Abstract
Protein arginine methyltransferases (PRMT) have been implicated in the regulation of transcription. They are recruited to promoters via interaction with transcription factors and exert their coactivator function by methylating arginine residues in histones and other chromatin proteins. Here, we employ an unbiased approach to identify novel target genes, which are under the control of two members of the enzyme family, PRMT1 and CARM1/PRMT4 (coactivator associated arginine methyltransferase 1). By using cDNA microarray analysis, we find that the siRNA-mediated single knockdown of neither CARM1 nor PRMT1 causes significant changes in gene expression. In contrast, double knockdown of both enzymes results in the deregulated expression of a large group of genes, among them the CITED2 gene. Cytokine-stimulated expression analysis indicates that transcriptional activation of CITED2 depends on STAT5 and the coactivation of both PRMTs. ChIP analysis identifies the CITED2 gene as a direct target gene of STAT5, CARM1 and PRMT1. In reporter gene assays, we show that STAT5-mediated transcription is cooperatively enhanced by CARM1 and PRMT1. Interaction assays reveal a cytokine-induced association of STAT5 and the two PRMTs. Our data demonstrate a widespread cooperation of CARM1 and PRMT1 in gene activation as well as repression and that STAT5-dependent transcription of the CITED2 gene is a novel pathway coactivated by the two methyltransferases.
INTRODUCTION
Protein arginine methylation is a covalent posttranslational modification carried out by a family of enzymes, the PRMTs (protein arginine methyltransferases), which are evolutionary conserved in eukaryotes from fungi to plants and mammals (1). In humans, the PRMT family consists of nine members (2,3). PRMTs use S-adenosylmethionine (SAM) as methyl donor and add one or two methyl groups onto the guanidino nitrogens of the arginine residue. The majority of PRMTs catalyse the formation of asymmetric dimethyl-arginine by transferring both methyl groups to the same nitrogen atom, whereas PRMT5 and PRMT7 generate symmetric dimethylation (2). Unmodified and monomethyl arginines can be either converted by the peptidylarginine deiminases PAD4 to citrulline or demethylated by the Jumonji domain-containing protein JMJD6 (4–6).
As reflected by the diversity of their substrates, PRMTs are implicated in the regulation of a plethora of cellular processes, among which regulation of chromatin-related processes has been well studied (2). Chromatin-related functions of PRMTs comprise regulation of DNA-repair, imprinting and gene expression. PRMT1 and PRMT6 influence nucleotide excision repair by modification of DNA polymerase β (7,8). PRMT7 plays a role in male-specific imprinting (9). Trancriptional regulation is conducted by several family members: PRMT1, CARM1/PRMT4 (coactivator associate arginine methyltransferase 1), PRMT5, and PRMT6 (10–14). To fulfil their function in gene expression the enzymes are recruited via interaction with transcription factors to target promoters, where they affect gene activity in a methyltransferase-dependent manner and modify histones and other chromatin-associated proteins. Promoter binding of PRMT5 associates with transcriptional repression (15,16), whereas CARM1 and PRMT1 have been identified as individual coactivators for a number of transcription factors. For example, CARM1 enhances NFkB- (17), MEF2- (18) and βCatenin/TCF-mediated gene expression (19) and PRMT1 is a coactivator for YY1 (20). Recent findings suggest that PRMT1 and CARM1 not only synergize in their transcriptional function with one another but also with histone acetyltransferases, e.g. in gene regulation by nuclear hormone receptors (NR) (21–23) and p53 (24).
Whereas the molecular impact of arginine methylation of histones is still obscure, arginine methylation of other non-histone chromatin proteins has been shown to control assembly or disassembly of coactivator and corepressor complexes. For example, methylation of the transcriptional coregulators CBP/p300 (25), SRC-3 (26,27), PGC-1 (28), NIP45 (29) and RIP140 (30) by CARM1, PRMT1, respectively modulates their interactions with other proteins. This is in agreement with the observation that methylation increases the hydrophobicity of the arginine and hence alters intra- as well as intermolecular interactions (1).
To study individual and cooperative functions of CARM1 and PRMT1 in gene expression in greater detail and to identify novel signalling pathways, which are transcriptionally regulated by these two enzymes, we carried out a screen for novel target genes. We established short interfering RNA (siRNA)-mediated single and double knockdowns for CARM1 and/or PRMT1 in HeLa cells and performed a cDNA microarray expression profiling. Whereas comparison of wild-type and single knockdown cells showed no differential expression pattern, a group of 46 genes were significantly deregulated in the CARM1 and PRMT1 double knockdown cells suggesting a widespread cooperation of the two enzymes in the control of gene expression. For the CITED2 gene, which was downregulated upon codepletion of both PRMTs in the microarray analysis, we found that its expression is activated by interleukin-4 (IL-4) stimulation in a STAT5-dependent manner. By means of ChIP (chromatin immuoprecipitation) analysis, reporter gene assays and interaction studies we show that CARM1 and PRMT1 are coactivators of the STAT5-mediated transcription of the CITED2 gene upon IL-4-stimulation.
MATERIALS AND METHODS
Cell lines and antibodies
Hela cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS, Gibco/BRL) at 37°C and 5% CO2. Cells were treated with 20 ng/ml interleukin-4 (IL-4, recombinant, Stratmann, Hamburg). The following antibodies were employed: anti-PRMT1 from Upstate (07-404), anti-PRMT1 generated in rabbits against recombinant human PRMT1 protein (aa 1–343), anti-CARM1 from Upstate (07-080), anti-CARM1 generated in rabbits against recombinant murine PRMT4 protein (aa 433–608), anti-H4 R3me2 from Upstate (07-213), anti-H3 R17me2 from Abcam (ab8284), anti-STAT5a/b from Santa Cruz, CA, USA (sc-835), anti-phospho Tyr694/699 STAT5 from Cell Signaling (9351), anti-STAT6 from Santa Cruz (sc-981), anti-phospho Tyr641 STAT6 from Cell Signaling (9361) and anti-β tubulin (MAB3408) from Chemicon, Billerica, MA, USA.
Plasmids
The pSG5-HA mCARM1 and pGEX-4T1 mCARM1 has been previously published (31). The pGEX-4T1 hPRMT1 was published by ref. (32) and pcDNA3.1 hPRMT1 by ref. (33). The pGL3 (STAT5-RE)6-luc was a gift from Iris Behrmann. The pECE-STAT5b was described by ref. (34).
siRNA transfections
The siRNA oligonucleotide duplexes were purchased from Dharmacon for targeting the human PRMT1, CARM1, STAT5 and STAT6 transcripts, respectively in HeLa cells. The siRNA against STAT5 does not distinguish between STAT5a and b. The siRNA sequences are (sense strand indicated): siPRMT1_1 5′-CGUGUAUGGCUUCGACAUG-3′, siPRMT1_2 5′-UCAAAGAUGUGGCCAUUAA-3′, siCARM1_1 5′-CAUGAUGCAGGACUACGUG-3′, siCARM1_2 5′-GGACAUGUCUGCUUAUUGC-3′, siSTAT5 5′-GCAGCAGACCAUCAUCCUG-3′, siSTAT6 5′-GAUGUGUGAAACUCUGAAC-3′ and siNON-targeting 5′-UUGAUGUGUUUAGUCGCUA-3′. siRNA duplexes (80 nM final concentration) were transfected with Oligofectamine (Invitrogen, Karlsruhe, Germany) for 2 days. Afterwards, cells were treated without or with stimulus (IL-4) and harvested for RNA or protein preparation.
cDNA microarray hybridization
Total RNA was prepared from siRNA-treated HeLa cells using the peqGOLD total RNA kit (PeqLab, Erlangen, Germany). One microgram of RNA was amplified using the MessageAmpTM II aRNA Amplification kit (Ambion, Austin, TX, USA). Amplified RNA measuring 0.8–2 μg were employed to synthesize Cy3 and Cy5-labelled cDNA using random nonamer primers. We used a cDNA array, which comprised the Human Sequence-verified cDNA Unigene Set gf200, gf201u and gf202 gene sets (ResGen/Invitrogen/Cat.No.97001.V) covering 11 552 sequence-verified cDNAs. Each experiment was performed as sandwich hybridization, i.e., instead of a coverslip, a second microarray slide was used. This provides a replicated measurement for each hybridization that can be used for quality control and to reduce technical variability. For each condition, we generated four sandwich experiments. For each spot, median signal and background intensities for both channels were obtained. The background-corrected ratio of the two channels was calculated and log2 transformed. To balance the fluorescence intensities for the two dyes, as well as to allow for comparison of expression levels across experiments, the raw data were standardized. We used the printtip-lowess normalization to correct for inherent bias on each chip (35). Expression data and gene annotations were stored in Array Express accession no. E-MEXP-1173 (http://www.ebi.ac.uk/arrayexpress/), which complies with MIAME (minimal information about a microarray experiment) guidelines. The R environment (http://www.r-project.org/) was used for gene filtering and normalization of the data.
