. Vol. 212. San Diego: Academic Press; 1992. pp. 155–180. [DOI] [PubMed] [Google Scholar]
- 23.Carpenter M, Divvela P, Pingoud V, Bujnicki J, Bhagwat AS. Sequence-dependent enhancement of hydrolytic deamination of cytosines in DNA by the restriction enzyme PspGI. Nucleic Acids Res. 2006;34:3762–3770. doi: 10.1093/nar/gkl545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daujotyte D, Vilkaitis G, Manelyte L, Skalicky J, Szyperski T, Klimasauskas S. Solubility engineering of the HhaI methyltransferase. Protein Eng. 2003;16:295–301. doi: 10.1093/proeng/gzg034. [DOI] [PubMed] [Google Scholar]
- 25.Sokolowska M, Kaus-Drobek M, Czapinska H, Tamulaitis G, Szczepanowski RH, Urbanke C, Siksnys V, Bochtler M. Monomeric restriction endonuclease BcnI in the apo form and in an asymmetric complex with target DNA. J. Mol. Biol. 2007;369:722–734. doi: 10.1016/j.jmb.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Russell DW. Molecular Cloning. 3rd. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 27.O'Gara M, Roberts RJ, Cheng X. A structural basis for the preferential binding of hemimethylated DNA by HhaI DNA methyltransferase. J. Mol. Biol. 1996;263:597–606. doi: 10.1006/jmbi.1996.0601. [DOI] [PubMed] [Google Scholar]
- 28.Daujotyte D, Serva S, Vilkaitis G, Merkiene E, Venclovas C, Klimasauskas S. HhaI DNA methyltransferase uses the protruding Gln237 for active flipping of its target cytosine. Structure. 2004;12:1047–1055. doi: 10.1016/j.str.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 29.Klimasauskas S, Roberts RJ. M.HhaI binds tightly to substrates containing mismatches at the target base. Nucleic Acids Res. 1995;23:1388–1395. doi: 10.1093/nar/23.8.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klimasauskas S, Szyperski T, Serva S, Wüthrich K. Dynamic modes of the flipped-out cytosine during HhaI methyltransferase-DNA interactions in solution. EMBO J. 1998;17:317–324. doi: 10.1093/emboj/17.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vilkaitis G, Dong A, Weinhold E, Cheng X, Klimasauskas S. Functional roles of the conserved threonine 250 in the target recognition domain of HhaI DNA methyltransferase. J. Biol. Chem. 2000;275:38722–38730. doi: 10.1074/jbc.m005278200. [DOI] [PubMed] [Google Scholar]
- 32.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE–enzymes and genes for DNA restriction and DNA modification. Nucleic Acids Res. 2007;35:D269–D270. doi: 10.1093/nar/gkl891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar S, Cheng X, Pflugrath JW, Roberts RJ. Purification, crystallization, and preliminary X-ray diffraction analysis of an M.HhaI-AdoMet complex. Biochemistry. 1992;31:8648–8653. doi: 10.1021/bi00151a035. [DOI] [PubMed] [Google Scholar]
- 34.Yang AS, Shen JC, Zingg JM, Mi S, Jones PA. HhaI and HpaII DNA methyltransferases bind DNA mismatches, methylate uracil and block DNA repair. Nucleic Acids Res. 1995;23:1380–1387. doi: 10.1093/nar/23.8.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen JC, Zingg JM, Yang AS, Schmutte C, Jones PA. A mutant HpaII methyltransferase functions as a mutator enzyme. Nucleic Acids Res. 1995;23:4275–4282. doi: 10.1093/nar/23.21.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pingoud V, Conzelmann C, Kinzebach S, Sudina A, Metelev V, Kubareva E, Bujnicki JM, Lurz R, Luder G, Xu SY, et al. PspGI, a type II restriction endonuclease from the extreme thermophile Pyrococcus sp.: structural and functional studies to investigate an evolutionary relationship with several mesophilic restriction enzymes. J. Mol. Biol. 2003;329:913–929. doi: 10.1016/s0022-2836(03)00523-0. [DOI] [PubMed] [Google Scholar]
- 37.Pestov DG, Dayn A, Siyanova EY, George DL, Mirkin SM. H-DNA and Z-DNA in the mouse c-Ki-ras promoter. Nucleic Acids Res. 1991;19:6527–6532. doi: 10.1093/nar/19.23.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kladde MP, Kohwi Y, Kohwi-Shigematsu T, Gorski J. The non-B-DNA structure of d(CA/TG)n differs from that of Z-DNA. Proc. Natl Acad. Sci. USA. 1994;91:1898–1902. doi: 10.1073/pnas.91.5.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Gara M, Horton JR, Roberts RJ, Cheng X. Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base. Nat. Struct. Biol. 1998;5:872–877. doi: 10.1038/2312. [DOI] [PubMed] [Google Scholar]
- 40.McLean MJ, Larson JE, Wohlrab F, Wells RD. Reaction conditions affect the specificity of bromoacetaldahyde as a probe for DNA cruciforms and B-Z junctions. Nucleic Acids Res. 1987;15:6917–6935. doi: 10.1093/nar/15.17.6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dayn A, Malkhosyan S, Duzhy D, Lyamichev V, Panchenko Y, Mirkin S. Formation of (dA-dT)n cruciforms in Escherichia coli cells under different environmental conditions. J. Bacteriol. 1991;173:2658–2664. doi: 10.1128/jb.173.8.2658-2664.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]