Abstract
Mammalian genomes contain numerous evolutionary harbored mobile elements, a part of which are still active and may cause genomic instability. Their movement and positional diversity occasionally result in phenotypic changes and variation by causing altered expression or disruption of neighboring host genes. Here, we describe a novel microarray-based method by which dispersed genomic locations of a type of retrotransposon in a mammalian genome can be identified. Using this method, we mapped the DNA elements for a mouse retrotransposon, intracisternal A-particle (IAP), within genomes of C3H/He and C57BL/6J inbred mouse strains; consequently we detected hundreds of probable IAP cDNA–integrated genomic regions, in which a considerable number of strain-specific putative insertions were included. In addition, by comparing genomic DNAs from radiation-induced myeloid leukemia cells and its reference normal tissue, we detected three genomic regions around which an IAP element was integrated. These results demonstrate the first successful genome-wide mapping of a retrotransposon type in a mammalian genome.
INTRODUCTION
Substantial portions of mammalian genomes are composed of interspersed repeats that have accumulated during evolution due to insertions of transposable elements (TEs), which play crucial roles in both shaping mammalian genomes and expanding genome size (1–4). Recently, researchers have proposed that in certain cases TEs have been incorporated into host genomes and have acquired specific functions, including the recruitment of coding sequences (5), regulatory sequences (6,7) and sequences for noncoding regulatory RNAs, such as micro RNAs (8–11).
TEs occasionally have deleterious effects on their host genome by causing genetic or epigenetic aberrations (12). In addition, TEs may increase genomic instability depending on their interspersed homologous nature, since a tract of nonallelic sequence homology have the potential to cause genomic alterations during both meiotic recombination and DNA repair processes (13). In mammalian genomes, most TEs are retroelements (RTEs), which mobilize indirectly via an RNA intermediate. Although most RTEs are immobile and thus considered to be fossil genetic elements, some have remained active, including a small subfamily of human L1 and Alu (2,14) and mouse intracisternal A-particle (IAP) and early transposon (ETn) elements (3,15). In germline cells, the mutation rate mediated by RTEs differs significantly among species. It has been reported that ∼0.1% and 10–12% of spontaneous mutations in human and mouse, respectively, may be due to TE insertions (15).
The genetic background of mice is considered to affect the frequency of retrotransposition judging from the apparent strain bias, which shows the association of insertions of IAP elements and ETn with C3H/He and A/J inbred mouse strains, respectively (15). We previously reported that higher levels of IAP transcripts, the source of cDNA for the integration, were observed in normal cells, particularly in hematopoietic cells, from C3H/He mice compared with inbred strains C57BL/6J and STS/A (16). Other studies have reported strain-specific insertions of IAPs as well as their significant effects on the transcription of neighboring genes (17–19). These findings suggest that there are considerable interstrain differences in both RTE integration sites and retrotransposition competence; however, it has been very difficult to comprehensively evaluate these interstrain differences because of the lack of a method to identify integration sites of numerous RTEs dispersed across a genome.
In various organisms, including microorganisms, plants and mammals, environmental genotoxic stresses are known to evoke genomic responses including activation of RTEs (20–22). For example, ionizing radiation increases the transcription level of IAPs in mouse cells (23) and activates L1 retrotransposition in cultured human cells (24). In addition, we have reported insertions of IAP elements in radiation-induced acute myeloid leukemia (AML) cells from a C3H/He mouse (25–27). Because there have been many reports of IAP insertions near cytokine genes (25,28–31) and cytokine receptor genes (32,33) in spontaneous and radiation-induced tumor cells, activation of these tumor-related genes by inserted IAP elements in somatic cells is likely to be involved in carcinogenesis. However, because it is difficult to simultaneously detect numerous RTEs, the efficiency with which each type of RTE can be activated in whole genomes in response to certain types of genotoxic stress remains poorly understood.
PCR-based methods have been previously developed to experimentally identify novel mobile element insertions in mammalian genomes. In these methods, certain types of RTEs together with their flanking regions are specifically amplified by PCR with various strategies in combination with techniques, such as anchored-PCR, suppression PCR and subtractive hybridization, and subsequently the resultant PCR fragments are displayed by fingerprinting (34–36) or characterized after cloning (37–40). These methods were successfully applied to identify many polymorphic and human-specific integration sites of RTEs. In these methods, however, information on genomic positions of the sites can be obtained only after determining the sequence of isolated PCR fragments or individual clones. Recently, novel microarray-based methods for the global mapping of TEs in yeast were developed (41,42). In the present study, we developed a similar microarray-based method for global mapping of RTEs in mouse, the genome of which is much larger and more complex than that of yeast. We mapped and compared the locations of major types of IAP elements in genomes from the inbred mouse strains C3H/He and C57BL/6J. In addition, we compared the locations of IAPs between genomic DNAs from radiation-induced myeloid leukemia cells and from its reference normal tissue. Consequently, we could detect many differences between the strains and identified three genomic regions around which an IAP element was integrated possibly during or after radiation-induced leukemogenesis.
MATERIALS AND METHODS
Genomic DNA samples
Normal genomic DNA from a C3H/He and C57BL/6J female mouse was extracted from the tail according to standard procedures using proteinase K and phenol–chloroform extraction. AML from C3H/He male mouse was induced by exposure to 0.5 Gy of cyclotron-derived 10 MeV fast neutrons at eight weeks old. Genomic DNA of leukemic cells was isolated from the spleen, while its reference normal genomic DNA was isolated from the tail of the same mouse. These experiments were conducted according to the legal regulations in Japan and in compliance with the institutional guidelines for the care of laboratory animals.
Generation of genomic DNA fragments
Two methods were used to generate genomic DNA fragments containing a common sequence at both ends: the degenerate oligonucleotide-primed PCR (DOP–PCR) method and digestion by restriction enzymes followed by linker ligation. Four degenerate primers were designed to amplify the flanking regions of the IAP element (Supplementary Figure S1A). The 5′ anchor sequences and six nucleotides from random sequences of these four primers were identical to the published 6MW primer (43). Each primer was used independently, and the PCR products were then combined, followed by purification using a QIAquick PCR Purification kit (Qiagen, Valencia, CA, USA). After an initial denaturation at 94°C for 3 min, the DOP–PCR reaction was as follows: 3 cycles of 94°C for 1 min, 30°C for 2.5 min, ramp at 0.1°C/s to 72°C, 72°C for 5 min; 2 cycles of 94°C for 1 min, 38°C for 2 min, ramp at 0.2°C/s to 72°C, 72°C for 5 min; 2 cycles of 94°C for 1 min, 45°C for 2 min, 72°C for 5 min; finally, 30 cycles of 94°C for 1 min, 62°C for 1.5 min, 72°C for 5 min and a final extension step of 72°C for 10 min. Reactions were performed in PTC-225 Tetrad thermocyclers (MJ Research, Cambridge, MA, USA). Digestion by restriction enzymes was carried out with the following three enzymes; BamHI, BglII and FbaI. These enzymes produce a common cohesive end and are resistant to cytosine methylation at the CpG site. Genomic DNA (1 μg) was digested by these enzymes independently or in combination with another enzyme. Subsequently, all digested fragments were mixed, purified using a QIAquick kit and ligated to a linker that was prepared by annealing the following two primers: 5′-phospholylated 5′-GATCCTCGAGTCGGGCTGTTTCC-3′ and nonphosphorylated 5′-GGAAACAGCCCGACTCGAG-3′. The 10 bases at the 3′ end of the latter primer, originally designed by Fiegler et al. (44), matched the 10 bases at the 5′ end of the DOP–PCR primers.