Reverse transcription (RT) and quantitative PCR (QPCR)
Total RNA from HeLa cells was isolated using the peqGOLD total RNA kit (PeqLab). Two micrograms of RNA were applied to RT by incubation with 0.5 μg oligo(dT)17 primer and 100 U M-MLV reverse transcriptase (Invitrogen). cDNA was analysed by QPCR, which was performed using SybrGreen (Bioline, Luckenwalde, Germany) and the Mx3000P real-time detection system (Stratagene, La Jolla, CA/USA). For RT–QPCR we used the following primers: hGAPDH forward 5′-AGCCACATCGCTCAGACAC-3′ and reverse 5′-GCCCAATACGACCAAATCC-5′, hCARM1 forward 5′-CACACCGACTTCAAGGACAA-3′ and reverse 5′-AAAAACGACAGGATCCCAGA-3′, hPRMT1 forward 5′-GAGAATTTTGTAGCCACCTTGG-3′ and reverse 5′-CCTGGCCACAGGACACTT-3′, hKRT8 forward 5′-AACGAATTTGTCCTCATCAAGAA-3′ and reverse 5′-GTTGATCTCGTCGGTCAGC-3′, hCITED2 forward 5′-TCACTTTCAAGTTGGCTGTCC-3′ and reverse 5′-CATTCCACACCCTATTATCATCTGT-3′. All amplifications were performed in triplicates using 0.5 μl cDNA per reaction. The triplicate mean values were calculated according to the ΔΔct quantification method (36) using the GAPDH gene transcription as reference for normalization. Standard deviation was calculated from the triplicates. Error bars are accordingly indicated. Gene transcription was expressed as fold increase in mRNA level, whereas the mRNA level in uninduced control (siNON-targeting) cells was equated 1 and all other values were expressed relative to this. The represented RT–QPCR assays were reproduced at least three times in independent experiments with the same results. Representative data sets are shown.
ChIP
Cells were crosslinked in the presence of 1% formaldehyde at 37°C for 10 min and harvested after twice washing with cold PBS. ChIP was performed as previously described (22). Immunoprecipitated and eluted DNA was purified with QIAquick columns (Qiagen, Hilden, Germany) and amplified by QPCR with the following primers: the CITED2 promoter region (nt −1324 to −1054, encompassing the STAT5 binding site) forward 5′-GCCCAGACCTGTGTTAGGGGTTT-3′ and reverse 5′-TGAGTAAGGCTGCTCTTGCTGGA-3′ and as control an intergenic promoter-free control region upstream of the CITED2 gene (nt −6835 to −6493) forward 5′-CTCAGAAGAGCCCAGTGTAGCA-3′ and reverse 5′-GGATGAGGTATGTTGGAAAGCAGA-3′. Amplifications were performed in triplicates and mean values were normalized to the values of chromatin enrichment by the control IgGs and expressed as fold control IgG, which was equated 1.
Reporter gene assay
HeLa cells were plated in 24-well dishes. After 24 h, cells were transiently transfected at a confluency of 50% with 2 μg of DNA per well (500 ng reporter plasmid and CMV-β-Gal, 300 ng of various expression plasmids filled up with mock plasmid DNA) using the Ca-Phosphat technique. Twenty-four hours post transfection medium was changed. Sixteen hours before harvesting, IL-4 stimulation was performed. Finally, 48 h post transfection cells were washed with PBS and subsequently harvested in passive lysis buffer (Promega, Madison, WI, USA). Luciferase assays were performed by adding 350 μl 2 mM ATP solution and 150 μl luciferin solution (0.2 mM in 25 mM glycylglycine pH 7.8) to 10 μl lysate. RLU (relative light units) were measured in a Berthold Lumat LB 9507 luminometer. To normalize for transfection efficiency β-Gal assays were conducted by incubation of 50 μl lysate with o-nitrophenyl β-d-galactopyranoside solution (400 μg of ONPG in 60 mM Na2HPO4/40 mM NaH2PO4 pH 8, 10 mM KCl, 1 mM MgCl2, 50 mM β-mercaptoethanol) at 37°C and subsequent determination of OD420 in the photometer. For normalization, the luciferase values (in RLU) were calculated relative to β-Gal values. Each transfection was performed in triplicates. Error bars show the standard deviation of the triplicates in a representative experiment. Experiments were repeated several times.
GST-pulldown experiments
GST, GST-CARM1 and GST-PRMT1 fusion proteins were expressed in and purified from Escherichia coli BL21 according to standard procedures. Two micrograms of each fusion protein immobilized on glutathione–agarose beads were blocked with bovine serum albumine (200 μg/ml) for 1 h at 4°C. In parallel, HeLa whole-cell extract was prepared after Ca-phosphat transfection of STAT5b using IPH buffer (50 mM Tris/HCl pH 8, 150 mM NaCl, 0.5% NP-40, 1 mM DTT) and precleared with glutathione beads. Subsequently, the blocked GST-fusion beads were incubated with 250 μg of the precleared cell extract for 2 h at 4°C. After intense washes of the beads in IPH buffer bound proteins were resolved by SDS–PAGE and analysed by anti-STAT5 Western Blot.
Immunoprecipitation analysis
Nuclear extracts were prepared from HeLa cell. Cells were washed in cold PBS and subsequently lysed in BufferA (10 mM HEPES–KOH, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.04% NP-40, 2 mM Na3VO4, 150 mM NaF) for 5 min. After centrifugation, the cytosolic components were removed. The remaining nuclear pellet was resolved in BufferB (20 mM HEPES–KOH, pH 7.9, 400 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5% NP-40, 2 mM Na3VO4, 150 mM NaF) and incubated under rotation for 20 min at 4°C. Debris was removed by centrifugation and the clear lysates were diluted 1:1 with Dilution Buffer (20 mM HEPES–KOH, pH 7.9, 0.5% NP-40). Five hundred micrograms of nuclear extract were incubated with 1–2 μg of the indicated antibodies at 4°C overnight and subsequently incubated with protein A and G sepharose (GE Health Care, München, Germany). After extensive washes in IPH buffer precipitates were analysed by SDS–PAGE and Western Blot.
RESULTS
Identification of novel target genes of CARM1 and PRMT1 by cDNA microarray analysis
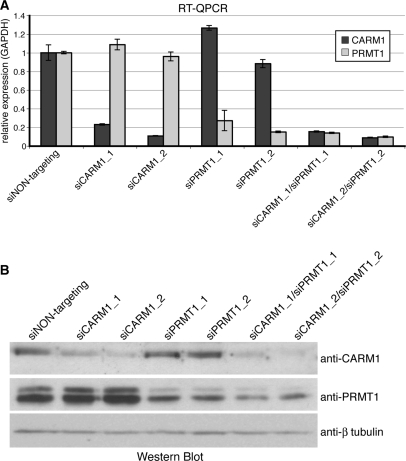
To identify novel transcriptional targets of CARM1 and/or PRMT1, we established single and double knockdowns using transient transfection of soluble double-stranded siRNAs to deplete one or both enzymes in HeLa cells. We employed two different siRNA sequences against each enzyme: siCARM1_1 or siCARM1_2 targeting CARM1 and siPRMT1_1 or siPRMT1_2 targeting PRMT1. Forty-eight hours post transfection, the endogenous expression of CARM1 and/or PRMT1 was efficiently suppressed on RNA (Figure 1A) and protein level (Figure 1B) with the aid of both alternative siRNAs in single as well as double knockdown experiments compared to control siRNA (siNON-targeting) transfection.
Figure 1.
