Abstract
Nonsense-mediated mRNA decay (NMD) is a quality control system that degrades mRNAs containing premature termination codons. Although NMD is well characterized in yeast and mammals, plant NMD is poorly understood. We have undertaken the functional dissection of NMD pathways in plants. Using an approach that allows rapid identification of plant NMD trans factors, we demonstrated that two plant NMD pathways coexist, one eliminates mRNAs with long 3′UTRs, whereas a distinct pathway degrades mRNAs harbouring 3′UTR-located introns. We showed that UPF1, UPF2 and SMG-7 are involved in both plant NMD pathways, whereas Mago and Y14 are required only for intron-based NMD. The molecular mechanism of long 3′UTR-based plant NMD resembled yeast NMD, whereas the intron-based NMD was similar to mammalian NMD, suggesting that both pathways are evolutionarily conserved. Interestingly, the SMG-7 NMD component is targeted by NMD, suggesting that plant NMD is autoregulated. We propose that a complex, autoregulated NMD mechanism operated in stem eukaryotes, and that despite aspect of the mechanism being simplified in different lineages, feedback regulation was retained in all kingdoms.
Keywords: EJC, NMD, SMG-7, UPF, VIGS
Introduction
Nonsense-mediated mRNA decay (NMD) is a eukaryotic quality control system that degrades mRNAs containing premature termination codons (PTC), thereby preventing the accumulation of potentially harmful truncated proteins. NMD also regulates the expression of many wild-type genes (Rehwinkel et al, 2006). NMD discriminates between PTC and authentic stop codons during translation. A stop codon is identified as a PTC if NMD cis elements are present downstream. This triggers the formation of a functional NMD complex on these mRNAs, which in turn target these transcripts for rapid degradation. In yeast and invertebrates, unusually long 3′UTRs can act as NMD cis elements (Gatfield et al, 2003; Amrani et al, 2004; Longman et al, 2007), whereas in mammals, 3′UTR-located introns are the predominant NMD cis elements (Nagy and Maquat, 1998).
UPF1, UPF2 and UPF3 are the core components of the functional NMD complex in yeast as well as animals. However, additional factors including SMG-1, SMG-5, SMG-6 and SMG-7 are also required for NMD in animals. Moreover, in mammals (but not invertebrates), specific components of the exon junction complex (EJC) also have a role in NMD. Two non-mutually exclusive NMD models are suggested. The faux UTR model proposes that the NMD complex is formed and the mRNA is quickly degraded if translation termination is aberrant. It suggests that translation termination of PTC-containing mRNAs is aberrant because their 3′UTR factors, which are required for efficient termination, are not properly positioned. Poly(A) binding protein (PABP) might be the most relevant 3′UTR factor, as its interaction with the terminating ribosome is important for normal translation termination (Amrani et al, 2004). If a long 3′UTR inhibits this interaction, the translation termination will be aberrant. In line with this model, PTC-containing yeast or Drosophila mRNAs are protected from NMD if PABP is tethered downstream of the PTC (Amrani et al, 2004; Behm-Ansmant et al, 2007). However, yeast NMD also targets PTC-containing non-polyadenylated mRNAs, suggesting that other 3′UTR factors also have a role in PTC definition (Meaux et al, 2008).
The second model explains intron-based NMD. When an intron is spliced, an EJC is deposited on the mRNA 20–25 nt upstream of the exon–exon junction (Le Hir et al, 2000). The mammalian EJC consists of four core (Y14, Mago, MLN51/BTZ and eIF4AIII) and many peripheral proteins including UPF3 and UPF2. During translation, the ribosomes displace EJC–UPF3–UPF2 complexes from the mRNA unless they reside downstream of the stop codon (Chang et al, 2007). During the termination of translation, a complex consisting of SMG-1, UPF1 and eukaryotic releasing factors (the SURF complex) binds to the ribosome. If an EJC–UPF2–UPF3 complex is associated with the 3′UTR, the UPF1 component of SURF binds UPF2 and SMG-1 phosphorylates UPF1 (Kashima et al, 2006). Phosphorylated UPF1 might recruit mRNA decay systems to the PTC-containing mRNA. Finally, three related proteins, SMG-5, SMG-6 and SMG-7, recruit a PP2A phosphatase to dephosphorylate UPF1 (Yamashita et al, 2005). As ribosomes also remove EJCs that are located downstream but in close proximity to a stop codon, mammalian transcripts are targeted for NMD only if a stop codon is situated >50 nt upstream of an exon–exon junction (Nagy and Maquat, 1998).
In yeast and humans, NMD complex formation results in decapping and deadenylation of mRNA harbouring a PTC, whereas in Drosophila it leads to the cleavage of aberrant mRNAs. Decapped and/or deadenylated mRNAs are degraded in the cytoplasm and/or in the P-bodies (Isken and Maquat, 2007).
Although little is known about plant NMD, it appears that plant NMD is more complex than the mammalian or the yeast NMD systems. Interestingly, in plants, both long 3′UTRs and 3′UTR-located introns could act as efficient NMD cis factors (Kertesz et al, 2006; Schwartz et al, 2006; Hori and Watanabe, 2007). In addition, the effect of long 3′UTRs is graded, with longer 3′UTRs triggering more efficient NMD. Intron-based plant NMD, like mammalian NMD, acts in a position-dependent manner. An intron located 28 nt downstream of a stop codon fails to destabilize the mRNA, whereas the same intron located 99 nt downstream of the stop codon elicits NMD (Kertesz et al, 2006). So far, only the core NMD trans factors (UPF1, UPF2 and UPF3) have been identified in plants. Mutants in the Arabidopsis UPF3 gene show a strong phenotype (Hori and Watanabe, 2005), whereas null mutants in UPF1 are lethal (Arciga-Reyes et al, 2006; Yoine et al, 2006). PTC-containing mRNAs were found to accumulate to high levels in both mutants. Recently, virus-induced gene silencing (VIGS) was used to characterize UPF2 in Nicotiana attenuata (Wu et al, 2007).
To better understand plant NMD, we identified and characterized novel NMD trans factors with respect to their involvement in long 3′UTR- and intron-based NMD pathways. To achieve these aims, we combined VIGS, a well-documented gene silencing approach in plants (Ratcliff et al, 2001; Valentine et al, 2004), with an agroinfiltration-based transient NMD assay (VIGS-NMD system). Using this system, we have shown that UPF1, UPF2 and SMG-7 are involved in both long 3′UTR-based NMD and intron-based NMD, whereas Mago and Y14 have a role only in intron-based NMD. This demonstrates that the two NMD pathways, despite having some degree of overlap, rely on pathway-specific regulatory genes. We also provide evidence that the mechanisms underlying eukaryotic NMD pathways are evolutionarily conserved. Finally, we demonstrate that plant NMD is feedback controlled, as expression of SMG-7, an NMD trans factor, is regulated by NMD.