Isolation of DNA fragments containing the sequences flanking the IAP-LTR
Genomic DNA fragments (200 ng) containing a common sequence at both ends were first mixed with a set of four biotin-labeled oligonucleotides (12.5 pmol each) in a 20 μl mixture containing 2 μl of 10 × LA Taq DNA polymerase buffer (Takara, Shiga, Japan) and 1.6 μl of 25 mM MgCl2. The mixture was heat-denatured at 95°C for 3 min and then annealed at 70°C for 5 min. After addition of 180 μl of STE solution (0.1 M NaCl, 10 mM Tris–HCl pH 8.0, 1 mM EDTA), the mixture was mixed with 50 μl of magnetic Dynabeads M-280 linked to streptavidin (Dynal Biotech ASA, Oslo, Norway) that had been prewashed and resuspended by 200 μl of Binding solution (Dynal kilobaseBINDER kit; Dynal Biotech ASA), followed by incubation at room temperature for 3 h with gentle mixing by rotation. The Dynabeads/DNA complex was washed once by Washing solution (Dynal kilobaseBINDER kit; Dynal Biotech ASA), followed by washing twice with 0.15 × SSC containing 0.05% SDS at 53°C for 15 min. DNA fragments containing the long terminal repeat (LTR) sequence were dissociated by incubating the beads in 10 mM Tris–HCl pH 8.0, 1 mM EDTA at 65°C for 10 min. The recovered DNA fragments originally prepared by DOP–PCR or digestion by restriction enzymes were both amplified by PCR using the primer, 5′-GGAAACAGCCCGACTCGAG-3′. The PCR products were applied to the secondary affinity-purification step in which a new set of biotin-labeled oligonucleotides targeted to a different segment within the LTR region was used. In this secondary affinity-purification procedure, temperatures of annealing and washing reactions were changed to 68 and 51°C, respectively. These two sets of biotin-labeled oligonucleotides were designed to anneal to the adjacent conserved sequences within 45 IAP elements, many of which were previously reported to be retrotransposed in tumor cells, as shown in Supplementary Figure S1.
Hybridization, imaging and data analysis
Fluorescence labeling of DNA, microarray hybridization and post hybridization wash were carried out according to the manufacturer's protocol (version 2) for oligonucleotide array-based comparative genomic hybridization (CGH) for genomic DNA analysis (Agilent Technologies, Santa Clara, CA, USA). Briefly, PCR products amplified from the recovered DNA fragments in the secondary affinity-purification procedures and original unprocessed genomic DNA were digested by Rsa I and Alu I, then fluorescently labeled with Cy3 and Cy5, respectively. Clean-up of digested or fluorescently labeled DNA was carried out with QIAprep Spin Miniprep kit (Qiagen) or Microcon YM-30 filters (Millipore, Billerica, MA, USA), respectively. After hybridization at 65°C for 40 h, microarrays were serially washed by Oligo aCGH wash buffer 1 (Agilent Technologies), Oligo aCGH Wash Buffer 2 (Agilent Technologies), acetonitrile and stabilization and drying solution (Agilent Technologies). Hybridized arrays were scanned using the Axon 4000B scanner (Axon Instruments, Burlingame, CA, USA) and processed using GenePix Pro 5.1 software (Axon Instruments). Fluorescence ratios were normalized so that the mean of the median ratios for all of the features was equal to 1.0. Three types of Agilent 244k-formatted microarrays were used: two were high-density mouse custom array-CGH microarrays consisting of more than 200 000 oligonucleotide probes designed for genomic region on chromosome 18 alone with a 0.21 kb median spacing (type 1) or for whole genomic region with a 7.8 kb median spacing (type 2), and the other one was high-density mouse promoter ChIP-on-chip microarrays (G4490A, Agilent), which covers −5.5 kb upstream to +2.5 kb downstream of the transcriptional start sites for ∼17 000 of the best-defined mouse transcripts (type 3). Most of the probes in the second custom array-CGH microarray were the same probes used for a catalog mouse CGH array (G4415A, Agilent), while there were additional 2433 probes. These additional probes were selected from the genomic regions of probable IAP-cDNA integrating regions, including tens located adjacent to the two representative IAPs described in Figure 2A and from genomic regions for T-cell receptors and immunoglobulin genes. Because these were not randomly selected probes, the data on these additional probes were not included in the scatter plot in Figure 3A, in which probable genomic regions that flank IAP-LTR between C3H/He and C57BL/6J strains were compared. The microarray data reported in this article have been deposited in the Gene Expression Ominibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE9572).
Figure 2.
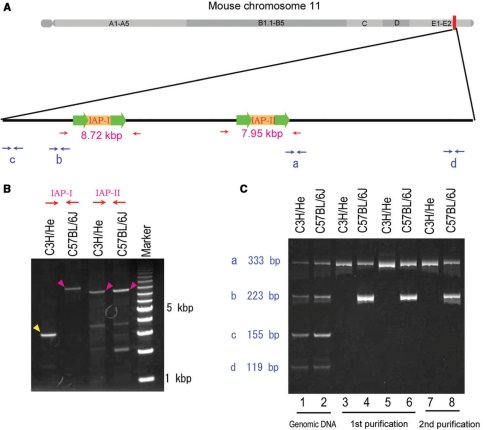
Monitoring the enrichment of DNA fragments containing the LTR sequences using multiplex PCR. (A) Diagram of a distal region of mouse chromosome 11. Relative positions of two IAP elements (IAP-I and IAP-II) and four PCR target sites (labeled as a–d) are illustrated. Arrows indicate PCR primers. (B) Long-distance PCR amplification of genomic regions surrounding IAP-I and IAP-II in (A) from C3H/He and C57BL/6J genomic DNA. Magenta arrowheads indicate PCR products with the length expected based on the genomic database, in which IAP elements were included, and the yellow arrowhead corresponds to the length without IAP insertion. (C) Multiplex PCR reaction was carried out on DNA from C3H/He (odd-numbered lanes) and C57BL/6J (even-numbered lanes), respectively. Templates were as follows: original genomic DNA (lanes 1 and 2), PCR-amplified DNA fragments originally prepared by protocol-I (lanes 3 and 4) and protocol-II (lanes 5 and 6) as shown in Figure 1C after the first round of affinity purification (lanes 3–6), and PCR-amplified DNA fragments after the second round of affinity purification (lanes 7 and 8).
Figure 3.
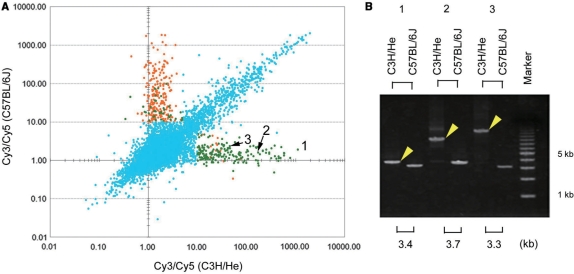
Comparison of probable genomic regions that flank IAP-LTR between C3H/He and C57BL/6J strains. (A) Scatter plots of the Cy3/Cy5 ratios for C3H/He (x-axis) and C57BL/6J (y-axis) genomes. The LTR sequence of IAP elements was present in the mouse genome database (NCBI m36.1) near the genomic regions corresponding to orange dots, but not to genomic regions corresponding to light green dots. For the light blue dots, we did not examine whether the LTR sequence of IAP elements was recorded as being present around the corresponding genomic regions in the database. (B) The presence of insertions around the regions corresponding to the probes with numbers from 1 to 3 was verified by PCR. PCR products (yellow arrowheads) that were longer than expected based on the database information (shown in the bottom of the figure in kbp) were detected only in the C3H/He genome.