Establishment of the CARM1/PRMT1 single and double knockdown in HeLa cells. (A) HeLa cells were transfected with siNON-targeting or two alternative siRNAs against CARM1 (siCARM1_1 or siCARM1_2) and/or two alternative siRNAs against PRMT1 (siPRMT1_1 or siPRMT1_2) for 48 h. Subsequently total RNA was analysed by RT–QPCR for CARM1 (dark grey bars) and PRMT1 transcription (bright grey bars) respectively normalized for GAPDH. (B) HeLa cells were treated with siRNAs as in (A). Forty-eight hours post transfection cells were harvested in SDS-lysis buffer and 50 μl of each sample were stained by Western Blot with the indicated antibodies.
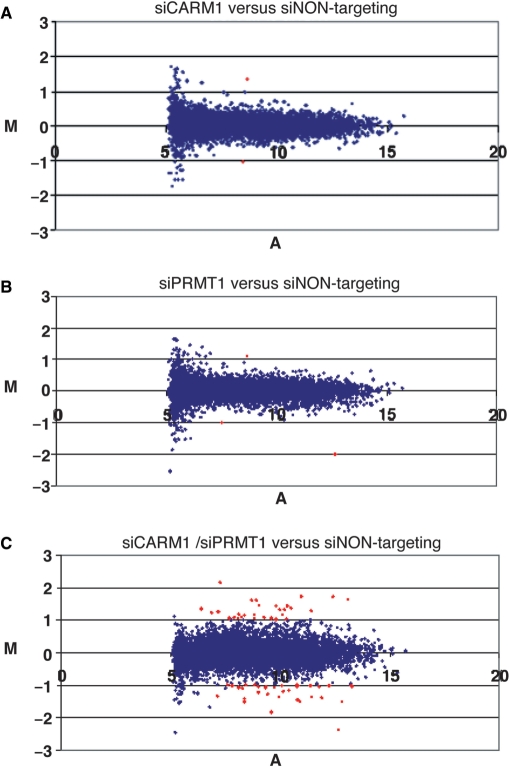
Subsequently we explored the gene expression profiles of these single or double PRMT-depleted HeLa cells relative to control (siNON-targeting transfected) cells by hybridization of a human cDNA microarray, which represents 11 552 human cDNAs. Microarray analysis was perfomed for both alternative siRNAs in duplicates and additionally in flip-colour experiments. As we obtained in total eight independent data sets for each gene and knockdown condition, mean log2 ratios (M-values) were calculated from replicates and used to compare the different conditions. To select for differentially expressed genes we used the significance analysis of microarrays (37) allowing a false-discovery rate (A-value) of ≥7% and a fold change (M-value) of at least 2 or a log2 of ≥1 or ≤−1, as indicated in the MA-scatterplots of Figure 2. Therefore, genes indicated by red-coloured spots were considered deregulated or differentially expressed. The MA-plot revealed no significant changes in the overall expression pattern for the CARM1 and PRMT1 single knockdown versus control knockdown cells (Figure 2A and B).
Figure 2.
Changes in the expression pattern of CARM1/PRMT1 single and double knockdown cells analysed by MA-scatterplot of the entire averaged microarray data set. The MA-scatterplot illustrates the distribution of log expression ratios (M-values) comparing single or double knockdown cells with siNON-targeting transfected (control) cells and the normalized fluorescence intensity values (A-values) for each of the 11 552 genes, which were spotted on the cDNA microarray. The presented results reflect the average of eight independent microarray hybridizations. Genes indicated by red-coloured spots were considered significantly deregulated, i.e. they displayed M-values ≥1 or ≤−1 (≥2-fold expression changes) and A-values ≥7%. (A) Scatterplot for CARM1 single knockdown versus control HeLa cells. (B) Scatterplot for PRMT1 single knockdown versus control HeLa cells. (C) Scatterplot for CARM1/PRMT1 double knockdown versus control HeLa cells.
Predominantly cooperative effects of CARM1 and PRMT1 on global transcription
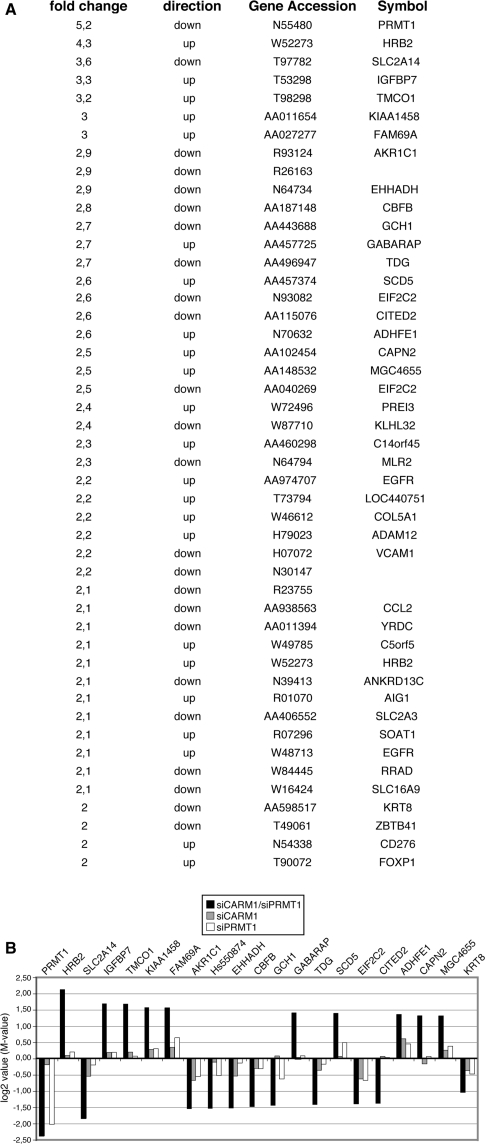
The MA-plot of double knockdown cells versus control cells identified 63 genes, which were >2-fold differentially expressed and were considered significantly deregulated (Figure 2C). Forty-six out of these deregulated genes exhibited a similar expression change when the two alternative siRNAs against CARM1 and PRMT1 were compared. Therefore the group of 46 genes, which are listed in Figure 3A, represent bona fide targets of both enzymes. Among them, 23 genes were downregulated and 23 genes were upregulated in their expression. PRMT1 itself was the strongest deregulated gene (5.2-fold downregulated, Figure 3A) and served as an internal control, whereas the CARM1 cDNA was not spotted on our array. We then analysed in detail the mean expression changes of the 20 strongest deregulated genes in double knockdown cells in comparison to their expression in single knockdown cells (Figure 3B). In agreement with the MA-plot analysis, we found weak differential expression for these genes in the single knockdown conditions (mostly below log2 value of 0.5) and stronger differential expression in the double knockdown conditions. A subset of the 46 deregulated targets was also validated by RT–QPCR from independent knockdown experiments (Supplementary Figure 1) confirming that single knockdown of either CARM1 or PRMT1 reduced the transcript levels of these genes, but double depletion of both enzymes caused a synergistically stronger effect. These results suggest a predominantly cooperative or redundant function of CARM1 and PRMT1 in global gene expression.
Figure 3.
Analysis of the by CARM1/PRMT1 double knockdown significantly deregulated genes. (A) List of the 46 deregulated genes from the microarray analysis. Fold change of expression, direction of deregulation, accession number and gene symbol are indicated for each gene. (B) Comparison of the expression changes (in log2 or M-values) of the 20 strongest deregulated genes and the KRT8 gene in the double knockdown cells with their expression in the single knockdown cells. Black bars indicated the average expression change in CARM1/PRMT1 double knockdown cells (versus control cells). Grey bars indicated the average expression change in CARM1 single knockdown cells (versus control cells). White bars indicated the average expression change in PRMT1 single knockdown cells (versus control cells).
Regulation of CITED2 by STAT5, CARM1 and PRMT1
In the following, we raised the question whether the differentially expressed genes of our microarray analysis might be transcriptional targets of CARM1 and PRMT1. Accordingly, we aimed to identify transcription factors, which might be responsible for recruitment of the two PRMTs to target gene promoters and might coregulate a subset of the identified genes. Such promoter analysis could point to novel signalling pathways that are cooperatively regulated by CARM1 and PRMT1. Therefore, we searched for conserved cis-regulatory elements in the 46 target gene promoters by using the TRANSFAC database on promoter segments conserved among human, mouse and rat (38). This analysis suggested the existence of potential binding sites of STAT5 and STAT6 (signal transducer and activator of transcription 5 and 6) transcription factors respectively in a subset of the downregulated genes, among them the CITED2 and KRT8 gene. No enrichment for transcription factor binding sites was found among the upregulated genes (data not shown).
