Abstract
The origin of the high pathogenicity of an emerging avian influenza H5N1 due to the –RRRKK– insertion at the cleavage loop of the hemagglutinin H5, was studied using the molecular dynamics technique, in comparison with those of the noninserted H5 and H3 bound to the furin (FR) active site. The cleavage loop of the highly pathogenic H5 was found to bind strongly to the FR cavity, serving as a conformation suitable for the proteolytic reaction. With this configuration, the appropriate interatomic distances were found for all three reaction centers of the enzyme-substrate complex: the arrangement of the catalytic triad, attachment of the catalytic Ser368 to the reactive S1-Arg, and formation of the oxyanion hole. Experimentally, the –RRRKK– insertion was also found to increase in cleavage of hemagglutinin by FR. The simulated data provide a clear answer to the question of why inserted H5 is better cleaved by FR than the other subtypes, explaining the high pathogenicity of avian influenza H5N1.
INTRODUCTION
Proteolytic activation of the hemagglutinin (HA) is essential for viral infectivity and for spread of the avian influenza virus through the host's body (1–3). This process is determined by a cleavage reaction at the HA cleavage site, a conserved arginine, by host proteases (4–6). Insertion of the –RRRKK– residues into the low pathogenic avian influenza (LPAI) cleavage site is known to potentially activate infectivity of viruses, i.e., the LPAI viruses, which then become high pathogenic avian influenza (HPAI) viruses, allowing highly virulent strains to be cleaved by furin (FR), an ubiquitously expressed protease. The goal of this study is to understand why FR cleaves the inserted hemagglutinin strains better than the noninserted strains.
The HA is synthesized initially as an inactive precursor (HA0) that is then activated through a controlled proteolytic cleavage by protease into HA1 and HA2 subunits. HA1 mediates virus binding to host-cell receptors whereas HA2 promotes the release of the viral RNA complexed with polymerase through membrane fusion (1–6). Without proteolysis, the fusion peptide cannot occur and therefore the virus is essentially noninfectious.
The HA cleavage site relies on the presence of basic amino acids and relates directly to influenza virus pathogenicity, i.e., the LPAI viruses possess a single arginine at the cleavage site, while the HPAI viruses containing the polybasic insertion upstream of the cleavage site of the H5 subtype have potential to cause devastating pandemics in the future (3,7). In addition, in vitro cleavage of a series of peptide substrates showed that the insertion of the –RRRKK– residues at cleavage site led to an increase in cleavage by FR protease (2,8,9).
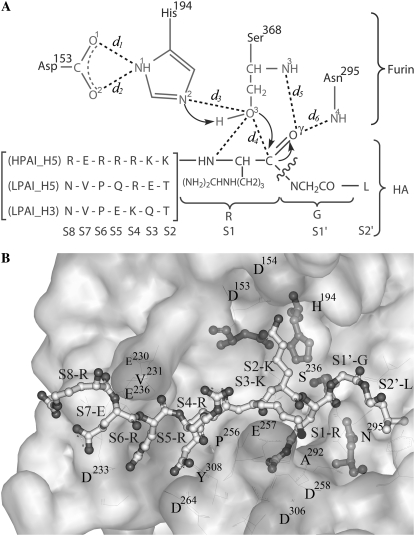
FR, a subtilisin-like serine endoprotease, seems to be a highly specific enzyme, cleaving pro-protein precursors at specific consensus sequence –RXK/RR–, usually to produce biologically active products (10–15). Based on the x-ray structure of mouse FR complexed with the dec-RVKR-cmk inhibitor, the catalytic ability of FR is considered to have originated from the catalytic triad residues (Ser368, His194, and Asp153) and the oxyanion hole (formed by the carboxamide nitrogens of Asn295 and backbone nitrogen of Ser368 to carbonyl oxygen of the inhibitor/substrate's centered arginine) at the active site (12). The proposed cleavage mechanism is shown in Fig. 1 (16,17).
FIGURE 1.
(A) Proposed cleavage mechanism of HA by furin and definitions of d1–d6. (B) Loop of HPAI H5 (ball and stick model) in the electrostatic surface of furin.
This work seeks the source of high pathogenicity in avian influenza A virus subtype H5 in comparison with LPAI subtypes H5 and H3. Three molecular dynamics simulations were carried out for the three complexes, HPH5–FR, LPH5–FR, and LPH3–FR. The investigation was focused to intra- and intermolecular interactions and geometries of the substrate-furin complex potentially involved to the cleavage mechanism.