Results
Roles of UPF1, UPF2 and UPF3 in plant NMD
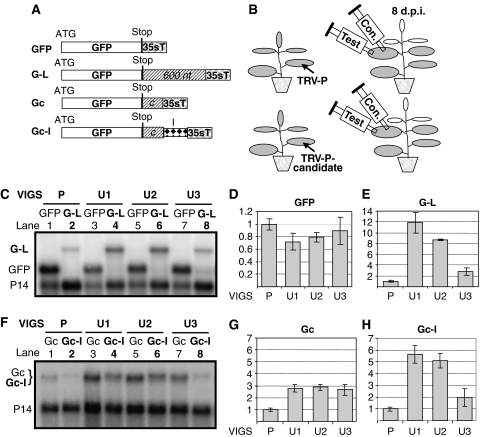
Previously, we described a GFP-based transient NMD assay (Kertesz et al, 2006). Here we have developed an approach (VIGS-NMD system) that allows rapid identification and characterization of NMD trans factors by combining our transient NMD assay with Tobacco rattle virus (TRV)-mediated VIGS (Ratcliff et al, 2001; Liu et al, 2002; Valentine et al, 2004). Following VIGS-mediated knockdown of putative plant NMD factors, NMD activity in silenced plants is assessed by transient expression of GFP-based NMD reporter constructs (Figure 1B). Transient NMD assay is based on agroinfiltration (Supplementary Figure 1) (Kertesz et al, 2006). Infiltration of Nicotiana benthamiana leaves with Agrobacteria expressing GFP leads to abundant GFP mRNA accumulation and strong green fluorescence. In contrast, agroinfiltration with NMD reporter constructs such as those encoding a GFP mRNA with an unusually long 3′UTR (G-L construct) or a GFP mRNA with an intron in the 3′UTR (Gc-I) leads to a weak green fluorescence and low mRNA levels because both G-L and Gc-I transcripts are targeted by NMD. Indeed, upon inhibition of NMD, infiltration of G-L or Gc-I leads to strong fluorescence and abundant transcript levels (Supplementary Figure 1). G-L differs from GFP by harbouring a 600 nt stuffer sequence in the 3′UTR region; thus, G-L transcripts are targeted by long 3′UTR-based NMD (Figure 1A). Gc-I, the reporter construct used to induce intron-based NMD, is also a GFP derivative, but this time containing an intron in the 3′UTR. This intron is separated from the stop codon by a 200 nt stuffer sequence. Gc-I is weakly targeted by the long 3′UTR-based NMD (due to the presence of 200 nt stuffer in the 3′UTR) and strongly targeted by the intron-based NMD. To separate the effect of the two NMD pathways on Gc-I, an intronless construct (Gc), derived from Gc-I, was engineered (Figure 1A). Importantly, although the processed mRNAs transcribed from Gc and Gc-I are identical, Gc-I-derived transcripts accumulate to much lower levels because, although both Gc and Gc-I are weak targets of long 3′UTR-based NMD, intron-based NMD targets only Gc-I (Figure 1F, compare lane 2 with 1; Supplementary Figure 1B) (Kertesz et al, 2006). Thus, the effect of intron-based NMD can be estimated by comparing Gc and Gc-I expression. To compare the expression of different constructs, each construct was co-infiltrated with Agrobacteria expressing P14 and then P14 mRNAs were used as normalization controls (see Figure 1C legend and Materials and methods) (Kertesz et al, 2006).
Figure 1.
Roles of UPFs in plant NMD. (A) Schematic representation of NMD constructs. G-L contains a 600 nt stuffer sequence cloned between the stop and the 35S terminator (35sT). Gc contains a 200 nt stuffer region. Gc-I is the same as Gc but contains an intron (I) between the c-stuffer and the terminator. (B) VIGS-NMD system. N. benthamiana plants are infected with TRV-P or a modified TRV-P harbouring a sequence from a candidate NMD trans factor (TRV-P-candidate). NMD activity of the silenced leaves is tested by agroinfiltration with control (con) and NMD test constructs. (C–E) Long 3′UTR-based NMD is inhibited in UPF-silenced plants. Leaves of P-silenced control (P) or U1-, U2- and U3-silenced test plants (U1, U2, U3) were infiltrated with G-L NMD test constructs (test constructs are shown as bold letters) or with GFP (as a control to show that agroinfiltration worked well in silenced plants). P14 was co-infiltrated with each construct. RNAs isolated at 3 d.p.i. were analysed in gel blot assays using P14 and GFP probes. GFP or G-L transcript levels were normalized to the corresponding P14 mRNA levels. Mean values were calculated from three independent experiments, and then these mean transcript levels were compared and graphically presented. (D, E) Mean values of GFP (D) or G-L mRNA levels (E) of P control leaves are taken as 1 and the corresponding transcript levels of U1-, U2- and U3-silenced leaves are shown relative to it. At NMD test constructs, numbers >1 indicate that the NMD is inhibited. s.d. is indicated by error bar. (F–H) Intron-based NMD activity in UPF-silenced plants. Leaves of silenced plants were infiltrated with Gc control or with Gc-I (bold letters) intron-based NMD test constructs. As processed Gc and Gc-I mRNAs are identical, they run to the same position on RNA gel blot. Gc and Gc-I mRNA levels were normalized and the mean values were calculated as described above. (G, H) Graphical representation of Gc or Gc-I expression in UPF-silenced plants relative to P-silenced control. Note that Gc and Gc-I expressions are similarly increased in U3-silenced leaves relative to P-silenced control, whereas in U1- or U2-silenced leaves Gc-I transcript levels are much more increased than Gc levels.
VIGS-mediated gene inactivation is based on the observation that viral infection triggers RNA silencing (RNAi) in plants. For instance, upon systemic infection of N. benthamiana by a TRV-VIGS vector harbouring a sequence from the phytoene desaturase (PDS) gene (TRV-P vector), silencing of the endogenous PDS mRNAs occurs, resulting in a characteristic leaf bleaching phenotype. To examine the role of UPF proteins in plant NMD, we silenced N. benthamiana UPF1, UPF2 and UPF3 by TRV-VIGS. Sequences from the putative N. benthamiana UPF1, UPF2 and UPF3 were cloned into the TRV-P vector. Plants were infected with TRV-P as a control, or with the modified TRV-P vectors containing sequences from UPF1, UPF2 or UPF3 (infected plants are referred to as P-, U1-, U2- and U3-silenced plants, respectively). In U1-silenced plants, both PDS and UPF1 will be knocked down, whereas in P-silenced control plants only PDS will be inactivated. Spread of the VIGS response can be visualized because PDS silencing leads to bleaching. In each silenced plant, leaf bleaching started at 7 days post inoculation (d.p.i.). RT–PCR confirmed that by 8 d.p.i. target mRNA levels were significantly reduced in the leaves of all silenced plants (data not shown). The long 3′UTR-based NMD activity of U1-, U2-, U3- and P-silenced plants was tested at 8 d.p.i. by infiltrating G-L NMD construct into the leaves. Infiltration of G-L into P-silenced leaves led to weak fluorescence, suggesting that long 3′UTR-based NMD operated efficiently in virus-infected plants (Figure 2A). If UPF1, UPF2 and UPF3 are involved in long 3′UTR-based NMD, this NMD pathway would be inhibited in U1-, U2- and U3-silenced plants and therefore G-L long 3′UTR-based NMD test construct would express to enhanced levels in these plants relative to the P-silenced controls. Enhanced G-L expression manifests in stronger fluorescence and in