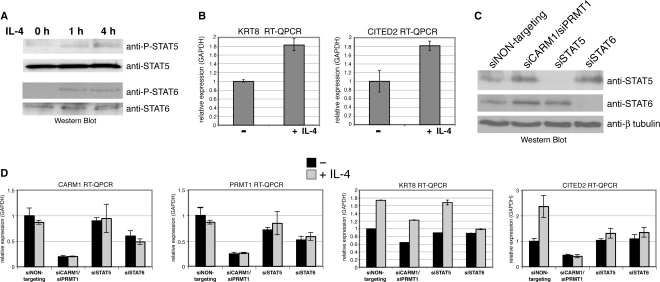
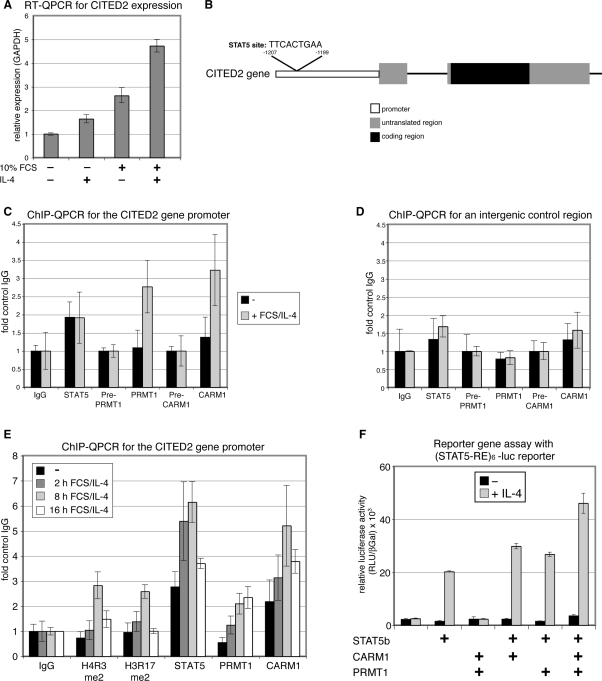
Given that the CITED2 gene expression was previously shown to be inducible upon treatment with various cytokines (39), we investigated whether the transcription of CITED2 and also KRT8 depends on STAT5 and/or STAT6. IL-4 is the canonical stimulus for STAT6 and has been reported to induce STAT6-dependent transcription in HeLa cells (40,41). Additionally, it was suggested that IL-4 stimulates STAT5 signalling in T-lymphocytes (42). Therefore, we treated HeLa cells with IL-4 for the indicated time course and analysed phosphorylation of STAT5 and STAT6 by Western Blot (Figure 4A). Upon IL-4 stimulus tyrosine phosphorylation of both transcription factors was detectable indicating that STAT5 and STAT6 signalling is activated under these conditions in HeLa. RT–QPCR of untreated and IL-4-treated HeLa cells revealed that the transcript levels of CITED2 and KRT8 were enhanced upon IL-4 exposure (Figure 4B). These results identified CITED2 and KRT8 as IL-4-inducible genes in HeLa cells.
Figure 4.
A subset of downregulated microarray targets is inducible by IL-4 treatment depending on the presence of STAT5, CARM1 and PRMT1. (A) HeLa cells were treated without (0 h) or with IL-4 for the indicated times (1 h and 4 h). Subsequently, cells were harvested in SDS-lysis buffer and 50 μl of each sample were analysed by Western Blot with the indicated antibodies. (B) HeLa cells were treated without (−) or with (+) IL-4 for 16 h. Subsequently, cells were harvested and total RNA was analysed by RT–QPCR for transcript levels of CITED2 and KRT8 normalized for GAPDH. (C) HeLa cells were transfected with siNON-targeting, siCARM1/siPRMT1, siSTAT5 or siSTAT6 for 24 h and harvested in SDS-lysis buffer. Fifty microlitres of each sample were analysed by Western Blot with the indicated antibodies. (D) HeLa cells were transfected with siNON-targeting, siCARM1/siPRMT1, siSTAT5 or siSTAT6 for 24 h. Subsequently, cells were treated without (−, black bars) or with (+, grey bars) IL-4 for 24 h. Total RNA was analysed by RT–QPCR for transcript levels of CARM1, PRMT1 and CITED2 normalized for GAPDH.
Next, we analysed whether the IL-4-mediated gene activation of CITED2 and KRT8 depends on the two PRMTs, STAT5 and STAT6, respectively. Thus, we transfected HeLa cells with NON-targeting siRNA, siCARM1/siPRMT1, siRNA against STAT5 or STAT6 and 24 h post transfection IL-4 treatment was performed. The effective knockdown was confirmed by Western Blot for STAT5 and STAT6 (Figure 4C) and by RT–QPCR for CARM1 and PRMT1 transcript levels (Figure 4D). In agreement with our microarray analysis, RT–QPCR revealed that the basal gene activity of KRT8 and CITED2 was reduced upon CARM1/PRMT1 knockdown (Figures 4D and 3B), which suggests that both genes are already activated under normal growth conditions (i.e. in the presence of FCS). In case of KRT8, the IL-4-induced expression was not influenced upon knockdown of CARM1/PRMT1 or STAT5, but diminished upon knockdown of STAT6 (Figure 4D) indicating that its IL-4-stimulated transcription might be regulated by STAT6 and not by STAT5 or the two PRMTs. In case of CITED2, the IL-4-induced expression was abolished upon knockdown of CARM1/PRMT1 and both STATs (Figure 4D). Given that STAT6 knockdown reduced the expression of CARM1 and PRMT1, STAT6 might exert its effect on CITED2 transcription indirectly by regulating the coactivator levels. STAT5 knockdown however did not influence the expression of the PRMTs (Figure 4D) suggesting that STAT5 might activate CITED2 expression directly on the promoter level. These results indicate that transcription of CITED2 in IL-4 signalling is regulated by STAT5, CARM1 and PRMT1.
CITED2 is a direct target gene of STAT5, CARM1 and PRMT1
To elucidate the potential coactivator function of CARM1 and PRMT1 for the STAT5 transcription factor at the CITED2 gene, we investigated whether CITED2 is a direct target gene of CARM1, PRMT1 and STAT5. Therefore, we analysed the recruitment of both PRMTs and STAT5 to the CITED2 gene promoter by ChIP. As we originally identified CITED2 as a PRMT1/CARM1 target in a microarray analysis with normal FCS growth conditions but without IL-4 stimulation, we asked whether FCS and IL-4 costimulation would cooperatively enhance CITED2 gene activation. Indeed, we found that transcript levels of CITED2 were stronger increased by costimulation of starved HeLa cells with FCS and IL-4 than by individual stimulation with FCS or IL-4 (Figure 5A). Therefore, we used for the ChIP analysis FCS/IL-4 costimulated cells versus unstimulated cells. We detected binding of STAT5 to a CITED2 gene promoter fragment encompassing the putative STAT5-binding site (Figure 5B and C). Interestingly, STAT5 recruitment was not enhanced upon FCS/IL-4 stimulation for 16 h (Figure 5C). In contrast, CARM1 and PRMT1 were not bound at the CITED2 gene promoter in unstimulated cells, but became recruited upon 16 h of FCS/IL-4 treatment coinciding with transcriptional activation of the gene (Figure 5A and C). Neither of the two PRMTs nor STAT5 were detected on an intergenic control fragment 6-kb uptream of CITED2 gene (Figure 5D).
Figure 5.
