Abstract
The eukaryotic Ccr4/Caf1/Not complex is involved in deadenylation of mRNAs. The Caf1 and Ccr4 subunits both potentially have deadenylating enzyme activity. We investigate here the roles of Ccr4 and Caf1 in deadenylation in two organisms that separated early in eukaryotic evolution: humans and trypanosomes. In Trypanosoma brucei, we found a complex containing CAF1, NOT1, NOT2 and NOT5, DHH1 and a possible homologue of Caf130; no homologue of Ccr4 was found. Trypanosome CAF1 has deadenylation activity, and is essential for cell survival. Depletion of trypanosome CAF1 delayed deadenylation and degradation of constitutively expressed mRNAs. Human cells have two isozymes of Caf1: simultaneous depletion of both inhibited degradation of an unstable reporter mRNA. In both species, depletion of Caf1 homologues inhibited deadenylation of bulk RNA and resulted in an increase in average poly(A) tail length.
INTRODUCTION
Degradation of eukaryotic mRNAs is critical in cellular homoeostasis. The first, rate-limiting step in degradation is usually removal of the poly(A) tail. Subsequently, the mRNA can be degraded in the 3′ → 5′ direction, probably by the exosome. Alternatively, the 5′ cap is removed by decapping enzymes and the mRNA is degraded in the 5′ → 3 direction. RNAs subject to rapid, regulated degradation, such as those bearing AU-rich instability elements, show accelerated deadenylation relative to more stable mRNAs (1,2). Saccharomyces cerevisiae has two deadenylation complexes, the Pan2/Pan3 complex and the Ccr4/Caf1/Not complex (3–5). Vertebrates have, in addition, the DAN/PARN enzyme (6). Evidence so far indicates that the Ccr4/Caf1/Not complex is the major eukaryotic deadenylase, while the other activities—PARN and the Pan2/Pan3 complex—play secondary or more specialized roles (1,7).
The central component of the Ccr4/Caf1/Not complex is Not1, which is conserved throughout eukaryotic evolution and serves as a scaffold for the attachment of the other subunits; it has been found to be essential in all organisms investigated so far. Not1p binds to Caf1p (8), which in turn binds to Ccr4p. A leucine-rich domain at the N-terminus of Ccr4p is required for binding to Caf1p in both yeast and humans and is conserved in the Ccr4 homologues from Drosophila and Caenorhabditis elegans (9–11). Other subunits of the complex are Not2, Not5, Not3, Not4, a ubiquitin ligase, Caf4p, Caf16p, Caf40p and Caf130p [see Table 1 and (12–14)]; it is not clear whether all of these subunits are associated simultaneously and their roles are unclear (15–18). In addition, the putative helicase Dhh1p interacts with complex both physically and genetically (19–21).
Table 1.
New plasmids used in this study
| Plasmid | Description | |
|---|---|---|
| pHD1534 | DHH1 RNAi | Part of the DHH1 ORF was amplified using forward primer 5′-gag aag atc taa gct tca cag tca agg act ttg cg-3′and reverse primer 5′-cgg aat tcg tcg acc caa tgc ggt gaa ggt aag t-3′, PCR product was cloned into p2T7TA blue (39) |
| pHD1535 | LCCR4 RNAi | Part of the CCR4-like ORF was amplified using forward primer 5′-gagaagatctgcatgccaaagcaatctgtggaagca-3′ and reverse primer 5′-cggaattcgtcgacccaaccaccaactcttcgtt-3′, PCR product was cloned into p2T7TA blue (39) |
| pHD1843 | CAF1 RNAi | Part of the CAF1 ORF was amplified using forward primer 5′-gaggatcsctctgcctcacagaggatgtg-3′ and reverse primer 5′-cgggatcccgaggaatcttcacagagcc-3′, PCR product was cloned into p2T7TA blue (39) |
| pHD1844 | NOT1 RNAi | Part of the NOT1 ORF was amplified using forward primer 5′-gaggatccgagctggtagaagtggacgc-3′ and reverse primer 5′-cgggatccacgactcgggaccacaatag-3′, PCR product was cloned into p2T7TA blue (39) |
| pHD1847 | GST-CAF1 | CAF1 ORF was amplified using forward primer 5′-gagagaattcatgcagtatggtggcgggac-3′ and reverse primer 5′-cgatgaattctcagctatgaccctttaccgc-3′ and cloned into pGEX-λ T (Eco RI digest) |
| pHD1849 | CAF1-TAP tag (C-terminal) | CAF1 ORF was amplified using forward primer 5′-aagcttatgatgcagtatggtggcggga-3′ and reverse primer 5′-gttaacgctatgaccctttaccgcttgtgt-3′and cloned into pHD 924 (Hind III, Hpa I digest) (34) |
| pHD1850 | LCCR4- TAP tag (C-terminal) | LCCR4 ORF was amplified using forward primer 5′-aagcttatggcacggcaccccaacacggc-3′ and reverse primer 5′-gttaactttgtacgacaaatcggcaatcaa-3′ and cloned into pHD 1337 (Hind III, Hpa I digest) (34) |
A variety of observations suggest that in yeast, Ccr4 has the major deadenylation activity, while Caf1 serves mainly to attach Ccr4 to the rest of the complex (22,23). Ccr4 (an exonuclease III) has 1000 times higher specific activity than Caf1, which resembles an RNase D (24,25). Neither protein is essential in yeast but deletion mutants show slow growth and a deadenylation defect (5,25–27). Critically, yeast Δcaf1 mutants can be complemented either by over-expression of Ccr4p, or by expression of a mutant mouse CAF1 lacking the active site (4,24); but human Ccr4, which is less catalytically active than the yeast homologue, cannot complement a yeast Δccr4 mutant (3).
Results from vertebrates and Drosophila, in contrast to those from yeast, suggest that Caf1 is more important in deadenylation than Ccr4. RNA interference (RNAi) targeting Caf1, but not Ccr4, in C. elegans resulted in an embryonic lethal phenotype (28,29). Drosophila cells depleted of Caf1 showed a marked increase in overall poly(A) length and a delay in the deadenylation of Hsp70 mRNA during the recovery from heat shock, whereas Ccr4 deletion mutants were viable and showed only slight defects in deadenylation of bulk or Hsp70 RNA (15). The relative roles of Caf1 and Ccr4 in mammalian deadenylation have not yet been resolved, essentially because of the presence of several potential homologues. Five human genes encode proteins with Ccr4-like catalytic domains, but only two—Ccr4a and Ccr4b—retain the leucine-rich repeat and associate with Caf1a (7). Over-expression of wild-type Ccr4a in mammalian cells destabilized the (normally very stable) beta-globin mRNA (7) and a reporter mRNA containing a premature termination codon. In contrast, over-expression of a catalytically inactive mutant Ccr4a, or siRNA treatment targeting Ccr4a, slightly delayed deadenylation of the mRNA containing a premature termination codon, but the effects were subtle (7). There are also two homologues of Caf1p, called Caf1a and Caf1b [(7,14,30) and see Table 1]. Mouse Caf1a is a processive deadenylase; Caf1a−/− mice are apparently healthy but males are sterile (24,31). The roles of Ccr4b and Caf1b have not been examined.
So far detailed experiments concerning the Ccr4/Caf1/Not complex have been performed on organisms in a single eukaryotic group—the Opisthokonta (32). The Kinetoplastid protists—which diverged very early from the Opisthokonta—are unique among eukaryotes in relying extremely heavily on mRNA degradation for the control of gene expression (33). Trypanosoma brucei is able to multiply in the mammalian bloodstream, as ‘bloodstream form’ trypomastigotes, and in the midgut of the Tsetse fly vector, as ‘procyclic form’ trypomastigotes. In bloodstream forms, constitutively expressed mRNAs have half-lives varying from 15 min to over an hour, while mRNAs which are specific to the Tsetse vector forms are mostly destroyed within 5 min; in the more slowly growing procyclic forms, constitutive mRNAs have rather longer half-lives but mRNAs specific to bloodstream forms are again destroyed with 5–10-min half-lives. The trypanosome exosome seems to play a minor role in mRNA degradation (34,35). In contrast, a putative 5′–3′ exonuclease, XRNA, is required for the very rapid degradation of developmentally unstable mRNAs and may operate on polyadenylated mRNAs (36). Milone et al. (37) previously demonstrated cap-independent deadenylation activity, which was inhibited by poly(A) binding protein, in extracts from the Kinetoplastid Leptomonas seymouri and deadenylation has also been demonstrated in vivo [see e.g. (36)].
In this paper, we test the roles of Caf1 and Ccr4 in mRNA degradation in both human cells and trypanosomes.
MATERIALS AND METHODS
Cell culture and plasmids
Procyclic or bloodstream form T. brucei stably expressing the tetracycline repressor (38,39), with or without T7 polymerase expression (39,40), were grown, transfected and clonally selected as described in ref. (41). Plasmids used in this study are described in Table 1. To analyse the degradation of a reporter mRNA the CAF1 RNAi line was transfected with pHD 1034 (42).
CAF1- and CCR4-TAP purification
Procyclic cells expressing the tet repressor [pHD 1313 (39)] were transfected with pHD 918 (tetracycline-inducible expression of the TAP-tag), pHD1849 (tetracycline inducible over-expression of TbCAF1-TAP) or pHD1850 (tetracycline inducible over-expression of TbLCCR4) (Table 1). The expression of the TAP tag alone, TAP-tagged TbCAF1 or TAP-tagged TbLCCR4 was induced by adding 1 µg/ml tetracycline and cells were harvested 20 h later. For tandem affinity purification 6 × 109 cells were used, with methodology as previously described (43). The whole preparation was run on a 10% SDS–page gel and stained with SyproRuby (Invitrogen). Bands which were not present in the TAP-only control were identified by mass spectrometry.
Intracellular detection of the TAP tag
106 trypanosomes were sedimented, re-suspended, then fixed in 4% paraformaldehyde (w/v) in 1× PBS for 18 min, sedimented again for 2 min, re-suspended in PBS, and allowed to settle down on a poly-lysine-coated glass surface for 25 min. The cells were then permeabilized with 0.2% (v/v) Triton X-100 (in 1× PBS) at room temperature for 20 min. Before staining, slides were blocked with 0.5% (w/v) gelatine for 20 min. To detect the TAP tag, cells were incubated with a 1:50 000 dilution of rabbit α-Protein A (Sigma) for 1 h and with a 1:500 dilution of the second antibody Alexa Fluor 594 goat α- rabbit IgG (Molecular probes) for 40 min. The kinetoplast and the nuclear DNA were then stained with 100 ng/ml DAPI/1× PBS for 10 min.