increased G-L mRNA levels. We found that infiltration of G-L resulted in dramatically enhanced fluorescence in U1- and U2-silenced leaves and in moderately enhanced fluorescence in U3-silenced leaves relative to the P-silenced control plants (Figure 2A). Moreover, G-L mRNAs accumulated to significantly higher levels in U1-, U2- and U3-silenced leaves relative to P-silenced leaves (Figure 1C, compare lanes 4, 6 and 8 with lane 2, and Figure 1E). In U1-, U2- and U3-silenced leaves, G-L mRNA levels were dramatically enhanced in U1- and U2-silenced leaves (12- and 9-fold), but only moderately in U3-silenced leaves (3-fold) compared with P-silenced plants (Figure 1E). These results are in line with previous findings that an endogenous PTC-containing mRNA (trypsin proteinase inhibitor), which is likely targeted by long 3′UTR-based NMD, overaccumulated markedly in U1- and U2-silenced N. attenuata plants but only slightly in U3-silenced plants (Wu et al, 2007). Our data suggest that all UPFs are required for long 3′UTR-based NMD in plants, and that UPF1 and UPF2 have a major role in this plant NMD pathway. However, we do not know whether UPF3 has only a minor role in long 3′UTR-based NMD or that plant NMD can still operate with reduced UPF3 levels (the role of UPF3 in plant NMD is discussed in Supplementary Text 1C). In a separate set of experiments, we studied the role of UPF proteins in intron-based NMD by infiltrating Gc control and Gc-I test constructs into the leaves of silenced plants. Consistent with our previous finding that all three UPFs are required for long 3′UTR-based NMD, we found that Gc, a weak target of long 3′UTR-induced NMD, was expressed to higher levels in U1-, U2- and U3-silenced leaves than in P-silenced plants (Figure 1F, compare lanes 3, 5 and 7 with lane 1, and Figure 1G). Importantly, infiltration of the Gc-I intron-based NMD reporter construct led to a very strong increase in fluorescence (Figure 2A) and Gc-I transcript levels in U1- and U2-silenced leaves (5.5- and 5-fold) relative to P-silenced control leaves (Figure 1F, compare lanes 4 and 6 with lane 2, and Figure 1H). These results suggest that UPF1 and UPF2 are also required for intron-based NMD. In contrast, it appears that intron-based NMD is not affected in U3-silenced leaves. A slight increase in Gc-I mRNA levels was found in U3-silenced leaves relative to P-silenced controls (Figure 1F, compare lane 8 with 2, and Figure 1H). Gc-I is targeted by both long 3′UTR-based NMD and intron-based NMD. As long 3′UTR-based NMD is less efficient in U3-silenced plants than in control leaves (Figure 1G), we conclude that the slight increase in Gc-I expression in U3-silenced leaves is due to the less efficient long 3′UTR-based NMD rather than reduced intron-based NMD activity. These data suggest that UPF3 is not required for intron-based NMD. However, as VIGS-mediated knockdown is not complete, we cannot exclude that UPF3 also has a role in intron-based NMD (see Discussion in Supplementary Text 1C).
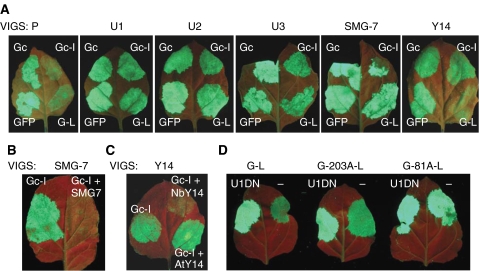
Figure 2.
UPF1, UPF2, UPF3, SMG-7 and Y14 are required for plant NMD. (A) Leaves of silenced control (P) and U1-, U2-, U3-, SMG-7- and Y14-silenced test plants (U1, U2, U3, SMG-7 and Y14) were infiltrated with GFP and Gc control constructs or were infiltrated with G-L or Gc-I NMD test constructs. UV pictures were taken at 3 d.p.i. Fluorescence of the infiltrated silenced plant should be compared with the corresponding patch on P control. Enhanced fluorescence indicates that the silenced gene is required for that type of NMD. (B) Complementation of SMG-7-silenced plants. Leaves of SMG-7-silenced plants were infiltrated with Gc-I or were co-infiltrated with Gc-I and with FLAG-tagged Arabidopsis SMG-7 (SMG-7). Complementation restores NMD activity leading to reduced Gc-I accumulation, which manifests in lowered fluorescence. (C) Y14 complementation. Y14-silenced leaves were infiltrated with Gc-I or were co-infiltrated with Gc-I and with either Arabidopsis (AtY14) or N. benthamiana Y14 (NbY14). Note that only NbY14 complemented the NMD deficiency of Y14-silenced leaf. (D) Position of poly(A) tail has a role in PTC definition. G-L control and PABP localization test constructs (G-203A-L, G-81A-L) were infiltrated or co-infiltrated with a dominant-negative UPF1 (U1DN) construct into N. benthamiana leaves.
A frequent concern is that VIGS might suppress off-target genes, thus leading to phenotypes that are not solely due to the inactivation of the target gene. To prove that altered NMD in U1- and U2-silenced leaves was a consequence of the specific silencing of the targeted UPF gene, we performed a complementation assay (for details, see Supplementary Text 1D). Leaves of U1- and U2-silenced plants were co-infiltrated with the Gc-I NMD reporter construct and a construct expressing Arabidopsis UPF1 or UPF2. It was found that expression of Arabidopsis UPF1 (but not UPF2) restored NMD activity in U1-silenced plants, whereas U2-silenced plants were complemented only by transiently expressed Arabidopsis UPF2 (Supplementary Figure 2). These data confirm that our VIGS-mediated knockdowns were gene specific.
Role of plant SMG-7 in NMD
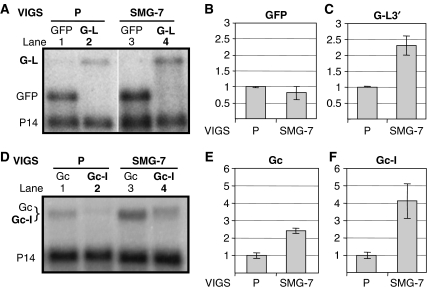
Phosphoregulation of UPF1 has a key role in animal NMD. SMG-1 phosphorylates UPF1, whereas three related proteins (SMG-5, SMG-6 and SMG-7) regulate UPF1 dephosphorylation. An SMG-1 orthologue was not found in Arabidopsis. However, Arabidopsis At5g19400 encodes a 14-3-3-like protein that is similar to SMG-5, SMG-6 and SMG-7 (Fukuhara et al, 2005). In animals, SMG-5 and SMG-6 contain a PIN domain, which is not found in SMG-7 (Glavan et al, 2006). As the predicted At5g19400 product does not have a PIN domain and because it is most similar to SMG-7 (Supplementary Figure 7), we refer to At5g19400 as SMG-7. To test whether SMG-7 is involved in plant NMD, SMG-7-silenced plants were established by infecting N. benthamiana plants with a modified TRV-P VIGS vector containing a segment of N. benthamiana SMG-7 cDNA, and NMD activity in SMG-7-silenced and control plants was tested as described. In SMG-7-silenced leaves, infiltration of both G-L and Gc-I led to strong GFP fluorescence (Figure 2A). Consistent with this, G-L (Figure 3A, compare lane 4 with 2, and Figure 3C) and Gc-I (Figure 3D, compare lane 4 with 2, and Figure 3F) transcripts were significantly increased in SMG-7-silenced plants relative to P-silenced control plants. Thus, we conclude that both long 3′UTR-based NMD and intron-based NMD are inhibited in SMG-7-silenced plants. A complementation assay confirmed that the observed NMD deficiency in SMG-7-silenced leaves was solely due to the inactivation of SMG-7 (Figure 2B; Supplementary Figure 3A–C).
Figure 3.