Long-distance PCR amplification of genomic regions
To amplify long genomic fragments, hot-start PCR was performed by combinational use of a LA-Taq DNA polymerase (Takara) and a wax barrier (AmpliWax PCR Gem 50, Takara). All designed primer sequences were checked for their specificity by BLAST search and are listed in Supplementary Table S1. PCR cycling conditions were set according to the manufacturer's recommendations. For amplification using the template DNA from a leukemia sample and its reference tissue, the PCR cycling number was set below 28 to prevent the one-sided amplification of shorter products, which hinder detection of longer one co-amplified with the same primers.
Multiplex PCR
Multiplex PCR was carried out using Multiplex PCR kit (Qiagen) in a 12.5 μl volume, in which 10 ng of template DNA was included. All primer sequences are listed in Supplementary Table S1. PCR cycling conditions were set according to the manufacturer's recommendations. The reaction was subjected to 26 cycles of amplification. Samples (2.5 μl) were resolved by 8% polyacrylamide gel electrophoresis, followed by staining with SYBR Green I (Takara) for 1 h.
Real-time PCR
Enrichment of the flanking regions of IAP-LTR was analyzed by real-time PCR and Syber-Green fluorescence using SYBER Premix Ex taq (Takara) on a Stratagene MX3000 real-time PCR instrument (Stratagene, La Jolla, CA, USA) for 45 two-step cycles, which was followed by a final dissociation stage to generate a melting curve. Genomic DNA and the PCR products after the first and the secondary affinity-purification step were diluted into 1 ng/μl and were analyzed in duplicate.
RESULTS
Outline of genome-wide mapping of IAP integration sites
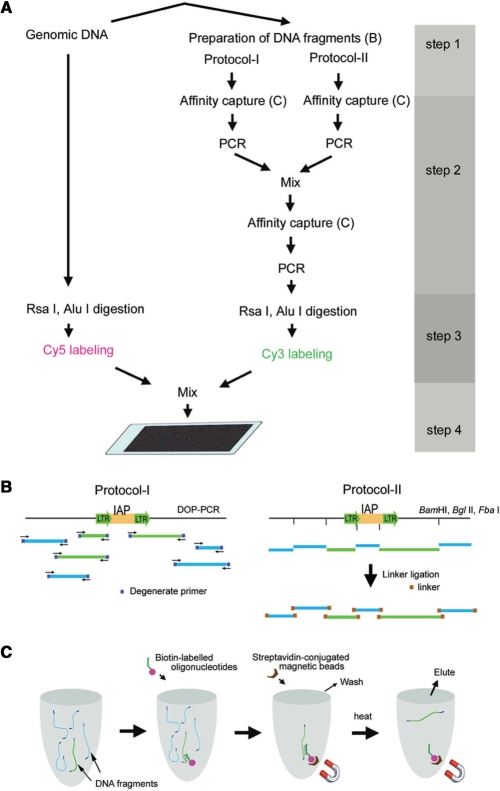
For the global identification of genomic regions containing the IAP-integrated sites, genomic DNA fragments containing sequences that flank IAP elements were isolated and hybridized to the microarray. Our protocol mainly consisted of the following four steps (Figure 1): (i) preparation of genomic DNA fragments (Figure 1B), (ii) affinity purification of DNA fragments containing the contiguous sequences covering the LTR region of IAP elements and their flanking regions (Figure 1C), (iii) differential fluorescent labeling of the affinity-captured DNA and original unprocessed DNA and (iv) hybridization on the high-density oligo microarrays.
Figure 1.
Outline of the procedure for isolating genomic DNA fragments containing sequences that flank IAP elements. (A) Two rounds of affinity purification followed by PCR amplification. The affinity-captured DNA and original unprocessed DNA were digested with Rsa I and Alu I and then fluorescently labeled with Cy3 and Cy5, respectively. The mixture of these labeled DNA fragments was used for hybridization on high-density oligo microarrays. (B) Genomic DNA fragments containing a common sequence at both ends were prepared by two different methods: the degenerate DOP–PCR method (protocol-I) and digestion by restriction enzymes followed by linker ligation (protocol-II). Light green and blue lines indicate the fragments in which the highly conserved sequences within LTRs of IAP elements are included or not included, respectively. (C) A schematic diagram of the steps for the affinity purification of DNA fragments containing the contiguous sequences covering the LTR region of the IAP element and its flanking region.
In the first step, genomic DNA fragments were prepared by two different methods: DOP–PCR (protocol-I) and the digestion by restriction enzymes followed by linker ligation (protocol-II). We used DNA fragments prepared by both methods because we considered that certain genomic regions flanking IAP elements would have an increased chance of generating longer DNA fragments by combining DNA fragments originally prepared by two different ways. In Protocol-I, DOP–PCR was carried out using four specifically designed degenerate primers with modified cycling conditions as described in Materials and Methods to amplify relatively long products of LTR-flanking regions. In Protocol-II, the digestion by restriction enzymes was carried out using the following three enzymes: BamHI, BglII and FbaI. These enzymes are insensitive to methylation at CpG sites and produce a common cohesive end. These enzymes were used both independently and in double-enzyme digestions to increase the chance of producing DNA fragments with variable sizes from an IAP-LTR-flanking region. We expected that this could increase the possibility that DNA fragments prepared by restriction enzyme digestion include DNA fragments with suitable sizes for the detection by the microarray analysis from as many IAP-LTR-flanking regions as possible.
In the second step, DNA fragments containing the LTR sequence were enriched. The desired DNA fragments were annealed to biotin-labeled oligonucleotides and subsequently captured by streptavidin-conjugated magnetic beads (Figure 1C). Both heat-dissociated DNA fragments originally prepared by Protocols-I and -II were separately amplified by PCR and mixed, followed by a similar secondary affinity-purification step in which a new set of biotin-labeled oligonucleotides was targeted to an adjacent segment within the LTR region. PCR products amplified from the recovered DNA fragments in the secondary affinity-purification procedure and the original unprocessed genomic DNA was digested by Rsa I and Alu I, and then labeled fluorescently with Cy3 and Cy5, respectively. Digestion by Rsa I and Alu I was carried out essentially according to the manufacturer's protocol (version 2) for oligonucleotide array-based CGH for genomic DNA analysis to minimize the hybridization between the genomic DNA fragments at the subsequent hybridization step on the high-density oligo microarrays.
