Abstract
We report here on the design, synthesis and evaluation of small molecule inhibitors of the interaction between a steroid receptor coactivator and estrogen receptor α. These inhibitors are based upon an amphipathic benzene scaffold whose hydrophobic face mimics the leucine-rich α-helical consensus sequence on the steroid receptor coactivators that interacts with a shallow groove on estrogen receptor α. Several of these molecules are among the most potent inhibitors of this interaction described to date, and they are active at low micromolar concentrations in both in vitro models of estrogen receptor action and in cell-based assays of estrogen receptor-mediated coactivator interaction and transcription.
INTRODUCTION
Estrogen receptor α (ERα), a ligand-activated transcription factor and member of the nuclear hormone receptor superfamily, is a well-validated target for the treatment of breast cancer, osteoporosis, and other endocrine disorders. It exerts its genomic effects by binding natural or synthetic estrogens, and then recruiting steroid receptor coactivators (SRCs) that regulate the magnitude of gene transcription. SRCs of the p160 class bind to the ER through “nuclear receptor (NR)-box” sequence motifs comprising two turns of an amphipathic α-helix and containing an LXXLL sequence (where L is leucine and X is any amino acid, though typically a polar one, e.g., His, Arg); these NR boxes bind to the C-terminal activation function 2 of ER-agonist complexes in a shallow hydrophobic groove formed by hydrophobic residues from helices 3, 4, 5, and 12.(1) Conventional ER antagonists, exemplified by the selective estrogen receptor modulator (SERM) tamoxifen, block this interaction indirectly: The large basic side chain of tamoxifen disrupts the folding of helix 12, causing it to move so that it blocks the recruitment of SRC-NR boxes to the ER.(1–3) Although many breast cancer patients benefit from tamoxifen therapy, resistance to this drug develops in the majority of cases; moreover, if tamoxifen treatment is not discontinued after onset of resistance, the drug can actually promote, rather than inhibit, tumor growth.(4)
We (5,6) and others (7–9) have envisioned a novel strategy to overcome this problem: Directly blocking the ER/SRC interaction with a small molecule, termed a “coactivator binding inhibitor” (CBI), that is capable of binding to the groove formed on the receptor surface. This approach is unique in that it allows for the activity of an agonist-bound ER to be inhibited directly, thereby potentially circumventing the need for SERM treatment and the risk of the development of tamoxifen resistance. Because protein-protein interactions typically occur over large surface areas, they have historically been viewed as difficult targets for inhibition by small molecules, but, auspiciously, there have been a number of recent advances in the field.(10–13) Due to the short, well-defined nature of the LXXLL interaction motif,(14) the ERα-SRC interaction seems to be a promising target for small molecule therapy. Toward that goal, we report here a series of amphipathic benzene CBIs obtained from de novo design that are active as inhibitors in cell-based assays of ERα-mediated transcription.
RESULTS AND DISCUSSION
Design and Synthesis
We have previously reported the trisubstituted pyrimidine A that inhibits the interaction of ERα and SRC1 NR Box II at mid-micromolar potency as assayed in a fluorescence polarization assay.(6) The three leucine residues of the LXXLL motif are sufficiently mimicked by the three alkyl substituents in A, but we were interested in synthesizing molecules of a more refined design: Ones that not only keep the 1,3,5-relationship of the alkyl substituents, but also are both more conformationally constrained in their positioning of leucine-mimicking sidechains and incorporate structural elements that mimic the amphipathic nature of the LXXLL α-helix (Figure 1). Benzenes exhibiting a substitution pattern of alternating hydrophobic and hydrophilic residues give rise to facially amphipathic molecules that have been used as tripodal receptors for metals and ions (15, 16) as well as generating supramolecular assemblies.(17, 18) Owing to the amphipathic nature of the NR-box helix (e.g., solvent exposed His and Arg residues in SRC1 Box II), as well as the success garnered in mimicking the NR-box Leu residues in our previously reported work in this area, we designed a series of hexa-substituted amphipathic benzenes with alternating hydrophobic groups to mimic Leu residues and with hydrophilic groups to increase solubility, to mimic the amphipathic nature of the peptide, and to allow interaction with the exposed solvent.
Figure 1.