RNAi
For RNAi in bloodstream forms, gene-specific fragments of ∼500 nt (44) were cloned into the vector p2T7TA blue (see Table 1). In this vector, the RNAi fragments are flanked by opposing T7 promoters with downstream tet operators. The resulting plasmids were linearized with Not I and transfected into bloodstream-form trypanosomes expressing the tet repressor (pHD 1313) and T7 RNA polymerase (pHD514) (39). Transfectants were selected in 10 µg/ml hygromycin and cloned by limiting dilution. RNAi was induced by adding tetracycline to the medium at a concentration of 1 µg/ml. Samples were taken after 18.5–26 h of RNAi induction. For procyclic forms, the entire CAF1 open reading frame was cloned into p2T7 and transfected into trypanosomes containing integrated plasmids pLEW29 and pLEW13 (40). RNAi was again induced by adding tetracycline to the medium at a concentration of 1 µg/ml.
Expression of GST-CAF1 in Escherichia coli
The E. coli strain BL21 was transformed with pHD1847 (Table 1). The bacteria were grown to an optical density (600 nm) of 2 in LB medium at 37°C. Expression of GST-CAF1 was induced with 1 mM IPTG and the bacteria were allowed to grow for 2 h more before harvesting and sonification in 1× PBS. Triton X-100 was next added to a final concentration of 1%. The sonicate was gently mixed for 30 min at 4°C, then cleared by centrifugation at 12 000g at 4°C for 10 min. The supernatant was incubated with glutathione–Sepharose 4B slurry (GE Healthcare) for 30 min at room temperature in a column. The slurry was washed three times with 1× PBS then incubated three times with 50 mM Tris–HCl, 10 mM reduced glutathione (pH8.0) for 10 min at room temperature to elute the fusion protein. The purity of the preparations was checked on SDS–page gels stained with colloidal Coomassie; a single dominant band was seen with two to three very faint additional bands. Unfortunately, serum from a rabbit immunized with purified TbCAF1-GST recognized the recombinant protein only with very poor sensitivity, and failed to detect CAF1 in trypanosome lysates. Affinity purification did not improve the sensitivity.
In vitro deadenylation assay
Ten picomoles of each RNA oligonucleotide substrate (see Figure 1B) (from MWG, Ebersberg) was labelled with 30 U T4 polynucleotide kinase and 50 µCi γ-32P ATP (GE Healthcare) at 37°C for 30 min. They were purified with Sephadex G-25 columns equilibrated with 10 mM Tris. For the assay, substrate (2 × 105 c.p.m.) was incubated with 50 pmol GST-CAF1 in 10 mM Tris–HCl and 1 mM magnesium acetate. The reaction was stopped by adding 1 volume formamide loading buffer (95% formamide, 0.025% xylene cyanol, 0.025% bromophenol blue, 18 mM EDTA, 0.025% SDS) at appropriate time points. The Decade Marker System (Ambion) was used to synthesize the 10-bp RNA ladder. The samples were run on 12% polyacrylamide/urea gels, which were then fixed, dried and analysed with a phosphorimager.
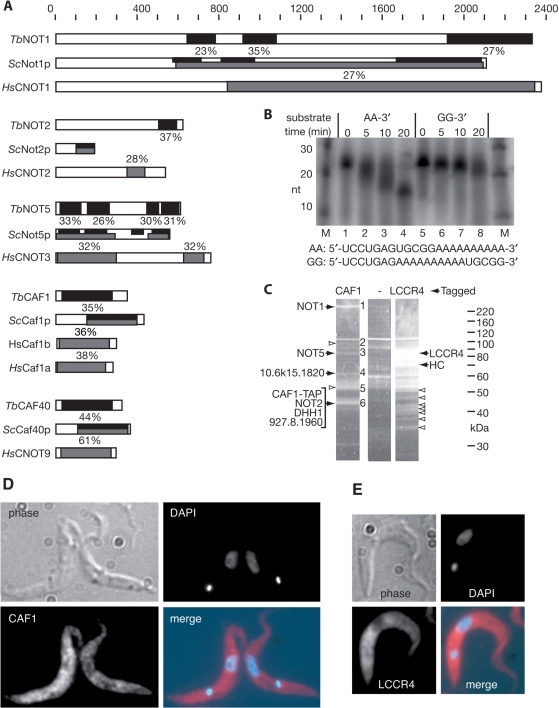
Figure 1.
(A) Diagrammatic representation of likely CAF1/NOT complex components from trypanosomes, aligned with the yeast and human homologues. The scale at the top represents amino acid residues. The black bars indicate regions of sequence similarity between the trypanosome and yeast proteins, and the percentage identical amino acids is shown, using output from GeneDB blastp using default settings. (The percentage similar amino acids was 1–5–2 times higher.) Grey bars similarly indicate the regions of sequence similarity between the human and yeast proteins, using the output of Blastp with NCBI database default settings. (B) Enzyme activity of recombinant TbCAF1. The oligonucleotides shown were 5′-radiolabelled; the samples without enzyme were incubated 20 min in buffer alone to exclude that the degradation observed was caused by contaminating RNases (lanes 1 and 5). Products were separated by denaturing gel electrophoresis. (C) Proteins associated with CAF1-TAP (left), CCR4L-TAP (right) or the TAP tag alone (centre) were subjected to purification on IgG and calmodulin columns, then separated by SDS–PAGE and stained with SyproRuby. The major potential complex components are indicated by solid arrows; other bands which were analysed and contained either tagged protein only, or highly abundant proteins which are probably contaminants are indicated by open arrows. Bands in the CAF1 lane are numbered on the right (identities in Table 3). HC: hypothetical protein conserved in all three trypanosomatids. (D) Location of TAP-tagged CAF1. Procyclic trypanosomes expressing CAF1-TAP were fixed and permeabilized, and the TAP tag was detected by immunofluorescence. The kinetoplast (small dot near the end of the parasite) and nuclear DNA (larger stain near the middle) were counter-stained with DAPI. A typical image is shown. Cells not expressing the TAP tag gave no fluorescence. (E) Location of TAP-tagged LCCR4. Details as for (D).
mRNA degradation assays and mRNA characterization
To analyse mRNA decay in trypanosomes, transcription was stopped with Actinomycin D at a final concentration of 10 µg/ml. Total RNA was either isolated by using peqGold Trifast (peqLab, Germany) or RNeasy mini kit (QIAGEN) according to the manufacturer's instructions.
To remove poly(A) tails, 170 pmol oligo d(T) were incubated with 10–12 µg total RNA at 42°C for 10 min. After that 5 U RNaseH (New England Biolabs) and RNaseH buffer were added and samples were incubated for 40 min at 37°C.
Levels of mRNAs were assessed by formaldehyde agarose or denaturing polyacrylamide gel electrophoresis, followed by blotting onto Nytran membranes (Amersham Biosciences) and hybridization with radioactive DNA (random priming) (Stratagene) or anti-sense RNA (MAXIscriptT7/T3, Ambion) probes. Signals were measured using a phosphorimager. All mRNA signals were normalized relative to the signal from an SRP (signal recognition particle) probe. Northerns were stripped by brief boiling in 0.1% SDS; the efficacy of stripping assessed by phosphorimager and any residual signals were left to decay as necessary before re-use. The half-lives were measured by plotting relative signal intensities using Kaleidograph (Synergy software), including only the segments of the time course which gave exponential decay curves (fitted with a linear correlation coefficient generally exceeding 0.9).
Poly(A) tail assay
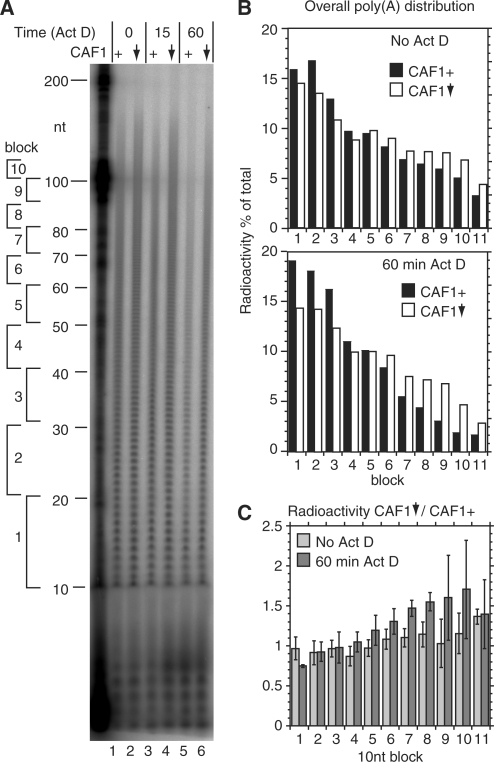
To induce CAF1 RNAi, bloodstream-form trypanosomes transfected with pHD1843 (Table 1) were cultured with 1 µg/ml tetracycline for 24 h. Ten micrograms per millilitre Actinomycin D was added to the cultures to stop transcription and samples taken at appropriate time points. Total RNA was isolated using the RNeasy mini kit (QIAGEN) according to the manufacturer's instructions. Five hundred nanograms of total trypanosome RNA or 1 µg of mammalian RNA, 20 U T4 RNA ligase (New England BioLabs) and 5 µCi pCp (GE Healthcare) in 50 mM Tris–HCl, 10 mM MgCl2, 10 mM DTT, 1 mM ATP and 10% (v/v) DMSO have been incubated at 37°C for 1.5 h to label the 3′-ends. After heat inactivation of the T4 RNA ligase, the labelled RNA was incubated with 2 µg RNase A, 5 U T1 (RNase A/T1 mix, Fermentas) and 20 µg yeast tRNA (Ambion) in 10 mM Tris–Cl, 2 mM MgCl2, 2 mM DTT, 0.2 mM ATP, 2% (v/v) DMSO, 300 mM NaCl and 5 mM EDTA at 37°C for 30 min. The poly(A) tails were purified by phenol/chloroform extraction and ethanol precipitation then separated on 8% polyacrylamide/urea gels. Markers used were the Decade RNA ladder (Ambion, see above), labelled with T4 polynucleotide kinase and 50 µCi γ-32P ATP, or an RNA ladder (Invitrogen), labelled with T4 RNA ligase and 5 µCi α-32P pCp. Bands were detected and quantified in the fixed and dried gels with a phosphoimager. For analysis, the poly(A) tails were subdivided into blocks of 10 nt, either by counting the bands (Figure 3) or using the labelled Decade marker (Figure 5). The ratios between the signals from the first block and the other blocks were determined.