Enrichment of DNA fragments
To monitor how DNA fragments containing the LTR sequence were effectively enriched, two representative genomic positions at the distal region of chromosome 11 were selected: one position contained an IAP element in both genomes of C3H/He and C57BL/6J inbred mice (IAP-II in Figure 2A and B), whereas another position contained an IAP element only in the C57BL/6J genome (IAP-I in Figure 2A and B). Based on the mouse genomic database, the LTR sequences at both ends of IAP-II contained a sequence identical to the one of the biotin-labeled oligonucleotides used in both the first and second affinity-purification procedures. However, the LTR sequences at both ends of IAP-I included at least one base difference from any of the biotin-labeled oligonucleotides (Supplementary Figure S1). Four pairs of PCR primers were designed based on four genomic regions surrounding the two IAP elements (Figure 2A). By simultaneous amplification of these four regions in a single reaction, similar amounts of all four PCR products were amplified from both the C3H/He and C57BL/6J DNA samples before purification (lanes 1 and 2 in Figure 2C, respectively). After the first (lanes 3–6 in Figure 2C) and the second (lanes 7 and 8 in Figure 2C) purification procedures, PCR products originating from the DNA sequences near IAP elements were visible whereas those from the sequences distal to the insertions were not visible (Figure 2B), indicating the effective enrichment of the desired DNA fragments. Furthermore, a 223-bp PCR product (b in Figure 2C) originating from the DNA sequence adjacent to an IAP insertion (IAP-I in Figure 2A) was only visibly amplified from the C57BL/6J DNA sample but not the C3H/He DNA sample. This result indicated that the affinity-purified DNAs reflected the difference between the genomic DNAs from the two different strains.
Microarray-based mapping of integration sites for IAP elements
We next examined whether dispersed integration sites of IAP elements could be mapped by the combinational use of the affinity-purified DNAs and the oligo microarrays. We expected that the microarray oligonucleotide probes that corresponded to flanking regions of IAP elements would generate high ratios of Cy3/Cy5 fluorescence intensities because Cy3-labeled DNA fragments, which included flanking regions of IAP elements, were enriched by the affinity capture procedure. DNA fragments containing the IAP element and the flanking sequences in C57BL/6J genomes were isolated in the same way as described above in two separate experiments. They were re-amplified by PCR, labeled fluorescently with Cy3, mixed with the original DNA fragments labeled with Cy5 and then hybridized to an Agilent high-density mouse custom array-CGH microarray that consisted of more than 200 000 oligonucleotide probes for genomic region on chromosome 18 alone with 0.33-kb average spacing. In this experiment, we diluted the original C57BL/6J genomic DNA and the PCR products after the first and the secondary affinity-purification step into 1 ng/μl and monitored the enrichment of five flanking regions of IAP-LTR by real-time PCR (Supplementary Table S2). The result indicated that the genomic DNA fragments near the IAP-LTR were recovered and more or less enriched by both or either protocol I and/or II, while the fragments far from the IAP-LTR were almost completely eliminated at least after two step purifications. In addition, in contrast to the efficient enrichment of an RS subtype of IAP elements, on which we previously detected retrotransposition events in AML cells of C3H/He mice (26), another highly repetitive SINE B1 sequence was diluted. This incomplete elimination of SINE B1 is probably due to the fact that many SINE B1 sequences are located near IAP elements.
We calculated each Cy3/Cy5 ratio for all probes on the array-CGH microarray and examined whether microarray features corresponding to the flanking regions of IAP-LTR showed high Cy3/Cy5 ratios. While the vast majority of the microarray features showed Cy3/Cy5 ratios <10 with the average ratio of 2.6 in both experiments (Supplementary Figure 2A), many features relatively close to IAP elements (∼2.0 kb) showed Cy3/Cy5 ratios higher than 10. The frequency of neighboring features showing higher Cy3/Cy5 ratios appeared to vary significantly depending on the distance from the IAP-LTR (Supplementary Figure S2B) and the subtype of IAP elements (Supplementary Table S3). While majority of the neighboring features to IAP-LTR classified as IAPLTR1, IAPLTR1a, IAPLTR2, IAPLTR2a and IAPLTR2b in the database (NCBI Build 36.1) showed higher ratios, those to IAP-LTR belonging to other groups rarely showed such high ratios. It is noteworthy that most of IAP elements reported to be activated in tumor cells (16,26,27) belong to the former groups and most of the LTRs in the latter groups are solo-LTR, which is composed of a single LTR region alone.
We next asked what extent the locations of IAP-LTR in the database and the microarray probes showing high Cy3/Cy5 ratios were correlated. In this analysis, we designated locations as candidate IAP-integrated regions if two or more consecutive features had Cy3/Cy5 ratios >10. As a result, clusters of features corresponding to 145 and 167 genomic regions in experiment 1 and 2, respectively, passed through this criterion and were considered to correspond to IAP-integrated genomic regions. Among them, about 70% (102 in experiment 1, 114 in experiment 2 and 100 in both experiments) corresponded to the locations of IAP-LTR in the database (NCBI Build 36.1).
Comparison of C3H/He and C57BL/6J genomes using oligo microarrays
We next examined whether our microarray-based method can be used to detect the difference in the localization of IAP elements. In this analysis, the affinity-purified DNAs described in Figure 2 were labeled and hybridized on two types of microarrays. One was an Agilent high-density mouse custom array-CGH microarray that consisted of more than 200 000 oligonucleotide probes with 6.4 kb average spatial resolution, including tens of probes located adjacent to the two representative IAPs described in Figure 2A. The other one was an Agilent high-density mouse promoter ChIP-on-chip microarray that covered −5.5 kb upstream to +2.5 kb downstream of the transcriptional start sites of ∼17 000 of the best-defined mouse transcripts.
Figure 3A shows a plot of each Cy3/Cy5 ratio for all probes on the array-CGH microarray for the C3H/He genome (x-axis) versus that for the C57BL/6J genome (y-axis). While the vast majority of the microarray features showed Cy3/Cy5 ratios less than 10 in both x and y axes, about 800 features showed higher ratios in both or either x and/or y axes. We expected that many of the features showing higher ratios in both x and y axes would correspond to regions around which IAP elements were located in both C3H/He and C57BL/6J genomes, whereas the features showing higher ratios in either x-or y-axis would correspond to regions around which IAP elements were located in either the C3H/He or C57BL/6J genome. For the features of which Cy3/Cy5 ratios were higher than 10 in one axis and less than 5 in another axis, the presence of IAP elements in LTR sequences around the corresponding genomic regions in the mouse genome database (NCBI 36.1) were examined. As a result, LTR sequence of IAP elements was recorded as being either present (features indicated by orange dots in Figure 3A) or absent (features indicated by green dots) within 2.5 kb of the genomic regions. The orange and green dots are distributed quite differently on the plot. For features having higher Cy3/Cy5 ratios only for the y-axis (C57BL/6J genome), 85.9% (225/262) of them were orange. In contrast, for features having higher Cy3/Cy5 ratios only for the x-axis (C3H/He genome), 8.4% (16/190) of them were orange. This stark contrast can be logically explained by the fact that the mouse genome database is largely based on the information from the C57BL/6J strain; hence, most of the IAP elements inserted into C3H/He, but not into the C57BL/6J genome, were not registered in the database. Of the three features showing relatively high Cy3/Cy5 ratios only for the C3H/He genome (indicated by numbers 1–3 in Figure 3A), we confirmed the presence of insertions around the corresponding regions only in the C3H/He genome by PCR (Figure 3B). Complete or partial sequencing analysis of these insertions indicated that the insertion corresponding to number 1 in Figure 3B contained IAP-LTR sequence alone (accession number AB425193) and the other two insertions contained IAP-LTR sequence (accession numbers AB425194, AB425195). Together with their sizes of insertions, these three insertions seemed to be solo IAP-LTR, a subtype of IAP element containing a deletion and a full-length IAP element, respectively.
