Abstract
This article compares 32 bacterial genomes with respect to their high transcription potentialities. The σ70 promoter has been widely studied for Escherichia coli model and a consensus is known. Since transcriptional regulations are known to compensate for promoter weakness (i.e. when the promoter similarity with regard to the consensus is rather low), predicting functional promoters is a hard task. Instead, the research work presented here comes within the scope of investigating potentially high ORF expression, in relation with three criteria: (i) high similarity to the σ70 consensus (namely, the consensus variant appropriate for each genome), (ii) transcription strength reinforcement through a supplementary binding site—the upstream promoter (UP) element—and (iii) enhancement through an optimal Shine-Dalgarno (SD) sequence. We show that in the AT-rich Firmicutes’ genomes, frequencies of potentially strong σ70-like promoters are exceptionally high. Besides, though they contain a low number of strong promoters (SPs), some genomes may show a high proportion of promoters harbouring an UP element. Putative SPs of lesser quality are more frequently associated with an UP element than putative strong promoters of better quality. A meaningful difference is statistically ascertained when comparing bacterial genomes with similarly AT-rich genomes generated at random; the difference is the highest for Firmicutes. Comparing some Firmicutes genomes with similarly AT-rich Proteobacteria genomes, we confirm the Firmicutes specificity. We show that this specificity is neither explained by AT-bias nor genome size bias; neither does it originate in the abundance of optimal SD sequences, a typical and significant feature of Firmicutes more thoroughly analysed in our study.
INTRODUCTION
This article addresses potentially high ORF expression related to σ70-like promoters, in bacterial genomes. In these genomes, a single enzyme, the RNA polymerase, is responsible for the synthesis of all RNA types. The core holoenzyme α2ββ′ is competent for transcribing a specific region of the DNA strand into an RNA molecule. However, transcription can only be initiated (at the so-called +1 transcription site) through a temporary biochemical complex. This complex is composed of the four previous subunits and of a protein, the σ factor, the primary one being σ70. As one of the simplest known bacterial models, Escherichia coli K-12 has been subjected to intensive research, especially with regard to transcription (1–8). Knowledge was therefore gained about the E. coli σ70 factor's; binding sites. Their consensuses are, respectively, TTGACA and TATAAT, in the 5′ to 3′ direction. The optimal fixation of the RNA polymerase requires that the site with the consensus TTGACA should be located between 35 bp and 30 bp or thereabouts upstream of the first transcribed nucleotide. This former site is thus called the −35 box. The Pribnow box, TATAAT, is called −10 box for similar reasons. These sites are separated by 15–21 bp in the known functional promoters, the canonical σ70 promoter being characterized by the optimal distance of 17 bp. Various methods and softwares devoted to the prediction of functional promoters in E. coli genome have been developped (9–12) (to restrain to a few examples). We do not mention here the numerous softwares designed to uncover a motif common to a set of biological sequences.
Not only is the RNA polymerase conserved through evolution in bacteria, but also there seems to be a single σ70 factor, responsible for housekeeping gene transcription, across the bacterial kingdom (13–14). Both points legitimate searches for σ70-like binding sites in other prokaryotic genomes (15–17). Furthermore, the number of complete prokaryotic genomes sequenced has increased at a high speed (594 in october 2007), which allows genome-wide computational investigations. In the domain of in silico analyses related to σ70 factor transcription, a reference contribution showed that σ70 promoter-like sequences are present throughout the kingdom of prokaryotic organisms (18). This former study demonstrated that the density of promoter-like sequences is high within regulatory regions, in contrast to coding regions and regions located between convergently transcribed genes. For instance, an average of 38 promoter-like sequences was computed for E. coli, within each 250 bp subregion located upstream of the start codon (SC).
In vivo, transcriptional regulations are known to compensate for promoter weakness (19–20). For example, Huerta and Collado-Vides established that more than 50% of experimentally verified promoters are not the promoters with the highest scores when scoring relies on the proximity to the canonical promoter, both in terms of consensus similarity and optimal bp distances between boxes (9). This statement was checked on the 111 promoters constituting a training set designed in a former work (15). On the other hand, in E. coli genome, it has been shown that mutations in the −10 box or the −35 box that bring the promoter sequence closer to the σ70 consensus tend to increase the strength of the promoter, and conversely, mutations decreasing homology to the σ70 consensus tend to lower the promoter strength (1). Thus, the more similar to the canonical σ70 promoter, the more potentially strong this promoter would be, with the noteworthy exception that the consensus promoters may actually be weak because RNA polymerase binds them so strongly that it cannot escape (21). Therefore, it is attractive to study and compare genomes from the point of view of potentially high transcription, allowing for mismatches, under a minimal similarity constraint. This large-scale comparative analysis is feasible through an in silico approach.
No computational method can capture the biological features and environmental conditions involved in vivo, to predict functional strong promoters. Besides, even for the most intensively studied prokaryotic genome, E. coli's, the available repositories of σ70 promoters do not provide annotations about promoter strength. The measurement of promoter activity in cellular or cell-free expression systems cannot be applied on a large scale. ChIP on chip assays allow the identification of transcription factor binding sites, under given environmental conditions, but high-throughput promoter strength measurement cannot be implemented using this technique. Thus, before such large-scale array experimentations may be conducted on the 32 genomes we are interested in, an in silico genome-comparative analysis focused on intrinsically high transcription potentiality is worth being performed.