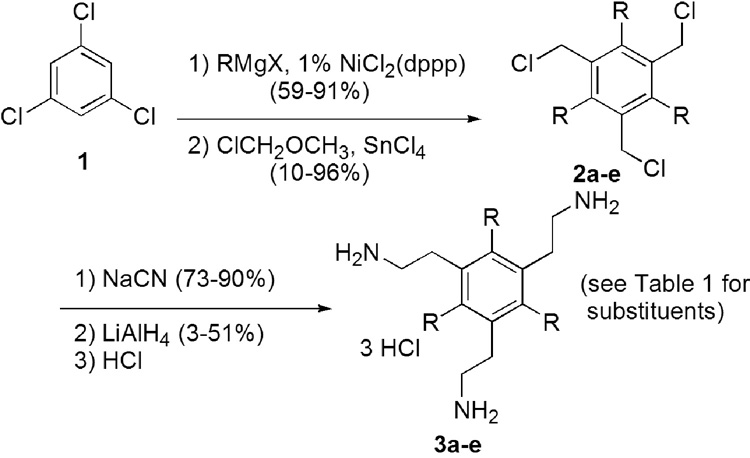
Hydrophobic groups were added at the 1, 3 and 5 positions of benzene by Kumada coupling of 1,3,5-trichlorobenzene (1) and an alkyl Grignard reagent. Exhaustive chloromethylation of the 1,3,5-trialkylbenzenes afforded persubstituted benzenes 2, which were further functionalized by nucleophilic substitution with cyanide. Reduction of these intermediates gave tris-ethylamines 3.(18)
In Vitro CBI and Ligand Binding Assays
We have developed a reliable time-resolved fluorescence resonance energy transfer (TR-FRET) assay (Figure 2a and 2b) to measure coactivator binding inhibition. Briefly, the ligand binding domain of ERα (residues 304–554; N-terminally His-tagged, with previously described C381,530S mutations that do not affect activity but leave one reactive cysteine) is site-specifically labeled (Cys417) (19) with biotin while bound to a nickel column during protein purification and subsequently tagged with a streptavidin-terbium complex. The SRC-3 nuclear receptor domain (NRD) (residues 627–829, which include all three NR-boxes) is non-specifically labeled through the four available cysteines using 5-iodoacetamidofluorescein. When the ER is bound with a high concentration of the agonist estradiol, fluorescein-SRC3 (FRET acceptor, λem = 520 nm) is recruited to the terbium/estrogen receptor complex (FRET donor, λem = 495 nm), and a high FRET signal is produced. Coactivator binding inhibitor activity is assayed by the ability of increasing concentrations of compound to compete for ER-SRC binding and disrupt the energy transfer. As shown in Table 1, the tris-ethylamines 3c and 3e exhibit, respectively, Kis of 1.7 and 2.1 µM, among the lowest reported for such inhibitors. Interestingly, n-pentyl-substituted 3c shows a more complete blockade of SRC-NRD binding than does the neo-hexyl substituted 3e in this assay.
Figure 2.

Table 1.
Summary of binding affinity and inhibitory potency data for aminoethyl-substituted CBIs
| R | TR-FRET assay (µM) | Reporter gene assay (µM) | Mammalian 2-hybrid assay (µM) | |
|---|---|---|---|---|
| NR Box II 15mer | n/a | 0.23 | n/a | n/a |
| 3a | propyl | >50 | >50 | >50 |
| 3b | butyl | 1.8 | 8.2 | 10.1 |
| 3c | pentyl | 1.7 | 3.2 | 3.2 |
| 3d | hexyl | 2.0 | 3.6 | 3.4 |
| 3e | neo-hexyl | 2.1 | 3.8 | 2.2 |
It is important to establish that these compounds act as coactivator binding inhibitors rather than conventional antagonists. When assayed in a radiometric competitive ligand binding assay with [3H]estradiol, 3e binds to the ligand binding pocket of ERα with an affinity approximately 1/1000 that of estradiol. Since the estradiol concentration in the FRET assays is 1 µM, the concentration of CBI needed to displace estradiol from its pocket and exert conventional antagonistic effects would be 1 mM, or roughly 550 times greater than the Ki measured for inhibition of coactivator binding by 3e This provides strong evidence that this compound is, in fact, working through a CBI mechanism.
Cell-Based Assays of Coactivator Binding and Transcription Inhibition
To further establish their activity, we have assayed these compounds in cell-based assays of ER-mediated transcription. In the first, a human endometrial cancer (HEC-1) cell line that expresses nuclear receptor coactivators but contains no endogenous ERα was transfected with a full-length ERα expression vector, an estrogen-responsive luciferase reporter gene plasmid (2ERE Luc), and pCMV β-galactosidase (β-gal; internal control). The transfected cells were incubated with two different concentrations of estradiol and increasing concentrations of CBI; luciferase activity was then measured. If the compound acts by a coactivator binding inhibition mechanism—that is, by directly competing with the SRCs—then a change in the concentration of estradiol should have no effect on the IC50 of the compound. As seen in Figure 2c and Table 1, both 3c and 3e have IC50 values near 3 µM in the presence of either 1 nM or 100 nM estradiol, indicating that the inhibition of transcription is, in fact, occurring by a coactivator binding inhibition mechanism The internal control β-galactosidase is unaffected except at the highest concentrations of 3c and 3e, indicating that the suppressive effect of the CBIs on transcription is not due to cell toxicity.