SMG-7 is required for both types of NMD. (A–C) Effect of SMG-7 silencing on long 3′UTR-based NMD. Silenced control (P) and SMG-7-silenced test plants (SMG-7) were infiltrated with GFP control and G-L NMD test constructs. (D–F) The effect of SMG-7 silencing on intron-based NMD. P- and SMG-7-silenced plants were infiltrated with Gc control or Gc-I NMD test constructs. Panels B, C, E and F are the graphical representations of these experiments. Expression of each construct in SMG-7-silenced plant is shown relative to P-silenced control.
Mago and Y14 are required only for intron-based NMD
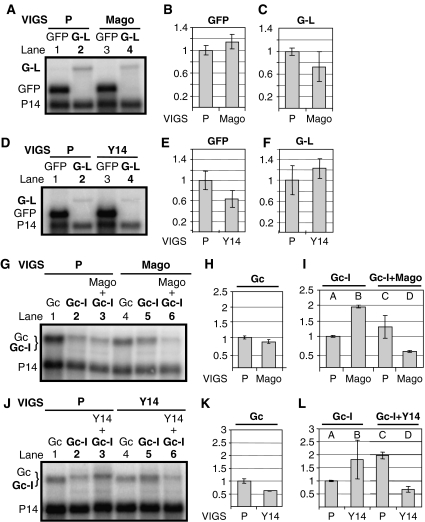
Intron-based mammalian NMD is mediated by the EJC. We hypothesized that intron-based plant NMD is also mediated by the EJC. However, only indirect evidence supports the existence of the EJC in plants. Homology searches found orthologues of Y14, Mago and eIF4AIII as putative EJC core components in Arabidopsis, but failed to identify orthologues of MLN51 (Pendle et al, 2005). We postulated that if the EJC has a role in plant NMD, Y14 and Mago would be required. To test this, NMD activity was tested in Mago- and Y14-silenced plants as described. As G-L mRNA levels did not increase in either Mago- or Y14-silenced leaves relative to controls (Figure 4A–F), we concluded that Mago and Y14 are not required for long 3′UTR-based NMD. In contrast, Gc-I mRNAs were more abundant in both Mago-silenced (Figure 4G, compare lane 5 with 2, and Figure 4I, compare column B with A) and Y14-silenced (Figures 2A and 4J, compare lane 5 with 2, and Figure 4L, compare column B with A) leaves than in control leaves. These data suggest that both Y14 and Mago are required for intron-based NMD. Complementation assays showed that transient expression of N. benthamiana Y14 restored the intron-based NMD activity of Y14-silenced leaves (Figures 2C and 4J, compare lane 6 with 5, and Figure 4L, compare column D with B), whereas expression of N. benthamiana Mago complemented the intron-based NMD activity of Mago-silenced leaves (Figure 4G, compare lane 6 with lane 5, and Figure 4I, compare column D with B). Thus, silencing of both Mago and Y14 was specific.
Figure 4.
Role of Mago and Y14 in plant NMD. (A–F) Mago and Y14 are not involved in long 3′UTR-based NMD. Leaves of P-silenced control (P) and Mago- or Y14-silenced test plants (Mago, Y14) were infiltrated with GFP or G-L long 3′UTR-based NMD test construct. (G–L) Effect of Mago and Y14 VIGS on intron-based NMD. Leaves of silenced plants were infiltrated with Gc control or Gc-I NMD test constructs. Gc expression is not enhanced in Mago- or Y14-silenced leaves (H and K). Gc-I expression is enhanced in both Mago- and Y14-silenced plants relative to the P-silenced control (I and L, compare column B with A). To carry out complementation assay, N. benthamiana Mago or Y14 was co-infiltrated with Gc-I test constructs into Mago- and Y14-silenced leaves (G and J, lanes 6). Complementation led to enhanced NMD activity, which manifested in reduced Gc-I expression (G, J, compare lanes 6 with 5; I, L, compare columns D with B).
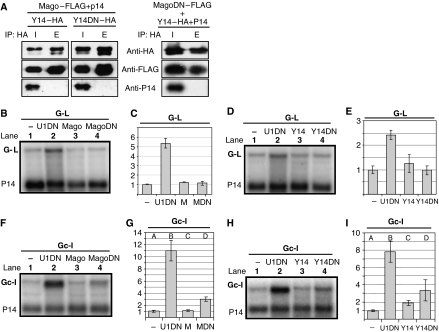
In animals, Y14–Mago and MLN51–eIF4AIII heterodimers form the tetrameric core of the EJC. Changing two conserved residues in human Y14 (L106E/R108E) leads to a mutant protein that is NMD defective in tethering assays. This mutant binds Mago but fails to incorporate into the EJC core (Fribourg et al, 2003), perhaps this incorporation requires a direct interaction between Y14R108 and eIF4AIII (Bono et al, 2006). Similarly, NMD-defective human Mago mutants were reported to bind Y14 but failed to incorporate into the EJC (Fribourg et al, 2003), likely because the residues affected in the mutants normally interact with MLN51. In plants, as in animals, Y14 and Mago form a strong heterodimer (Park and Muench, 2007). If plant Y14–Mago heterodimers act in NMD as components of a tetrameric EJC, and similar interactions stabilize the EJC in both plants and mammals, overexpression of plant mutants corresponding to those described above could inhibit NMD in a dominant-negative manner by depleting the cognate heterodimer pair. To test this, corresponding mutations were introduced into N. benthamiana Y14 (L125E/R127E) and Mago (KF21-22EA/KN46-47DA) (henceforth referred to as Y14DN and MagoDN, respectively). First, co-immunoprecipitation (co-IP) assays were carried out to test if the mutant proteins retained the ability to form heterodimers with their wild-type partners (Figure 5A). HA-tagged Y14DN (Y14DN–HA) was co-infiltrated with a FLAG-tagged Mago (Mago–FLAG) and with P14 (as a negative control) into N. benthamiana leaves. Interactions were studied by HA co-IP assays. Both Y14DN–HA and Mago–FLAG were easily detected in the HA precipitate, suggesting that Y14DN bound Mago in planta. As P14 was present in the input but could not be detected in the precipitate, we concluded that the IP was specific. Similar co-IP assays showed that MagoDN also retained the ability to form a complex with wild-type Y14 (Figure 5A). Next, we studied the effect of overexpression of the mutant proteins on NMD by co-infiltrating them with G-L and Gc-I NMD test constructs. It was found that expression of MagoDN or Y14DN did not affect long 3′UTR-based NMD (Figure 5B–E). In contrast, expression of either MagoDN or Y14DN inhibited intron-based NMD (Figure 5F and H, compare lane 4 with 1, and Figure 4G and I, compare columns D with A). These data also suggest that Y14 and Mago are required only for intron-based plant NMD. Moreover, these results suggest that both Y14 and Mago operate in plant NMD as components of the EJC and that similar interactions stabilize the EJC core complex in plants and mammals.
Figure 5.
Overexpression of a mutant Mago or Y14 inhibits intron-based NMD. (A) Interactions of N. benthamiana Mago and Y14. HA-tagged Y14 (Y14–HA) or a Y14 mutant (Y14DN–HA) was co-infiltrated with a FLAG-tagged Mago (Mago–FLAG) or a mutant Mago (MagoDN–FLAG). P14 was co-infiltrated with each sample. At 3 d.p.i., proteins were extracted and HA co-IP was carried out. Input (I) and elutes of precipitate (E) were analysed by western blotting. P14 probe was used as negative controls. E is 20 × concentrated relative to the input, thus equal signal strength at I and E means that IP efficiency was ∼5%. (B–I) Overexpression of Y14 and Mago dominant-negative mutants inhibits intron-based NMD but does not affect long 3′UTR-based NMD. N. benthamiana leaves were infiltrated with G-L (B–E) or Gc-I (F–I) NMD test construct (−) or were co-infiltrated with a dominant-negative UPF1 (U1DN), a wild-type Y14 or a Mago, and a mutant Y14 (Y14DN) or a Mago (MagoDN). Note that the wild-type Y14 (but not Mago) showed mild dominant-negative effect on intron-based NMD, whereas both Y14DN and MagoDN showed strong dominant-negative effect on intron-based NMD.