For the features located within four regions adjacent to the two representative IAP elements described in Figure 2A, which were arranged to be included in our custom microarray, the Cy3/Cy5 ratios for C3H/He and C57BL/6J genomes were plotted while maintaining the spatial relationships between the position of each probe and IAP element (Supplementary Figure S3). For features positioned around IAP-I in Figure 2A, high Cy3/Cy5 ratios were observed only for some probes relatively close (∼2.0 kb) to both ends of the IAP element only in the C57BL/6J strain, suggesting successful LTR-dependent enrichment of flanking DNA fragments of IAP elements by the affinity capture procedure. As for features positioned relatively close to both ends of IAP-II in Figure 2A, however, Cy3/Cy5 ratios were considerably lower compared with those for IAP-I, indicating inefficient enrichment of flanking DNA fragments. This difference in enrichment efficiency between IAP-I and IAP-II might be due to the aforementioned sequence variations within the LTR regions or to another factor associated with the method used to prepare DNA fragments containing the LTR sequence.
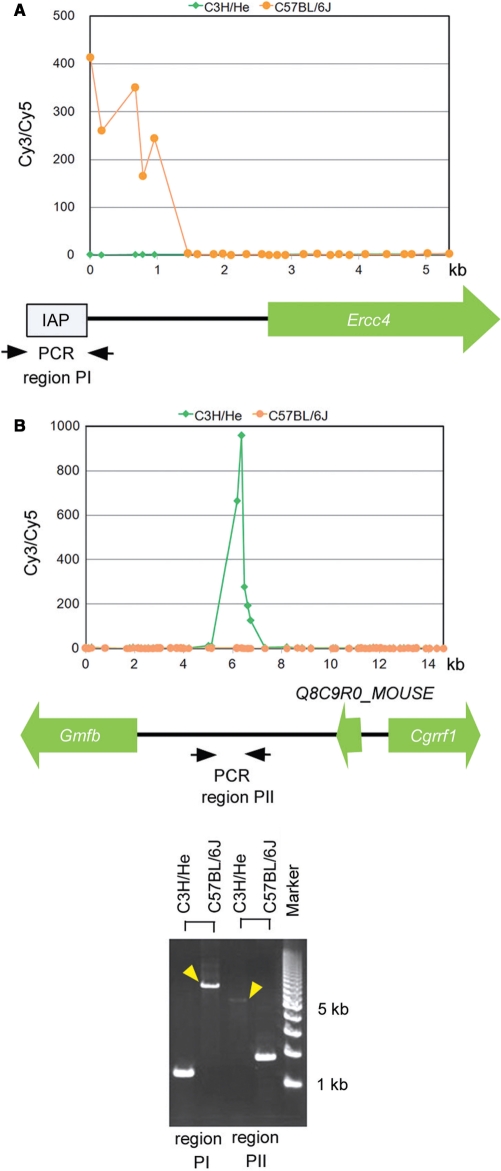
To search in depth for IAP elements located around the promoter regions, similar hybridization experiments were carried out on the Agilent mouse promoter ChIP-on-chip microarray. Because the average spatial resolution was very high (∼200 bp) around the promoter regions in this microarray, we designated locations as candidate IAP-integrated regions if two or more consecutive features had Cy3/Cy5 ratios >10. As a result, clusters of features corresponding to 147 genomic regions passed through this criterion and were considered to correspond to IAP-integrated genomic regions located near the promoter or intragenic regions, of which 91 were considered to contain an IAP element only in the C3H/He or C57BL/6J genome (Supplementary Table S4). For all of these regions, we asked whether LTR sequences of IAP elements were recorded around the corresponding genomic regions in the database. Of 97 candidate regions in which IAP elements were judged to be inserted in the C57BL/6J genome, 69 (71%) corresponded to the genomic regions located near the LTR insertion recorded in the database. In contrast, for 50 candidate regions in which IAP elements were judged to be inserted only in the C3H/He genome, only 1 (2%) corresponded to the genomic position located near the LTR insertion recorded in the database. For the two promoter regions in which IAP elements were present in either the C3H/He or C57BL/6J genome alone, we used PCR to confirm that the insertions around the corresponding region were located only in the genome of each respective mouse strain (Figure 4). These comparative results between C3H/He and C57BL/6J indicated that there are considerable strain-differences in the localization of IAP elements and that multiple differences can be detected using our microarray-based method.
Figure 4.
Differential insertions at promoter regions observed between genomes from C3H/He and C57BL/6J mice. (A and B) Cy3/Cy5 ratios for C3H/He (light green dots) and C57BL/6J (orange dots) genomes were plotted separately to maintain the spatial relationships between the position of each probe and gene. Gene names and their directions are illustrated below the graphs. (C) The presence of insertions around regions PI and PII shown in (A) and (B) was verified by PCR. Strain-dependent long PCR products (yellow arrowheads) indicated the presence of insertions around regions PI and PII only in the C57BL/6J or C3H/He genome alone.
Insertions in a genome of radiation-induced leukemic cells
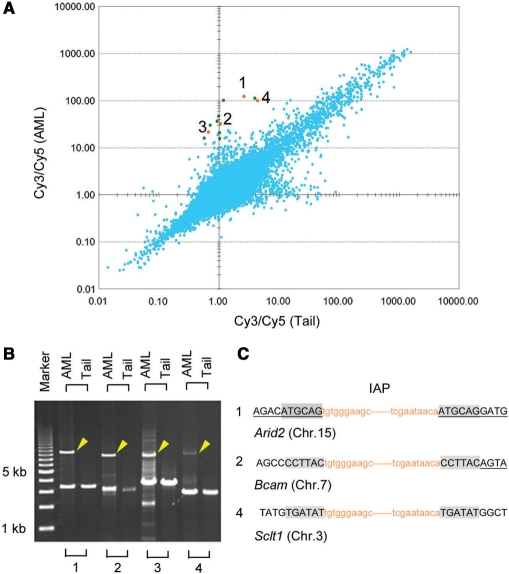
Based on the successful genome-wide survey of retrotransposon integration sites to compare the germlines of mouse strains by this method, we tried to expand the method to analyze genomic aberrations in tumor cells. From a C3H/He mouse that had acquired radiation-induced leukemia, a DNA sample was prepared from each of leukemia cells and germline cells isolated from the spleen and tail, respectively. Figure 5A shows a plot of each Cy3/Cy5 ratio for all features on the array-CGH microarray for the leukemia genome (y-axis) versus that for the reference normal tail genome (x-axis). For the 12 features showing relatively large ratios only for the leukemia genome, the presence of inserted sequences was examined by PCR. In addition to a PCR product of a expected size from the database that were commonly amplified from both AML and normal DNA, another longer PCR product was detected only for AML DNA in four cases, suggesting the presence of inserted sequences (indicated by numbers 1–4 in Figure 5A and B).
Figure 5.