In our work, we intentionally focus on the subset of putative strong σ70 promoters already potentially favoured by the presence of an optimal Shine-Dalgarno (SD) sequence (GGAGG). The presence of the SD sequence has been ascertained for a large number of bacteria (22) and it was established that the extent to which a SD sequence is conserved relates to its translation efficiency (23). Besides, our study also puts emphasis on strength transcription reinforcement through the upstream promoter (UP) element presence. The UP element is an enhancer for transcription and thus for ORF expression (24–25). In about 3% of E. coli promoters, an UP element has been identified upstream of the −35 region, conferring additional strength to the promoter. The high conservation of the domain of the alpha subunit of the RNA polymerase involved in the interaction with the UP element suggests that the UP element consensus should be valid throughout the bacterial kingdom. To our knowledge, in addition to E. coli genome, the UP element has been experimentally identified in Bacillus subtilis (26), Vibrio natriegens (27) and Bacillus stearothermophilus (28). UP elements were previously taken into account by PlatProm algorithm (29); to our knowledge, the only other work devoted to in silico identification of σ70 promoter-like sequences harbouring an UP element is by M. Dekhtyar, A. Morin and V. Sakanyan (Sakanyan, personal communication.).
In this article, we perform a comparison of the frequencies observed for the putative strongest promoters over 32 bacterial genomes. We distinguish two strength levels, depending on the relaxation allowed with respect to the canonical σ70 promoter, and combine them with either mandatory or optional UP element presence. Thus, we perform four genome-comparative studies. We discuss the statistical significance of our results through comparisons with randomly generated genomes, highlighting and elucidating the specific case of Firmicutes.
SYSTEMS AND METHODS
Genome analysis upon request
For each genome studied, BacTrans2 (http://www.sciences.univ-nantes.fr/lina/bioserv/BacTrans2/) takes as an input the Fasta genome sequence provided by GenBank (http://www.ncbi.nlm.nih. gov/genomes/lproks.cgi) together with the corresponding genome annotation. For each gene encoding a protein, the tool first extracts the subregion spanning to 350 nucleotides upstream of SC's; first nucleotide. Then, occurrences of the σ70 promoter binding sites are searched for under constraints relative to: (i) bp distances between binding sites or distances between binding sites and translation signals playing the role of ‘anchors’ and (ii) the maximal number of mismatches allowed with respect to each consensus. In GenBank files, the only location annotation available is that of the SC. Hence, for each gene, the SC is considered a right anchor and each region upstream of SC is scanned to retrieve in priority the structured motif [UP element] <3-18> [ − 35box] <15-20> [ − 10box] <10-200> [SD] <2-10> [SC] (described in the 5′ to 3′ direction), where SD denotes the Shine-Dalgarno sequence and [box1] < dmin − dmax > [box2] states the minimal and maximal bp distances allowed between the two boxes concerned. Actually, the full motif identification is performed in the 3′ to 5′ direction, successively considering each possible occurrence of the current box as a right anchor. In the absence of any UP element, the structured motif [ − 35box] <15-20> [ − 10box] <10-200> [SD] <2-10> [SC] is looked for.
For each genome, the consensuses used have been adapted from E. coli σ70 promoter, relying on the work of Huerta and co-workers (18). These authors first identified a pair of Position-Specific Scoring Matrices (PSSMs), corresponding to the −35 and −10 boxes, associated with an interval of minimal and maximal bp distances, best describing E. coli σ70 functional promoters (see latter reference, Matrix_18_15_13_2_1.5 in Figure 2). Second, for any genome other than E. coli, they normalized the frequencies of the pair of E. coli PSSMs, using the a priori nucleotide probabilities characterizing this genome. Then, they relied on the normalized PSSM pair, to identify a set of promoter-like sequences within each genome. Finally they computed the −10 and −35 consensuses for each genome. In our study, for each genome, the consensuses retained are the subsequences of the consensuses of Huerta and co-workers, corresponding to the locations of the canonical TTGAC and TATAAT E. coli consensuses. We were careful to set accordingly the optimal bp distance between the −10 and the −35 boxes. As a result, the two −10 consensus TATAAT and TAAAAT have been used, respectively, for 20 and 12 genomes; TTGAC, TTGAA and TTTAA were the three −35 consensuses used to scan 6, 18 and 8 genomes, respectively (see Supplementary Appendix 1). A value of 200 bp was chosen for the maximal distance between SC and SD; it was selected on the basis of the average 5′UTR region's; length (50 or thereabouts, with variations between 0 and 200). The UP consensus used is that of E. coli, AAAWWTWTTTTNNAAAA (The genuine UP element has NN and NNN, respectively, as 5′ and 3′ termini).
Figure 2.
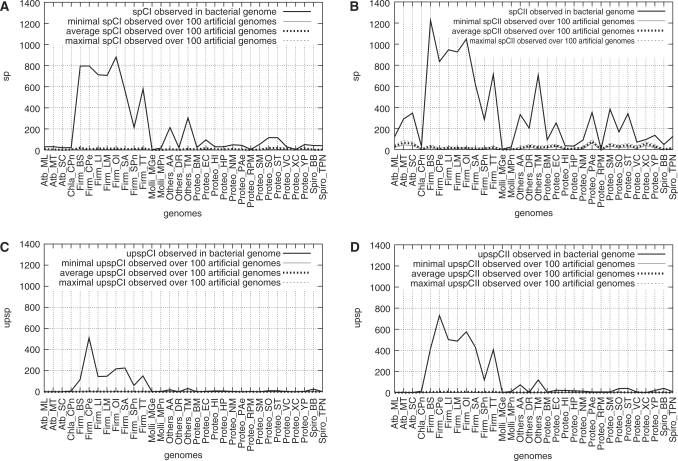
Observed bacterial genome values versus minimal, average and maximal values observed over 100 similarly AT-rich genomes generated at random, for sp and upsp, respectively, under four constraint sets. See Figure 1 for definition of sp and upsp, and for genome abbreviations. See text, Subsection “Genome analysis upon request” for the definition of CI and CII constraints. (A): CI, UP element optional; (B): CII, UP element optional; (C): CI, UP element required; (D): CII, UP element required.