METHODS
Initial structure of individual protein and substrates
The crystal structure of FR with bound dec-RVKR-cmk inhibitor (12) used as receptor model for MD simulations was obtained from Protein Data Bank (PDB), code: 1P8J. This study covers the loop of HA substrate features of subtypes H5 (both HPAI and LPAI) and H3 (known as LPAI). The sequence of the HPAI subtype H5 isolated during the 2006 influenza outbreaks in Thailand was taken from Genbank LOCUS ID ABK13784 (A/chicken/Thailand/PC-168/2006(H5N1)) (18). The sequence of the LPAI subtype H5 was obtained from Protein Data Bank (PDB entry code: 1JSM) (19).
The initial model for the HPAI_H5 loop (RERRRKKRGL) was built up using the sequence alignments and the atomic coordinates of the x-ray structure (residues 322–331: NVPEKQTQGL) of the HA0 of H3 (PDB entry code: 1HA0) (20) and dec-RVKR-cmk inhibitor of FR (12) as a template, by using the homology module in the Insight II program (Accelrys, San Diego, CA) (21). The Needleman-Wunsch algorithm (22) was used for pairwise alignment to identify the correspondence region. The coordinates of the structurally conserved regions (SCRs) of the template —in which amino acid sequences in the template and the model are identical—were copied to be those of the model. In addition to the similar SCRs, only the backbone coordinates were replicated and the side chain atoms were added in a manner that preserves the residue type of the model protein.
For LPAI subtype H5, the initial structure of the cleavage loop (NVPQRETRGL) (19) was constructed using the backbone atoms of the HPH5 loop built previously. The side chains were generated by the LEaP module of AMBER 7 (23). With this module, the H3 loop above was mutated at residue 329 from glutamine (Q) to arginine (R).
Initial structure of enzyme-substrate complexes
The cleavage loop of HA complexed with FR was generated by the following procedures (Fig. 1): i), all HA heavy atoms of S1–S4 were superimposed with the crystal structure of the dec-RVKR-cmk inhibitor whereas the HA backbone atoms of S5–S8 and S1′–S2′ were superimposed with the HA0 loop of H3. The coordinates of S1–S4 residues were changed according to the –RVKR– coordinates of inhibitor leading to a newly formed conformation of the HA loop; ii), this loop was then placed manually into the FR active site by superimposition between the –R_KR– residues of the HA loop and the dec-RVKR-cmk with a creation of the furin-substrate complex; iii), the complex was minimized by keeping the S1–S4 residues and the FR fixed; and iv), finally, three steps of the restrained MD simulations at 298 K were carried out for relaxing the modeled systems with the restrain factors of 10, 5, and 2.5 kcal mol−1·Å2 for 200 ps, 200 ps, and 200 ps, respectively. Therefore, the conformation of each cleavage loop was adapted (from the initial model) to fit better with the FR cavity. The last snapshot obtained from restrained MD procedure was used as the starting structure of the substrate-enzyme complex for the next MD simulations with all atoms allowed to move freely for 2 ns.
Molecular dynamics simulations
Three MD simulations for the HA cleavage loop complexed with furin, HPH5–FR, LPH5–FR, and LPH3–FR, were carried out where their initial structures were generated as described above. The simulated systems were neutralized by 6, 11, and 11 Na+ ions and solvated by TIP3P water molecules leading to total atoms of 54,500, 56,663, and 56,667 for the HPH5–FR, LPH5–FR, and LPH3–FR complexes, respectively. The dimensions of the obtained simulation boxes for the three systems are 86 Å × 90 Å × 88 Å. The periodic boundary condition with the NPT ensemble was used. Energy minimization and MD simulations were carried out using the SANDER module of AMBER 7 (23) with the Cornell force field (24). A Berendsen coupling time of 0.2 ps was used to maintain the temperature and standard pressure of the system (25). The SHAKE algorithm (26) was applied to constrain all bonds involving hydrogens. The simulation time step of 2 fs was used. All MD simulations were run with a 12 Å residue-based cutoff for nonbonded interactions and the particle mesh Ewald method was used for an adequate treatment of long-range electrostatic interactions (27). The MD simulations were carried out for 2.0 ns where the production phase starts from 0.75 ns to 2.0 ns. The convergences of energies, temperature, pressure, and global root mean-square displacement (RMSD) were used to verify the stability of the systems. The MD trajectories were collected every 0.2 ps. Analysis of all MD trajectories, i.e., RMSD, distances, hydrogen bonds, etc., were carried out using the Ptraj, CARNAL, and MMGB/SA modules of AMBER 7.