The distance between stop and PABP has a role in plant PTC definition
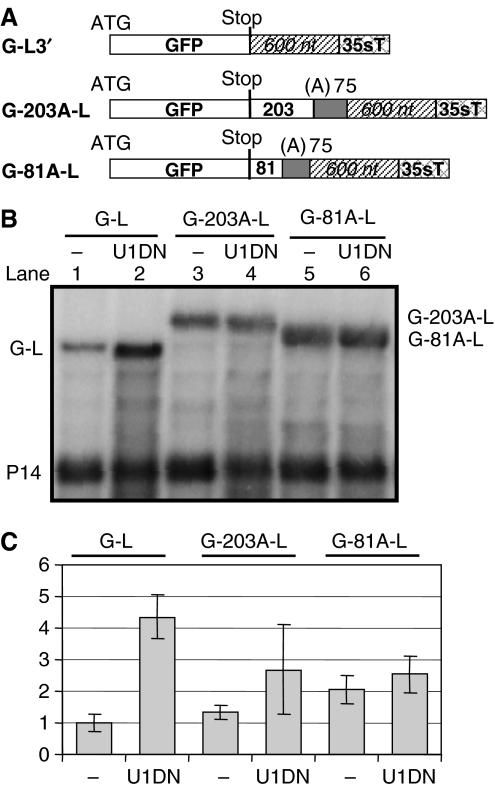
Interactions between PABP and terminating ribosomes stimulate translation termination (Ivanov et al, 2008). In yeast and Drosophila, an NMD complex is assembled and the mRNA is degraded if a long 3′UTR prevents interactions between PABP and terminating ribosomes (Amrani et al, 2004; Behm-Ansmant et al, 2007). We hypothesized that long 3′UTR-based plant NMD is mechanistically similar to yeast or Drosophila NMD. If this is true, artificial localization of PABP downstream of a PTC could protect plant mRNAs with long 3′UTR from NMD by allowing PABP to interact with ribosomes terminating at PTCs. To test this, we generated two constructs (G-81A-L and G-203A-L) by inserting 75 adenosine (A) residues into the G-L NMD reporter construct between the stop codon and the long 3′UTR. These constructs differ only in the length of the spacer that separates the stop codon from the 75A stretch (respectively 81 and 203 nt in the G-81A-L and G-203A-L constructs; Figure 6A). As 75A stretches acted as an efficient binding platform for PABP in yeast and Drosophila (Dower et al, 2004; Behm-Ansmant et al, 2007), we reasoned that plant PABP could also bind to 75A stretches. To study the effect of artificial PABP binding on mRNA levels, expression of the G-81A-L and G-203A-L constructs was compared with a G-L control. We found that G-81A-L (but not G-203A-L) expressed to significantly increased levels relative to the G-L control (Figures 2D and 6B, compare lane 5 with 1, and Figure 6D). Moreover, G-81A-L mRNA levels were not significantly increased in NMD-deficient tissues in co-infiltration experiments with the dominant-negative UPF1 mutant (U1DN) construct, whereas expression of G-L controls was strongly increased in NMD-deficient tissues (Figures 2D and 6B, compare lane 6 with 5 and lane 2 with 1, and Figure 6C). Therefore, we concluded that G-81A-L transcripts were not targeted by NMD. Thus, artificial localization of PABP to the close proximity of a PTC protects mRNAs from long 3′UTR-activated NMD. These data suggest that in plants, as in yeast and Drosophila, the distance between the stop codon and the PABP has an important role in PTC definition.
Figure 6.
Role of PABP position in plant PTC definition. (A) Constructs used for artificial PABP localization assays. (B, C) G-L control and test (G-203A-L, G-81A-L) constructs were infiltrated or were co-infiltrated with a dominant-negative UPF1 (U1DN) into N. benthamiana leaves. RNA gel blot was hybridized with P14 and GFP probes.
Conserved interactions of eukaryotic UPF proteins
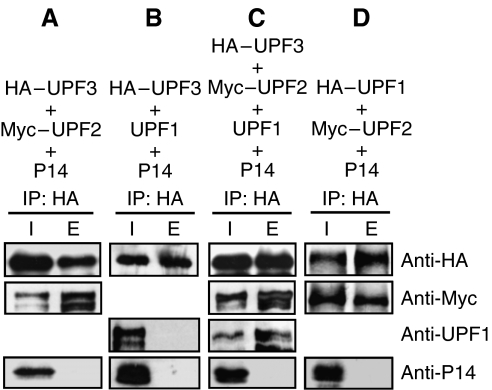
The interactions between UPF proteins are a conserved feature in fungi and animals, where UPF2 connects UPF3 and UPF1 (He et al, 1997; Serin et al, 2001). To map the interactions of plant UPF proteins, we carried out co-IP assays. HA-tagged Arabidopsis UPF3 (HA–UPF3) was co-infiltrated with a Myc-tagged Arabidopsis UPF2 (Myc–UPF2) and with P14 negative control. HA antibody precipitated both HA–UPF3 and Myc–UPF2, indicating that UPF3 bound UPF2 in planta. P14 was present only in the input (Figure 7A), indicating that IP was specific. Similar assays revealed that HA–UPF1 also bound Myc–UPF2 (Figure 7D). To test if plant UPF2 connects UPF1 and UPF3, we compared the interactions of UPF3 and UPF1 in the absence and presence of UPF2. As Figure 7B shows, HA–UPF3 failed to immunoprecipitate with UPF1 in the absence of UPF2. However, when UPF1, Myc–UPF2 and HA–UPF3 were co-infiltrated, all three proteins could be detected in the HA IP fraction (Figure 7C). These data suggest that the plant UPF2, like yeast and mammalian UPF2, connects UPF3 and UPF1. Therefore, we conclude that the interactions of UPF proteins are conserved in all eukaryotic kingdoms.
Figure 7.
Interactions between UPF proteins. (A–D) Interactions were analysed by HA co-IP assays. N. benthamiana leaves were co-infiltrated with Arabidopsis UPF1, UPF2 and UPF3 in different combinations. P14 was co-infiltrated with each mixture. Elute (E) is 20 × concentrated relative to the input (I).
In mammals, Staufen selectively destabilizes mRNAs by recruiting UPF1 to Staufen bound transcripts (Kim et al, 2007). We showed that tethering of plant UPF1 to either the 5′UTR or 3′UTR of an mRNA results in a strong reduction of mRNA levels (Kertesz et al, 2006). Therefore, it is possible that mRNA binding plant proteins that can bind UPF1 also destabilize their target mRNAs by recruiting UPF1. To isolate UPF1-interacting plant proteins, a yeast two-hybrid screen was carried out using UPF1 as bait. The strongest interactor corresponded to the C-terminal 439 amino acids of UPF2 (C-UPF2). Co-IP assays showed that UPF1 could also bind C-UPF2 in planta (data not shown). These data suggest that other UPF1 interactions could also be physiologically relevant (Supplementary Figure 4). For instance, the second strongest interactor is a putative non-LTR retrotransposon. In mammals, many transcripts derived from transposons and retroviral elements are NMD regulated. It is tempting to speculate that the same is true in plants. However, the biological relevance of UPF1 interactions remained to be determined.