CARM1 and PRMT1 are recruited in an IL-4-dependent manner to the endogenous CITED2 gene promoter and cooperatively coactivate STAT5 in an IL-4-dependent manner. (A) Starved HeLa cells (0.1% FCS) were treated without (−) or with (+) IL-4 and/or 10% FCS for 16 h. Subsequently, cells were harvested and total RNA was analysed by RT–QPCR for transcript levels of CITED2 normalized for GAPDH. (B), Scheme of the CITED2 gene illustrating the location of the putative STAT5-binding site within the promoter. (C and D) Starved HeLa cells (0.1% FCS) were treated without (−, black bars) or with 10% FCS/IL-4 (+, grey bars) for 16 h. Subsequently, cells were harvested and subjected to ChIP analysis using antibodies against STAT5 (corresponding control antibody: IgG), PRMT1 (corresponding control antibody: preimmune-PRMT1) and CARM1 (corresponding control antibody: preimmune-CARM1). Immunoprecipitated DNA was analysed in triplicates by QPCR with primers for the CITED2 gene promoter encompassing the STAT5-binding site (in C) and an intergenic control region 6-kb upstream of the CITED gene (in D). Mean values were expressed as fold control IgG, which was equated 1. (E) Starved HeLa cells (0.1% FCS) were treated without (−, black bars) or with 10% FCS/IL-4 for 2, 8 and 16 h. Subsequently, cells were harvested and subjected to ChIP analysis using control antibodies (IgG), antibodies against STAT5, H4 R3me2, H3 R17me2, PRMT1 and CARM1. Immunoprecipitated DNA was analysed in triplicates by QPCR with primers for the CITED2 gene promoter as in C. (F) HeLa cells were transiently transfected with a (STAT5-RE)6-luciferase (luc) reporter gene constructs and a CMV-β-galactosidase reporter (the latter for normalization) in the absence or presence of expression constructs for STAT5b, CARM1 and PRMT1. Twenty-four hours after transfection medium was changed and cells were treated without (−, black bars) or for 16 h with IL-4 (+, grey bars). Subsequently, cells were harvested and assayed for luciferase activity. Values given are the mean of triplicate measurements and expressed relative to the β-galactosidase activity. Equal protein expression was confirmed by Western Blot (data not shown).
To investigate whether STAT5 is indeed constitutively bound to the CITED2 gene promoter, we performed ChIP analysis of uninduced, 2, 8 and 16 h FCS/IL-4-stimulated HeLa cells. As shown in Figure 5E, recruitment of STAT5 was already weakly detectable in the uninduced state, peaked after 8 h of induction and was barely visible after 16 h in agreement with Figure 5C. These findings suggest that promoter association of STAT5 is an early and transient event subsequent to its activation. Similar to STAT5, CARM1 recruitment to the CITED2 promoter was stimulus dependent and reached its maximum after 8 h of FCS/IL-4 treatment, whereas PRMT1 remained associated up to the 16 h time point (Figure 5E). To address the question whether the histone arginine methyltransferase activity of the two PRMTs is involved in their coactivation function at the CITED2 gene, we performed ChIP analysis with antibodies recognizing the modification site of PRMT1 (H4 R3me2) and CARM1 (H3 R17me2) in histones (43,44). Coinciding with the recruitment of PRMT1 and CARM1, we found increased H4 R3 dimethylation and H3 R17 dimethylation at the 8 h induction time point (Figure 5E). These data indicate that CITED2 is a direct target gene of IL-4-induced coactivation by CARM1 and PRMT1 and that the histone arginine methyltransferase activity of the two PRMTs might be required for gene activation.
Coactivation of STAT5-mediated transcription by CARM1 and PRMT1
To confirm the coactivator function of CARM1 and PRMT1 for the STAT5 transcription factor in an independent and more general experimental setting, we employed reporter gene assays using a reporter constructs with multimerized STAT5-response elements of the β-casein promoter cloned upstream of the luciferase gene (STAT5-RE)6-luc (45,46). HeLa cells were transfected with the (STAT5-RE)6-luc reporter in the absence and presence of STAT5b expression plasmid and IL-4 stimulus, respectively. The basal activity of the luciferase reporter was stimulated by 10-fold depending on the over-expression of STAT5b and IL-4 stimulus (Figure 5E). Overexpression of either CARM1 or PRMT1 resulted in a slight increase of the STAT5b/IL-4-dependent luciferase activity, whereas cotransfection of both PRMTs clearly enhanced the STAT5b/IL-4-stimulated reporter activity >2-fold (Figure 5E). These results suggest that CARM1 and PRMT1 coactivate STAT5-regulated transcription in a cytokine-dependent and cooperative manner.
Cytokine-enhanced interaction between STAT5 and the two PRMTs
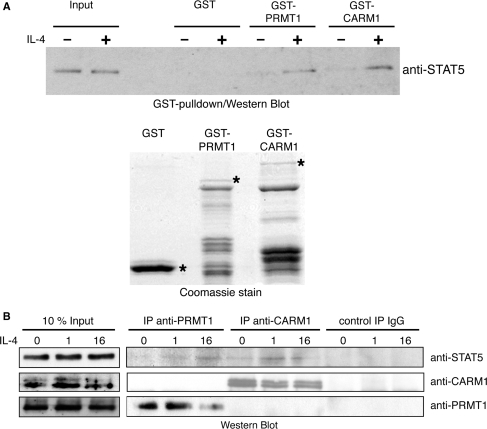
Given that STAT5, CARM1 and PRMT1 appear to collaborate in transcriptional activation and overlap in their promoter recruitment to a common target gene, we investigated whether CARM1 and PRMT1 are able to interact with STAT5b and whether this interaction is altered by a transcriptional stimulus like IL-4. Therefore, we first performed GST pulldown experiments with the aid of GST (as control), GST-PRMT1 or GST-CARM1 immobilized on agarose beads (Figure 6A, bottom panel) in the presence of unstimulated and IL-4-stimulated HeLa cell extracts overexpressing STAT5b. Subsequently, the pulldowns were analysed by anti-STAT5 Western Blot. As shown in Figure 6A (top panel), CARM1 and PRMT1 weakly interacted with exogenous STAT5b before stimulation and revealed enhanced interaction with STAT5b in IL-4-stimulated cell extracts. Similar results were gained in a second experimental approach by coimmunoprecipitation. Endogenous PRMT1 and CARM1 respectively were immunoprecipitated from unstimulated or IL-4-stimulated HeLa nuclear extracts. Subsequently, the precipitates were probed for the presence of endogenous STAT5 protein by Western Blot analysis. As shown in Figure 6B, both PRMTs interacted with STAT5 in vivo. PRMT1 precipitates contained STAT5 only subsequent to IL-4 stimulation, whereas CARM1–STAT5 interaction was already weakly detectable in unstimulated extacts and was enhanced upon IL-4 stimulation. These results indicate that CARM1 and PRMT1 interact stimulus dependent with STAT5 corroborating the direct role of these three proteins in the transcriptional control of the CITED2 gene in response to IL-4 stimulation.
Figure 6.
CARM1 and PRMT1 interaction with STAT5 is enhanced by IL-4. (A) GST alone (control), GST-CARM1 and GST-PRMT1 were expressed in E. coli and purified. Equal amounts of these GST-fusions, as indicated in the Coomassie-stained SDS-gel (bottom panel, full-length protein bands marked with asterisks), coupled to glutathione beads were used in pulldown experiments in the presence of HeLa whole-cell extract overexpressing STAT5b and treated without or with IL-4 for 16 h. Interactions between the proteins were detected by anti-STAT5 Western Blot (top panel). One percent input of HeLa whole-cell extract is shown. (B) HeLa cells were left unstimulated or stimulated with IL-4 for 1 or 16 h. Subsequently, nuclear extracts were prepared and subjected to immunoprecipitation with antibodies against PRMT1, CARM1 and as control IgG. Immunoprecipitates were resolved by SDS–PAGE and analysed by Western Blot analysis with the indicated antibodies to confirm the immunoprecipitation of PRMT1 and CARM1 (as control) and to investigate STAT5 coimmunoprecipitation. Ten percent input (50 μg) of HeLa cell extract is shown.