RESULTS AND DISCUSSION
Overall enzyme-substrate structure
To monitor the stability of the three simulated systems, HPH5–FR, LPH5–FR, and LPH3–FR, RMSD of the structures obtained during the 2 ns MD simulations relative to the initial structure of all heavy atoms in the substrate-enzyme complex and the 10 residues of HA substrate were evaluated and plotted in Fig. 2. It can be seen from the plot of RMSD plots versus simulation time that all three systems were found to reach equilibrium at 0.75 ns.
FIGURE 2.
RMSDs relative to the initial structure for all heavy atoms of the substrate-furin complexes (black) and the HA loops (gray) for the three systems studied: HPH5–FR, LPH5–FR, and LPH3–FR.
Enzyme-substrate conformation
The main results refer to the schematic representation shown in Fig. 1 where the conformation structures of the complexes are described in terms of d1–d6 (Fig. 3) in which the reaction coordinates involved in the acylation in three regions, the catalytic triad of FR, nucleophilic attack, and oxyanion hole, were described by the d1–d3, d4, and d5–d6, respectively (Fig. 1). Accordingly, percentage H-bond occupations were evaluated using the CARNAL module in AMBER7 based on the following criteria: i), proton donor-acceptor distance ≤3.5 Å; and ii), donor-H-acceptor bond angle ≥120°. The results were summarized in Table 1.
FIGURE 3.
Distributions of the d1–d6 distances defined in Fig. 1 for the three simulated systems, sampling from 0.75 to 2.0 ns in MD simulations.
TABLE 1.
Percentage of hydrogen bond according to the d1–d3 and d5–d6 distances defined in Fig. 1 for the three simulated systems
| Distance | H-bond | HPH5–FR | LPH5–FR | LPH3–FR |
|---|---|---|---|---|
| d1 | D153_O1…HN1_H194 | 100 | 100 | 0 |
| d2 | D153_O2…HN1_H194 | 96 | 75 | 32 |
| d3 | H194_N2…HO3_S368 | 81 | 90 | 98 |
| d5 | S1-R_ Oγ … HN3_ S368 | 100 | 86 | 15 |
| d6 | S1-R_ Oγ … HN4_N295 | 34 | 6 | 34 |
| S368_O3…HN_S1-R | 71 | 10 | 0 |
At the catalytic triad (Asp153, His194, and Ser368) region of the HPH5–FR, the simulated distances of d1 = d2 = d3 = 2.85 Å (Fig. 3) are comparable to those found experimentally for the inhibitor-furin complex of 2.53 Å, 3.10 Å, and 2.95 Å, respectively (12). This indicates that the three catalytic residues, Asp153, His194, and Ser368, are in the configuration suitable for initiating the nucleophilic reaction. The situation is different for the bound LPAI, H5, and H3, hemagglutinin loops, where only d3 of both systems and d1 of LPH5–FR fall within the range whereby the reaction can take place.
The nucleophilic attack is determined by the distance between the O3-hydroxyl oxygen of Ser368 and the carbonyl carbon of the S1-Arg, d4 in Fig. 1. The d4 bond-making distances were found to be at 3.05 Å, 3.45 Å, and 3.75 Å for the HPH5–FR, LPH5–FR, and LPH3–FR systems, respectively (Fig. 3). In addition to a short d4, the detected sharp and narrow peak was found only in the HPH5–FR complex, signifying the rigidity of the complex that thus serves as a more appropriate configuration for the nucleophilic attack. In contrast, for the other two complexes the d4 peak shows a broad distribution and takes place at significantly longer distances.