Figure 3.
CAF1 depletion inhibits deadenylation in bloodstream trypanosomes. Total RNA was purified from cells with inducible CAF1 RNAi, cultured without tetracycline (CAF1+, lanes 1, 3 and 5) or with tetracycline (24 h) to deplete CAF1 (downward arrow, lanes 2, 4 and 6). Actinomycin D was then added for the times indicated. RNA was 3′-labelled then digested with RNases A and T1. (A) Phosphorimager image showing radiolabelled poly(A) tails separated by denaturing gel electrophoresis. To the left are the positions of markers. Further to the left we have indicated blocks of 10 a residues. Block 1 is the group from 11–20 As; block 2 from 21–30 As, and so on. (B) Quantitation of results from the gel shown in (A) (no Actinomycin and 60-min time point). For each lane, the amount of radioactivity in each block was plotted relative to the total radioactivity in the lane. Black bars: CAF1+; open bars: CAF1-depleted. (C) Results of three independent experiments, shown as arithmetic mean and standard deviation. We first generated data like that shown in (B) for all three experiments. We then divided the number for the CAF1-depleted cells by the number obtained for the CAF1+ cells. For block 1, the numbers are automatically 1. For blocks 2–11, numbers >1 indicate that the CAF1-depleted cells have more radioactivity in the poly(A) tails than do the normal cells. Here it is clear that the differences are stronger for the poly(A) tails above 70 nt, especially after Actinomycin D treatment.
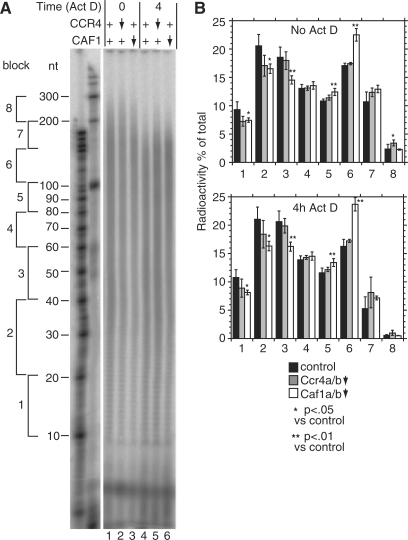
Figure 5.
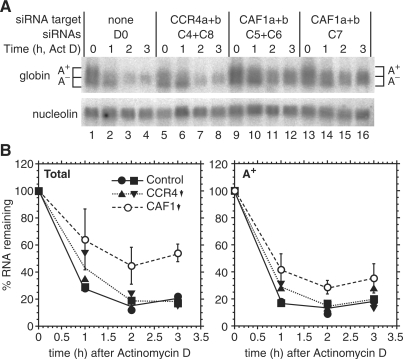
CAF1 depletion inhibits deadenylation in human cells. HTGM5 cells were transfected with siRNAs to deplete CAF1 or CCR4 (downward arrow) or with control siRNAs (+). The effect of the siRNAs on their targets is illustrated in Figure 6. Samples were taken either with, or without 4 h Actinomycin D treatment. Purified RNA was 3′-labelled then digested with RNases A and T1. (A) Phosphorimager image showing radiolabelled poly(A) tails separated by denaturing gel electrophoresis. To the left are the positions of markers. Further to the left we have indicated blocks of 20–50 A residues, as in Figure 5. (B) Quantitation of results from the gel shown in (A). For each individual lane, the amount of radioactivity each block was calculated relative to the total radioactivity in the lane. The results are expressed as arithmetic mean ± standard deviation. Black bars: control siRNA, five independent experiments; grey bars: Ccr4a+b-depleted, siRNAs C4+C8, three experiments; open bars: Caf1a+b-depleted, siRNAs C5+C6, four experiments. Upper panel: steady state, values for lanes 1, 2 and 3 (in that order); lower panel: after 4 h Actinomycin D, values for lanes 4, 5 and 6. P-values are for a 2-tailed students t-test (unequal variance) and show the probability that the poly(A) tail signals for Ccr4- or Caf1-depleted cells were the same as those of the control.
Human cell lines and siRNA
HTGM5 cells were derived from human HT1080 fibrosarcoma cells (45). Cells were transfected twice with siRNA duplexes (100 nM) over a period of 96 h using Lipofectamine 2000 (Invitrogen). siRNAs were designed using published recommendations (46) and purchased from Ambion. The following target sequences (sense strand) were chosen:
control siRNA (D0): 5′-GCAUUCACUUGGAUAGUAA-3′; Ccr4a siRNA (C4): 5′-CAAGACGGCUGCUGAACUA-3′;
Ccr4b siRNA (C8): 5′-UGACAGCGCUGCACCTAAA-3′;
Caf1a siRNA (C5): 5′-UUACGACUUUGGCUACUUA-3′;
Caf1b siRNA (C6): 5′-GUGCUCAGUUACAGUUAUA-3′;
Caf1a+b siRNA (C7): 5′-UGAAGAGCUGCAAAAAUCU-3′
The efficiency of siRNA knockdown was determined by real-time PCR.
Quantitation of globin-ARE mRNA
SiRNA-transfected HTGM5 cells were treated with Actinomycin D (5 µg/ml; Sigma) for indicated periods of time, and cytoplasmic RNA was extracted according to ref. 47. Twenty micrograms of RNA were resolved by 1.5% agarose/formaldehyde gel electrophoresis as described previously (48). For Figure 6, β-globin and nucleolin mRNA were detected with digoxigenin-labelled RNA probes (Roche). Templates were generated by PCR using primers G1000 and G1001 for the b-globin probe Bex12, and primers G83 and G1009 for the nucleolin probe. The results were quantitated by scanning densitometry. Oligonucleotides used were:
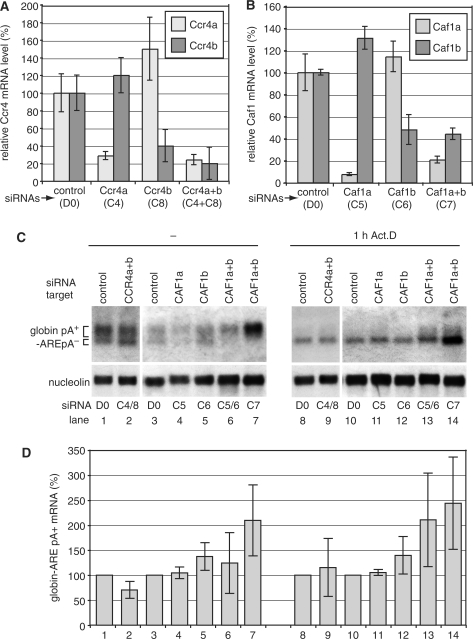
Figure 6.
Depletion of Caf1a and Caf1b inhibits deadenylation of an ARE-containing β-globin reporter mRNA in human cells. (A) HTGM5 cells were transfected over a period of 5 days with siRNAs targeting Ccr4a and Ccr4b; or (B) Caf1a and Caf1b. The knockdown efficiencies were measured at the mRNA expression level by quantitative PCR. The graphs show the mean and standard deviation from three assays in a single experiment; the siRNA transfection and the measurements were also repeated once (for Ccr4b) or three times (all other siRNAs) with very similar results. (C) Cytoplasmic RNA was prepared from siRNA-transfected cells with or without prior Actinomycin D treatment. Northern blots were hybridized with a globin probe (upper panel), and with a nucleolin probe as loading control (lower panel). (D) Quantitation of the poly(A)+ portion of globin-ARE mRNA shown in (C); mean and standard deviation from three (lanes 1, 2, 8 and 9) or four (lanes 3–7, 10–14) independent experiments.
G83: 5′-TTACAAAGTCACTCAGGATG-3′;
G1000: 5′-GTGCATCTGTCCAGTG-3′;
G1001: 5′-GCCGATTTAGGTGACACTATAGAATACCCTGAAGTTCTC-3′;
G1009: 5′-GCCGATTTAGGTGACACTATAGAATACTTAGCGTCTTCG-3′.
For Figure 7, the probes were 32P-labelled and hybridization was as described for trypanosome mRNA; signals were quantitated using the phosphorimager.
Figure 7.
Depletion of Caf1a and Caf1b inhibits degradation of the ARE-containing β-globin reporter mRNA. Transfection with siRNAs and RNA preparation was as for Figure 5, except that samples were taken 1, 2 and 3 h after addition of Actinomycin D. The experiment was done twice with similar results. (A) Northern blot for the first experiment. To quantitate the globin mRNA, the lower portion of the globin signal in lane 1 was designated as poly(A)–; the upper portion, which had the same area as the lower one, was designated poly(A)+ as indicated. Identical areas were then used for all of the other lanes, aligning the bottom of the quantitation box just beneath the signal in each case. (B) Quantitation for both experiments. Results for the control and Ccr4a+b knockdown are shown separately, as indicated in the legend within the graph; the lines are drawn through the arithmetic mean. Results for the siRNAs C5+6 and C7 (CAF1 RNAi) were indistinguishable. These results were therefore pooled, to generate a mean and standard deviation from a total of four measurements (2× C5+6 plus 2× C7).
RESULTS
Trypanosome homologues of CCR4/CAF1/NOT complex subunits
We searched the trypanosome genome database with the amino-acid sequences of the yeast and human Ccr4/Caf1/Not complexes, extracted the possible homologues and confirmed their identities by reciprocal BLAST analysis on the yeast genome database or the EMBL database. We found possible candidates (reciprocal-blast positive) for Caf1, Not1, Ccr4, Not2, Not5 and Caf40 (Figure 1A and Table 2). The matches for Caf4 and Caf16 were restricted to functional domains, so their status is uncertain. Not3, Not4 and Caf130 appeared to be absent.
