Abstract
The osmotic response element-binding protein (OREBP), also known as tonicity enhancer-binding protein (TonEBP) or NFAT5, is the only known osmo-sensitive transcription factor that mediates cellular adaptations to extracellular hypertonic stress. Although it is well documented that the subcellular localization and transactivation activity of OREBP/TonEBP are tightly regulated by extracellular tonicity, the molecular mechanisms involved remain elusive. Here we show that nucleocytoplasmic trafficking of OREBP/TonEBP is regulated by the dual phosphorylation of Ser-155 and Ser-158. Alanine scanning mutagenesis revealed that Ser-155 is an essential residue that regulates OREBP/TonEBP nucleocytoplasmic trafficking. Tandem mass spectrometry revealed that Ser-155 and Ser-158 of OREBP/TonEBP are both phosphorylated in living cells under hypotonic conditions. In vitro phosphorylation assays further suggest that phosphorylation of the two serine residues proceeds in a hierarchical manner with phosphorylation of Ser-155 priming the phosphorylation of Ser-158 and that these phosphorylations are essential for nucleocytoplasmic trafficking of the transcription factor. Finally, we have shown that the pharmacological inhibition of casein kinase 1 (CK1) abolishes the phosphorylation of Ser-158 and impedes OREBP/TonEBP nuclear export and that recombinant CK1 phosphorylates Ser-158. Knockdown of CK1α1L, a novel isoform of CK1, inhibits hypotonicity-induced OREBP/TonEBP nuclear export. Together these data highlight the importance of Ser-155 and Ser-158 in the nucleocytoplasmic trafficking of OREBP/TonEBP and indicate that CK1 plays a major role in regulating this process.
The osmotic response element-binding protein (OREBP),3 also known as TonEBP, belongs to the Rel family of transcription factors that includes NFκB and nuclear factor of activated T cells (NFAT). Members of this family contain a Rel homology domain for DNA binding (1, 2). Because the Rel homology domain of OREBP/TonEBP shares significant homology to that of other NFAT isoforms (NFAT1-4), it was independently identified by homology screening and named NFAT5 (3). However, unlike NFAT1-4, OREBP/TonEBP lacks the calcineurin binding domain that plays a critical role in regulating the subcellular localization and activity of the NFATs (4). Although NFAT-1, -2, and -4 regulate the immune response as well as the development of heart, bone, thymus, and the nervous system (5, 6), OREBP/TonEBP plays a pivotal role in activating adaptive cellular responses to extracellular hypertonic stress (2, 7).
OREBP/TonEBP is the only known osmo-sensitive transcription factor in mammalian cells. Upon activation by hypertonic stress, it induces the expression of a battery of osmoprotective genes, including aldose reductase, the betaine/γ-aminobutyric acid transporter, and the Na+-dependent myoinositol transporter, through binding to a cognate cis-acting element known as the osmotic response element or the tonicity-responsive enhancer (8-11) in the promoter region of these genes. Consequently, there is an increase in the level of intracellular organic osmolytes, including sorbitol, betaine, and myoinositol (12), that are required for the preservation of cell volume and osmolality without perturbing macromolecular structure and function (13, 14). Consistent with its putative role in osmoprotection, OREBP/TonEBP-deficient mice or transgenic mice ectopically expressing dominant-negative OREBP/TonEBP in epithelial cells of collecting tubules develop renal atrophy (15, 16). More recently, OREBP/TonEBP has also been implicated in lymphoid (17) and lens (18) development, myoblast migration and differentiation (19), and in cancer metastasis (20).
Earlier studies have established that extracellular tonicity regulates OREBP/TonEBP through altering its subcellular distribution and transcriptional activity. Hypertonic stress promotes nuclear translocation and activates its enhancer activity. In contrast, hypotonicity induces OREBP/TonEBP nuclear export (1, 21-25). These studies have suggested that OREBP/TonEBP is subjected to bi-directional regulation in response to changes in tonicity. Furthermore, a number of kinases have been implicated in this process. Nevertheless, the molecular mechanisms that control the subcellular localization and activity of OREBP/TonEBP remain largely unknown. Previously, we have identified and characterized three domains of OREBP/TonEBP that regulate its subcellular localization (26). These domains include a nuclear localization signal (NLS), a canonical nuclear export sequence (NES), and a novel auxiliary export domain (AED). The NES, in conjunction NLS, regulates nucleocytoplasmic shuttling of OREBP/TonEBP under isotonic conditions, whereas the AED is indispensable for its nuclear export under both isotonic and hypotonic conditions. Here we identify the mechanism underlying the AED-mediated nuclear export of OREBP/TonEBP. We show that Ser-155 in the AED, together with Ser-158, plays a critical role in regulating OREBP/TonEBP nuclear export. Ser-155 and Ser-158 are both phosphorylated in response to hypotonicity. Our data further reveal that phosphorylation proceeds in a hierarchical manner with Ser-155 phosphorylation priming the phosphorylation of Ser-158. Furthermore, we show that casein kinase 1 (CK1) directs the phosphorylation of Ser-158. Our findings thus provide an important insight into the mechanism of the nucleocytoplasmic trafficking of OREBP/TonEBP in response to extracellular tonicity.