Regulation of plant NMD
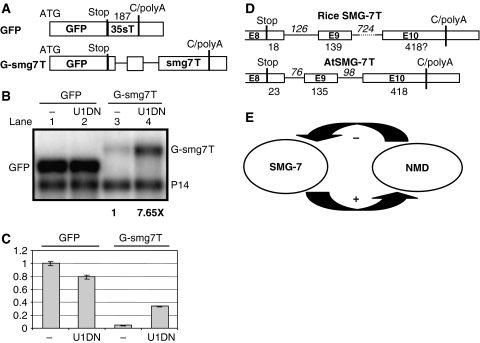
It is not known what proportion of the plant transcriptome is affected by NMD. However, as plant NMD could target mRNAs harbouring 3′UTR longer than 300 nt (Kertesz et al, 2006; Schwartz et al, 2006) and because ∼18% of Arabidopsis 3′UTRs are longer than 300 nt (data not shown), it is likely that NMD might modify the expression of many plant genes. Moreover, NMD might also target other types of transcript, for instance mRNAs with 3′UTR-located introns or mRNAs with an ORF in the 5′ UTR region. Thus, we hypothesize that plant NMD should be strictly regulated. As the simplest regulatory system would be a feedback control, we searched the gene structure of NMD trans factors for potential NMD cis elements. We found that the 3′UTR of Arabidopsis SMG-7 is exceptionally long (576 nt) and that it contains two introns 23 and 158 nt downstream of the stop codon (Figure 8D). Thus, SMG-7 mRNA might be targeted by NMD. Indeed, our qRT–PCR analysis revealed that SMG-7 mRNA was overexpressed (4- to 6-fold) in Arabidopsis UPF3 mutants (data not shown). In line with these data, microarray analysis showed that SMG-7 was moderately (two-fold) overexpressed in a missense UPF1 mutant (Yoine et al, 2006). These data suggest that Arabidopsis SMG-7 is downregulated by NMD.
Figure 8.
SMG-7 is downregulated by NMD. (A) The terminator of Arabidopsis SMG-7 gene triggers NMD. Schematic representation of the constructs. C/polyA shows the annotated polyadenylation sites. Introns are shown as thin lines. (B, C) GFP and G-smg7T test construct were infiltrated (lanes 1 and 3) or were co-infiltrated (lanes 2 and 4) with a dominant-negative UPF1 (U1DN) into N. benthamiana leaves. Bold numbers below the blot show the effect of U1DN co-infiltration on G-smg7T expression. (C) Graphical representation of the expression of each construct relative to GFP. (D) The structure of Arabidopsis and rice SMG-7 3′UTRs is conserved. (E) Model of autoregulation of plant NMD.
To test if NMD-mediated downregulation of SMG-7 transcripts is conserved, we measured the N. benthamiana SMG-7 mRNA levels in different silenced leaves. As expected, SMG-7 transcripts accumulated to low levels in SMG-7-silenced leaves (0.3-fold relative to a P-silenced control sample), showing that SMG-7 silencing was effective. Importantly, SMG-7 transcript levels were strongly increased in U1- and U2-silenced leaves (6.5- and 8-fold, respectively) and moderately enhanced in U3-silenced leaves (2-fold) but were not altered in Y14- and Mago-silenced samples (Supplementary Figure 5A). These data suggest that N. benthamiana SMG-7 mRNAs are negatively regulated by long 3′UTR-based NMD. The 3′UTR region of N. benthamiana SMG-7 was cloned and sequenced. It was found to resemble the 3′UTR of Arabidopsis SMG-7 on the basis of its length (>600 nt) and the presence of two introns in similar positions (data not shown). Moreover, we found that the structure (but not the sequence) of the SMG-7 3′UTRs is conserved within flowering plants as the 3′UTRs of SMG-7 orthologues of other dicot and a monocot species were also exceptionally long and contained two introns in similar positions (Supplementary Figure 6 and Figure 8D). As very long 3′UTRs and introns located within 3′UTRs are rare in plants, it is likely that the extreme length as well as the presence and position of SMG-7 3′UTR introns are under strong selection. It is tempting to speculate that the SMG-7 3′UTR structure is conserved because it can act as a sensor for NMD and ‘measure' the intensity of long 3′UTR-based NMD. Moreover, it is possible that in cells where long 3′UTR-based NMD is impaired, the SMG-7 3′UTR can sense the intensity of intron-based NMD. Our results show that SMG-7 transcripts are downregulated by NMD and suggest that SMG-7 is a direct target of NMD. To prove experimentally that SMG-7 is a direct target, we replaced the terminator of the GFP construct with the terminator of Arabidopsis SMG-7 (G-smg7T) (Figure 8A), and then compared the expression of G-smg7T in normal tissues and tissues in which NMD was inactivated by coexpressing a dominant-negative UPF1 mutant. Although G-smg7T was weakly expressed in wild-type leaves, G-smg7T expression was dramatically increased in NMD-deficient tissues (Figure 8B and C; Supplementary Figure 5B). These data show that the smg7T terminator triggers NMD. Therefore, we propose that in Arabidopsis, SMG-7 mRNAs are directly targeted by NMD. As SMG-7 is required for both types of plant NMD, the findings that Arabidopsis SMG-7 transcripts are targeted by NMD suggest that plant NMD is feedback controlled. Intense NMD would reduce SMG-7 expression and thus lead to a weakening in NMD activity (Figure 8E).
Discussion
Previously, we have found that two NMD pathways operate in plants, one degrades mRNAs with long 3′UTRs, whereas the second pathway eliminates mRNAs with 3′UTR-located introns. Here, we have characterized several plant NMD trans factors. Our data reveal an unexpected conservation of eukaryotic NMD pathways. Moreover, these data suggest that a similar autoregulatory circuit controls NMD activity in plants, fungi and animals.
Conservation of eukaryotic NMD pathways
VIGS-NMD assays (advantages of this system are summarized in Supplementary Text 1D) demonstrate that plant UPF1, UPF2, UPF3 and SMG-7 are required for long 3′UTR-based NMD. As these proteins are also required for long 3′UTR-based NMD in yeast and animals, and because interactions between UPF proteins are conserved within eukaryotes, we propose that the mechanism of long 3′UTR-based NMD is similar in most eukaryotes. In addition, artificial binding of PABP downstream of a PTC protects mRNAs from NMD in plants, Drosophila and yeast. Therefore, we suggest that in plants, as in yeast or Drosophila, the interaction of PABP with the terminating ribosome stimulates translation termination, and that inefficient termination leads to NMD-mediated mRNA decay.
UPF1, UPF2, SMG-7, Y14, Mago, and possibly UPF3, are components of intron-based plant NMD. Importantly, in plants, as in mammals, Y14 and Mago are required only for intron-based NMD. Taken together with the finding that 3′UTR-located introns elicit NMD in a position-dependent manner in plants as well as in mammals, these results suggest that the mechanism of plant and mammalian intron-based NMD is similar. We propose that the plant splicing machinery also deposits an EJC upstream of the exon–exon boundary, and that 3′UTR-located plant EJCs can induce NMD-mediated mRNA degradation. It would be interesting to test if 3′UTR-associated plant EJCs, like mammalian EJCs, have an active role in NMD, or if plant EJCs promote NMD in a passive manner, for example by hindering steric interactions between PABP and the terminating ribosome.