For each binding site, minimal similarity is described through a maximal number of mismatches allowed. Notation (err(UP),err( − 35box),err( − 10box)) specifies the maximal numbers of mismatches allowed with regard to the UP element, the −35 box and the −10 box, respectively. Given this notation, two mismatch constraints are retained in our study; they are described as follows: (4,2,1) and (4,3,2). From now on, the two mismatch constraints (4,2,1) and (4,3,2) will be, respectively, denoted CI and CII. CI is more stringent than CII. Finally, four configurations will be considered in our analysis: CI, UP element required; CII, UP element required; CI, UP element optional; CII, UP element optional. The requirement of a greatest specificity for the −10 box compared to the −35 box is modeled after observations relative to functional σ70 promoters.
Hereafter, we denote sp the number of strong σ70 promoter-like sequences obtained from a given genome, when the presence of the UP element is optional. Similarly we define upsp when the UP element is required. From now on, we will refer to spCI, spCII, upspCI and upspCII.
Scoring function used
In the sequel, err(b) denotes the number of mismatches observed with respect to the consensus box b; d1 denotes the bp distance observed between the −35 box and the −10 box; d2 denotes the bp distance observed between the UP element and the −35 box. The score is calculated as follows: score = 0.60err( − 10box) + 0.40err( − 35box)+ t1 + err(UP) + t2, where t1 = 0 if d1 belongs to [17–19] else t1 = 5*d1, and t2 = 0 if d2 ranges in interval [6–8] else t2 = 3*d2. When no UP element can be identified, the score is merely computed as: score = penalty+ 0.60err( − 10box) + 0.40err( − 35box) + t1. The penalty value is set in order to systematically favour a candidate with an UP element within the regulatory region. This scoring function takes into account the specificity increase of the −10 box with respect to the −35 box. The choice of the coefficients 0.6 and 0.4 may be debatable. The most important point remains that the ratio between these coefficients be consistent with the behaviour of RNA polymerase as observed through functional promoters. Besides, we wished to emphasize the UP element weight, in the case when two promoter candidates harbour an UP-like element. Therefore, we assigned a value of 1 to the coefficient of the UP element. Finally, BacTrans2 outputs 0 or 1 putative SP per gene encoding a protein. The scoring function is one of the six major differences with the approach by Dekhtyar et al. (V. Sakanyan, personal communication). For an enumeration of the differences, the reader is referred to https://hal.archives-ouvertes.fr/hal-00153303/en/.
Comparison with randomly generated genomes
For each bacterial genome considered in this study, we compare the sp value (respectively, upsp value) observed with respect to the corresponding value expected on average for a similarly AT-rich genome generated at random. This latter artificial genome is only constrained to have the same following characteristics as the prokaryotic genome considered: same total number of genes coding for proteins and same proportions of A, C, T and G nucleotides in the 350 nucleotide-long region upstream of the SC. Due to the high bp distance allowed between the −10 box and the SD sequence (200), and the numbers of mismatches allowed, the calculation of the theoretical expected value would not be tractable. Thus, for each genome, and under the four conditions studied, we computed the minimum, maximum, mean and standard deviation for sp and upsp values, over 100 such randomly generated genomes. Scanning the largest batch of genomes (1400 artificial genomes) required no more than two days and a half under CII conditions. To evaluate whether two distributions are statistically different when the latter are not of the Gaussian type and when their variances are not in the same order of magnitude, we relied on the Wilcoxon test. The H0 hypothesis is stated as follows: the populations from which the two distributions are taken have identical median values. This test first ranks all n1 + n2 values from both distributions (n1 and n2) combined, then sums the ranks on each distribution, ws being the smallest sum and ws′ being computed as n1(n1 + n2 + 1) − ws. If either ws or ws′ is smaller than the theoretical value mentioned in Wilcoxon tables for n1 and n2 and an a priori level of significance, then hypothesis H0 is rejected. We also computed the Z-score as the absolute difference between the number of SPs obs observed in the prokaryotic genome and the average number Memp of promoters computed from the 100 artificial genomes, divided by the standard deviation σemp computed over these 100 latter genomes: Z-score = ∣ obs − Memp ∣ / σemp, where obs is an spCI value (respectively, spCII, upspCI, upspCII value). Again, statistical significance will be discussed, this time, with respect to several Z-score thresholds.
RESULTS AND DISCUSSION
Are potentially strong σ70 promoter-like sequences frequent?
The 32 genomes compared belong to ten Firmicutes, 13 Proteobacteria, 3 Actinobacteria, 2 Spirochaetales, 1 Chlamydia and 3 other taxa outside latter phyla. We draw the reader's; attention to the case of small genomes: Borrelia burgdorferi (0.91 Mbp), Chlamydophila pneumoniae (1.22 Mbp), Mycoplasma genitalium (0.58 Mbp), Mycoplasma pneumoniae (0.81 Mbp), Rickettsia prowazekii (1.11 Mbp) and Treponema pallidum nichols (1.13 Mbp). All previous six species are either obligate intracellular pathogens, symbionts or animal commensal parasites and have undergone massive gene decay, as well as numerous genomic rearrangements. The presence of functional σ70 promoters is disputable in these genomes. Hereafter the two Firmicutes M. genitalium and M. pneumoniae will be referred to as Mollicutes. Nevertheless, except for R. prowazekii, these genomes were investigated in the reference work of Huerta and co-workers (18). We will follow this line, taking great care regarding the discussion. The total number of genes g encoding proteins in a genome and the size of this genome are proven to be correlated over the 32 genomes studied (linear correlation coefficient: 0.93). To escape the size bias when comparing genomes, we define the percentage p1 (p1 = 100 × sp/g). The top section of Figure 1 (A and B) depicts the variations of sp values and p1 percentages through genomes (also see Supplementary Data, Appendix 2). For illustration, the output files relative to E. coli genome are provided (see Supplementary Data, Appendix 3).