EXPERIMENTAL PROCEDURES
Plasmid Constructs and Chemicals—The original OREBP cDNA clone KIAA0827 was a gift from Dr. Takahiro Nagase (Kazusa DNA Research Institute, Japan). FLAG-OREBP-(1-581)-Δ1-131 was constructed as described previously (26). GST-OREBP-(146-167) was derived by in-frame insertion of double-stranded oligonucleotides corresponding to amino acid residues 146-167 of OREBP between the BamHI and XhoI restriction sites of pGEX4T-1 (GE Healthcare). Point mutants of FLAG-OREBP-(1-581)-Δ1-131 and GST-OREBP-(146-167) were made using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) using the corresponding parental constructs as templates. All the constructs were verified by sequencing. CKI-7 was obtained from United States Biological.
Cell Culture and Transfection—HeLa cells (American Type Culture Collection, Manassas, VA) were maintained in minimal essential medium supplemented with 10% fetal bovine serum, 1 mm sodium pyruvate, 2 mml-glutamine (300 mosmol/kg H2O). For subcellular localization studies, cells grown in 6-well plates to 50% confluence were transfected with 0.6 μg of the plasmids expressing the proteins using GeneJuice (Novagen) according to the manufacturer's instructions. Transfected cells were incubated for 16-24 h in complete growth medium before switching to medium with different osmolality. Hypotonic (260 mosmol/kg H2O), isotonic (300 mosmol/kg H2O), or hypertonic (500 mosmol/kg H2O) medium was prepared by supplementing 10% fetal bovine serum, 1 mm sodium pyruvate, 2 mm l-glutamine and different amounts of NaCl to NaCl-deficient minimal essential medium (Invitrogen) to the desired osmolality. Medium osmolality was measured by the Vapro® vapor pressure osmometer (Wescor).
Immunoaffinity Purification, Multidimensional Protein Identification Technology (MudPIT), Mass Spectrometry, and Data Base Searching—20 × 106 HeLa cells were transfected with FLAG-OREBP-(1-581)-Δ1-131. Twenty four hours after transfection, cells were treated with hypotonic medium (260 mosmol/kg H2O) for 90 min and lysed with SDS lysis buffer (50 mm Tris-Cl, pH 6.8, 2% SDS, 5% glycerol). Cell lysates were diluted 20-fold by dilution buffer (20 mm Tris-Cl, pH 7.6, 150 mm NaCl, 1 mm EDTA, 1 mm Na3VO4, 1 mm β-glycerol phosphate, 1% Triton X-100, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitors) and concentrated by ultrafiltration. The recombinant protein was purified by affinity chromatography using anti-FLAG affinity resin (Sigma).
The purified protein was precipitated by 20% trichloroacetic acid. The precipitate was washed twice with acetone, dried, and resuspended in 8 m urea and 100 mm Tris-HCl, pH 8.5. The solubilized protein was reduced by the addition of tris(2-carboxyethyl)phosphine to 5 mm, followed by the carboxyamidomethylation of cysteines using 10 mm iodoacetamide. The concentration of urea was then diluted 2-fold (to 4 m) by the addition of an equal volume of 100 mm Tris-HCl, pH 8.5. Sequencing grade endoproteinase Lys-C (Roche Diagnostics) and modified trypsin (Promega) was added at ∼1:50 enzyme to substrate ratio (w/w) and incubated at 37 °C for 4 and 12-16 h, respectively. The resulting peptides were extracted with 5% formic acid and re-dissolved into buffer A (5% acetonitrile with 0.1% formic acid) prior to the liquid chromatography-MS/MS analysis.