We also measured the effect of the CBIs in a mammalian two-hybrid assay in which HEC-1 cells were transfected with pFR luciferase, pCMV β-gal, Gal4-SRC-1NRD, and ERαDEF-VP16. The cells were then treated with estradiol and CBI, and assayed for luciferase activity. As shown in Figure 2d and Table 1, 3c and 3e inhibit the interaction between ERα and SRC-1 with an IC50 of approximately 2–3 µM. The β-galactosidase controls, carried out as above, also support a mechanism not invoked by general cellular toxicity (See Figure 2d). A 100-fold increase in estradiol concentration also resulted in superimposable inhibition curves for the two compounds (data not shown).
In conclusion, we have synthesized a series of amphipathic benzenes that directly inhibit the interaction of ERα and steroid receptor coactivators in both in vitro and cell-based experiments. These molecules build on and further establish the relatively simple 1,3,5-trisubstituted pharmacophore that we have previously described, (6) and which could be useful in designing inhibitors of other helix-groove interactions. To date, these compounds are among the most potent inhibitors and the few known to work in a cellular context. Their extensive evaluation in transient transfection assays confirms their ability to interrupt estrogen receptor/coactivator interactions in a cellular context and paves the way for more advanced biological assays using cellular breast cancer models. Thus, they are promising probes of an alternative mechanism of estrogen action that could be used to explore new directions for controlling hormone-dependent breast cancer.
METHODS
Luciferase Reporter Gene Assay
Human endometrial cancer (HEC-1) cells were maintained in culture as described and transfected in 24 well plates.(20) A mixture of HBSS (50 µL/well), Holo-transferrin (Sigma T1408) (20 µL/well), and lipofectin (Invitrogen #18292-011) (5 µL/well) were incubated at room temperature for 5 minutes. The DNA mixture was made by adding 200 ng of pCMVβ-galactosidase as internal control, 500 ng of the estrogen responsive reporter gene plasmid 2ERE Luc, and 100 ng of full-length ER alpha expression vector with 75 µL HBSS per well and, after addition to the first mixture, allowed to incubate for 20 minutes at room temperature. The cell media was changed to Opti-MEM (350 µL/well) and 150 µL of the transfection mixture was added to each well. The cells were incubated at 37 °C in a 5% CO2 containing incubator for 6 h. The medium was then replaced with fresh medium containing 5% charcoal-dextran-treated calf serum and the desired concentrations of ligands. Reporter gene activity was assayed at 24 h after ligand addition. Luciferase activity, normalized for the internal control β-galactosidase activity, was assayed as described.(20)
In the initial screen, antagonist activity is determined at four concentrations, ranging from 20 µM to 0.6 µM, in the presence of 10−9 M estradiol (E2). Upon validation that compounds do act as antagonists, mechanism of action was examined by repeating the compound titration in the presence of both 10−7 and 10−9 M E2. Changing the concentration of E2 100-fold should not change the coactivator binding inhibitor IC50 determined in the reporter gene assay, because its mode of action is through direct displacement of coactivator, not competition for estradiol binding.