Comparison of candidate genomic regions flanking IAP-LTRs between genomic DNA from a radiation-induced myeloid leukemia and its reference normal tissue. (A) Scatter plots of Cy3/Cy5 ratios for DNA obtained from reference normal tail tissue (x-axis) or radiation-induced myeloid leukemia cells (y-axis). The presence of insertion sequences was examined by PCR for the 12 dots that showed relatively large Cy3/Cy5 ratios only for the myeloid leukemia genome. For four orange dots (numbered 1–4, panel A), PCR products that were longer than expected based on the database information (yellow arrowheads, B) were detected only in the genomic DNA from radiation-induced myeloid leukemia cells by PCR. For features 1, 2 and 4, sequence analysis at the insertion junctions showed the duplication of six bases of genomic sequence (shaded, C). Lowercase orange letters indicate the terminal sequences of inserted IAP elements. Capital letters with or without underlines correspond to genomic sequences of exons and introns, respectively, of the gene indicated below.
To ascertain the presence of LTR sequences in these four long PCR products, these fragments were purified from an agarose gel and were used as templates for the amplification of the sequences containing the junction boundary at both ends of the insertions. In these secondary PCR reactions, two types of primers, IAP-RM1 and IAP-s57F2, designed for the conserved sequences among IAP elements (Supplementary Figure S1), were used in combination with a specific primer that was designed for sequences adjacent to the insertions at both ends. Except for the number. 3 fragment in Figure 5B, we obtained clear PCR products (data not shown) and determined the boundary sequences. Consequently, IAP elements were found to be inserted within an exon of Arid2 (AT rich interactive domain 2), the region near the intron–exon boundary of Bcam (Basal cell adhesion molecule), and an intron of Sclt1 (sodium channel and clathrin linker 1), corresponding to features 1, 2 and 4, respectively. Each of these insertions showed a characteristic duplication of six bases of target sequence (Figure 5C), strongly suggesting that the insertions were due to IAP element retrotransposition.
DISCUSSION
In recent years, extensive studies on genetic variations, such as single nucleotide polymorphisms (SNPs) and copy number variations (CNVs), have been performed to investigate their association with disease susceptibility in humans (45,46). Mice harbor many SNPs and CNVs, similar to those observed among humans (47,48). Genotypic differences among mouse strains that result from SNPs or CNVs may be responsible for certain interstrain phenotypic differences. Although substantial portions of mammalian genomes are composed of interspersed repeats derived from insertions of TEs, information is limited on the differences in genomic locations of TEs among humans and mouse strains. In the present study, we report the development of a novel microarray-based method for global mapping of IAP elements in the mouse genome. In each analysis of the normal genomes of C3H/He and C57BL/6J mice using array-CGH oligo microarrays, we identified hundreds of probable IAP cDNA–integrated genomic regions. Because the median probe spacing of the array-CGH oligo microarray (7.8 kb) was longer than the typical length of LTR-flanking sequences enriched effectively in our protocol (which varied between 0.5–2 kb depending on the region), we only detected less than half of the insertion sites. Indeed, it was previously calculated that there are about 1000 or more copies of IAP elements per haploid murine genome (49,50). Nonetheless, we identified about 200 probable IAP cDNA–integrated genomic regions that were considered to be present either in the C3H/He or C57BL/6J genome, but not both. Similarly, among about 150 insertions located in close proximity to the promoter regions, which were detected using a mouse promoter ChIP-on-chip microarray, about two-thirds of them were present either in the C3H/He or C57BL/6J genome. These results indicate that there are considerable strain differences in the locations of IAP elements. This observation is consistent with a recent report by Horie et al. (19), in which 6 of 11 transcripts, induced by the presence of an IAP element, were found to be derived from insertions differentially distributed between C57BL/6J and 129 mouse strains.
These interstrain differences may be partially responsible for certain interstrain phenotypic differences because insertions of IAP elements may alter the normal expression of neighboring gene(s) in a variety of ways. Insertion of IAP elements around the promoter region may perturb epigenetic regulation (51), and a cryptic promoter in the IAP can induce ectopic expression of neighboring genes (15). In addition, some retrotransposons probabilistically escape epigenetic silencing, and they are responsible for the variable expressivity of adjacent alleles—termed metastable epialleles (52). On the other hand, insertion of IAP elements within introns can cause transcriptional aberrations by causing premature polyadenylation, aberrant splicing or ectopic transcription (15). It would be interesting to analyze how the insertion sites of IAP elements we detected around promoter regions either in the C3H/He or C57BL/6J genome affect the expression of neighboring genes.
By comparing IAP cDNA–inserted genomic positions between a radiation-induced AML and its reference normal tissue from the same mouse, we found there were at least three sites in which IAP elements were inserted only in the AML genome, suggesting the occurrence of multiple retrotransposition events in one tumor during or after radiation-induced leukemogenesis. Because the average spatial resolution of the array-CGH oligo microarray was larger than the typical length of LTR-flanking sequences enriched effectively in our protocol (as mentioned above), there may be more undetected insertions. Furthermore, subsequent analysis of partial sequences of three inserted IAP elements (accession numbers AB365179, AB365180, AB365181) revealed that they were classified into the IΔ1 subtype, which is a minor population consisting of internally deleted IAP elements, but predominantly transposed in AML cells [Ishihara et al. (27), accession numbers of AB099819 and AB099821]. In addition, the partial sequences of these three inserted IAP elements had mutual variations, indicating that these inserted IAP elements were not derived from one common active IΔ1 IAP element. These insertions within Arid2, Bcam and Sclt1 have the potential to cause gene disruption or aberrant splicing. Because we could find no reports suggesting the association of the aberrations in these genes with tumorigenesis, these integrations might not be directly involved in leukemogenesis. However, the successful identification of three IAP cDNA–integrated intragenic regions in radiation-induced tumor cells demonstrate the potential usefulness of this method to identify critical responsive genes for tumorigenesis and to a better understanding of the mechanisms of retrotransposition in radiation-induced tumor cells.
There were a number of false-positive probes using our method as was also seen in two recently developed microarray-based methods for the global mapping of TEs in yeast (41,42). In both of these yeast TE-mapping methods, likely true positives were judged to show positive hybridization signals at consecutive features within the corresponding genomic regions. We could apply a similar criterion in the analysis with the microarrays which focused on chromosome 18 or promoter regions. However, about 30% of the genomic regions showing positive hybridization signals at consecutive features did not correspond to the genomic regions located near the LTR insertion recorded in the database and thus should be defined as false positives. However, since we could not find any significant sequence similarities within many of these genomic regions to the sequence of biotin-labeled primers, a part of these probable false positives might be true positives, such as ones derived from differences in the integration sites between substrains or individuals or from retrotransposition events in some somatic cells included in the tissue from which genomic DNA was isolated. We also found that there were many false-negative features. Although we could map majority of several types of IAP elements on chromosome 18, some features positioned relatively close to the targeted types of IAP-LTR did not show high Cy3/Cy5 ratios. These false-negative features were possibly caused by several ways through uneven enrichment or amplification of DNA fragments containing the LTR sequence. We expect that the reliability of our method will benefit from improvements in certain experimental procedures that would yield more uniform enrichment and amplification of IAP flanking sequences; these improvements include more accurate sequences of biotin-labeled oligonucleotides, optimization of hybridization conditions and an enhanced method of preparing DNA fragments containing the LTR sequence. In addition, a plot of each Cy3/Cy5 ratio for all features in Supplementary Figure S2 showed broader distribution of the plots than the similar plots in Figures 3 and 5. We consider that this is related to the fact that two samples were prepared and enriched independently in the former experiment, while the two samples to be compared in Figures 3 and 5 were processed simultaneously in one experiment. This suggested the importance of simultaneous processing in the experiment to identify the difference between two different samples. Furthermore, by altering the sequence of biotinylated oligonucleotides used during affinity purification, we expect that our method will be applicable to global mapping of other types of retrotransposons and of multiple dispersed sequences in mammalian genomes.