Peptide mixtures were enriched by the TiO2 method (27) and pressure-loaded onto a biphasal column, which consists of a strong cation exchange material back-to-back with reversed phase material inside fused silica capillaries (collectively known as multidimensional protein identification technology, Mud-PIT) (28-30), and were eluted onto an analytical reversed phase column in the front with a salt gradient of ammonium acetate (5-100%). Data-dependent tandem mass spectrometry (MS/MS) analysis was performed in an LTQ mass spectrometer (ThermoFisher, San Jose, CA). One full MS spectrum was acquired followed by five MS/MS events, sequentially generated on the first to the fifth most intense ions selected from the full MS spectrum. MS scan functions and high pressure liquid chromatography solvent gradients were controlled by the Xcalibur data system (ThermoFisher). Tandem mass spectra were searched against the NCBI human data base on December 12, 2006, with the sequence of OREBP/TonEBP added, using the SEQUEST (29) algorithm, where phosphorylation at serine, threonine, and tyrosine were differentially considered. The validity of peptide/spectrum matches was assessed using the SEQUEST-defined parameters, cross-correlation score (XCorr), and normalized difference in cross-correlation scores (DeltaCn). Spectra/peptide matches were only retained if they had a DeltaCn of at least 0.08 and minimum XCorr of 1.8 for +1, 2.5 for +2, and 3.5 for +3 spectra. DTASelect (31) with phosphorylated modification as described (29) was used to select and sort peptide/spectrum matches passing this criteria set. Proteins were considered detected if they were identified by at least three spectra passing all of the selection criteria or with at least 10% sequence coverage.
Immunofluorescence Microscopy—Cells were washed three times with PBS and fixed with 4% w/v paraformaldehyde for 15 min at 4 °C. The cells were permeabilized with absolute methanol for 2 min at room temperature. Primary antibodies against FLAG (Sigma) were used. The fluorescein isothiocyanate (FITC)-conjugated anti-rabbit antibody (Chemicon International, Temecula, CA) was used as secondary antibodies. To visualize the nuclei, the cells were stained with 4,6-diamidino-2-phenylindole (DAPI) (Sigma). The fluorescence images were viewed with a Zeiss Axiovert 200 M fluorescence microscope and analyzed with AxioVision software. Quantification of fluorescence signal in different subcellular compartment was conducted as we have described (26).
Live Cell Microscopy and Time-lapse Imaging—HeLa cells transiently expressed GFP reporter fusion proteins were viewed with an inverted Zeiss Axiovert 200 M fluorescence microscope. A heated chamber perfused with CO2 was used to incubate the cells at 37 °C. After 30 min, original growth medium was removed and replaced with pre-warmed hypotonic growth medium. Time-lapse monitoring was initiated within 5 min. Images were taken at 30-min intervals and were analyzed using AxioVision software.
Preparation of Cytoplasmic and Nuclear Fractions—Cytoplasmic and nuclear fractions were purified as described (8). Briefly, 1 × 106 subconfluent HeLa cells were treated with hypotonic (260 mosmol/kgH2O), isotonic (300 mosmol/kg H2O), or hypertonic (500 mosmol/kg H2O) growth medium for the indicated time and then scraped into hypotonic, isotonic, or hypertonic PBS, respectively. Cells were centrifuged for 1 min at 4 °C, and the pellet was resuspended in 1 ml of cold buffer B (10 mm HEPES-KOH, pH 7.9, at 4 °C, 1.5 mm MgCl2, 10 mm KCl, 0.05% Nonidet P-40, 0.5 mm DTT, and 0.5 mm PMSF). Cells were disrupted using a Dounce homogenizer and were centrifuged for 10 min at 4000 rpm at 4 °C. The supernatant was collected as the cytoplasmic fraction. The pellet was washed with buffer B, resuspended in 100 μl of cold buffer C (20 mm HEPES-KOH, pH 7.9, 25% glycerol, 420 mm NaCl, 1.5 mm MgCl2, 0.2 mm EDTA, 0.5 mm DTT, and 0.5 mm PMSF), and incubated for 15 min on ice. Cellular debris was removed by centrifugation for 30 min at 13,000 rpm, at 4 °C, and the supernatant was collected as the nuclear extract. The extract was diluted with 200 μl of modified buffer D (20 mm HEPES, pH 7.9, 0.05 m KCl, 0.2 mm EDTA, 0.5 mm DTT, 0.5 mm PMSF, and 20% glycerol). The quality of nuclear and cytoplasmic fractions preparations was verified by analysis of α-tubulin (cytoplasmic) and TFIIB (nucleus), respectively.