Table 2.
Components of the yeast CCR4/CAF1/NOT complex, with trypanosome, human and Drosophila homologues
| Yeast | Trypanosome protein | Trypanosome GeneDB number | Humana | Drosophila CG number |
|---|---|---|---|---|
| Caf1/Pop2 | TbCAF1 | 927.6.600 | CNOT7/hCaf1a | 5684 |
| CNOT8/hCaf1b | ||||
| Ccr4 | None foundb | CNOT6/hCCR4a | 31137 | |
| CNOT6L/hCCR4b | ||||
| Not1 | TbNOT1 | 10.70.6450 | CNOT1 | 1884 |
| Not2 | TbNOT2 | 927.6.850 | CNOT2 | 2138 |
| Not3 | – | – | CNOT3S | |
| Not4 | – | – | CNOT4 N/S/L | 31716 |
| Not5 | TbNOT5 | 927.3.1920 | CNOT3L | 8426 |
| Caf4 | unclearc | Unclearc | ||
| Caf16 | – | (927.6.2810)d | ||
| Caf40 | TbCAF40 | 927.4.410 | CNOT9/hCAF40 | 14213 |
| Caf130 | ?Tb10.6k15.1820e | CNOT10 | – |
aHuman subunit names are preceded by either ‘C’ or ‘h’ (14). Ccr4a is Acc# NM_015455, Ccr4b, Acc# NM_144571, Caf1a, Acc# NM_013354, Caf1b, Acc# NM_004779.
bThe work in this article indicates that Tb927.4.2430 is not a functional homologue of Ccr4.
cSeveral possible homologues, none of which is convincing.
dUncertain.
eCaf130 has a molecular weight of 130 kDa while the Tb10.6k15.1820 protein is only 35 kDa.
We first compared the trypanosome CAF1 protein with homologues from multiple eukaryotic genomes. We found Caf1 homologues in all genomes searched, including Schizosaccharomyces pombe and the very early-branching protists Trichomonas (six copies) and Giardia. All catalytic residues are conserved in all homologues apart from that of S. cerevisiae (see Supplementary Figure 8 for the alignment). The first essential region (starting at residue 188 of the consensus) has the sequence DTEF in all homologues except S. cerevisiae (STEF). The Asp at residue 369 was universally conserved. At position 421, the conserved HQAGSD was changed to TTTGGQ only in S. cerevisiae. This supports the previous suggestion that the low deadenylase activity seen for S. cerevisiae Caf1 is not the norm (25).
To check the enzymatic activity of TbCAF1, we expressed the protein as a fusion with glutathione S-transferase and assayed its ability to digest a 5'-labelled oligonucleotide substrate. TbCAF1 shortened a substrate containing 3'-oligo(A), but not internal oligo(A) (Figure 1B), confirming that it is a deadenylase.
The best trypanosome match for Ccr4 was a predicted protein which contained the conserved C-terminal exonuclease domain, but lacked the leucine-rich repeats which are required both for interaction with CAF1 (11) and for deadenylase activity (10). In evolutionary trees, the position of this sequence varied depending on the method used (data not shown). It did not convincingly belong to any family but clearly did not branch with the confirmed Ccr4 proteins, so we named it LCCR4 (Like-Ccr4).
Trypanosome CAF1 is associated with NOT1, NOT2 and NOT5
In order to find out whether trypanosomes contain a Ccr4/Not1/Caf1 complex, we expressed TAP-tagged versions of TbCAF1 and TbLCCR4 in trypanosomes. Expression was tetracycline inducible; 3–5 days after tetracycline addition to the TbCAF1-TAP cells mild growth inhibition (no more than 50% increase in division time) was apparent (data not shown). After two steps of affinity purification, associated proteins were identified by mass spectrometry (Figure 1C). All bands excised from the TbLCCR4-TAP purification contained predominantly TbLCCR4 sequence. In addition, we identified abundant proteins which routinely contaminate such preparations, such as tubulin, HSP70 variants, ribosomal proteins and glycolytic enzymes, whose specific association is virtually impossible to verify biochemically. The only other proteins found were Tb08.10J17.920 (annotated as PIF1 DNA repair and recombination helicase) and Tb10.6k15.1430 (hypothetical protein conserved in other Kinetoplastids) (Figure 1C, both found in the band marked as ‘HC’). There was no evidence for any association of TbLCCR4 with NOT1 or CAF1, as expected from the absence of the leucine-rich repeat.
Proteins co-purifying with TbCAF1-TAP gave four major bands (Figure 1C). The band (1) at above 220 kDa contained TbNOT1. Judging from the peptide coverage (Table 3), the major protein in band 3 was TbNOT5. Band 4 contained a 61-kDa protein (encoded by Tb10.6k15.1820), which has a TPR repeat and gives a poor match (P = 0.6 and above) to various CNOT10 homologues. Band 5 was broad and contained various proteins of 37–48 kDa, including TbDHH1, TbNOT2, TbCAF1 and a protein of unknown function encoded by Tb927.8.1960 (Table 3). Band 2 was very faint and contained an aminopeptidase which may be a contaminant; other likely contaminants included actin and a ubiquitin-like protein which was also identified in purified RNA polymerase II (49). The TbCAF40 protein was not detected, and neither was the possible TbCAF16. When we immunoprecipitated TbDHH1 with specific antibody (50) we also found TbNOT1 as the major specifically associated protein (See Supplementary Figure 9). Additional work will be required to confirm the association of the other potential subunits. Since there was no evidence for association of either TbLCCR4, or any other trypanosome CCR4-like protein, with components of the NOT1/CAF1 complex we speculated that TbCAF1 might be the active deadenylating enzyme in the complex.
Table 3.
Mass spectrometry of proteins associated with TAP-tagged CAF1
| Band | GeneDB number | Protein function (known or putative) | Score | Peptides | Coverage | Molecular weight | Migration |
|---|---|---|---|---|---|---|---|
| 1 | Tb10.70.6450 | NOT1 | 3389 | 65 | 33 | 259 | >220 |
| 2 | Tb927.2.280 (and related)a | Retroposon hotspot protein (several similar predicted proteins) | 251 | 7 | 8 | 91 | 90-100 |
| 2 | Tb11.02.1070 | Aminopeptidase | 57 | 3 | 3 | 98 | 100 |
| 3 | Tb927.3.1920 | NOT3/5 | 815 | 19 | 29 | 67 | 85 |
| 3 | Tb11.02.1600 | Ubiquitin-like | 92 | 2 | 3 | 67 | 85 |
| 4 | Tb10.6k15.1820 | TPR repeat protein, similar to CNOT10/Caf130 | 578 | 11 | 21 | 61 | 65 |
| 4 | Tb11.02.1860 | Hypothetical conserved, PF02622, domain of unknown function | 105 | 2 | 4 | 52 | 65 |
| 5 | Tb927.1.2340a | Alpha tubulin | 975 | 20 | 5 | 50 | 50 |
| 5 | Tb927.1.2330a | Beta tubulin | 614 | 14 | 28 | 50 | 50 |
| 6 | Tb927.6.600 | CAF1 | 196 | 7 | 16 | 39 | 45 |
| 6 | Tb927.6.850 | NOT2 | 172 | 4 | 8 | 37 | 45 |
| 6 | Tb10.70.3290 | DHH1 | 52 | 2 | 4 | 46 | 45 |
| 6 | Tb09.211.0620 | Actin A | 135 | 3 | 11 | 42 | 45 |
| 6 | Tb927.8.1960 | Hypothetical conserved | 714 | 20 | 43 | 48 | 45 |
The overall score, number of different peptides identified, and % coverage of the amino acid sequence are shown.
aBands 2 and 5 predominantly consisted of proteins found frequently as contaminants.
The intra-cellular locations of TbCAF1-TAP (Figure 1D) and TbLCCR4-TAP (Figure 1E) were analysed by immunofluorescence: both appeared to be predominantly—but not exclusively—in the cytoplasm. Although the distribution of CAF1 was somewhat uneven, clear punctate staining indicative of association with RNA storage or degradation granules was not evident; this is as expected since, in trypanosomes, such structures can be seen by immunofluorescence only after stress [(50,51) and our unpublished data]. A polyclonal antibody to CAF1 unfortunately had a low titre and did not detect the protein in trypanosome lysates (data not shown).
TbCAF1, TbNOT1 and TbDHH1 are required for bloodstream trypanosome growth
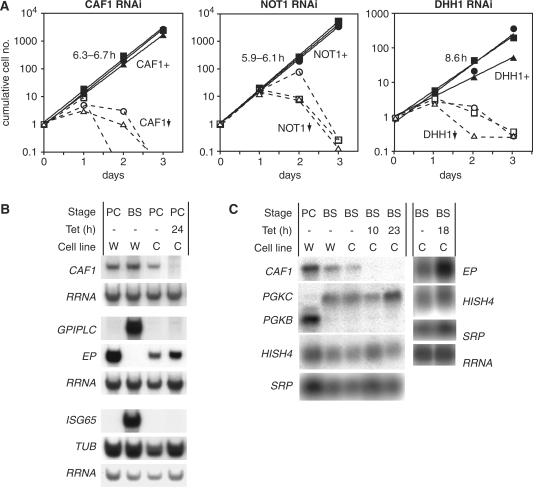
The roles of TbCAF1, TbNOT1, TbDHH1 and TbLCCR4 in bloodstream trypanosomes were probed by tetracycline-inducible RNAi. Depletion of TbCAF1, TbNOT1 or TbDHH1 RNA severely inhibited trypanosome growth with death at days 2 or 3 after tetracycline addition (Figure 2A). Reduction of RNA was verified by northern blotting (shown for CAF1 in Figure 2B and C, not shown for NOT1 and DHH1) and, for DHH1, by western blotting and immunoprecipitation. Notably, despite the lethal effect, the DHH1 protein was depleted only by 50% (data not shown and Supplementary Figure 9). TbLCCR4 RNAi, in contrast, had no effect on growth, but we were unable to detect the mRNA even in wild-type cells so we could not be sure that the RNAi had worked (data not shown). Similar experiments in procyclic trypanosomes gave the same results: growth of TbCAF1 or TbDHH1 RNAi cells ceased 1 day after tetracycline addition, and numbers started to decrease after 2 days, whereas growth of TbLCCR4 RNAi cells was unaffected over 6 days of incubation with tetracycline (data not shown).