Figure 1.
Frequencies of genes harbouring a putative strong promoter (SP), under four constraint sets, in 32 prokaryotic genomes. See text, Subsection “Genome analysis upon request” for the definition of CI and CII constraints. (A) and (B): UP element optional; (C) and (D): UP element required. Along the x-axis, the following phyla and groups are encountered: Actinobacteria, Chlamydia, Firmicutes (among which Mollicutes), “Others” group, Proteobacteria, Spirochaetales. (A) y-axis: number of genes harbouring a SP (sp); (B) y-axis: ratio p1 of genes harbouring a SP (sp) to the total number of genes encoding proteins in the genome (g), p1 = 100 × sp / g; (C) y-axis: number of genes identified with an UP element harboured in the SP (upsp); (D) y-axis: ratio p2 of the number of genes with an UP element in the SP (upsp) to the number of genes with a SP (sp), p2 = 100 × upsp / sp.
As a first result, we check that the number of putative strong promoters identified increases when constraints are relaxed from CI to CII. Secondly, we observe that for the AT-rich genomes of Firmicutes, putative SPs are over-represented under the two constraints CI and CII. This differentiates Firmicutes from all other genomes studied. Nonetheless, among Firmicutes, the numbers of SPs may differ in high proportions (1 to 4 under CI and CII constraints); Streptococcus pneumoniae is always characterized by the lowest value whereas B. subtilis, Oceanobacillus ihenyensis and Clostridium perfringens happen to show peaks depending on the constraint. The differentiation between Firmicutes and other genomes holds for p1 percentage. The non-Firmicutes genomes pointed out by the highest p1 percentages (over 5%) are Aquifex aeolicus, Thermotoga maritima and B. burgdorferi. Thirdly, a more thorough examination shows that the genomes with the highest numbers of genes (g) are not necessarily those with the highest numbers of putative strong promoters (sp). The percentage p1 is variable and no linear correlation can be shown to exist between sp and g. More comments are provided in Supplementary Appendix 4, including a brief report about investigating the nature of genes associated with putative SPs.
The high AT-richness of Firmicutes could justifiably be suspected to yield these high numbers of σ70 promoter-like sequences. Indeed, we show that AT-content does not interfere much with p1: over the 32 genomes, the linear correlation coefficient between p1CI and AT-content is 0.52; the correlation coefficient between p1CII and AT-content is equal to 0.30, which was expected indeed under relaxed constraints allowing more blurred occurrences of the σ70 promoter model. When we take into account all bacteria but Firmicutes, such coefficients go down to 0.26 (CI) and −0.14 (CII), respectively. When the 10 AT-richest genomes are considered (Firmicutes), the coefficients are 0.27 and 0.20, respectively. Anyway, in the latter case, 10 is a borderline value regarding correlation analysis validity.
Are potentially strong σ70 promoter-like sequences harbouring an UP-like element frequent?
We now define percentage p2 as follows: p2 = 100 × upsp/sp. The bottom section of Figure 1 (C and D) depicts the variations of upsp and p2 among the 32 micro-organisms, under CI and CII constraints (also see Supplementary Data, Appendix 2). The output files relative to E. coli genome are provided (see Supplementary Data, Appendix 5).
Again, detailed complements to the present paragraph may be found in Supplementary Appendix 4. We first show that the differentiation between Firmicutes and other genomes holds, but it is more subdued for p2 percentage than for p1 percentage. Secondly, we observe that σ70 promoter-like sequences of relatively ‘lesser quality’ (constraint CII) are more frequently associated with an UP-like element than sequences of ‘better quality’ (constraint set CI) (Figure 1(C and D)): the ratio p2CII / p2CI is calculable for 24 genomes and its average is 2.13; the average computed for all Firmicutes but Mollicutes is 2.07. Thirdly, we show that some genomes characterized by a low number of strong promoters show in contrast a high (p2) percentage of them harbouring an UP element, whatever the constraint (see Supplementary Appendix 4 for more details).
We calculate a correlation coefficient between p2CI and AT-content of 0.84 when all 32 genomes are considered; the correlation between p2CII and AT-content is similarly high (0.87). A high correlation is still observed when Firmicutes are not taken into account (0.82 and 0.86, respectively). In contrast with the case when no UP element was required, the 10 Firmicutes clearly show a correlation between p2 and AT-content (0.87 and 0.65, respectively). As expected, a stronger correlation is observed for p2 with respect to p1, since 7 out of the 17 nucleotides of the UP element consensus are nucleotides A, 5 are nucleotides T and 3 are A or T (W).
We now recapitulate the results obtained regarding AT-richness influence on p1 and p2: (i) depending on the species considered, AT-richness interferes but moderately so long as the UP element is not considered (p1); (ii) on the contrary, AT-content and percentage p2 are highly correlated. A pending question is then: does AT-richness alone entail high upspCI and upspCII values? To answer this question, we will in particular compare Firmicutes’ genomes with similarly AT-rich genomes generated at random.