DISCUSSION
Using an unbiased microarray approach, we identified novel target genes directly or indirectly regulated by CARM1 and/or PRMT1, which have previously been linked to transcriptional control. We could demonstrate that single knockdown of neither CARM1 nor PRMT1 significantly changed the global expression pattern in HeLa cells, whereas double knockdown of both enzymes resulted in >2-fold deregulation of 46 genes. One explanation for this finding is that deregulated gene expression upon codepletion of both PRMTs demonstrates additive effects of the single knockdowns. Indeed, we found evidence that CARM1 and PRMT1 cooperatively regulate the CITED2 gene. However, we cannot exclude that some targets might be redundantly regulated by the two PRMTs. For example, neither CARM1 nor PRMT1 single knockdown affected RRAD expression (Supplementary Figure 1), whereas other targets are at least slightly deregulated under single knockdown conditions. Since siRNA-mediated knockdown was efficient (Figure 1) but certainly incomplete, residual PRMT-activity might complicate the discrimination of cooperatively from redundantly regulated target genes. Additionally, 48 h of siRNA treatment, which we employed in the microarray analysis to circumvent secondary effects, might not be sufficient to allow complete depletion of the corresponding methylation marks and to reveal a clear synergism of the two enzymes in gene expression.
In general, our data suggest a more widespread cooperation and overlap of target genes between CARM1 and PRMT1 than so far reported in the literature. Most studies investigated the individual coregulator function of either CARM1 or PRMT1 on distinct transcription factors and signalling pathways besides two examples in the literature, which describe synergism between the two enzymes in p53- and NR-mediated transcription (21,24). Given that the two PRMTs possess non-overlapping substrate specificity and their recruitment to p53 and NR target gene promoters is temporally separated (11,24,43,47), it was suggested that they fullfil non-redundant functions in p53- and NR-signalling. PRMT1 is targeted very early during the initial phase of transcriptional activation to these target gene promoters, whereas CARM1 subsequently appears at the promoters triggering late coactivation events and also disassemby of coactivator complexes (24,26,27). As our aim was to identify novel target genes and pathways regulated by the two arginine methyltransferases, we did not specifically stimulate steroid-hormone signalling in our experiments. Furthermore p53 signalling is inactive in HeLa cells because of E6 expression from human papillomavirus (48). Therefore, we did not expect to identify any cooperatively regulated target genes of these two known pathways in our screen.
Interestingly, among the 46 deregulated target genes of CARM1 and PRMT1, genes were downregulated as well as upregulated upon depletion of the enzymes indicating that the two enzymes cooperate in activation as well as repression of gene expression. Arginine methylation of histones by the two enzymes, i.e. R17 in histone H3 by CARM1 and R3 in histone H4 by PRMT1, have up to now exclusively been linked to transcriptional activation (11,43), whereas arginine methylation of non-histone chromatin proteins was also found to be involved in transcriptional repression. For example, arginine methylation of the histone acetyltransferase CBP/p300 by CARM1 has been reported to occur at various sites. Methylation of the KIX domain in the N-terminus of CBP/p300 by CARM1 was shown to inhibit its interaction with the CREB transcription factor and hence abolishes cAMP-activated gene expression (25). Other reports find methylation of CBP/p300 within its C-terminal interaction surface for the p160 proteins, which abolishes interaction between CBP/p300 and GRIP1 (49). In contrast, methylation of the central part of CBP/p300 by CARM1 enhances the coactivation function of the HAT in NR-mediated transcription (50). The p160 family members SRC-1 and SRC-3 are regulated as well by CARM1-dependent methylation, as they become methylated in their glutamine-rich region. This methylation event associates with decreased protein stability of p160, disassembly of the p160-CBP/p300 coactivator complex and accordingly reduction of the transcriptional activity (26,27). Modification of non-histone chromatin proteins by PRMT1 is also linked to transcriptional activation and repression. The NR transcription factor HNF-4 becomes methylated in its DNA-binding domain by PRMT1, which enhances DNA-binding affinity of HNF-4 and subsequent coactivation (51). In contrast, the elongation factor Spt5 is methylated by PRMT1, which regulates its interaction with RNA polymerase II and causes transcriptional pausing (52). Taking the literature into account, which describes mechanisms for transcriptional regulation either by CARM1 or by PRMT1, it appears plausible that the enzymes might not only cooperate in activation but also in repression of transcription.
PRMTs possess many other cellular interaction partners and substrates besides histones and non-histone chromatin proteins, for example the RNA-binding protein HuD, whose methylation by CARM1 influences p21 mRNA stability (53). Therefore, some of our differentially expressed microarray targets might be regulated indirectly and/or posttranscriptionally by the two PRMTs, e.g. on the level of RNA stability, protein turnover, cellular localization or other mechanisms. Further work has to be done to dissect transcriptional from non-transcriptional targets identified in our microarray approach.
As transcriptional cooperation of CARM1 and PRMT1 is mediated by sequence-specific transcription factors, we searched for common transcription factor binding site in the 46 deregulated targets. Promoter analysis suggested that a subset of downregulated genes might be regulated by STAT5 and/or STAT6. STATs are downstream targets of various cytokines and lipophilic hormones, like erythropoetin and prolactin (40,54). Cytokine stimulation and subsequent phosphorylation of STATs by cytokine receptor-associated kinases results in nuclear translocation of STATs and activation of a variety of genes involved in the control of proliferation, apoptosis and differentiation. Although IL-4 is the canonical stimulus for STAT6-activated transcription (40), it was previously reported to activate also STAT5 signalling (42). IL-4 treatment induces the expression of two microarray targets, namely CITED2 and KRT8. CARM1/PRMT1 codepletion and STAT5 depletion, respectively abolished IL-4 induced expression of CITED2, whereas KRT8 expression was unaffected under these conditions. This observation identified CITED2 as a CARM1/PRMT1 and STAT5 target in IL-4 signalling. In agreement with this observation, the CITED2 gene, which encodes a nuclear protein interacting with CBP/p300 and interfering with hypoxia-driven transcription, was previously shown to be transcriptionally activated upon cytokine treatment (39,55) and was identified as a STAT5 target gene (56). As knockdown of STAT6 inhibited the expression of CARM1 and PRMT1, we suggest that STAT6 regulates CITED2 gene expression indirectly. In agreement with the finding that IL-4 induced KRT8 expression depends neither on CARM1, PRMT1 nor STAT5, but on STAT6, we could not detect coactivator function of both PRMTs using a STAT6-dependent reporter gene (data not shown). Instead, we detected cooperative coactivation of IL-4 induced STAT5-mediated transcription by CARM1 and PRMT1 in reporter gene assays. As we found that PRMT1/CARM1 did not coactivate the transcript levels of other well-known STAT5 target genes, like Cis and βCasein (data not shown), we conclude that the two PRMTs are no general coactivators of STAT5, but act on a subset of STAT5 targets, e.g. CITED2.
ChIP analysis identified CITED2 as a direct target gene of CARM1 and PRMT1, as we could detect the recruitment of both coregulators to the gene promoter subsequent to FCS/IL-4 stimulation. Similarly, promoter association of STAT5 was increased by the FCS/IL-4 stimulus and presented an early and transient event. Furthermore, in GST-pulldown and endogenous coimmunoprecipitation experiments STAT5 interacted with both PRMTs in an IL-4-dependent manner. Altogether, these data suggest that CARM1 and PRMT1 cooperatively enhance STAT5-dependent transcriptional activation of CITED2 on the promoter level in IL-4 signalling and that the IL-4 stimulus enforces the interaction of the two coactivators with STAT5 and their promoter recruitment.
Previous reports proposed that arginine methylation plays an important role in cytokine signalling either on the level of the cytokine receptor or on the level of the STAT transcription factors (57–59). Although it was suggested that STAT1 and STAT6 are themselves substrates of arginine methylation and that this modification event enhances their coactivation function (58,59), arginine methylation of STAT transcription factors is still under debate (60,61). Based on our ChIP data, in which we detected increased H4 R3 and H3 R17 dimethylation at the CITED2 gene promoter upon FCS/IL-4 stimulation coinciding with the promoter recruitment of the two PRMTs, it seems likely that CARM1 and PRMT1 regulate STAT5-dependent transcription due to histone methylation. Whether arginine methylation of other promoter-bound proteins by PRMT1 and/or CARM1 is also required in the coactivation process of the CITED2 gene should be addressed in the future. Interestingly, STAT5 uses a similar set of coactivator proteins as p53 and NR transcription factors, for example p160 proteins, p300/CBP (62,63) and PRMTs, as we show here. This suggests that a common coactivator complex consisting of a p160 family member, histone acetyltransferase and arginine methyltransferase activities might cooperatively contribute to gene activation by various transcription factors.