Figure 2.
CAF1, DHH1 and NOT1 are essential for bloodstream trypanosome growth. (A) Trypanosomes were transfected with plasmids designed for tetracycline-inducible RNAi targeting CAF1, DHH1 or NOT1 and three independent cell lines were selected. Growth without tetracycline (filled symbols, continuous lines, CAF1+, NOT1+, DHH1+ cells) is illustrated as a cumulative exponential curve (log scale, initial number divided by 105) and the division time is indicated. Growth in the presence of tetracycline (added at time = 0) is shown using open symbols and dashed lines; the depletion of CAF1, DHH1 or NOT1 is indicated by the downward arrow. The three different symbols represent three different cell lines in each case. (B) Effect of CAF1 depletion on the levels of various RNAs in procyclic trypanosomes. The CAF1 mRNA was reduced to about 10% of normal after RNAi induction. Total glyoxal-treated RNA was separated on agarose gels, blotted and hybridized with the probes shown on the left. PC: procyclic; BS: bloodstream; tet: tetracycline. Cell lines are: W-cells expressing tet repressor and T7 polymerase; C-cells with inducible CAF1 RNAi. The experiments were performed in duplicate. Quantitation using the rRNA as a loading control revealed no reproducible effects of CAF1 depletion on any mRNA, apart from EP which increased 1.2–3.1-fold. (C) Effect of CAF1 depletion on the levels of various RNAs in bloodstream-form trypanosomes. Details as for (B) except that formaldehyde-agarose gels were used.
Effect of TbCAF1 depletion on mRNA abundance
Figure 2B and C show the effects of TbCAF1 depletion on the abundances of various trypanosome mRNAs. First, we looked at the abundances of some mRNAs which do not show developmental regulation, encoding Histone H4 (HISH4), actin (ACT) and Tubulin (TUB) (35,36,52,53). The abundances of all of these mRNAs were unaffected by TbCAF1 depletion (Figure 2B and C; for ACT see later in Figure 4; note that any minor variations visible in the blots shown were not reproducible). In addition, we looked at mRNAs which are developmentally regulated: Cytosolic phosphoglycerate kinase (PGKB) is expressed predominantly in procyclic forms (PC) (Figure 2C), while the mRNAs encoding glycosomal phosphoglycerate kinase (PGKC) (Figure 2C), glycosyl phosphatidylinositol phospholipase C (GPIPLC) (Figure 2B), and the membrane protein ISG65 (Figure 2B) are found exclusively in bloodstream forms (BS) (54–57). By northern blotting, we did not detect any defect in developmental regulation of these transcripts in TbCAF1-depleted cells (Figure 2B and C). Results from quantitative PCR analysis also revealed no change in PGKB mRNA levels (Stewart,M., ZMBH, data not shown). The only transcript which increased slightly in abundance after TbCAF1 depletion was that encoding EP procyclin mRNA in bloodstream forms (Figure 2C). This mRNA is exceptional because it is produced by RNA polymerase I, and both transcription and degradation are subject to developmental regulation; since it is possible that growth arrest alone might mimic the first stages of differentiation, and thereby affect regulation of EP expression, these mRNAs will not be discussed further here.
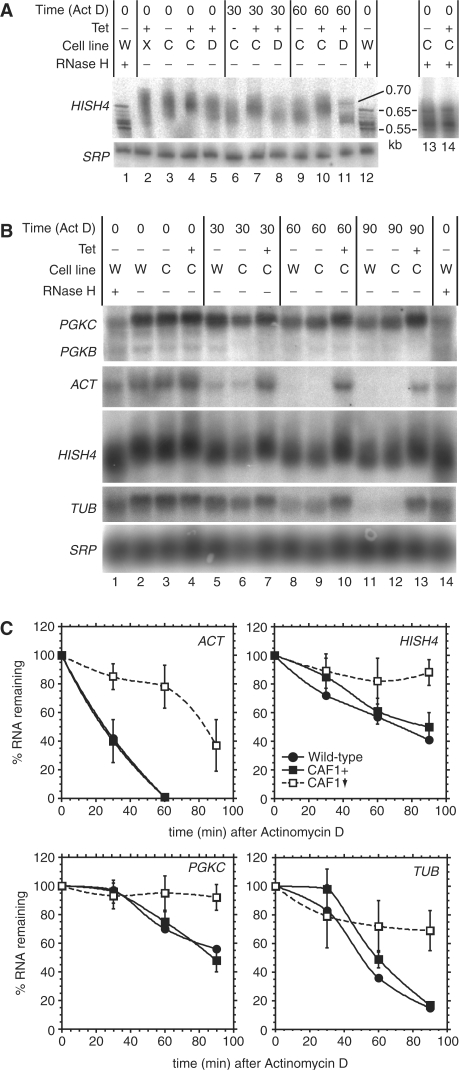
Figure 4.
CAF1 depletion inhibits degradation of ACT, PGKC, TUB and HISH4 mRNAs in trypanosomes. (A) Bloodstream trypanosomes expressing repressor alone (cell line W) or lines with inducible RNAi targeting XRNA (cell line X, lane 2), CAF1 (cell line C) or DHH1 (cell line D) were grown without (–) or with (+) tetracycline for 24 h as shown. Actinomycin D was then added for the times indicated (times include the centrifugation time). RNA was isolated and separated on a denaturing polyacrylamide gel. For lanes 1, 12, 13 and 14 the mRNA was treated with oligo d(T) and RNase H to remove the poly(A) tails. The histone H4 (HISH4) mRNA and the signal recognition particle RNA control (SRP) were detected after northern blotting. (B) As in (A), but separation on an agarose gel. RNA was detected using various probes, as indicated: PGKC: glycosomal phosphoglycerate kinase; ACT: actin; TUB: alpha tubulin; HISH4 (usually hybridized in that order, with intermediate stripping as required). (C) Results for three independent experiments [as in (B)] were quantitated, using SRP as the loading control, and results plotted as mean ± standard deviation (four experiments). Black circles: wild-type cells (two experiments only, arithmetic mean only shown); Black squares: inducible RNAi line without tetracycline (CAF1+); Open squares: inducible RNAi line with tetracycline (CAF1 down, with downward arrow).
TbCAF1is involved in deadenylation of many trypanosome RNAs
To find out if TbCAF1 was important for poly(A) shortening in trypanosomes, we measured the overall poly(A) tail distribution in total RNA. Briefly, total RNA was extracted, labelled at the 3′ end, then digested with RNases A and T1, which together cleave downstream of G, C and U. The remaining labelled poly(A) was separated on a polyacrylamide gel and visualized by phosphorimaging. Results are shown in Figure 3 [note that these preparations are for total cellular RNA, so the poly(A) tails seen will include the short tails present on nuclear RNAs that are destined for degradation (58)]. In normal cells, poly(A) tails ranged from 10 to over 100 nt in length (Figure 3A, lane 2). To quantitate the poly(A) tail lengths, we compared the radioactivity in tails specific lengths with the total radioactive signal, taking blocks of 10 nt (Figure 3B). Cells depleted of CAF1 showed a very slight, but reproducible decrease in the proportion of tails under 40 nt with a corresponding shift towards longer tail lengths (Figure 3C). One hour after addition of Actinomycin D, in the cells with a normal CAF1 level, there was a lower poly(A) tail signal overall and the signal was concentrated at shorter lengths (Figure 3A, lane 6, and Figure 3B). In cells with CAF1 RNAi (Figure 3A, lane 3), poly(A) tails were clearly longer than in the normal cells. This difference was evident 15 min after Actinomycin D treatment and became more pronounced after 60 min (Figure 3B). As a control we did similar experiments after DHH1 depletion; no increase in poly(A) tail lengths was seen (data not shown).
To further quantitate the effects of TbCAF1 depletion, we compared poly(A) tail lengths from three different experiments. First, the amount of radioactivity in the different 10-nt length ranges was divided by the total amount. Next, the value for CAF1 RNAi cells was divided by the value for the normal cells. This confirmed that 60 min after addition of Actinomycin D, the CAF1 RNAi cells had more 70–110-nt poly(A) tails than normal cells (Figure 3C, blocks 7–10, dark bars). These results suggest that CAF1 is involved in deadenylation of many trypanosome mRNAs.
TbCAF1 is involved in constitutive degradation of trypanosome mRNAs
To assess the effects of CAF1 depletion on the decay of individual mRNAs in bloodstream-form trypanosomes, we incubated them with Actinomycin D. Figure 4A shows results for HISH4. Trypanosome transcription is polycistronic. During processing, a 39-nt spliced leader RNA is added to the 5′ end of each mRNA, the position usually being determined by a polypyrimidine tract. Polyadenylation of the ‘upstream’ mRNA follows, in a reaction that is coupled to splicing of the downstream transcript. Polyadenylation occurs not at a specific single site, but at various positions between 70 and 140-nt upstream of the polypyrimidine tract (59,60). From the genome sequence, there are 8 HISH4 open reading frames organized in a tandem repeat; for the first seven genes the maximum predicted length of the mRNA [without the 39-nt spliced leader and the poly(A) tail] is about 600 nt. For the last gene of the cluster, the prediction is for about 500 nt. Deadenylated HISH4 mRNA migrated as 6 bands between 0.55 and 0.65 kb (Figure 4A, lanes 1 and 12) and the polyadenylated mRNA migrated between 0.775 and 0.575 kb, suggesting a maximum poly(A) tail length of about 100 nt (Figure 4A, lane 3). The choice of poly(A) site was unaffected by CAF1 depletion (Figure 4A, compare lanes 13 and 14). As previously demonstrated (36), depletion of the 5′->3′ exonuclease XRNA resulted in an increase of shorter HISH4 mRNA species (Figure 4A, lane 2). Depletion of DHH1 had no effect (Figure 4A, lane 5), but CAF1 RNAi resulted in a clear increase in the average mRNA size, as expected for an inhibition of deadenylation (Figure 4A, lane 4). Thirty minutes after addition of Actinomycin D, shortening of the HISH4 mRNA was clearly visible (Figure 4A, lane 6); this was unaffected by depletion of DHH1 (lane 8) but inhibited by CAF1 RNAi (lane 7). This difference persisted after 60 min Actinomycin D. Notably, deadenylation was not complete (compare lane 11 with lane 12); the results suggested that it ceased when 25–50 nt of poly(A) remained. A poly(A) of 27 nt is probably sufficient to bind a single molecule of poly(A) binding protein (61); further deadenylation presumably leads to linear RNA which lacks poly(A) binding protein and is susceptible to rapid degradation.