Finally, the normalized ratio ρ of the number of promoter-like sequences (associated with an optimal SD) to the number of genes harbouring an optimal SD sequence has been calculated under all four conditions (see Supplementary Appendix 4, Table 4.1). The first observation drawn from Table 4.1 is that CII conditions do not entail any selection, thus leading to the conclusion that CII conditions alone are not adequate for potentially strong promoter description. Moreover, interestingly, under all three other conditions (CI and CII, UP element required; CI, UP element optional), this normalized ρ ratio is always significantly higher in Firmicute genomes than in non-Firmicute genomes. Therefore, we have indisputably confirmed the existence of a meaningful bias for frequencies of σ70 promoter-like sequences associated with optimal SDs, in Firmicute genomes.
Comparing observations in bacterial genomes with expectations in randomly generated genomes
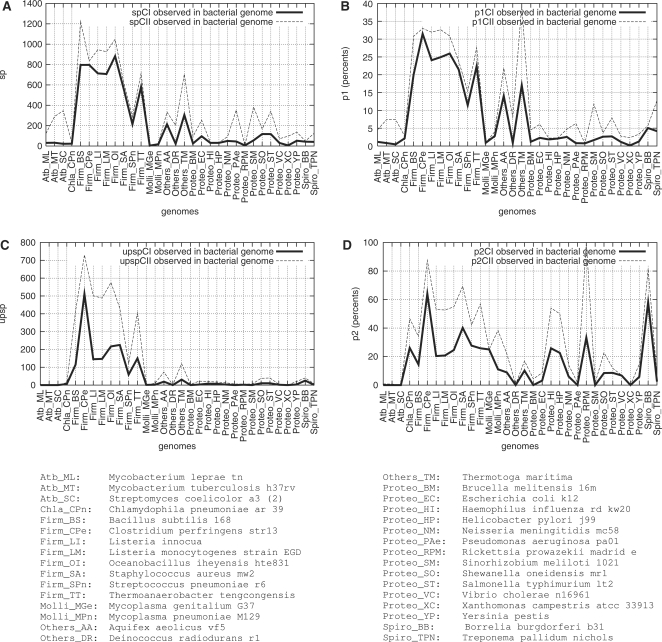
For each genome, we compare the frequency of putative SPs with that obtained for a similarly AT-rich ‘average’ genome generated at random (Figure 2). For comparison purposes, a common scale is used in the four pictures of Figure 2 (The reader interested in details is referred to Supplementary Data, Appendix 6, for a magnification relative to artificial genomes’ results).
We start our analysis focusing on the CI case. Figure 2A (CI) shows that strong σ70 promoter-like sequences are significantly more frequent in Firmicutes genomes than in corresponding artificial genomes. From now on, we distinguish the two Mollicutes from the other eight Firmicutes. Given as quadruplets (minimum, maximum, average, standard deviation), Z-scores are as follows: Firmicutes except Mollicutes: (81.3, 308.5, 193.0, 66.1); Proteobacteria: (1.0, 32.4, 16.0, 9.5). We check that the eight Firmicutes’ Z-scores are above threshold 140, except for Listeria monocytogenes (81.3). Concerning the 12 large Proteobacteria genomes studied, 10 have their Z-scores above threshold 7, among which 6 have their Z-scores above threshold 15. In particular, the Z-score obtained for E. coli genome is 21.7.
When restraining our examination to the 26 species with large genomes, under condition CI, we observe that 24 genomes have their Z-scores over threshold 7, among which 15 have their Z-scores over threshold 15 and finally 10 Z-scores exceed threshold 80. For a detailed description relative to spCII, upspCI and upspCII values (Figure 2; B, C and D), the reader is referred to Tables 6.1 through 6.4 in Supplementary Appendix 6. Table 6.3 focuses on E. coli. We recapitulate the main results and conclusions in the following paragraph.
First, we confirm that, except for the slightly more subdued case of L. monocytogenes, Firmicutes clearly show a specific trend, with Z-scores above thresholds 160, 100 and 150, respectively, under CII condition (UP optional), and CI and CII conditions (UP required). Yet, under all four conditions, the Z-scores calculated for L. monocytogenes stay rather high (they range in interval [69,93]). Secondly, relaxing the constraint from CI to CII entails no decrease of the Z-score (see Supplementary Appendix 6, Table 6.1). At first sight, this is not a trivial result, as the opposite was expected instead. But CII condition alone has been shown to be under-constrained. Therefore, no valid information can be drawn from the Z-scores, in this case. Besides, the number of putative SPs harbouring an UP element, observed in the average random genome under CI condition, drastically decreases down to 0 for 26 species out of 32. Under this latter condition, it is obvious that both observed and expected upspCI distributions strongly differ from one another. More rigorously, and more generally, the Wilcoxon test successively performed on p1CI, p2CI and p2CII allows us to conclude that the difference between observed values and values expected by chance is statistically significant under all three conditions, for the 0.05 threshold. Thus, the σ70 promoter-like sequences retrieved in bacterial genomes are not due to mere chance. Additionally, Table 6.2 in Supplementary Appendix 6 enables evaluation of the statistical significance for each non-Firmicute genome with respect to the Z-score thresholds 7, 15 and 80. Table 6.4 recapitulates the number of large genomes for which statistical significance is ascertained with regard to these thresholds: at least half of them under CI condition, for threshold 15, which we consider a high threshold; nearly all of them for threshold 7. Finally, since similarly AT-rich average genomes generated at random are far from yielding such high frequencies as those observed for the eight corresponding Firmicutes genomes, AT-richness is clearly not the reason for the Firmicutes specificity.