In summary, our microarray data present a first unbiased study, which globally searches for individual and overlapping target genes of CARM1 and PRMT1 and unravels a more general cooperation in transcriptional regulation of the two enzymes than so far expected. Further, we identified IL-4-stimulated STAT5-dependent CITED2 expression as a novel pathway coregulated by the two PRMTs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Iris Behrmann and Michael Lohoff for providing us reagents. We are grateful to Iris Behrmann, Martin Eilers and Alexander Brehm for critical reading the article and valuable suggestions. We thank Inge Pelz and Karin Theis for technical assistance. We appreciate the help of members of the U.M.B. laboratory and Martin Eilers laboratory during work progress. This work was supported by the DFG (Deutsche Forschungsgemeinschaft), by funding from Land Hessen and from BMBF (Bundesministerium für Bildung und Forschung) to U.M.B.
Conflict of interest statement. None declared.
REFERENCES
- 1.Boisvert FM, Chenard CA, Richard S. Protein interfaces in signaling regulated by arginine methylation. Sci STKE. doi: 10.1126/stke.2712005re2. 2005 re2. [DOI] [PubMed] [Google Scholar]
- 2.Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol. Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Cook JR, Lee JH, Yang ZH, Krause CD, Herth N, Hoffmann R, Pestka S. FBXO11/PRMT9, a new protein arginine methyltransferase, symmetrically dimethylates arginine residues. Biochem. Biophys. Res. Commun. 2006;342:472–481. doi: 10.1016/j.bbrc.2006.01.167. [DOI] [PubMed] [Google Scholar]
- 4.Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318:444–447. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
- 5.Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, Yamada M, Schneider R, Gregory PD, Tempst P, Bannister AJ, et al. Histone deimination antagonizes arginine methylation. Cell. 2004;118:545–553. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y, et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306:279–283. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- 7.El-Andaloussi N, Valovka T, Toueille M, Steinacher R, Focke F, Gehrig P, Covic M, Hassa PO, Schar P, Hubscher U, et al. Arginine methylation regulates DNA polymerase beta. Mol. Cell. 2006;22:51–62. doi: 10.1016/j.molcel.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 8.El-Andaloussi N, Valovka T, Toueille M, Hassa PO, Gehrig P, Covic M, Hubscher U, Hottiger MO. Methylation of DNA polymerase beta by protein arginine methyltransferase 1 regulates its binding to proliferating cell nuclear antigen. FASEB J. 2007;21:26–34. doi: 10.1096/fj.06-6194com. [DOI] [PubMed] [Google Scholar]
- 9.Jelinic P, Stehle JC, Shaw P. The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol. 2006;4:e355. doi: 10.1371/journal.pbio.0040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hyllus D, Stein C, Schnabel K, Schiltz E, Imhof A, Dou Y, Hsieh J, Bauer UM. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 2007;21:3369–3380. doi: 10.1101/gad.447007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst P, et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- 12.Chen D, Huang SM, Stallcup MR. Synergistic, p160 coactivator-dependent enhancement of estrogen receptor function by CARM1 and p300. J. Biol. Chem. 2000;275:40810–40816. doi: 10.1074/jbc.M005459200. [DOI] [PubMed] [Google Scholar]
- 13.Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell. Biol. 2004;24:9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boulanger MC, Liang C, Russell RS, Lin R, Bedford MT, Wainberg MA, Richard S. Methylation of Tat by PRMT6 regulates human immunodeficiency virus type 1 gene expression. J. Virol. 2005;79:124–131. doi: 10.1128/JVI.79.1.124-131.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fabbrizio E, El Messaoudi S, Polanowska J, Paul C, Cook JR, Lee JH, Negre V, Rousset M, Pestka S, Le Cam A, et al. Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep. 2002;3:641–645. doi: 10.1093/embo-reports/kvf136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, Surani MA. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat. Cell. Biol. 2006;8:623–630. doi: 10.1038/ncb1413. [DOI] [PubMed] [Google Scholar]
- 17.Covic M, Hassa PO, Saccani S, Buerki C, Meier NI, Lombardi C, Imhof R, Bedford MT, Natoli G, Hottiger MO. Arginine methyltransferase CARM1 is a promoter-specific regulator of NF-kappaB-dependent gene expression. EMBO J. 2005;24:85–96. doi: 10.1038/sj.emboj.7600500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen SL, Loffler KA, Chen D, Stallcup MR, Muscat GE. The coactivator-associated arginine methyltransferase is necessary for muscle differentiation: CARM1 coactivates myocyte enhancer factor-2. J. Biol. Chem. 2002;277:4324–4333. doi: 10.1074/jbc.M109835200. [DOI] [PubMed] [Google Scholar]
- 19.Koh SS, Li H, Lee YH, Widelitz RB, Chuong CM, Stallcup MR. Synergistic coactivator function by coactivator-associated arginine methyltransferase (CARM) 1 and beta-catenin with two different classes of DNA-binding transcriptional activators. J. Biol. Chem. 2002;277:26031–26035. doi: 10.1074/jbc.M110865200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rezai-Zadeh N, Zhang X, Namour F, Fejer G, Wen YD, Yao YL, Gyory I, Wright K, Seto E. Targeted recruitment of a histone H4-specific methyltransferase by the transcription factor YY1. Genes Dev. 2003;17:1019–1029. doi: 10.1101/gad.1068003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee YH, Koh SS, Zhang X, Cheng X, Stallcup MR. Synergy among nuclear receptor coactivators: selective requirement for protein methyltransferase and acetyltransferase activities. Mol. Cell. Biol. 2002;22:3621–3632. doi: 10.1128/MCB.22.11.3621-3632.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagner S, Weber S, Kleinschmidt MA, Nagata K, Bauer UM. SET-mediated promoter hypoacetylation is a prerequisite for coactivation of the estrogen-responsive pS2 gene by PRMT1. J. Biol. Chem. 2006;282:27242–27250. doi: 10.1074/jbc.M605172200. [DOI] [PubMed] [Google Scholar]
- 23.Daujat S, Bauer UM, Shah V, Turner B, Berger S, Kouzarides T. Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr. Biol. 2002;12:2090–2097. doi: 10.1016/s0960-9822(02)01387-8. [DOI] [PubMed] [Google Scholar]
- 24.An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 25.Xu W, Chen H, Du K, Asahara H, Tini M, Emerson BM, Montminy M, Evans RM. A transcriptional switch mediated by cofactor methylation. Science. 2001;294:2507–2511. doi: 10.1126/science.1065961. [DOI] [PubMed] [Google Scholar]
- 26.Feng Q, Yi P, Wong J, O’Malley BW. Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol. Cell. Biol. 2006;26:7846–7857. doi: 10.1128/MCB.00568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naeem H, Cheng D, Zhao Q, Underhill C, Tini M, Bedford MT, Torchia J. The activity and stability of the transcriptional coactivator p/CIP/SRC-3 are regulated by CARM1-dependent methylation. Mol. Cell. Biol. 2007;27:120–134. doi: 10.1128/MCB.00815-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teyssier C, Ma H, Emter R, Kralli A, Stallcup MR. Activation of nuclear receptor coactivator PGC-1alpha by arginine methylation. Genes Dev. 2005;19:1466–1473. doi: 10.1101/gad.1295005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mowen KA, Schurter BT, Fathman JW, David M, Glimcher LH. Arginine methylation of NIP45 modulates cytokine gene expression in effector T lymphocytes. Mol. Cell. 2004;15:559–571. doi: 10.1016/j.molcel.2004.06.042. [DOI] [PubMed] [Google Scholar]