Expression of GST-tagged Fusion Proteins in Escherichia coli—The GST-OREBP-(146-167) and its point mutants were transformed into E. coli BL21(DE3) bacteria (Invitrogen) to produce GST fusion proteins. Expression was induced with 1 mm isopropyl β-d-1-thiogalactopyroanoside for 2 h at 37 °C. Bacteria were harvested, lysed in lysis buffer (PBS with 5% glycerol, 100 mm MgCl2, 1 mm PMSF, 1 mm DTT, 1 mg/ml lysozyme, 0.2 units/ml DNase I, and Complete protease inhibitors), and centrifuged to remove debris. GST fusion proteins were purified from lysates with glutathione-Sepharose 4 Beads (GE Healthcare). The size and purity of the protein preparations were verified by SDS-PAGE and Coomassie Blue staining.
In Vitro Phosphorylation Assay—Whole cell, nuclear, or cytoplasm extracts (15 μg) obtained from HeLa cells, or recombinant CK1α1 (Invitrogen), were incubated with GST proteins (10 μg) in a kinase buffer containing 20 mm HEPES KOH, pH 7.5, 100 μm ATP, 10 mm β-glycerol phosphate, 30 mm magnesium chloride, 1 mm DTT, and 2 μCi of [γ-32P]ATP. After incubation for 20 min at 30 °C, the reactions were terminated by the addition of SDS sample buffer, and the proteins were separated by SDS-PAGE. Substrate phosphorylation was analyzed by autoradiography. For experiments involving CKI-7, 150 μm CKI-7 (United States Biological) or the same volume of ethanol (solvent) was added to the reaction mixture 10 min before the addition of ATP and [γ-32P]ATP.
RNA Interference-mediated Knockdown of CK1s—HeLa cells were transfected with 100 nm CK1 isoform-specific (CK1α1L, CK1α1, CK1-γ3, CK1-ε, and CK1-δ) or nontargeting control siRNAs (Qiagen) by DharmaFECT1 (Dharmacon). After 48 h, cells were transfected with FLAG-OREBP-(1-581)-Δ1-131. After another 24 h, cells were subjected to total RNA isolation or immunofluorescence microscopy.
Quantitative Real Time Reverse Transcription-PCR—Total RNA was prepared using TRIzol (Invitrogen) according to the manufacturer's instructions. cDNA synthesis and reverse transcription-PCR were carried out using SuperScript™ III Platinum CellsDirect two-step quantitative reverse transcription-PCR kit with SYBR® Green (Invitrogen). Each RNA sample was amplified in triplicate for the genes of interest and β-actin for normalization on a Mx3000P® quantitative PCR system. The primers for detecting CK1α1L (CSNK1A1L), CK1α1 (CSNK1A1), CK1-γ3, CK1-ε, and CK1-δ were obtained from Qiagen.
RESULTS
Ser-155 Located in the AED of OREBP/TonEBP Plays a Critical Role in Hypotonicity-induced Nuclear Export—Our earlier study showed that the transactivation domain of OREBP/TonEBP is not required for nucleocytoplasmic trafficking, whereas the NES (amino acids 5-15) and AED (amino acids 132-156) are involved in this process (Fig. 1A). The NES is mainly responsible for the nucleocytoplasmic shuttling of OREBP/TonEBP under isotonic conditions, and the AED plays an indispensable role in the nuclear export of the transcription factor under both isotonic and hypotonic conditions (26). To identify the critical amino acid residues responsible for hypotonicity-induced nuclear export in this domain, we carried out alanine scanning mutagenesis on selected residues in the AED, using the FLAG-OREBP-(1-581)-Δ1-131 plasmid as template. This plasmid contains a mutant OREBP/TonEBP protein devoid of NES that is preferentially localized to the nucleus under isotonic conditions but remains responsive for hypotonicity-induced nuclear export signaling (26). All the amino acid residues in the AED that were potential candidates for post-translational modifications, including serine (Ser-134, Ser-145, and Ser-155), threonine (Thr-135 and Thr-140), tyrosine (Tyr-143), lysine (Lys-137), and arginine (Arg-138 and Arg-156), were evaluated (Fig. 1A). The FLAG-tagged OREBP mutant constructs were transiently expressed in HeLa cells, and their subcellular localization in response to hypotonic conditions was determined. In agreement with our previous findings (26), indirect immunofluorescence analysis using anti-FLAG antibodies revealed that wild-type FLAG-OREBP-(1-581)-Δ1-131 resided in the nucleus under isotonic conditions and was localized to the cytoplasm in response to hypotonic challenge (Fig. 1B). All the alanine mutants remained responsive to hypotonic challenge except for the alanine substitution at Ser-155 (S155A) (Fig. 1B), indicating that this amino acid plays an important role in nuclear exclusion of OREBP/TonEBP. Western blotting analysis showed that all the protein mutants were correctly expressed (Fig. 1C). In addition, all the mutants were expressed at levels similar to the WT protein, suggesting that alanine substitution did not result in protein misfolding, which may have targeted the protein for proteasome degradation.