Results for more mRNAs are shown in Figure 4B, with quantitation in Figure 4C. In normal cells, degradation of TUB and PGKC mRNAs started after a delay of 30 min; decay of HISH4 and ACT mRNAs was faster. Depletion of CAF1 strongly inhibited the degradation of all four RNAs and inhibition of poly(A) tail shortening was evident. From these results, we concluded that degradation of constitutively expressed trypanosome mRNAs depends on CAF1. The results of experiments using a reporter mRNA produced by RNA polymerase I indicated that the effects of CAF1 depletion on mRNA degradation were not linked to transcription by RNA polymerase II (Supplementary Figure 10).
Depletion of CAF1 inhibits deadenylation in human cells
The clear importance of TbCAF1 in trypanosomes, combined with observations in the literature, suggested to us that Caf1 might be important in mammalian cells. We therefore repeated the poly(A) tail length measurements for cytoplasmic RNA in HTGM5 cells, which are derived from the human HT1080 fibrosarcoma cell line (45). A typical autoradiogram shown in Figure 5A and quantitations are shown in Figure 5B and C. The overall distributions of poly(A) tail lengths cannot be compared, since Figure 3 shows total trypanosome RNA and Figure 5 is cytoplasmic mammalian RNA. Nevertheless, in the sample from HTGM5 cells there were substantially more poly(A) tails over 100 nt than in trypanosomes (compare Figure 5A, lane 1 with Figure 3A, lane 1). For human RNA, 29% of poly(A) tails were shorter than 40 nt, and 5.6% were in the 10–20-nt range (Figure 5B). In another recent analysis of human poly(A) tail lengths, done by solution hybridization, up to 25% of expressed genes appeared to have <30 nt of poly(A) tail on at least a quarter of transcripts (62).
To deplete human Ccr4 and Caf1, we targeted the two paralogues that have previously been identified in the human genome: Ccr4a, Ccr4b, Caf1a and Caf1b (Table 2) (30,63). The mRNA depletion was slightly more efficient for Ccr4 (Figure 6A) than for Caf1 (Figure 6B). Although depletion of Ccr4a and Ccr4b resulted in a very slight increase in average poly(A) lengths the differences were generally not statistically significant (Figure 5A, lane 2, and Figure 5B). Caf1a and Caf1b depletion (Figure 5A, lane 3), in contrast, caused a shift towards longer lengths that was particularly marked in the 100–150-nt range (Figure 5B, block 6). After a 4-h-Actinomycin D treatment, deadenylation occurred in both the control (lane 4) and Ccr4-depleted cells (lane 5), but was inhibited by Caf1 RNAi (Figure 5A, lane 6); in the Caf1-depleted cells, the proportion of short tails was less than in the control (Figure 5B, blocks 1–3) and those in the 100–150-nt range were again markedly more abundant (Figure 5B, block 6). These results confirmed that although Ccr4 may contribute, as previously suggested (7), depletion of Caf1 has a stronger effect on deadenylation of bulk mRNA in human HTGM5 cells. Since Caf1 depletion presumably results in dissociation of Ccr4 from the complex, the effects we see should reflect a failure to recruit both Ccr4 and Caf1 to susceptible mRNAs.
Depletion of Caf1 inhibits degradation of an unstable mammalian mRNA
In mammalian cells, the rapid degradation of mRNAs containing an AU-rich element (ARE) is initiated by poly(A) tail shortening (2). We therefore analysed the effects of Caf1 depletion in human HTGM5 cells that stably express a β-globin reporter gene with the ARE of granulocyte-macrophage colony-stimulating factor in the 3′UTR (45). Knocking down Ccr4a and Ccr4b (Figure 6C, lanes 1and 2) did not affect expression the of the globin-ARE reporter mRNA. The amount of poly(A)+ globin-ARE mRNA was quantified (Figure 6D) and the result suggested that neither Ccr4a nor Ccr4b are limiting factors in the deadenylation of the ARE-containing reporter mRNA.
We then tested the effect of knocking down Caf1a and Caf1b, either alone or in combination. This was done either using two different, gene-specific siRNAs, or by using a single siRNA (C7) that targets a conserved region in both mRNAs. The same total amounts of siRNA were used throughout; the knockdown was most effective for Caf1a (Figure 6B). At steady state, knocking down Caf1a had no effect (Figure 6C and D, lane 4). Targeting Caf1b (lane 5), or both Caf1s together (lanes 6 and 7), gave a small increase in poly(A)+ globin-ARE mRNA. The effects were more pronounced after treatment with Actinomycin D for 1 hour. Knocking down Caf1b alone gave a 1.4-fold increase in poly(A)+ globin-ARE mRNA (lane 12); despite the relatively low siRNA efficiency, whereas knocking down both together resulted in a 2- to 2.5-fold increase (lanes 13 and 14). In each experiment, after the 1-h Actinomycin D treatment the mRNA level decreased more in the control cells than in the cells with Caf1a+b RNAi.
The results so far suggested that Caf1a and Caf1b are redundant, and play a role in degradation of the globin-ARE mRNA. We next analysed the effects of Caf1a+b RNAi on degradation in more detail. Results are shown in Figure 7. The globin-ARE mRNA decayed with a half-life of <1 h, and Ccr4a+b siRNA treatment had very little effect on degradation; in contrast, Caf1a+b siRNA treatment delayed deadenylation and allowed persistence of 50% of the mRNA 2–3 h after Actinomycin D treatment (Figure 6B). This confirmed that depletion of Caf1a+b inhibited globin-ARE mRNA degradation.
DISCUSSION
The composition of the trypanosome CAF1/NOT complex
Our results indicate that trypanosomes have a complex which contains TbCAF1, TbNOT1, TbNOT2, TbNOT5, TbDHH1 and possibly two more subunits, encoded by Tb10.6k15.1820 (a possible equivalent of Caf130) and Tb927.8.1960. We do not yet know whether possible Caf16 and Caf40 homologues are associated with the complex, but if so, they must be sub-stoichiometric.
In yeast, mammalian cells and Drosophila, a Ccr4 subunit is an integral part of the complex. The only possible homologue of Ccr4 in trypanosomes (and other Kinetoplastids) lacked the leucine-rich domain that is required for interaction with Caf1 and for deadenylase activity, and did not co-purify with the complex. Therefore, all evidence suggests that trypanosomes and other Kinetoplastids lack an equivalent to Ccr4. This absence might indicate that the ancestral complex lacked a Ccr4 subunit. To investigate this possibility we looked for Ccr4 homologues, defined as predicted proteins including the leucine-rich repeat in addition to the exonuclease domain, in the six major eukaryotic groups (32). Yeasts and animals, which contain Ccr4, belong in one group, the Opisthokonta; Plantae (higher plants, algae) also have Ccr4. We could not find any complete genomes from the Rhizaria. Trypanosomes are currently usually grouped in the Excavata, with Giardia intestinalis and Trichomonas vaginalis, but molecular evidence for this group is lacking (32). Giardia has no Ccr4 but Trichomonas has a homologue. We found no likely Ccr4 in Tetrahymena or Plasmodium (Chromalveolata) or in Dictyostelium and Entamoeba (Amoebozoa). From this limited dataset, therefore, it seems unlikely that Ccr4 was a component of the ancestral Caf1/Not complex.
The roles of CAF1 in deadenylation
Deadenylation in eukaryotes has mostly been studied focusing on transcripts which are relatively unstable, either because they contain destabilizing sequences in the 3′-UTR, or because the open reading frame contains a premature termination codon. In experiments with mammalian cells, stable transcripts have half-lives of many hours; their degradation has so far only been analysed on an individual basis, using inducible promoters. A transgene mRNA with a beta-globin 3′-UTR, for example, decreased only by 10% over the first 10 h after transcriptional shut-off (64). In contrast, the previous report that CAF1 depletion results in an increase in the average poly(A) tail length in Drosophila cells (15) implies that Drosophila CAF1 plays a major role in bulk mRNA deadenylation.
To determine the overall contribution of Caf1 and Ccr4 homologues to deadenylation we analysed the overall distribution of poly(A) tail lengths. In both trypanosomes and human cells, a reduction in CAF1 (Caf1a+b) activity increased the average lengths of poly(A) tails and inhibited deadenylation (Figures 3 and 5). In human cells, Ccr4 depletion had, at best, a marginal effect on poly(A) tail lengths. The overall poly(A) tail distributions in these figures cannot be compared, since the experiments were done with total trypanosome RNA, but with cytoplasmic human RNA. Nevertheless, it was notable that the longest poly(A) tails were much shorter in trypanosomes than in mammals. Perhaps this is an intrinsic property of the trypanosome polyadenylation complex, but we favour another explanation: that at steady state, a large proportion of trypanosome mRNAs is undergoing degradation. Even ‘stable’ mRNAs in trypanosomes have much shorter half-lives than are generally seen in mammalian cells—compare for example trypanosome PGKC and TUB with mammalian cyclophilin RNA—and polycistronic transcription creates numerous unwanted mRNAs which are rapidly destroyed.
We also examined the effects of Caf1 depletion on deadenylation and decay of individual mRNAs. When we depleted CAF1 in trypanosomes, the four mRNAs studied—PGKC, HISH4, TUB and ACT—were deadenylated more slowly and were stabilized (Figure 4). They did not, paradoxically, show an increase in abundance relative to rRNA, which is very stable and should be unaffected by the RNAi. The simplest explanation is that inhibition of bulk mRNA decay caused feedback inhibition of transcription by RNA polymerase II. This could result from a specific regulatory mechanism; alternatively, ribonucleotides produced by mRNA degradation might be a significant source of substrate for mRNA synthesis. In other eukaryotes also, inhibition of mRNA degradation does not always affect mRNA abundance (45,65–68); for example, in yeast lacking Ccr4p, there is no general increase in the abundances of unstable mRNAs, and although mRNAs encoding ribosomal proteins are markedly stabilized their steady-state levels are little affected (67). Thus, this sort of feedback regulation may be widespread, although the mechanism(s) are unknown.