- 30.Mostaqul Huq MD, Gupta P, Tsai NP, White R, Parker MG, Wei LN. Suppression of receptor interacting protein 140 repressive activity by protein arginine methylation. EMBO J. 2006;25:5094–5104. doi: 10.1038/sj.emboj.7601389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 32.Scott HS, Antonarakis SE, Lalioti MD, Rossier C, Silver PA, Henry MF. Identification and characterization of two putative human arginine methyltransferases (HRMT1L1 and HRMT1L2) Genomics. 1998;48:330–340. doi: 10.1006/geno.1997.5190. [DOI] [PubMed] [Google Scholar]
- 33.Balint BL, Szanto A, Madi A, Bauer UM, Gabor P, Benko S, Puskas LG, Davies PJ, Nagy L. Arginine methylation provides epigenetic transcription memory for retinoid-induced differentiation in myeloid cells. Mol. Cell. Biol. 2005;25:5648–5663. doi: 10.1128/MCB.25.13.5648-5663.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ellis L, Clauser E, Morgan DO, Edery M, Roth RA, Rutter WJ. Replacement of insulin receptor tyrosine residues 1162 and 1163 compromises insulin-stimulated kinase activity and uptake of 2-deoxyglucose. Cell. 1986;45:721–732. doi: 10.1016/0092-8674(86)90786-5. [DOI] [PubMed] [Google Scholar]
- 35.Yang YH, Dudoit S, Luu P, Speed TP. Normalization for cDNA microarray data. In: Bittner ML, Chen Y, Dorsel AN, Dougherty ER, editors. Microarrays: Optical Technologies and Informatics. Vol. 4266. Bellingham, WA, USA: Proceedings of SPIE; 2001. [Google Scholar]
- 36.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharan R, Ovcharenko I, Ben-Hur A, Karp RM. CREME: a framework for identifying cis-regulatory modules in human-mouse conserved segments. Bioinformatics. 2003;19(Suppl. 1):i283–i291. doi: 10.1093/bioinformatics/btg1039. [DOI] [PubMed] [Google Scholar]
- 39.Sun HB, Zhu YX, Yin T, Sledge G, Yang YC. MRG1, the product of a melanocyte-specific gene related gene, is a cytokine-inducible transcription factor with transformation activity. Proc. Natl Acad. Sci. USA. 1998;95:13555–13560. doi: 10.1073/pnas.95.23.13555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levy DE, Darnell J.E., Jr. Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell. Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 41.Yang J, Aittomaki S, Pesu M, Carter K, Saarinen J, Kalkkinen N, Kieff E, Silvennoinen O. Identification of p100 as a coactivator for STAT6 that bridges STAT6 with RNA polymerase II. EMBO J. 2002;21:4950–4958. doi: 10.1093/emboj/cdf463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lischke A, Moriggl R, Brandlein S, Berchtold S, Kammer W, Sebald W, Groner B, Liu X, Hennighausen L, Friedrich K. The interleukin-4 receptor activates STAT5 by a mechanism that relies upon common gamma-chain. J. Biol. Chem. 1998;273:31222–31229. doi: 10.1074/jbc.273.47.31222. [DOI] [PubMed] [Google Scholar]
- 43.Bauer UM, Daujat S, Nielsen SJ, Nightingale K, Kouzarides T. Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep. 2002;3:39–44. doi: 10.1093/embo-reports/kvf013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, Zhang Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell. 2001;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- 45.Friedrich K, Kammer W, Erhardt I, Brandlein S, Sebald W, Moriggl R. Activation of STAT5 by IL-4 relies on Janus kinase function but not on receptor tyrosine phosphorylation, and can contribute to both cell proliferation and gene regulation. Int. Immunol. 1999;11:1283–1294. doi: 10.1093/intimm/11.8.1283. [DOI] [PubMed] [Google Scholar]
- 46.Moriggl R, Gouilleux-Gruart V, Jahne R, Berchtold S, Gartmann C, Liu X, Hennighausen L, Sotiropoulos A, Groner B, Gouilleux F. Deletion of the carboxyl-terminal transactivation domain of MGF-Stat5 results in sustained DNA binding and a dominant negative phenotype. Mol. Cell. Biol. 1996;16:5691–5700. doi: 10.1128/mcb.16.10.5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 48.Talis AL, Huibregtse JM, Howley PM. The role of E6AP in the regulation of p53 protein levels in human papillomavirus (HPV)-positive and HPV-negative cells. J. Biol. Chem. 1998;273:6439–6445. doi: 10.1074/jbc.273.11.6439. [DOI] [PubMed] [Google Scholar]
- 49.Lee YH, Coonrod SA, Kraus WL, Jelinek MA, Stallcup MR. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc. Natl Acad. Sci. USA. 2005;102:3611–3616. doi: 10.1073/pnas.0407159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chevillard-Briet M, Trouche D, Vandel L. Control of CBP co-activating activity by arginine methylation. EMBO J. 2002;21:5457–5466. doi: 10.1093/emboj/cdf548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barrero MJ, Malik S. Two functional modes of a nuclear receptor-recruited arginine methyltransferase in transcriptional activation. Mol. Cell. 2006;24:233–243. doi: 10.1016/j.molcel.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kwak YT, Guo J, Prajapati S, Park KJ, Surabhi RM, Miller B, Gehrig P, Gaynor RB. Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Mol. Cell. 2003;11:1055–1066. doi: 10.1016/s1097-2765(03)00101-1. [DOI] [PubMed] [Google Scholar]
- 53.Fujiwara T, Mori Y, Chu DL, Koyama Y, Miyata S, Tanaka H, Yachi K, Kubo T, Yoshikawa H, Tohyama M. CARM1 regulates proliferation of PC12 cells by methylating HuD. Mol. Cell. Biol. 2006;26:2273–2285. doi: 10.1128/MCB.26.6.2273-2285.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wittig I, Groner B. Signal transducer and activator of transcription 5 (STAT5), a crucial regulator of immune and cancer cells. Curr. Drug Targets Immun. Endocr Metabol. Disord. 2005;5:449–463. doi: 10.2174/156800805774912999. [DOI] [PubMed] [Google Scholar]
- 55.Leung MK, Jones T, Michels CL, Livingston DM, Bhattacharya S. Molecular cloning and chromosomal localization of the human CITED2 gene encoding p35srj/Mrg1. Genomics. 1999;61:307–313. doi: 10.1006/geno.1999.5970. [DOI] [PubMed] [Google Scholar]
- 56.Bakker WJ, van Dijk TB, Parren-van Amelsvoort M, Kolbus A, Yamamoto K, Steinlein P, Verhaak RG, Mak TW, Beug H, Lowenberg B, et al. Differential regulation of Foxo3a target genes in erythropoiesis. Mol. Cell. Biol. 2007;27:3839–3854. doi: 10.1128/MCB.01662-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abramovich C, Yakobson B, Chebath J, Revel M. A protein-arginine methyltransferase binds to the intracytoplasmic domain of the IFNAR1 chain in the type I interferon receptor. EMBO J. 1997;16:260–266. doi: 10.1093/emboj/16.2.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mowen KA, Tang J, Zhu W, Schurter BT, Shuai K, Herschman HR, David M. Arginine methylation of STAT1 modulates IFNalpha/beta-induced transcription. Cell. 2001;104:731–741. doi: 10.1016/s0092-8674(01)00269-0. [DOI] [PubMed] [Google Scholar]
- 59.Chen W, Daines MO, Hershey GK. Methylation of STAT6 modulates STAT6 phosphorylation, nuclear translocation, and DNA-binding activity. J. Immunol. 2004;172:6744–6750. doi: 10.4049/jimmunol.172.11.6744. [DOI] [PubMed] [Google Scholar]
- 60.Meissner T, Krause E, Lodige I, Vinkemeier U. Arginine methylation of STAT1: a reassessment. Cell. 2004;119:587–589. doi: 10.1016/j.cell.2004.11.024. discussion 589–590. [DOI] [PubMed] [Google Scholar]
- 61.Komyod W, Bauer UM, Heinrich PC, Haan S, Behrmann I. Are STATS arginine-methylated? J. Biol. Chem. 2005;280:21700–21705. doi: 10.1074/jbc.C400606200. [DOI] [PubMed] [Google Scholar]
- 62.Pfitzner E, Jahne R, Wissler M, Stoecklin E, Groner B. p300/CREB-binding protein enhances the prolactin-mediated transcriptional induction through direct interaction with the transactivation domain of Stat5, but does not participate in the Stat5-mediated suppression of the glucocorticoid response. Mol. Endocrinol. 1998;12:1582–1593. doi: 10.1210/mend.12.10.0180. [DOI] [PubMed] [Google Scholar]
- 63.Litterst CM, Kliem S, Marilley D, Pfitzner E. NCoA-1/SRC-1 is an essential coactivator of STAT5 that binds to the FDL motif in the alpha-helical region of the STAT5 transactivation domain. J. Biol. Chem. 2003;278:45340–45351. doi: 10.1074/jbc.M303644200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.