Mammalian Two-Hybrid Assay
The plasmid pM- SRC-1NRD was constructed by releasing the insert from pVP16-SRC-1NRD(21) with EcoRI and HindIII and subcloning into the pM vector that contains the Gal4 DNA binding domain (Clontech) between EcoRI and HindIII sites. ERDEF was subcloned into pVP16 vector (Clontech) by digestion with EcoRI and MluI of pM-ERDEF, and insertion into pVP16 vector between EcoRI and MluI sites.(21)
Human endometrial cancer (HEC-1) cells were maintained in culture as described and transfected in 24 well plates using lipofectin.(20) HEC-1 cells were plated at 2 × 104 per well in 24-well plates and transfected 24 h later with 1 µg of pFR-Luc (Stratagene), 0.2 µg of pCMVβ-gal, 0.2 µg of pM- SRC-1NRD and 0.2 µg of pVP16-ERDEF. At 8 h after transfection, cells were treated with ligand or control vehicle. Cells were harvested 24 h after ligand treatment, and cell extracts were prepared. β-Galactosidase activity and luciferase activity were assayed as described.(20)
TR-FRET CBI Assay
Purified biotin-ERα-417 and fluorescein-SRC3-NRD were used in the time-resolved FRET assays. A portion (5 µL) of a stock solution of ERα-417 (8 nM), estradiol (4 µM), and LanthaScreen™ Streptavidin-Terbium (Invitrogen) (2 nM) in TR-FRET buffer (20 mM Tris, pH 7.5, 0.01% NP40, 50 mM NaCl) was placed in separate wells of a black 96-well Molecular Devices HE high efficiency microplate (Molecular Devices, Inc.). In a second 96-well Nunc polypropylene plate (Nalge Nunc International, Rochester, NY), a 0.02 M solution of the coactivator binding inhibitor was serially diluted in a 1:10 fashion into DMF Each concentration of coactivator binding inhibitor was then diluted 1:10 into TR-FRET buffer, and 10 µL of this solution or vehicle was added to the stock estrogen receptor α solution in the 96-well plate. After a two-minute incubation, 5 µL of 200 nM fluorescein-SRC3-NRD was added to each well. This mixture was allowed to incubate for 1 h at room temperature in the dark TR-FRET was measured using an excitation filter at 340/10 nm, and emission filters for terbium and fluorescein at 495/20 and 520/25 nm, respectively. The final concentrations of the reagents were as follows: ERα-417 (2 nM), streptavidin-terbium (0.5 nM), estradiol (1 µM), coactivator binding inhibitor (0–1 mM), SRC3-NRD (50 nM).(6)
General Procedure for the Preparation of 1,3,5-trialkylbenzenes
1,3,5-Trichlorobenzene and [1,3-bis(diphenylphosphino)propane]dichloronickel(II) were combined with diethyl ether. The alkyl Grignard reagent was added, and the solution began to reflux and turned from orange to brown The reaction was maintained at reflux overnight. After being cooled to room temperature, the reaction mixture was poured into cold 1 M HCl. The aqueous layer was exhaustively extracted with ether. The combined organic extracts were dried over MgSO4, filtered and concentrated in vacuo to an oil, which was purified through a short silica gel column, using hexanes as the eluant.
General Procedure for the Preparation of 1,3,5-tris(chloromethyl)-2,4,6-trialkylbenzene Derivatives
1,3,5-Trialkylbenzene (0.60 mmol) was dissolved in methylene chloride (2.3 mL). Upon cooling to 0 °C, chloromethyl methyl ether (420 µL, Aldrich) was added, followed by the slow addition of tin (IV) chloride (1 mL). The reaction was then stirred at 0 °C for three hours, and then overnight at room temperature After quenching with water at 0 °C and extracting exhaustively with chloroform, the organic layer was dried with sodium sulfate, filtered and then concentrated to give a green foam. The product was recrystallized from absolute ethanol.
General Procedure for the Preparation of 1,3,5-tris(cyanomethyl)-2,4,6-trialkylbenzene Derivatives
Chloromethyl intermediate 2 (0.21 mmol) was dissolved in acetone (5.6 mL). Sodium cyanide (0.94 mmol) was added, followed by water (2.3 mL). To the thick mixture was added acetone (0.3 mL). The reaction was stirred at reflux for 17 hours. After cooling to room temperature, the reaction mixture was poured over ice and filtered to isolate the product.
General Procedure for the Preparation of 1,3,5-tris(aminoethyl)-2,4,6-trialkylbenzene Derivatives
Nitrile intermediate (0.22 mmol) was dissolved in 3 mL THF. Lithium aluminum hydride (1 mL 1 M in THF, 1.00 mmol) was added, and the solution was refluxed overnight. As the reaction progressed, the color changed from yellow to red. The THF was removed under vacuum, and 3 mL ether was added in its place. The reaction was quenched by adding sequentially 40 µL water, 40 µL 3 M NaOH, and 120 µL water. The slurry was filtered and the filtrate was dried over MgSO4, filtered and concentrated to a yellow semi-solid. The amine was taken up in dry CH2Cl2 and was converted to the HCl salt by the addition of 1 M HCl in ether The precipitated product was collected by filtration after cooling to −15 °C.
Supplementary Material
Scheme 1.
ACKNOWLEDGMENTS AND DEDICATION
This paper is dedicated to Professor E. J. Corey in honor of his 80th birthday. We thank J. Amin for assistance in performing biochemical assays. Support through a grant from National Institutes of Health (PHS R37 DK15556) is gratefully acknowledged. J. Gunther received support from a David Robertson Fellowship and the National Institutes of Health (NRSA 1 F30 ES016484-01 and NRSA 5 T32 GM070421). We are grateful to A. Rodriguez for helpful suggestions regarding this project.