Depletion of CAF1 in trypanosomes also had no effect on the abundances of four highly unstable mRNAs—PGKC, GPIPLC and ISG65 in procyclic forms and PGKB in bloodstream forms. We have previously shown that depletion of XRNA does, in contrast, cause an increase in the steady-state levels of such mRNAs (36). Perhaps for these mRNAs, the CAF1-independent, XRNA-dependent mechanism of mRNA decay predominates; but since bulk mRNA decay is not much affected, transcription continues unabated.
In human cells, depletion of both Ccr4 homologues did not affect the abundance (Figure 6) or degradation (Figure 7) of an unstable mRNA that contains an ARE. In contrast, simultaneous depletion of Caf1b and Caf1a inhibited degradation of the ARE mRNA (Figures 6 and 7). We note that interpretation of these results is not straightforward. All of our experiments were subject to the inevitable weakness of RNAi: that the depletion of the target mRNA was not complete. Thus, residual deadenylase activity in CAF1- (or Caf1-)depleted cells might be attributable either to remaining Caf1, or to alternative deadenylases or both. Similarly, the absence of an effect of Ccr4a/b depletion on ARE mRNA decay does not necessarily mean that the proteins have a minor role, but could merely be due to persistence of sufficient protein to give wild-type function. In addition, after Ccr4a+b RNAi, cells retain a Caf1–Not complex. In contrast, since Ccr4 is linked to Not1 via Caf1, depletion of Caf1 is expected to result in cells with a Not complex lacking both Ccr4 and Caf1. Even if Ccr4 persists in the cells, its recruitment to ARE mRNA may depend on attachment to Not1. Our experiments therefore do not allow us to tell whether a complex with Ccr4 activity, but without Caf1 activity, such as is found in yeast, would be fully functional in mammalian deadenylation. Nevertheless, the clear effects after depletion of both Caf1 homologues indicate that Caf1a and Caf1b have redundant but important roles in ARE-mediated mRNA decay.
In vitro decay studies had previously shown that another deadenylase, PARN, can promote the deadenylation of an ARE-containing RNA (69). Future studies will be required to reveal whether this is also true in vivo, and whether PARN may cooperate with Caf1a/b. The analysis of nonsense-mediated mRNA decay in mouse NIH3T3 fibroblasts suggested that a reporter mRNA containing a premature termination codon (PTC) is initially deadenylated by Pan2 to a length of about 110 adenosine residues, followed by a second phase during which Ccr4a/b digests the remaining poly(A) tail (7). Since the contribution of Caf1a/b was not addressed in this study, it is not clear whether Caf1a/b also participates in the deadenylation of a PTC-containing mRNA.
The fact that RNAi targeting CAF1 in T. brucei was lethal, with clear effects on deadenylation, shows that in trypanosomes other deadenylases are not able to provide essential CAF1 functions. Since our experiments gave similar results for organisms from entirely different branches of eukaryotic evolution, we suggest that CAF1 may be a major deadenylase in eukaryotes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (DFG Cl112/9) (A.S.) and the Wellcome Trust (L.E. and M.C.). We thank Ute Leibfried (ZMBH) and Jochen Kreth (DKFZ) for technical assistance. G.S. was supported by a Young Investigators Grant from the Helmholtz Association of German Research Centres. The page charges were paid from the Grundausstattung (State of Baden Württemberg) of the Clayton laboratory.
Conflict of interest statement. None declared.
REFERENCES
- 1.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 2.Chen CY, Xu N, Shyu AB. mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol. Cell. Biol. 1995;15:5777–5788. doi: 10.1128/mcb.15.10.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J, Chiang Y.-C, Denis CL. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 2002;21:1414–1426. doi: 10.1093/emboj/21.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tucker M, Staples RR, Valencis-Sanchez MA, Muhlrad D, Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 6.Körner CG, Wormington M, Muckenthaler M, Schneider S, Dehlin E, Wahle E. The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes. EMBO J. 1998;17:5427–5437. doi: 10.1093/emboj/17.18.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, Shyu AB. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- 8.Bai Y, Salvadore C, Chiang YC, Collart MA, Liu HY, Denis CL. The CCR4 and CAF1 proteins of the CCR4-NOT complex are physically and functionally separated from NOT2, NOT4, and NOT5. Mol. Cell. Biol. 1999;19:6642–6651. doi: 10.1128/mcb.19.10.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Draper M, Liu H.-Y, Nelsbach A, Mosley S, Denis C. CCR4 is a glucose-regulated transcription factor whose leucine-rich repeat binds several proteins important for placing CCR4 in its proper promoter context. Mol. Cell. Biol. 1994;14:4522–4531. doi: 10.1128/mcb.14.7.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark LB, Viswanathan P, Quigley G, Chiang YC, McMahon JS, Yao G, Chen J, Nelsbach A, Denis CL. Systematic mutagenesis of the leucine-rich repeat (LRR) domain of CCR4 reveals specific sites for binding to CAF1 and a separate critical role for the LRR in CCR4 deadenylase activity. J. Biol. Chem. 2004;279:13616–13623. doi: 10.1074/jbc.M313202200. [DOI] [PubMed] [Google Scholar]
- 11.Dupressoir A, Morel A.-P, Barbot W, Loireau M.-P, Corbo L, Heidmann T. Identification of four families of yCCR4- and Mg2+-dependent endonuclease-related proteins in higher eukaryotes, and characterization of orthologs of yCCR4 with a conserved leucine-rich repeat essential for hCAF1/hPOP2 binding. BMC Genomics. 2001;2:9. doi: 10.1186/1471-2164-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Albert T, Hanzawa H, Legtenberg Y.IA, deRuwe MJ, van der Heuvel F.AJ, Collart MA, Boelens R, Timmers M.HT. Identification of a ubiquitin–protein ligase subunit within the CCR4-NOT transcription repressor complex. EMBO J. 2002;21:355–364. doi: 10.1093/emboj/21.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russell P, Benson J, Denis C. Characterization of mutations in NOT2 indicates that it plays an important role in maintaining the integrity of the CCR4 – NOT Complex. J. Mol. Biol. 2002;322:27–39. doi: 10.1016/s0022-2836(02)00707-6. [DOI] [PubMed] [Google Scholar]
- 14.Collart M, Timmers H. The eukaryotic Ccr4-Not complex: a regulatory platform Integrating mRNA metabolism with cellular signaling pathways? Prog. Nucleic Acid Res. Mol. Biol. 2004;77:289. doi: 10.1016/S0079-6603(04)77008-7. [DOI] [PubMed] [Google Scholar]
- 15.Temme C, Zaessinger S, Meyer S, Simonelig M, Wahle E. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J. 2004;23:2862–2871. doi: 10.1038/sj.emboj.7600273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okazaki N, Okazaki K, Watanabe Y, Kato-Hayashi M, Yamamoto M, Okayama H. Novel factor highly conserved among eukaryotes controls sexual development in fission yeast. Mol. Cell. Biol. 1998;18:887–895. doi: 10.1128/mcb.18.2.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsukahara K, Yamamoto H, Okayama H. An RNA binding protein negatively controlling differentiation in fission yeast. Mol. Cell. Biol. 1998;18:4488–4498. doi: 10.1128/mcb.18.8.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, Rappsilber J, Chiang YC, Russell P, Mann M, Denis CL. Purification and characterization of the 1.0 MDa CCR4-NOT complex identifies two novel components of the complex. J. Mol. Biol. 2001;314:683–694. doi: 10.1006/jmbi.2001.5162. [DOI] [PubMed] [Google Scholar]
- 19.Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hata H, Mitsui H, Liu H, Bai Y, Denis CL, Shimizu Y, Sakai A. Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics. 1998;148:571–579. doi: 10.1093/genetics/148.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maillet L, Collart MA. Interaction between Not1p, a component of the Ccr4-not complex, a global regulator of transcription, and Dhh1p, a putative RNA helicase. J. Biol. Chem. 2002;277:2835–2842. doi: 10.1074/jbc.M107979200. [DOI] [PubMed] [Google Scholar]
- 22.Goldstrohm AC, Seay DJ, Hook BA, Wickens M. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J. Biol. Chem. 2006;282:109–114. doi: 10.1074/jbc.M609413200. [DOI] [PubMed] [Google Scholar]
- 23.Ohn T, Chiang Y.-C, Lee D, Yao G, Zhang C, Denis C. CAF1 plays an important role in mRNA deadenylation separate from its contact to CCR4. Nucleic Acids Res. 2007;35:3002–3015. doi: 10.1093/nar/gkm196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Viswanathan P, Ohn T, Chiang YC, Chen J, Denis CL. Mouse CAF1 can function as a processive deadenylase/3′-5′-exonuclease in vitro but in yeast the deadenylase function of CAF1 is not required for mRNA poly(A) removal. J. Biol. Chem. 2004;279:23988–23995. doi: 10.1074/jbc.M402803200. [DOI] [PubMed] [Google Scholar]
- 25.Daugeron MC, Mauxion F, Seraphin B. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 2001;29:2448–2455. doi: 10.1093/nar/29.12.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakai A, Chibazakura Y, Shimizu Y, Hishinuma F. Molecular analysis of the POP2 gene, a gene required for glucose repression of Saccharomyces cerevisiae. Nucleic Acids Res. 1992;20:6227–6233. doi: 10.1093/nar/20.23.6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denis CL, Malvar T. The CCR4 gene from Saccharomyces cerevisiae is required for both nonfermentative and spt-mediated gene expression. Genetics. 1990;124:283–291. doi: 10.1093/genetics/124.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molin L, Puisieux A. C. elegans homologue of the Caf1 gene, which encodes a subunit of the CCR4-NOT complex, is essential for embryonic and larval development and for meiotic progression. Gene. 2005;358:73–81. doi: 10.1016/j.gene.2005.05.023. [DOI] [PubMed] [Google Scholar]