At the oxyanion hole region of the FR active site (d5 and d6 in Fig. 1), the hole is generally formed by the backbone nitrogen of Ser368 and the carboxamide nitrogen of Asn295, which specifically binds with the carbonyl group of the S1 reacting residue (12). In Fig. 3, the d5 distance of 2.95 Å for the HPH5–FR (solid line) is slightly shorter than that of 3.15 Å for the LPH5–FR (dashed line) with the presence of a strong hydrogen bond to Ser368, 100% and 86% occupations (d5 in Table 1), respectively. The percentage occupation of 15% for the LPH3–FR confirms a very weak interaction and highly flexible of the complex. This conclusion was also confirmed by a broad distribution of d5 distance varying from 3.00 Å to 5.85 Å (Fig. 3). In terms of d6 distance, the distribution plot for the HPH5–FR complex shows two apparent peaks centered at 2.85 Å and 5.45 Å where the sharper first peak indicated a preferential interaction. For LPAI systems, the d6 distance is detected at 3.05 Å in H3 whereas this distance is significantly longer in H5. This means that interaction between S1 residue and Asn295 was almost lost in the LPH5–FR system. The observed intermolecular distances agree very well with the percentage pair of hydrogen bond through the d5 and d6 in which those values (d5 = 100% and d6 = 34%) for the HPH5–FR are higher than those (d5 = 86% and d6 = 6%) for the LPH5–FR. On the contrary, bonding via these two distances almost disappear for the LPH3–FR complex. Note that the experimental d5 and d6 distances for the inhibitor-furin complex are 3.34 Å and 2.78 Å, respectively (12).
H-bonds of the hemagglutinin loop
To assess to molecular interaction at the cleavage loop of hemagglutinin, percentage, and number of hydrogen bond between each HA residue and the active site residues of FR, were evaluated and plotted in Fig. 4. The criteria used are the same as those mentioned earlier. Here, the experimentally detected hydrogen bonds for the inhibitor-furin complex (12) were also given for comparison, the residue with a box around the amino acid label. Interest was focused on S1-Arg of HA and FR's flat groove—a cavity formed by residues Pro256, Asp258, Ala292, and Asp306. The latter was found to play an important role in facilitating the cleavage reaction by forming hydrogen bonds to the guanidinium side chain (Fig. 1) of P1-Arg of the inhibitor (12) (Si denotes a residue of the substrate whereas Pi represents one of the inhibitors, where i = 1, 2, 3, …) (28). Strong hydrogen bonds between the four flat groove residues and S1-Arg were highly detected for the HPH5–FR complex (S1 in Fig. 4). The cleavage reaction in the HPH5–FR system was also promoted by the other hydrogen bonds from S1-Arg to Ser368 (two bonds, one of which is d5 in Fig. 1) and Ser253 (one bond) of FR with almost 100% occupation. These observations are strongly supported by the experimentally detected hydrogen bonds also shown in Fig. 1. This is in contrast with LPH5–FR in which no hydrogen bond with the flat groove was detected and only a single bond with the catalytic Ser386 (d5 in Fig. 1) was formed. The LPH3–FR loop shows much less interaction with the FR residues, forming no hydrogen with either the flat groove or the catalytic Ser368 residue.
FIGURE 4.
Percent occupations of hydrogen bonds between furin and the ten HA residues (defined in Fig. 1) where the residues with a box around the label represent experimentally detected bonds for the inhibitor-furin complex.
Some comments could be made concerning the two hydrogen bonds formed between the reacting S1-Arg and the catalytic Ser386 of HPH5–FR (Fig. 4). Besides the bond defined by d5, another bond is S368_O3…NH_S1-Arg (71% occupation). This hydrogen bond that has never been found in the oxyanion hole region of any other HA subtype or in the inhibitor-furin complex (12), could be a reason for the shortening and rigidity (narrow and sharp peaks shown in Fig. 3) of the intermolecular d4 distance, which could directly facilitate the nucleophilic reaction in HPH5–FR.
Considering the role of the –RRRKK– insertion (Fig. 1), more hydrogen bonds and a higher percentage occupation between the S2-S6 residues of HA and the surrounding residues of FR (Fig. 4) were found for HPH5–FR in comparison with the two LPAI systems. This means that the –RRRKK– insertion can directly help to hold the substrate in place.
Taking into account all the hydrogen bond data given above, the hemagglutinin loop of HPH5–FR is observed to bind much more tightly into the catalytic site of FR than the LPH5–FR and LPH3–FR systems.