In summary, the present results demonstrate for the first time the successful genome-wide mapping of integration sites of a type of retrotransposon in a mammalian genome. Using this method, we located many differences in probable IAP-cDNA integration regions between the C3H/He and C57BL/6J genomes. In addition, we identified three genomic regions around which an IAP element likely retrotransposed in the AML genome. Although the method has certain limitations at present, these successful applications indicate that our method will be useful not only for the study of IAP elements themselves but also for screening candidate responsive genes for the phenotypic changes and variations due to the movement and positional diversity of IAP elements.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank the Division of Animal Facility staff for help with the laboratory analysis and maintenance of animals. We also thank Dr Akihiro Shima for his encouragement throughout the course of research. The work was supported in part by grants from Aomori Prefecture, Japan, an LRI grant from Japan Chemical Industry Association (Grant 2007CC03-01), a grant from Ground-Based Research Announcement for Space Utilization Research from Japan Space Forum (Grant 17-031-18) and a grant-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (18510053). Funding to pay the Open Access publication charges for this article was provided by Institute for Environmental Sciences.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kidwell MG. Transposable elements and the evolution of genome size in eukaryotes. Genetica. 2002;115:49–63. doi: 10.1023/a:1016072014259. [DOI] [PubMed] [Google Scholar]
- 2.Deininger PL, Batzer MA. Mammalian retroelements. Genome Res. 2002;12:1455–1465. doi: 10.1101/gr.282402. [DOI] [PubMed] [Google Scholar]
- 3.Deininger PL, Moran JV, Batzer MA, Kazazian H.H., Jr. Mobile elements and mammalian genome evolution. Curr. Opin. Genet. Dev. 2003;13:651–658. doi: 10.1016/j.gde.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 4.Kazazian HH., Jr. Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 5.Volff JN. Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays. 2006;28:913–922. doi: 10.1002/bies.20452. [DOI] [PubMed] [Google Scholar]
- 6.Britten RJ. DNA sequence insertion and evolutionary variation in gene regulation. Proc. Natl Acad. Sci. USA. 1996;93:9374–9377. doi: 10.1073/pnas.93.18.9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van de Lagemaat LN, Landry JR, Mager DL, Medstrand P. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet. 2003;19:530–536. doi: 10.1016/j.tig.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Smalheiser NR, Torvik VI. Mammalian microRNAs derived from genomic repeats. Trends Genet. 2005;21:322–326. doi: 10.1016/j.tig.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 9.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 10.Piriyapongsa J, Jordan IK. A family of human microRNA genes from miniature inverted-repeat transposable elements. PLoS ONE. 2007;2:e203. doi: 10.1371/journal.pone.0000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piriyapongsa J, Marino-Ramirez L, Jordan IK. Origin and evolution of human microRNAs from transposable elements. Genetics. 2007;176:1323–1337. doi: 10.1534/genetics.107.072553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hedges DJ, Batzer MA. From the margins of the genome: mobile elements shape primate evolution. Bioessays. 2005;27:785–794. doi: 10.1002/bies.20268. [DOI] [PubMed] [Google Scholar]
- 13.Hedges DJ, Deininger PL. Inviting instability: transposable elements, double-strand breaks, and the maintenance of genome integrity. Mutat. Res. 2007;616:46–59. doi: 10.1016/j.mrfmmm.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian H.H., Jr. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl Acad. Sci. USA. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maksakova IA, Romanish MT, Gagnier L, Dunn CA, van de Lagemaat LN, Mager DL. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet. 2006;2:e2. doi: 10.1371/journal.pgen.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishihara H, Tanaka I, Furuse M, Tsuneoka K. Increased expression of intracisternal A-particle RNA in regenerated myeloid cells after X irradiation in C3H/He inbred mice. Radiat. Res. 2000;153:392–397. doi: 10.1667/0033-7587(2000)153[0392:ieoiap]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 17.Chang-Yeh A, Mold DE, Brilliant MH, Huang RC. The mouse intracisternal A particle-promoted placental gene retrotransposition is mouse-strain-specific. Proc. Natl Acad. Sci. USA. 1993;90:292–296. doi: 10.1073/pnas.90.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banno F, Kaminaka K, Soejima K, Kokame K, Miyata T. Identification of strain-specific variants of mouse Adamts13 gene encoding von Willebrand factor-cleaving protease. J. Biol. Chem. 2004;279:30896–30903. doi: 10.1074/jbc.M314184200. [DOI] [PubMed] [Google Scholar]
- 19.Horie K, Saito ES, Keng VW, Ikeda R, Ishihara H, Takeda J. Retrotransposons influence the mouse transcriptome: implication for the divergence of genetic traits. Genetics. 2007;176:815–827. doi: 10.1534/genetics.107.071647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wessler SR. Turned on by stress. Plant retrotransposons. Curr. Biol. 1996;6:959–961. doi: 10.1016/s0960-9822(02)00638-3. [DOI] [PubMed] [Google Scholar]
- 21.Kimura RH, Choudary PV, Schmid CW. Silk worm Bm1 SINE RNA increases following cellular insults. Nucleic Acids Res. 1999;27:3380–3387. doi: 10.1093/nar/27.16.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagan CR, Sheffield RF, Rudin CM. Human Alu element retrotransposition induced by genotoxic stress. Nat. Genet. 2003;35:219–220. doi: 10.1038/ng1259. [DOI] [PubMed] [Google Scholar]
- 23.Faure E, Emanoil-Ravier R, Champion S. Induction of transcription from the long terminal repeat of the intracysternal particles type A (IAP) by X-irradiation. Arch. Physiol. Biochem. 1997;105:183–189. doi: 10.1076/apab.105.2.183.12926. [DOI] [PubMed] [Google Scholar]
- 24.Farkash EA, Kao GD, Horman SR, Prak ET. Gamma radiation increases endonuclease-dependent L1 retrotransposition in a cultured cell assay. Nucleic Acids Res. 2006;34:1196–1204. doi: 10.1093/nar/gkj522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka I, Ishihara H. Unusual long target duplication by insertion of intracisternal A-particle element in radiation-induced acute myeloid leukemia cells in mouse. FEBS Lett. 1995;376:146–150. doi: 10.1016/0014-5793(95)01262-2. [DOI] [PubMed] [Google Scholar]
- 26.Ishihara H, Tanaka I. Detection and cloning of unique integration sites of retrotransposon, intracisternal A-particle element in the genome of acute myeloid leukemia cells in mice. FEBS Lett. 1997;418:205–209. doi: 10.1016/s0014-5793(97)01383-5. [DOI] [PubMed] [Google Scholar]