- 29.Maeda I, Kohara Y, Yamamoto M, Sugimoto A. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr. Biol. 2001;11:171–176. doi: 10.1016/s0960-9822(01)00052-5. [DOI] [PubMed] [Google Scholar]
- 30.Albert TK, Lemaire M, van Berkum NL, Gentz R, Collart MA, Timmers HT. Isolation and characterization of human orthologs of yeast CCR4-NOT complex subunits. Nucleic Acids Res. 2000;28:809–817. doi: 10.1093/nar/28.3.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berthet C, Morera A.-M, Asensio M.-J, Chauvin M.-A, Morel A.-P, Dijoud F, Magaud J.-P, Durand P, Rouault J.-P. CCR4-associated factor CAF1 Is an essential factor for spermatogenesis. Mol. Cell. Biol. 2004;24:5808–5820. doi: 10.1128/MCB.24.13.5808-5820.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parfrey L, Barbero E, Lasser E, Dunthorn M, Bhattacharya D.EA. Evaluating support for the current classification of eukaryotic diversity. PLoS Genet. 2006;2:e220. doi: 10.1371/journal.pgen.0020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clayton C, Shapira M. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol. Biochem. Parasitol., 2007;156:93–101. doi: 10.1016/j.molbiopara.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 34.Estévez AM, Lehner B, Sanderson CM, Ruppert T, Clayton C. The roles of inter-subunit interactions in exosome stability. J. Biol. Chem. 2003;278:34943–34951. doi: 10.1074/jbc.M305333200. [DOI] [PubMed] [Google Scholar]
- 35.Haile S, Estévez AM, Clayton C. A role for the exosome in the initiation of degradation of unstable mRNAs. RNA. 2003;9:1491–1501. doi: 10.1261/rna.5940703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li C-H, Irmer H, Gudjonsdottir-Planck D, Freese S, Salm H, Haile S, Estévez AM, Clayton CE. Roles of a Trypanosoma brucei 5′->3′ exoribonuclease homologue in mRNA degradation. RNA. 2006;12:2171–2186. doi: 10.1261/rna.291506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milone J, Wilusz J, Bellofatto V. Characterization of deadenylation in trypanosome extracts and its inhibition by poly(A)-binding protein Pab1p. RNA. 2004;10:448–457. doi: 10.1261/rna.5180304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biebinger S, Wirtz LE, Clayton CE. Vectors for inducible over-expression of potentially toxic gene products in bloodstream and procyclic Trypanosoma brucei. Mol. Biochem. Parasitol. 1997;85:99–112. doi: 10.1016/s0166-6851(96)02815-0. [DOI] [PubMed] [Google Scholar]
- 39.Alibu VP, Storm L, Haile S, Clayton C, Horn D. A doubly inducible system for RNA interference and rapid RNAi plasmid construction in Trypanosoma brucei. Mol. Biochem. Parasitol. 2004;139:75–82. doi: 10.1016/j.molbiopara.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 40.Wirtz E, Hoek M, Cross G.AM. Regulated processive transcription of chromatin by T7 RNA polymerase in Trypanosoma brucei. Nucleic Acids Res. 1998;26:4626–4634. doi: 10.1093/nar/26.20.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clayton CE, Estévez AM, Hartmann C, Alibu VP, Field M, Horn D. Down-regulating gene expression by RNA interference in Trypanosoma brucei. Methods Mol. Biol. 2005;309:39–60. doi: 10.1385/1-59259-935-4:039. [DOI] [PubMed] [Google Scholar]
- 42.Quijada L, Hartmann C, Guerra-Giraldez C, Drozdz M, Irmer H, Clayton CE. Expression of the human RNA-binding protein HuR in Trypanosoma brucei induces differentiation-related changes in the abundance of developmentally-regulated mRNAs. Nucleic Acids Res. 2002;30:1–11. doi: 10.1093/nar/gkf577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Estévez A, Kempf T, Clayton CE. The exosome of Trypanosoma brucei. EMBO J. 2001;20:3831–3839. doi: 10.1093/emboj/20.14.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Redmond S, Vadivelu J, Field MC. RNAit: an automated web-based tool for the selection of RNAi targets in Trypanosoma brucei. Mol. Biochem. Parasitol. 2003;128:115–118. doi: 10.1016/s0166-6851(03)00045-8. [DOI] [PubMed] [Google Scholar]
- 45.Stoecklin G, Mayo T, Anderson P. ARE-mRNA degradation requires the 5'–3' decay pathway. EMBO Rep. 2005;7:72–77. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall W, Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 47.Gough N. Rapid and quantitative preparation of cytoplasmic RNA from small numbers of cells. Anal. Biochem. 1988;173:93–95. doi: 10.1016/0003-2697(88)90164-9. [DOI] [PubMed] [Google Scholar]
- 48.Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby W.FC, Blackwell TK, Anderson P. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004;23:1313–1324. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Das A, Li H, Liu T, Bellofatto V. Biochemical characterization of Trypanosoma brucei RNA polymerase II. Mol. Biochem. Parasitol. 2006;150:201–210. doi: 10.1016/j.molbiopara.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 50.Hartmann C, Benz C, Brems S, Ellis L, Luu V.-D, Stewart M, D’Orso I, Busold C, Fellenberg K, Frasch A.CC, et al. The small trypanosome RNA-binding proteins TbUBP1 and TbUBP2 influence expression of F box protein mRNAs in bloodstream trypanosomes. Eukaryot. Cell. 2007;6:1964–1978. doi: 10.1128/EC.00279-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cassola A, De Gaudenzi J, Frasch A. Recruitment of mRNAs to cytoplasmic ribonucleoprotein granules in trypanosomes. Mol. Microbiol. 2007;65:655–670. doi: 10.1111/j.1365-2958.2007.05833.x. [DOI] [PubMed] [Google Scholar]
- 52.Hotz H-R, Hartmann C, Huober K, Hug M, Clayton CE. Mechanisms of developmental regulation in Trypanosoma brucei: a polypyrimidine tract in the 3'-untranslated region of a trypanosome surface protein mRNA affects RNA abundance and translation. Nucleic Acids Res. 1997;25:3017–3025. doi: 10.1093/nar/25.15.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hug M, Carruthers V, Sherman D, Hartmann C, Cross G.AM, Clayton CE. A possible role for the 3′-untranslated region in developmental regulation in Trypanosoma brucei. Mol. Biochem. Parasitol. 1993;61:87–96. doi: 10.1016/0166-6851(93)90161-p. [DOI] [PubMed] [Google Scholar]
- 54.Colasante C, Robles A, Li C.-H, Schwede A, Benz C, Voncken F, Guilbride DL, Clayton C. Regulated expression of glycosomal phosphoglycerate kinase in Trypanosoma brucei. Mol. Biochem. Parasitol. 2007;151:193–204. doi: 10.1016/j.molbiopara.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 55.Webb H, Burns R, Ellis L, Kimblin N, Carrington M. Developmentally regulated instability of the GPI-PLC mRNA is dependent on a short-lived protein factor. Nucleic Acids Res. 2005;33:1503–1512. doi: 10.1093/nar/gki298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ziegelbauer K, Overath P. Identification of invariant surface glycoproteins in the bloodstream stage of Trypanosoma brucei. J. Biol. Chem. 1992;267:10791–10796. [PubMed] [Google Scholar]
- 57.Ziegelbauer K, Multhaup G, Overath P. Molecular characterisation of two invariant surface glycoproteins specific for the bloodstream stage of Trypanosoma brucei. J. Biol. Chem. 1992;267:10797–10803. [PubMed] [Google Scholar]
- 58.Cristodero M, Clayton C. Trypanosome MTR4 in involved in ribosomal RNA processing. Nucleic Acids Res. 2007;35:7023–30. doi: 10.1093/nar/gkm736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liang X, Haritan A, Uliel S, Michaeli S. Trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot. Cell. 2003;2:830–840. doi: 10.1128/EC.2.5.830-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benz C, Nilsson D, Andersson B, Clayton C, Guilbride DL. Messenger RNA processing sites in Trypanosoma brucei. Mol. Biochem. Parasitol. 2005;143:125–134. doi: 10.1016/j.molbiopara.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 61.Baer B, Kornberg R. Repeating structure of cytoplasmic poly(A)-ribonucleoprotein. Proc. Natl Acad. Sci. USA. 1980;77:1890–1892. doi: 10.1073/pnas.77.4.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meijer H, Bushell M, Hill K, Gant T, Willis A, Jones P, de Moor C. A novel method for poly(A) fractionation reveals a large population of mRNAs with a short poly(A) tail in mammalian cells. Nucleic Acids Res. 2007;35:e132. doi: 10.1093/nar/gkm830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wagner E, Clement S, Lykke-Andersen J. An unconventional human Ccr4-Caf1 deadenylase complex in nuclear Cajal bodies. Mol. Cell. Biol. 2007;27:1686. doi: 10.1128/MCB.01483-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Dijk E, Schilders G, Pruijn G. Human cell growth requires a functional cytoplasmic exosome, which is involved in various mRNA decay pathways. RNA. 2007;13:1027–1035. doi: 10.1261/rna.575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muhlrad D, Parker R. Recognition of yeast mRNAs as “nonsense containing” leads to both inhibition of mRNA translation and mRNA degradation: implications for the Control of mRNA decapping. Mol. Biol. Cell. 1999;11:3971–3978. doi: 10.1091/mbc.10.11.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5' to 3' mRNA decay pathways in yeast. Mol. Cell. 2003;12:1439–1452. doi: 10.1016/s1097-2765(03)00446-5. [DOI] [PubMed] [Google Scholar]
- 67.Grigull J, Mnaimneh S, Pootoolal J, Robinson M, Hughes T. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol. Cell. Biol. 2004;24:5534–5547. doi: 10.1128/MCB.24.12.5534-5547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lai W, Parker J, Grissom S, Stumpo D, Blackshear P. Novel mRNA targets for Tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP-deficient fibroblasts. Mol. Cell. Biol. 2006;26:9196–9208. doi: 10.1128/MCB.00945-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lai WS, Kennington EA, Blackshear PJ. Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Mol. Cell. Biol. 2003;23:3798–3812. doi: 10.1128/MCB.23.11.3798-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.