Per residue enzyme-substrate interactions
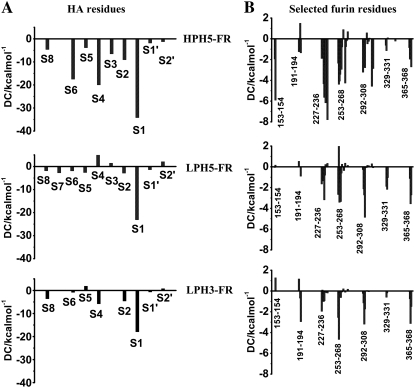
To seek the fundamental basis of substrate-furin interactions, the interaction energy between individual FR residue and the three HA loops and vice versa were calculated using the decomposition energy module of AMBER 7. Plots of the decomposition energies (DC) of the HA residues, as well as the selected FR residues that are located within 5 Å from the 10 residues of HA loop, were shown in Fig. 5. The overall DCs of FR residues were observed in the following order: HPH5–FR ≫ LPH5–FR ∼ LPH3–FR (Fig. 5 B). For the HA loops, the DCs for the residues of HPH5–FR are significantly lower than for the corresponding residues of LPH5–FR and LPH3–FR (Fig. 5 A). This is especially true for the S1, S4, and S6 residues. The calculated results are consistent with experimental results for the inhibitor-furin complex, which stated that P1, P4, and P6 residues of the inhibitor were observed to interact strongly with FR (11,29). In addition, these two P4 and P6 residues are required to dramatically increase the cleavage reaction (30). The DCs for the FR and HA residues agree very well with the hydrogen bond data shown (Fig. 4) and discussed above.
FIGURE 5.
Decomposition energies (DC) of (A) 10 HA residues (defined in Fig. 1), and (B) selected furin residues within 5 Å of those 10 HA residues.
Solvation structure
Hydrophilicity that is determined by the ligand solution in the cavity region of enzyme-substrate complex is known to play an essential role in the catalytic mechanism of biological systems. To seek this information, the plot of the radial distribution functions (RDFs, gxy(r))—the probability of finding the particle of type y in a spherical radii, r, around the particle of type x—was evaluated. Here, the RDFs from the acceptor atoms of FR pocket (O1, O2:Asp153; N1, N2:His194; N4:Asn295, and N3, O3:Ser368) and the reactive HA residue (Oγ:S1-Arg) (see label in Fig. 1), to oxygen atom of water was calculated. The results for the three simulated systems are shown in Fig. 6 together with the corresponding running integration numbers up to the first minimum of the RDF plot.
FIGURE 6.
Radial distribution functions, g(r), from acceptor atoms of furin catalytic residues and HA loops (see Fig. 1 for definition) to oxygen atoms of water molecules for the three simulated systems, HPH5–FR, LPH5–FR, and LPH3–FR complexes.
The RDFs for all oxygen atoms, O1, O2, O3, and Oγ, of the LPH3–FR complex show sharp and narrow first peaks at ∼2.9 Å with the integration number (coordination number, CN) of 1.5, 1, 1 and 2 water molecules, respectively (Fig. 6). The CN denotes the number of water molecules that locate in the first hydration shell around the central atom. This is in contrast for the LPH5–FR complex where only O2 can be accessed by water molecule whereas no water was detected around the four oxygen atoms of HPH5–FR.
The RDFs centered on N1 and N4 atoms of the three systems are almost similar, the first broad peaks at ∼3.8 Å and sharp peaks at ∼3 Å, respectively. Note that, N3 atom of only LPH3–FR was solvated by water molecule. Interest is focused to N2 atom of His194 where the catalytic water was observed and proposed to play role in the reaction mechanism of serine protease (14). In good agreement with our simulated results, one water (CN = 1) was detected to preferentially coordinate (first peak centered at ∼2.9 Å) to N2 atom of only for the HPH5–FR complex. This is not the case for the other two low-pathogenic systems, LPH3–FR and LPH5–FR, where less water, 0.5 and 0.2 were, respectively, was found (Fig. 6, dashed and dotted lines). In addition, distance to the LPH3-FR water molecule of ∼3.45 Å is significantly longer than that of the HPH5–FR complex.
Taking into account the solvation data shown above, the number of overall water molecules around the selected central atoms are in the following order: LPH3–FR > LPH5–FR > HPH5–FR. The obtained data indicates that more water molecules can take place in the catalytic pocket of FR in the complex whose substrate does not fit well with the FR active site, giving a subsequent result of more cavities or less binding between substrate and enzyme. This suggestion agrees very well with the conformational structures of the complexes (Fig. 3) and the hydrogen bond data (Fig. 4) shown and discussed previously.