UPF1 phosphoregulation is required for NMD in yeast and animals (Yamashita et al, 2005; Wang et al, 2006). UPF1 phosphorylation could also be important for both types of plant NMD. Mammalian SMG-7 binds phosphorylated UPF1 by means of its 14-3-3 phosphoserine binding domain and recruits (with SMG-5) a PP2A phosphatase to UPF1 (Fukuhara et al, 2005). Moreover, SMG-7 might remobilize phosphorylated UPF1-bound mRNAs to P-bodies (Unterholzner and Izaurralde, 2004). As SQ phosphorylation motifs are present in plant UPF1 (Dalmay et al, 2001), and because residues required for phosphoserine binding are conserved in plant SMG-7 (Supplementary Figure 7) (Fukuhara et al, 2005), we suggest that plant UPF1 is phosphoregulated and that SMG-7 binds phosphorylated UPF1. Plant SMG-7 might regulate UPF1 dephosphorylation or/and remobilize phosphorylated UPF1-containing mRNP complexes into P-bodies.
Evolution of eukaryotic NMD systems
It is proposed that long 3′UTR-based NMD operated in stem eukaryotes (the common ancestors of extant eukaryotes) and has evolved into a more complex intron-based NMD in vertebrates (Rehwinkel et al, 2006). An alternative theoretical model has suggested that in stem eukaryotes, intron-based NMD already existed and that it was mediated by the EJC (Lynch and Kewalramani, 2003). As we found that in plants, which are outgroup relatives to fungi and animals, both long 3′UTR- and intron-based NMD operated, we proposed that both NMD systems functioned in stem eukaryotes (Kertesz et al, 2006). This model predicts that the mechanisms of both NMD pathways are conserved between plants and animals. In agreement with this prediction, our results suggest that plant long 3′UTR-based NMD is mechanistically similar to yeast or Drosophila NMD (Figure 6), and that plant intron-based NMD, like mammalian NMD, is mediated by the EJC (Figures 4 and 5). These data support the model that in stem eukaryotes, an unexpectedly complex NMD system operated and that this system was simplified in certain lineages. For instance, splicing and NMD could have become uncoupled in intron-loss dominated lineages.
Here, we extend our NMD evolution model by proposing that NMD was already feedback controlled in stem eukaryotes. We found that SMG-7 is required for plant NMD and that plant SMG-7 is directly regulated by NMD (Figures 3 and 8). 14-3-3-like SMG proteins are essential for NMD in animals and the orthologue of SMG-7 promotes yeast NMD (Luke et al, 2007). Although NMD regulates different genes in yeast, Drosophila and mammals, 14-3-3-like SMGs are the striking exceptions. Mammalian SMG-5 (Chan et al, 2007), Drosophila SMG-5 and SMG-6 (Rehwinkel et al, 2005) and the yeast SMG-7 (Dahlseid et al, 2003) are overexpressed in NMD-deficient lines. Thus, SMG-7-related NMD factors are downregulated by NMD in plants, fungi and animals. Therefore, we propose that SMG-7 was already required for NMD in stem eukaryotes and that it was also controlled by NMD. This feedback could be important in regulating NMD activity, thereby protecting target mRNAs from hyperactive NMD. We believe that this SMG-mediated NMD autoregulation has been retained during the evolution of eukaryotes.
Materials and methods
Plasmid constructs
For agroinfiltrations, genes were cloned into Bin61S vector. P14, GFP, Gc, G-L, Gc-I, UPF1 and U1DN have been described. G-L and Gc-I plasmids were previously named as GFP-abc and GFPcLS+ (Kertesz et al, 2006). Details of clonings are presented in Supplementary Text 1. Bin-tag vectors are derivatives of pPILY-MESHI vectors (Ferrando et al, 2000). A modified binary TRV2 VIGS vector (Liu et al, 2002) was obtained by subcloning a fragment from pPK20.2b (Valentine et al, 2004). GFPc3 encompassing the 2b ORF, a duplicated copy of the CP subgenomic promoter upstream of GFPc3, was inserted into the binary TRV RNA2 vector pYL156 (Jens Tilsner, personal communication) to give pBinTRV-2b-GFP. To obtain TRV-P vector, the GFP region of pBinTRV-2b-GFP was replaced with a 319 nt from N. benthamiana PDS. N. benthamiana VIGS fragments were RT–PCR generated and cloned into TRV-P. Alignments of VIGS sequences with corresponding Arabidopsis sequences are shown in Supplementary Text 1E.
Agroinfiltration assays
Agroinfiltrations and GFP detections have been described previously (Silhavy et al, 2002). Cultures were mixed before infiltration. The final OD600 of each culture was 0.4 (except P14, which was 0.2) in the mixture. P14, a viral silencing suppressor, was co-infiltrated with each construct; thus, it prevented the activation of Agrobacterium-induced RNA silencing, which could have degraded the transgenic mRNAs (Kertesz et al, 2006). Moreover, P14 mRNAs were used as internal controls for RNA studies and P14 protein was used as a negative control in co-IP assays.
VIGS and complementation
To trigger VIGS, leaves of 21-day-old N. benthamiana were co-infiltrated with a mixture of three Agrobacterium cultures. One expressed P14, the second expressed TRV RNA1 (BINTRA6 vector) (Ratcliff et al, 2001), whereas the third expressed TRV-P VIGS vector or its derivatives. To complement VIGS plants, NMD constructs were co-infiltrated with a culture expressing the complementation factor (for details see Supplementary Text 1D).
Co-immunoprecipitation assay
Co-IP was carried out as described (Baumberger and Baulcombe, 2005) except G-25 separation was omitted. Anti-HA affinity matrix (Roche) was used for IPs. Rabbit polyclonal antibodies (anti-UPF1 and anti-P14) or monoclonal antibodies (anti-c-Myc peroxidase (Roche), anti-HA peroxidase (Roche), anti-FLAG M2-peroxidase (Sigma)) were used for chemiluminescence protein detections according to the manufacturer's instructions (ECL, Amersham-Biosciences).
Yeast two-hybrid screen
pGBKT7 (Clontech) was used to prepare GAL4 binding domain–UPF1 fusions for bait. Two-hybrid cDNA library was prepared from mRNA isolated from cell suspension of Arabidopsis Columbia ecotype, and cloned into Clontech pACT2 vector.
RT–PCR assays and RNA gel blot analysis
RNA methods and quantifications have been described previously (Silhavy et al, 2002; Kertesz et al, 2006). RT–PCR was carried out with QIAGEN OneStep RT–PCR Kit. For qRT–PCR, RNA was isolated with the RNeasy Plant Mini kit (Qiagen) and first strand was synthesized using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative PCR was performed with RotorGene RG-3000 PCR and analysed with RotorGene software (Corbett Research). Power SYBR Green PCR Master Mix (Applied Biosystems) was used for reactions. The expression of SMG-7 was normalized to actin or ubiquitin.
Supplementary Material
Supplementary Information
Supplementary Figures
Supplementary Figure Legends
Acknowledgments
We are grateful to Dr E Izzauralde (EMBL, Heidelberg, Germany) for pAdh-(A)75-Hhr, to Dr Cs Koncz (MPIZ Koln, Germany) for tag plasmids and to Dr D Baulcombe (University of Cambridge, UK) and Dr SP Dinesh-Kumar (University of Yale, USA) for TRV vectors. We thank Dr G Ingram (University of Edinburgh, UK) for critical reading of the manuscript. We thank M Magna and G Sotirios for creating Bin-tag and G-smg7T constructs. This work was supported by grants from the OTKA (K60102 and 60160) and the NKTH (4/064/2004, ‘Búzakalász'). F Nagy was supported by Howard Hughes Medical Institute International Research Scholar Grant. D Silhavy was supported by the Bolyai Scholarship. Z Mérai is a graduate student of ELTE ‘Classical and Molecular Genetics PhD Program'.