- 27.Ishihara H, Tanaka I, Wan H, Nojima K, Yoshida K. Retrotransposition of limited deletion type of intracisternal A-particle elements in the myeloid leukemia Clls of C3H/He mice. J. Radiat. Res. 2004;45:25–32. doi: 10.1269/jrr.45.25. [DOI] [PubMed] [Google Scholar]
- 28.Ymer S, Tucker WQ, Sanderson CJ, Hapel AJ, Campbell HD, Young IG. Constitutive synthesis of interleukin-3 by leukaemia cell line WEHI-3B is due to retroviral insertion near the gene. Nature. 1985;317:255–258. doi: 10.1038/317255a0. [DOI] [PubMed] [Google Scholar]
- 29.Blankenstein T, Qin ZH, Li WQ, Diamantstein T. DNA rearrangement and constitutive expression of the interleukin 6 gene in a mouse plasmacytoma. J. Exp. Med. 1990;171:965–970. doi: 10.1084/jem.171.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tohyama K, Lee KH, Tashiro K, Kinashi T, Honjo T. Establishment of an interleukin-5-dependent subclone from an interleukin-3-dependent murine hemopoietic progenitor cell line, LyD9, and its malignant transformation by autocrine secretion of interleukin-5. EMBO J. 1990;9:1823–1830. doi: 10.1002/j.1460-2075.1990.tb08307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leslie KB, Lee F, Schrader JW. Intracisternal A-type particle-mediated activations of cytokine genes in a murine myelomonocytic leukemia: generation of functional cytokine mRNAs by retroviral splicing events. Mol. Cell Biol. 1991;11:5562–5570. doi: 10.1128/mcb.11.11.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugita T, Totsuka T, Saito M, Yamasaki K, Taga T, Hirano T, Kishimoto T. Functional murine interleukin 6 receptor with the intracisternal A particle gene product at its cytoplasmic domain. Its possible role in plasmacytomagenesis. J. Exp. Med. 1990;171:2001–2009. doi: 10.1084/jem.171.6.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kono T, Doi T, Yamada G, Hatakeyama M, Minamoto S, Tsudo M, Miyasaka M, Miyata T, Taniguchi T. Murine interleukin 2 receptor beta chain: dysregulated gene expression in lymphoma line EL-4 caused by a promoter insertion. Proc. Natl Acad. Sci. USA. 1990;87:1806–1810. doi: 10.1073/pnas.87.5.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Badge RM, Alisch RS, Moran JV. ATLAS: a system to selectively identify human-specific L1 insertions. Am. J. Hum. Genet. 2003;72:823–838. doi: 10.1086/373939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy AM, Carroll ML, Kass DH, Nguyen SV, Salem AH, Batzer MA, Deininger PL. Recently integrated human Alu repeats: finding needles in the haystack. Genetica. 1999;107:149–161. [PubMed] [Google Scholar]
- 36.Sheen FM, Sherry ST, Risch GM, Robichaux M, Nasidze I, Stoneking M, Batzer MA, Swergold GD. Reading between the LINEs: human genomic variation induced by LINE-1 retrotransposition. Genome Res. 2000;10:1496–1508. doi: 10.1101/gr.149400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buzdin A, Khodosevich K, Mamedov I, Vinogradova T, Lebedev Y, Hunsmann G, Sverdlov E. A technique for genome-wide identification of differences in the interspersed repeats integrations between closely related genomes and its application to detection of human-specific integrations of HERV-K LTRs. Genomics. 2002;79:413–422. doi: 10.1006/geno.2002.6705. [DOI] [PubMed] [Google Scholar]
- 38.Buzdin A, Ustyugova S, Gogvadze E, Lebedev Y, Hunsmann G, Sverdlov E. Genome-wide targeted search for human specific and polymorphic L1 integrations. Hum. Genet. 2003;112:527–533. doi: 10.1007/s00439-002-0904-2. [DOI] [PubMed] [Google Scholar]
- 39.Mamedov I, Batrak A, Buzdin A, Arzumanyan E, Lebedev Y, Sverdlov ED. Genome-wide comparison of differences in the integration sites of interspersed repeats between closely related genomes. Nucleic Acids Res. 2002;30:e71. doi: 10.1093/nar/gnf071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mamedov IZ, Arzumanyan ES, Amosova AL, Lebedev YB, Sverdlov ED. Whole-genome experimental identification of insertion/deletion polymorphisms of interspersed repeats by a new general approach. Nucleic Acids Res. 2005;33:e16. doi: 10.1093/nar/gni018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wheelan SJ, Scheifele LZ, Martinez-Murillo F, Irizarry RA, Boeke JD. Transposon insertion site profiling chip (TIP-chip) Proc. Natl Acad. Sci. USA. 2006;103:17632–17637. doi: 10.1073/pnas.0605450103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gabriel A, Dapprich J, Kunkel M, Gresham D, Pratt SC, Dunham MJ. Global mapping of transposon location. PLoS Genet. 2006;2:e212. doi: 10.1371/journal.pgen.0020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Telenius H, Carter NP, Bebb CE, Nordenskjold M, Ponder BA, Tunnacliffe A. Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics. 1992;13:718–725. doi: 10.1016/0888-7543(92)90147-k. [DOI] [PubMed] [Google Scholar]
- 44.Fiegler H, Carr P, Douglas EJ, Burford DC, Hunt S, Scott CE, Smith J, Vetrie D, Gorman P, Tomlinson IP, et al. DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes Chromosomes Cancer. 2003;36:361–374. doi: 10.1002/gcc.10155. [DOI] [PubMed] [Google Scholar]
- 45.Farrall M, Morris AP. Gearing up for genome-wide gene-association studies. Hum. Mol. Genet. 2005;14(Spec No. 2):R157–R162. doi: 10.1093/hmg/ddi273. [DOI] [PubMed] [Google Scholar]
- 46.McCarroll SA, Altshuler DM. Copy-number variation and association studies of human disease. Nat. Genet. 2007;39:S37–S42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
- 47.Frazer KA, Eskin E, Kang HM, Bogue MA, Hinds DA, Beilharz EJ, Gupta RV, Montgomery J, Morenzoni MM, Nilsen GB, et al. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature. 2007;448:1050–1053. doi: 10.1038/nature06067. [DOI] [PubMed] [Google Scholar]
- 48.Cho EK, Tchinda J, Freeman JL, Chung YJ, Cai WW, Lee C. Array-based comparative genomic hybridization and copy number variation in cancer research. Cytogenet. Genome Res. 2006;115:262–272. doi: 10.1159/000095923. [DOI] [PubMed] [Google Scholar]
- 49.Lueders KK, Kuff EL. Sequences associated with intracisternal A particles are reiterated in the mouse genome. Cell. 1977;12:963–972. doi: 10.1016/0092-8674(77)90161-1. [DOI] [PubMed] [Google Scholar]
- 50.Ono M, Cole MD, White AT, Huang RC. Sequence organization of cloned intracisternal A particle genes. Cell. 1980;21:465–473. doi: 10.1016/0092-8674(80)90483-3. [DOI] [PubMed] [Google Scholar]
- 51.Whitelaw E, Martin DI. Retrotransposons as epigenetic mediators of phenotypic variation in mammals. Nat. Genet. 2001;27:361–365. doi: 10.1038/86850. [DOI] [PubMed] [Google Scholar]
- 52.Rakyan VK, Blewitt ME, Druker R, Preis JI, Whitelaw E. Metastable epialleles in mammals. Trends Genet. 2002;18:348–351. doi: 10.1016/s0168-9525(02)02709-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.