Another lead is thoroughly examined to attempt to explain the Firmicutes difference. Due to the lack of space, we refer the reader to Tables 6.6 and 6.7 in Supplementary Appendix 6. We demonstrate therein that the Firmicutes difference is neither explained by genome size bias. Summarizing, in this section, we have characterized the statistical significances for all genomes, under four conditions of stringency, and with respect to three Z-score thresholds. We have proven the existence of a specificity for Firmicutes (large) genomes with regard to our definition of potentially high transcription. Moreover, this specificity is neither an artefact due to high AT-richness nor to differences in gene numbers between genomes.
Discussing the Firmicutes case
To explain the fact that putative strong σ70 promoters appear much more frequently in Firmicutes than in other bacteria, including—paradoxically—E. coli, we recall that we adopted the consensus GGAGG. In E. coli, GGAGG is a very strong SD sequence; more frequent SDs are the submotifs GGAA, GGAG, GAGG, AGGA and AAGG (30, 23). On the other hand, ribosomes from many Gram-positive bacteria depend much more stringently upon a strong SD interaction for initiation (31). For instance, in B. subtilis genome, most SD sequences are close to the consensus sequence AAAGGAGG (32). This, we suggest, could be the reason for the abundance of putative SPs in Firmicutes genomes. This point has been investigated further. We show that the percentage pbact of genes associated with an optimal SD sequence ranges between 2.21% and 39.8% for the 26 large genomes. Immediately behind T. maritima, which shows the highest ratio, the eight large Firmicutes genomes rank first with respect to this pbact ratio ([15.3%, 32.6%]). The percentages prand expected for similarly AT-rich genomes generated at random have been calculated. The calculus is described in Supplementary Appendix 7. The pbact and prand distributions are proven statistically different through a Wilcoxon test (threshold 0.05). Furthermore, the correlation coefficient between prand and AT-richness is −0.97, over the 32 artificial genomes. This high negative value was expected, since the optimal SD sequence is enriched with four G nucleotides. In contrast, the correlation coefficient between pbact and AT-richness is low when computed over the 32 bacterial genomes (0.22). This point argues in favour of the biological significance of such GGAGG sequences in the close neighbourhood of SCs. Moreover, regarding this criterion, the Wilcoxon test also ascertains the statistical significance of the difference between the eight Firmicutes and the 18 other species with large genomes. This difference is reflected by the Z-scores. Z-scores range in interval [3.2, 363.9] when all genomes are considered (mean: 86.9, standard deviation: 103.1). The Z-scores calculated for the eight large Firmicutes genomes range between 86.8 (S. pneumoniae) and 363.9 (C. perfringens). When all large genomes but Firmicutes’ are considered, the mean and standard deviation are, respectively, equal to 41.8 and 40.0. Outside the Firmicutes taxon, T. maritima and A. aeolicus are the only two bacteria showing as outstanding Z-scores as Firmicutes (respectively, 168.7 and 106.2). Again, we emphasize that both previous genomes are also characterized with high AT percentages (54.6% and 57.6%), which confirms a bias for the presence of optimal SD sequences in some genomes.
Anyway, such bias exists for all genomes. For example, in the light of the previous explanation, we now explain the scarcity of putative SPs associated with optimal SD sequences, in E. coli, through the low pbact percentage of 6.2% observed. Though, the percentage expected is 0.9%. The bias measured through the Z-score is 37.9. Therefore, this point suggests that even in E. coli, hazard would only contribute for 15% (0.9/6.2) to yield false positive optimal SD sequences. Finally, considering the criteria retained in our analysis (high intrinsic transcription potentiality combined with strong SD interaction), we conclude that Firmicutes would appear as genomes more favoured by nature, especially with respect to other similarly AT-rich genomes.
Putative strong promoters versus experimentally verified functional promoters in E. coli genome
In vivo, activation by various factors is ascertained to compensate for promoter weakness. However, it is not known whether some functional promoters might also be intrinsic strong promoters. So far, data compilations relative to experimentally verified functional promoters are only available for E. coli genome, through two repositories, RegulonDB and PromEC (33–34). Therefore, we could compare the putative strong promoters identified by BacTrans2 software in E. coli genome with known E. coli functional promoters. For this purpose, we compiled our own σ70 promoter dataset from 5.8 RegulonDB release (september 2007, http://regulondb.ccg.unam.mx/data/PromoterSet.txt.) and PromEC database (http://margalit.huji.ac.il/). We checked that E. coli known functional promoters are intrinsically weaker than all putative SPs retrieved by our software BacTrans2, which was expected (see Supplementary Appendix 8).
Experimental verification of putative strong promoters identified in T. maritima genome
The hyperthermophilic model T. maritima has been intensively studied (35–36). In the context of a former study, the activity of 13 putative strong promoters harbouring an UP element has been measured in E. coli cell free extracts (37). The present work thereby benefits from these experimentations. The protocol used is described in Supplementary Appendix 9. Seven putative strong promoters harbouring an UP element identified by BacTrans2 were thus tested. Four were identified under the most constrained condition CI (TM1016, TM0373, TM0477, TM1667). The other three were identified under CII condition (TM0032, TM1429, TM1780). All of them promote protein synthesis, indicating that they are all functional promoters. Moreover, except TM0032, all provided a higher protein yield than that of the well-studied pTac promoter. TM0477 has been shown to be twice as strong as others regarding protein yield. Therefore, six potentially strong promoters among the seven tested do really favour high expression in E. coli cell free extracts.