References
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A (2004) A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118 [DOI] [PubMed] [Google Scholar]
- Arciga-Reyes L, Wootton L, Kieffer M, Davies B (2006) UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J 47: 480–489 [DOI] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC (2005) Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA 102: 11928–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E (2007) A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J 26: 1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono F, Ebert J, Lorentzen E, Conti E (2006) The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell 126: 713–725 [DOI] [PubMed] [Google Scholar]
- Chan WK, Huang L, Gudikote JP, Chang YF, Imam JS, MacLean JA II, Wilkinson MF (2007) An alternative branch of the nonsense-mediated decay pathway. EMBO J 26: 1820–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF (2007) The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 76: 51–74 [DOI] [PubMed] [Google Scholar]
- Dahlseid JN, Lew-Smith J, Lelivelt MJ, Enomoto S, Ford A, Desruisseaux M, McClellan M, Lue N, Culbertson MR, Berman J (2003) mRNAs encoding telomerase components and regulators are controlled by UPF genes in Saccharomyces cerevisiae. Eukaryot Cell 2: 134–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T, Horsefield R, Braunstein TH, Baulcombe DC (2001) SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J 20: 2069–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower K, Kuperwasser N, Merrikh H, Rosbash M (2004) A synthetic A tail rescues yeast nuclear accumulation of a ribozyme-terminated transcript. RNA 10: 1888–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando A, Farras R, Jasik J, Schell J, Koncz C (2000) Intron-tagged epitope: a tool for facile detection and purification of proteins expressed in Agrobacterium-transformed plant cells. Plant J 22: 553–560 [DOI] [PubMed] [Google Scholar]
- Fribourg S, Gatfield D, Izaurralde E, Conti E (2003) A novel mode of RBD-protein recognition in the Y14–Mago complex. Nat Struct Biol 10: 433–439 [DOI] [PubMed] [Google Scholar]
- Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E (2005) SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell 17: 537–547 [DOI] [PubMed] [Google Scholar]
- Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E (2003) Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J 22: 3960–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glavan F, Behm-Ansmant I, Izaurralde E, Conti E (2006) Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J 25: 5117–5125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Brown AH, Jacobson A (1997) Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol Cell Biol 17: 1580–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Watanabe Y (2005) UPF3 suppresses aberrant spliced mRNA in Arabidopsis. Plant J 43: 530–540 [DOI] [PubMed] [Google Scholar]
- Hori K, Watanabe Y (2007) Context analysis of termination codons in mRNA that are recognized by plant NMD. Plant Cell Physiol 48: 1072–1078 [DOI] [PubMed] [Google Scholar]
- Isken O, Maquat LE (2007) Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev 21: 1833–1856 [DOI] [PubMed] [Google Scholar]
- Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE (2008) Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J 27: 736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S (2006) Binding of a novel SMG-1–Upf1–eRF1–eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev 20: 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz S, Kerenyi Z, Merai Z, Bartos I, Palfy T, Barta E, Silhavy D (2006) Both introns and long 3′-UTRs operate as cis-acting elements to trigger nonsense-mediated decay in plants. Nucleic Acids Res 34: 6147–6157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Furic L, Parisien M, Major F, DesGroseillers L, Maquat LE (2007) Staufen1 regulates diverse classes of mammalian transcripts. EMBO J 26: 2670–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ (2000) The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J 19: 6860–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP (2002) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 30: 415–429 [DOI] [PubMed] [Google Scholar]
- Longman D, Plasterk RH, Johnstone IL, Caceres JF (2007) Mechanistic insights and identification of two novel factors in the C. elegans NMD pathway. Genes Dev 21: 1075–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B, Azzalin CM, Hug N, Deplazes A, Peter M, Lingner J (2007) Saccharomyces cerevisiae Ebs1p is a putative ortholog of human Smg7 and promotes nonsense-mediated mRNA decay. Nucleic Acids Res 35: 7688–7697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Kewalramani A (2003) Messenger RNA surveillance and the evolutionary proliferation of introns. Mol Biol Evol 20: 563–571 [DOI] [PubMed] [Google Scholar]
- Meaux S, van Hoof A, Baker KE (2008) Nonsense-mediated mRNA decay in yeast does not require PAB1 or a Poly(A) tail. Mol Cell 29: 134–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy E, Maquat LE (1998) A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci 23: 198–199 [DOI] [PubMed] [Google Scholar]
- Park NI, Muench DG (2007) Biochemical and cellular characterization of the plant ortholog of PYM, a protein that interacts with the exon junction complex core proteins Mago and Y14. Planta 225: 625–639 [DOI] [PubMed] [Google Scholar]
- Pendle AF, Clark GP, Boon R, Lewandowska D, Lam YW, Andersen J, Mann M, Lamond AI, Brown JW, Shaw PJ (2005) Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol Biol Cell 16: 260–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff F, Martin-Hernandez AM, Baulcombe DC (2001) Technical advance. Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J 25: 237–245 [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Raes J, Izaurralde E (2006) Nonsense-mediated mRNA decay: target genes and functional diversification of effectors. Trends Biochem Sci 31: 639–646 [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E (2005) Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA 11: 1530–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz AM, Komarova TV, Skulachev MV, Zvereva AS, Dorokhov Iu L, Atabekov JG (2006) Stability of plant mRNAs depends on the length of the 3′-untranslated region. Biochemistry (Mosc) 71: 1377–1384 [DOI] [PubMed] [Google Scholar]
- Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE (2001) Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol Cell Biol 21: 209–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy D, Molnar A, Lucioli A, Szittya G, Hornyik C, Tavazza M, Burgyan J (2002) A viral protein suppresses RNA silencing and binds silencing-generated, 21-to 25-nucleotide double-stranded RNAs. EMBO J 21: 3070–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Izaurralde E (2004) SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell 16: 587–596 [DOI] [PubMed] [Google Scholar]
- Valentine T, Shaw J, Blok VC, Phillips MS, Oparka KJ, Lacomme C (2004) Efficient virus-induced gene silencing in roots using a modified tobacco rattle virus vector. Plant Physiol 136: 3999–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Cajigas IJ, Peltz SW, Wilkinson MF, Gonzalez CI (2006) Role for Upf2p phosphorylation in Saccharomyces cerevisiae nonsense-mediated mRNA decay. Mol Cell Biol 26: 3390–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Kang JH, Hettenhausen C, Baldwin IT (2007) Nonsense-mediated mRNA decay (NMD) silences the accumulation of aberrant trypsin proteinase inhibitor mRNA in Nicotiana attenuata. Plant J 51: 693–706 [DOI] [PubMed] [Google Scholar]
- Yamashita A, Kashima I, Ohno S (2005) The role of SMG-1 in nonsense-mediated mRNA decay. Biochim Biophys Acta 1754: 305–315 [DOI] [PubMed] [Google Scholar]
- Yoine M, Ohto MA, Onai K, Mita S, Nakamura K (2006) The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signalling in Arabidopsis. Plant J 47: 49–62 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Figures
Supplementary Figure Legends