Footnotes
Supporting Information Available: This material is free via the Internet. Full details of the synthesis and characterization of compounds 3a–e and 2a–e can be found in the supporting information.
REFERENCES
- 1.Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 2.Brzozowski AM, Pike ACW, Dauter Z, Hubbard RE, Bonn T, Engstrom O, Ohman L, Greene GL, Gustafsson JA, Carlquist M. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 3.Pike ACW, Brzozowski AM, Walton J, Hubbard RE, Thorsell AG, Li YL, Gustafsson JA, Carlquist M. Structure. 2001;9:145–153. doi: 10.1016/s0969-2126(01)00568-8. [DOI] [PubMed] [Google Scholar]
- 4.Schiff R, Massarweh SA, Shou J, Bharwani L, Arpino G, Rimawi M, Osborne CK. Cancer Chemother. Pharmacol. 2005;56 Suppl 1:10–20. doi: 10.1007/s00280-005-0108-2. [DOI] [PubMed] [Google Scholar]
- 5.Zhou HB, Collins ML, Gunther JR, Comninos JS, Katzenellenbogen JA. Bioorg. Med. Chem. Lett. 2007;17:4118–4122. doi: 10.1016/j.bmcl.2007.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriguez AL, Tamrazi A, Collins ML, Katzenellenbogen JA. J. Med. Chem. 2004;47:600–611. doi: 10.1021/jm030404c. [DOI] [PubMed] [Google Scholar]
- 7.Becerril J, Hamilton AD. Angew. Chem. Int. Ed. Engl. Vol. 46. 2007. pp. 4471–4473. [DOI] [PubMed] [Google Scholar]
- 8.Shao D, Berrodin TJ, Manas E, Hauze D, Powers R, Bapat A, Gonder D, Winneker RC, Frail DE. J. Steroid Biochem. Mol. Biol. 2004;88:351–360. doi: 10.1016/j.jsbmb.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Norris JD, Paige LA, Christensen DJ, Chang C-Y, Huacani MR, Fan D, Hamilton PT, Fowlkes DM, McDonnell DP. Science. 1999;285:744–746. doi: 10.1126/science.285.5428.744. [DOI] [PubMed] [Google Scholar]
- 10.Arkin MR, Wells JA. Nat. Rev. Drug Discovery. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 11.Pagliaro L, Felding J, Audouze K, Nielsen SJ, Terry RB, Krog-Jensen C, Butcher S. Curr. Opin. Chem. Biol. 2004;8:442–449. doi: 10.1016/j.cbpa.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Yin H, Hamilton AD. Angew. Chem. Int. Ed. Engl. 2005;44:4130–4163. doi: 10.1002/anie.200461786. [DOI] [PubMed] [Google Scholar]
- 13.Wang S, Yang D, Lippman ME. Semin Oncol. 2003;30:133–142. doi: 10.1053/j.seminoncol.2003.08.015. [DOI] [PubMed] [Google Scholar]
- 14.Heery DM, Kalkhoven E, Hoare S, Parker MG. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 15.Voo JK, Lam KC, Rheingold AL, Riordan CG. J. Chem. Soc. Dalton Trans. 2001:1803–1805. [Google Scholar]
- 16.Yun SG, Kim YO, Kim D, Kim HG, Ihm H, Kim JK, Lee CW, Lee WJ, Yoon J, Oh KS, Yoon J, Park SM, Kim KS. Org. Lett. 2003;5:471–474. doi: 10.1021/ol0273203. [DOI] [PubMed] [Google Scholar]
- 17.Grawe T, Schrader T, Zadmard R, Kraft A. J. Org. Chem. 2002;67:3755–3763. doi: 10.1021/jo025513y. [DOI] [PubMed] [Google Scholar]
- 18.Kilway KV, Siegel JS. Tetrahedron. 2001;57:3615–3627. [Google Scholar]
- 19.Tamrazi A, Carlson KE, Daniels JR, Hurth KM, Katzenellenbogen JA. Mol. Endocrinol. 2002;16:2706–2719. doi: 10.1210/me.2002-0250. [DOI] [PubMed] [Google Scholar]
- 20.Sun J, Meyers MJ, Fink BE, Rajendran R, Katzenellenbogen JA, Katzenellenbogen BS. Endocrinology. 1999;140:800–804. doi: 10.1210/endo.140.2.6480. [DOI] [PubMed] [Google Scholar]
- 21.Martini PG, Delage-Mourroux R, Kraichely DM, Katzenellenbogen BS. Mol. Cell. Biol. 2000;20:6224–6232. doi: 10.1128/mcb.20.17.6224-6232.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.