CONCLUSION
Our work contributes to shedding new light on potentially high ORF expression in prokaryotic genomes, focusing on potentially high transcription combined with the presence of an optimal SD sequence. Our approach also puts emphasis on transcription initiation potentially enhanced through UP-like elements. In itself, this latter feature introduces originality with respect to other genome-comparative studies devoted to bacterial promoters. Moreover, genomes were compared in a rather unusual way, that is on the basis of their frequencies of intrinsically SP candidates, upstream of genes coding for proteins. Besides, our analysis clearly departs from other works, since it considers four different conditions of stringency and discusses in each framework the statistical significance of the presence of σ70 promoter-like sequences. Under all four conditions, we identified the species showing statistically significant differences between the bacterial genome and an average similarly AT-rich genome generated at random. Thus, specific features typical for E. coli promoters were used to extract promoter-like signals from other genomes and statistically significant differences were revealed on the basis of this approach. In particular, Firmicutes would appear as genomes more favoured by nature with respect to other genomes, including the cases when an UP-like element is required. A rigorous discussion allowed us to dismiss AT-richness and genome size bias as determining factors to explain the Firmicutes specificity. We have shown that this specificity is neither explained by the typical abundance of optimal SD sequences in Firmicutes' large genomes, thus revealing another Firmicute bias, unknown so far. Besides, so far, the UP element has been identified by experimentation in four genomes. Thus, our comparative study also brings novel knowledge about the statistical significance of the presence of putative σ70 promoters enhanced with an UP-like element, in various genomes.
The generic software platform BacTrans2 currently provides such putative strong promoters for 45 genomes. These data may be of interest to select a subset of promoters for experimental characterization and possible further use in biotechnological applications. In this latter field, inserting in cellular or cell-free expression systems regulatory regions including promoters enhanced with an UP element and an optimal SD sequence may be advocated, instead of inserting artificial binding sites in a synthetic sequence. A more thorough study of high translation potentiality related to high transcription potentiality in prokaryotic genomes is attractive and is currently under work. Finally, BacTrans2's genericity allows the user to analyse genomes with respect to any other super-motif consisting of three or four boxes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
S. Demey was supported by the Pays de la Loire Region (“Postgenomics and Technological Innovations” C.P.E.R. program) and by Ouest-Genopole consortium (National Network of Genopoles). The authors are thankful to V. Sakanyan for valuable comments and critically reading the manuscript. They wish to thank the anonymous reviewers for their constructive remarks. Thanks are also due to J. Bourdon for insightful discussions. Funding to pay the Open Access publication charges for this article was provided by the Pays de la Loire Region (Bioinformatics Research Project - BIL).
Conflict of interest statement. None declared.
REFERENCES
- 1.Hawley DK, McClure WR. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;25:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harley CB, Reynolds RP. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987;15:2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collado-Vides J, Magasanik B, Gralla JD. Control site location and transcriptional regulation in Escherichia coli. Microbiol. Rev. 1991;55:371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lisser S, Margalit H. Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res. 1993;21:1507–1516. doi: 10.1093/nar/21.7.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fenton MS, Lee SJ, Gralla JD. Escherichia coli promoter opening and -10 recognition: Mutational analysis of sigma70. EMBO J. 2000;19:1130–1137. doi: 10.1093/emboj/19.5.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- 7.Pager MS, Helmann JD. The sigma 70 family of sigma factors. Genome Biol. 2003;4:203. doi: 10.1186/gb-2003-4-1-203. 1–203.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herring CD, Raffaelle M, Allen TE, Kanin EI, Landick R, Ansari AZ, Palsson BO. Immobilization of Escherichia coli RNA polymerase and location of binding sites by use of chromatin immunoprecipitation and microarrays. J. Bacteriol. 2005;187:6166–6174. doi: 10.1128/JB.187.17.6166-6174.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huerta AM, Collado-Vides J. Sigma70 promoters in Escherichia coli: Specific transcription in dense regions of overlapping promoter-like signals. J. Mol. Biol. 2003;17:261–278. doi: 10.1016/j.jmb.2003.07.017. [DOI] [PubMed] [Google Scholar]
- 10.Eskin E, Gelfand M, Pevzner P. Genome-wide analysis of bacterial promoter regions. Pac. symp. on Biocomput. 2003;8:29–40. [PubMed] [Google Scholar]
- 11.Bulyk ML, McGuire AM, Masuda N, Church GM. A motif co-occurrence approach for genome-wide prediction of transcription-factor-binding sites in Escherichia coli. Genome Res. 2004;14:201–208. doi: 10.1101/gr.1448004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shultzaberger RK, Chen Z, Lewis KA, Schneider TD. Anatomy of Escherichia coli σ70 promoters. Nucleic Acids Res. 2007;35:771–788. doi: 10.1093/nar/gkl956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wosten MM. Eubacterial sigma-factors. FEMS Microbiol. Rev. 1998;22:127–150. doi: 10.1111/j.1574-6976.1998.tb00364.x. [DOI] [PubMed] [Google Scholar]
- 14.Mittenhuber G. An inventory of genes encoding RNA polymerase sigma factors in 31 completely sequenced eubacterial genomes. J. Mol. Microbiol. Biotechnol. 2002;4:77–91. [PubMed] [Google Scholar]