CONCLUSIONS
The technique of molecular dynamics simulations has been applied to look for detailed information focused on the intra- and intermolecular interactions and geometries of the three substrate-furin complexes, HPH5–FR, LPH5–FR, and LPH3–FR. The results obtained from this study indicate that the –RRRKK– insertion in the HPH5–FR (two arginines at S4 and S6 positions in particular) helps to directly hold the HA cleavage loop in place by forming many strong hydrogen bonds between the residues of HA and FR. This data is well supported by the corresponding strong-decomposition interaction energies and leads consequently to the accumulation of less water molecules accessible to the FR cavity. An active conformation of the HPH5–FR complex suitable for the acylation reaction by FR was formed. In conclusion, the results from this study provide a clear answer to the question of why HPAI hemagglutinin H5 is better cleaved by FR than the other two HA subtypes.
Acknowledgments
P.D. thanks the Sandwich Ph.D. Program and T. R. thanks the Post Doctoral Program from the Commission on Higher Education. The Computer Center for Advanced Research and the Computational Chemistry Unit Cell, Faculty of Science, Chulalongkorn University provided research facilities, software packages, and computing times.
This work was supported by the Thailand Research Fund.
Editor: Ron Elber.
References
- 1.Horimoto, T., and Y. Kawaoka. 2001. Pandemic threat posed by avian Influenza A viruses. Clin. Microbiol. Rev. 14:129–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horimoto, T., and Y. Kawaoka. 1995. The hemagglutinin cleavability of a virulent avian influenza virus by subtilisin-like endoproteases is influenced by the amino acid immediately downstream of the cleavage site. Virology. 210:466–470. [DOI] [PubMed] [Google Scholar]
- 3.Horimoto, T., and Y. Kawaoka. 2005. Influenza: lessons from past pandemics, warnings from current incidents. Nat. Rev. Microbiol. 3:591–600. [DOI] [PubMed] [Google Scholar]
- 4.Steinhauer, D. A. 1999. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology. 258:1–20. [DOI] [PubMed] [Google Scholar]
- 5.Cross, K. J., L. M. Burleigh, and D. Steinhauer. 2001. Mechanisms of cell entry by influenza virus. Expert Rev. Mol. Med. 6:1–18. [DOI] [PubMed] [Google Scholar]
- 6.Stevens, J., O. Blixt, T. M. Tumpey, J. K. Taubenberger, J. C. Paulson, and I. A. Wilson. 2006. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 312:404–410. [DOI] [PubMed] [Google Scholar]
- 7.Neumann, G., and Y. Kawaoka. 2006. Host range restriction and pathogenicity in the context of influenza pandemic. Emerg. Infect. Dis. 12:881–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basak, A., M. Zhong, J. S. Munzer, M. C. Tien, and N. G. Seidah. 2001. Implication of the proprotein convertases furin, PC5 and PC7 in the cleavage of surface glycoproteins of Hong Kong, Ebola and respiratory syncytial viruses: a comparative analysis with fluorogenic peptides. Biochem. J. 353:537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stieneke-Gröber, A., M. Vey, H. Angliker, E. Shaw, G. Thomas, C. Roberts, H. D. Klenk, and W. Garten. 1992. Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin-like endoprotease. EMBO J. 11:2407–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergeron, F., R. Ledue, and R. Day. 2000. Subtilase-like pro-protein convertases: from molecular specificity to therapeutic applications. J. Mol. Endocrinol. 24:1–22. [DOI] [PubMed] [Google Scholar]
- 11.Holyoak, T., C. A. Kettner, G. A. Petsko, R. S. Fuller, and D. Ringe. 2004. Structural basis for differences in substrate selectivity in kex2 and furin protein convertases. Biochemistry. 43:2412–2421. [DOI] [PubMed] [Google Scholar]
- 12.Henrich, S., A. Cameron, G. P. Bourenkov, R. Kiefersauer, R. Huber, I. Lindberg, W. Bode, and M. E. Than. 2003. The crystal structure of the proprotein processing proteinase furin explains its stringent specificity. Nat. Struct. Biol. 10:520–526. [DOI] [PubMed] [Google Scholar]
- 13.Thomas, G. 2002. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Mol. Cell. Biol. 3:753–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Molloy, S. S., P. A. Bresnahan, S. H. Leppla, K. R. Klimpel, and G. Thomas. 1992. Human furin is a calcium-dependent serine endoprotease that recognizes the sequence Arg-X-X-Arg and efficiently cleaves anthrax toxin protective antigen. J. Biol. Chem. 267:16396–16402. [PubMed] [Google Scholar]
- 15.Hosako, M., M. Nagahama, W.-S. Kim, T. Watanabe, K. Hatsuzawa, J. Ikemizu, K. Murakami, and K. Nakayama. 1991. Arg-X-Lys/Arg-Arg motif as a signal for precursor cleavage catalyzed by furin within the constitutive secretory pathway. J. Biol. Chem. 266:12127–12130. [PubMed] [Google Scholar]
- 16.Ishida, T., and S. Kato. 2003. Theoretical perspectives on the reaction mechanism of serine proteases: the reaction free energy profiles of the acylation process. J. Am. Chem. Soc. 125:12035–12048. [DOI] [PubMed] [Google Scholar]
- 17.Hedstrom, L. 2002. Serine protease mechanism and specificity. Chem. Rev. 102:4501–4523. [DOI] [PubMed] [Google Scholar]
- 18.Chutinimitkul, S., T. Songserm, A. Amonsin, S. Payungporn, K. Suwannakarn, S. Damrongwatanapokin, A. Chaisingh, B. Nuansrichay, T. Chieochansin, A. Theamboonlers, and Y. Poovorawan. 2007. New strain of influenza A virus (H5N1), Thailand. Emerg. Infect. Dis. 13:506–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ha, Y., D. J. Stevens, J. J. Skehel, and D. C. Wiley. 2002. H5 avian and H9 swine influenza virus hemagglutinin structures: possible origin of influenza subtypes. EMBO J. 21:865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen, J., K. L. Lee, D. A. Steinhauer, J. Stevens, J. J. Skehel, and D. C. Wiley. 1998. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell. 95:409–417. [DOI] [PubMed] [Google Scholar]
- 21.Accelrys Software Inc. Insight II Environment. 2006. Accelrys Software, San Diego.
- 22.Durbin, R., S. Eddy, A. Krogh, and G. Mitchison. 2004. Needleman-Wunsch Algorithm. Cambridge University:1–3.
- 23.Case, D. A., J. W. Pearlman, and T. E. Caldwell. J. C. III, W. S.Wang, C. L. Ross, T. A. Simmerling, K. M. Darden, R. V. Merz, A. L. Stanton, J. J. Cheng, M. Vincent, V. Crowley, H. Tsui, R. J. Gohlke, Y. Radmer, J. Duan, I. Pitera, G. L. Massova, U. C. Seibel, P. K. Singh, and P. A. Kollman. 2002. AMBER 7. University of California, San Francisco, CA.
- 24.Cornell, W. D., P. Cieplak, C. I. Bayly, I. R. Gould, K. M. Merz, D. M. Ferguson, D. C. Spellmeyer, J. W. C. T. Fox, and P. A. Kollman. 1995. A second generation forcefield for the simulation of proteins, nucleic-acids, and organic-molecules. J. Am. Chem. Soc. 117:5179–5197. [Google Scholar]
- 25.Berendsen, H. J. C., J. P. M. Postma, W. F. V. Gunsteren, and A. DiNola. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690. [Google Scholar]
- 26.Ryckaert, J. P., G. Ciccotti, and H. J. C. Berendsen. 1997. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 23:327–341. [Google Scholar]
- 27.York, D. M., T. A. Darden, and L. G. Pedersen. 1993. The effect of long-range electrostatic interactions in simulations of macromolecular crystals: a comparison of the Ewald and truncated list methods. J. Chem. Phys. 99:8345–8348. [Google Scholar]
- 28.Schechter, I., and A. Berger. 1968. On the active site of proteases. 3. Mapping the active site of papain; specific peptide inhibitors of papain. Biochem. Biophys. Res. Commun. 32:898–902. [DOI] [PubMed] [Google Scholar]
- 29.Rockwell, N. C., D. J. Krysan, T. Komiyama, and R. S. Fuller. 2002. Precursor processing by kex2/furin proteases. Chem. Rev. 102:4525–4548. [DOI] [PubMed] [Google Scholar]
- 30.Nakayama, K. 1997. Furin: a mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem. J. 327:625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]