- 15.Gralla J, Collado-Vides J. Organization and function of transcription regulatory elements. In: Neidhart FC, Curtiss R, Ingraham J, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella, Cellular and Molecular Biology. Vol. 57. Washington, D.C: American Society for Microbiology; 1996. pp. 1232–1246. [Google Scholar]
- 16.Li H, Rhodius V, Gross C, Siggia ED. Identification of the binding sites of regulatory proteins in bacterial genomes. Proc. Natl Acad. Sci. USA. 2002;99:11772–11777. doi: 10.1073/pnas.112341999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez-Antonio A, Collado-Vides J. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 2003;6:482–489. doi: 10.1016/j.mib.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 18.Huerta AM, Francino MP, Morett E, Collado-Vides J. Selection for unequal densities of sigma70 promoter-like signals in different regions of large bacterial genomes. PLoS Genet. 2006;2:e185. doi: 10.1371/journal.pgen.0020185. doi:10.1371/journal.pgen.0020185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gross CA, Chan C, Dombroski A, Gruber T, Sharp M, Tupy J, Young B. The functional and regulatory roles of sigma factors in transcription. Cold Spring Harb. Symp. Quant. Biol. 1998;63:141–155. doi: 10.1101/sqb.1998.63.141. [DOI] [PubMed] [Google Scholar]
- 20.Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2004;2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- 21.Ellinger T, Behnke D, Bujard H, Gralla JD. Stalling of Escherichia coli RNA polymerase in the +6 to +12 region in vivo is associated with tight binding to consensus promoter elements. J. Mol. Biol. 1994;239:455–465. doi: 10.1006/jmbi.1994.1388. [DOI] [PubMed] [Google Scholar]
- 22.Osada Y, Saito R, Tomita M. Analysis of base-pairing potentials between 16S rRNA and 5′ UTR for translation initiation in various prokaryotes. Bioinformatics. 1999;15:578–581. doi: 10.1093/bioinformatics/15.7.578. [DOI] [PubMed] [Google Scholar]
- 23.Ma J, Campbell A, Karlin S. Correlation between Shine-Dalgarno sequence and gene features such as predicted expression levels and operon structure. J. Bacteriol. 2002;184:5733–5745. doi: 10.1128/JB.184.20.5733-5745.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ross W, Gosink KK, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse RL. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 25.Estrem ST, Ross W, Gaal T, Chen ZW, Niu W, Ebright RH, Gourse RL. Bacterial promoter architecture: Subsite structure of UP elements and interactions with the carboxy-terminal domain of the RNA polymerase alpha subunit. Genes Dev. 1999;13:2134–2147. doi: 10.1101/gad.13.16.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fredrick K, Caramori T, Chen YF, Galizzi A, Helmann JD. Promoter architecture in the flagellar regulon of Bacillus subtilis: High-level expression of flagellin by the sigma δ RNA polymerase requires an upstream promoter element. Proc. Natl Acad. Sci. USA. 1995;92:2582–2586. doi: 10.1073/pnas.92.7.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aiyar SE, Gaal T, Gourse RL. rRNA promoter activity in the fast-growing bacterium Vibrio natriegens. J. Bacteriol. 2002;184:1349–1358. doi: 10.1128/JB.184.5.1349-1358.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savchenko A, Weigel P, Dimova D, Lecocq M, Sakanyan V. The Bacillus stearothermophilus argCJBD operon harbours a strong promoter as evaluated in Escherichia coli cells. Gene. 1998;212:167–177. doi: 10.1016/s0378-1119(98)00174-7. [DOI] [PubMed] [Google Scholar]
- 29.Ozoline ON, Deev AA. Predicting antisense RNAs in the genomes of Escherichia coli and Salmonella typhimurium using promoter-search algorithm PlatProm. J. Bioinf. Comput. Biol. 2006;4:443–454. doi: 10.1142/s0219720006001916. 16819794. [DOI] [PubMed] [Google Scholar]
- 30.Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Ann. Rev. Biochem. 1988;57:199–233. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- 31.Roberts MW, Rabinowitz JC. The effect of Escherichia coli ribosomal protein S1 on the translational specificity of bacterial ribosomes. J. Biol. Chem. 1989;264:2228–2235. [PubMed] [Google Scholar]
- 32.Rocha EPC, Danchin A, Viari A. Translation in Bacillus subtilis: Roles and trends of initiation and termination, insights from a genome analysis. Nucleic Acids Res. 1999;27:3567–3576. doi: 10.1093/nar/27.17.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salgado H, Gama-Castro S, Peralta-Gil M, Diaz-Peredo E, Sanchez-Solano F, Santos-Zavaleta A, Martinez-Flores I, Jimenez-Jacinto V, Bonavides-Martinez C, et al. RegulonDB (Version 5.0): Escherichia coli K-12 transcriptional regulatory network. operon organization, and growth conditions Nucleic Acids Res. 2006 doi: 10.1093/nar/gkj156. 34(Database issue): D394–D397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hershberg R, Bejerano G, Santos-Zavaleta A, Margalit H. PromEC: An updated database of Escherichia coli mRNA promoters with experimentally identified transcriptional start sites. Nucleic Acids Res. 2001;29:277. doi: 10.1093/nar/29.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morin A, Huysveld N, Braun F, Dimova D, Sakanyan V, Charlier D. Hyperthermophilic Thermotoga arginine repressor binding to full-length cognate and heterologous arginine operators and to half-site targets. J. Mol. Biol. 2003;332:537–53. doi: 10.1016/s0022-2836(03)00951-3. [DOI] [PubMed] [Google Scholar]
- 36.Braun F, Marhuenda FB, Morin A, Guevel L, Fleury F, Takahashi M, Sakanyan V. Similarity and divergence between the RNA polymerase alpha subunits from hyperthermophilic Thermotoga maritima and mesophilic Escherichia coli bacteria. Gene. 2006;380:120–126. doi: 10.1016/j.gene.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 37.Sakanyan V, Dekhtyar M, Morin A, Braun F, Modina L. Method for the identification and isolation of strong bacterial promoters. European patent application. 2003 3290203.3. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.