FIGURE 1.
Subcellular localization of OREBP/TonEBP mutants. A, schematic illustration of OREBP/TonEBP. AD1 and AD2, putative transcriptional activation domains; DBD, DNA-binding domain. The amino acid sequence of the AED has been boxed. Residues that were subjected to alanine-scanning mutagenesis are in boldface. B, representative fluorescence images of fixed HeLa cells expressing recombinant OREBP/TonEBP. Cells were transfected with plasmids containing the indicated alanine substitution. Subsequently, cells were either treated with isotonic or hypotonic medium for 90 min, respectively. FLAG-tagged recombinant protein was visualized after fixation with a FLAG antibody and a FITC-labeled secondary antibody, counterstained with DAPI, and analyzed by fluorescence microscopy. Images were examined using a ×63 objective lens. Scale bar, 20 μm. Data shown are representative of at least three independent transfections for each construct. C, expression of FLAG-tagged OREBP proteins as determined by Western blot analysis. Protein expression was detected using anti-FLAG and anti-β-actin antibodies.
Mapping the AED Phosphorylation Sites—The above data suggested that the hypotonicity-induced nuclear export of OREBP/TonEBP may be associated with Ser-155 phosphorylation. To identify the potential phosphorylated residue(s), recombinant OREBP/TonEBP was purified from HeLa cells using affinity chromatography using the anti-FLAG antibody and was analyzed by mass spectrometry. Purified protein was doubly digested with trypsin and Lys-C, and the resultant peptides were subjected to MudPIT (28-32) separation and analysis via mass spectrometry. Fig. 2 shows a representative MS/MS spectrum corresponding to the fragment ions derived from a peptide parent ion dually phosphorylated at Ser-155 and Ser-158. The MS/MS spectrum was consistent with serine sites modified by its dominated y-ions series. The observed y5-(pSCQDG) ion (m/z 646.3) and y8-(pSRMp-SCQDG) (m/z 1100.4) and their corresponding neutral loss of water peaks y5-H2O and y8-H2O confirmed the location of phosphorylated modified serine residues. The identity of the modified peptide was subsequently validated by performing a SEQUEST (29) and DTASelect (31) search with a predefined differential mass shift of +80 for serine. This approach led to the unambiguous identification of the FLAG-tagged OREBP with a sequence coverage of 56% and demonstrated phosphorylation on Ser-155 and Ser-158. The identified phosphorylated sites were further validated using the DeBunker algorithm (33), a software tool for automatic validation of phosphopeptide identifications from tandem mass spectrometry.
FIGURE 2.
Mass spectrometric analysis of recombinant OREBP. MS/MS spectrum of the modified Ser-155 and Ser-158 quadruple charged peptide, AAAYPSTPKRHTVLYISPPPEDLLDNS*RMS*CQDG (precursor ion m/z 989.72, S* corresponds to a phosphorylated serine, with a +80 mass shift). The y ions, which are at the specific modified sites, and their corresponding water loss peaks, are in red.
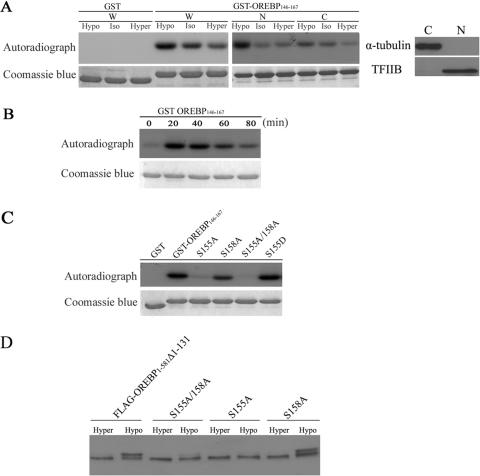
Ser-155 Phosphorylation “Primes” Ser-158 Phosphorylation—We next sought to determine whether Ser-155 and Ser-158 could be phosphorylated in vitro. Protein substrates were prepared by bacterial expression of cDNA constructs encoding amino acid residues 146-167 of OREBP, which encompasses Ser-155 and Ser-158 (there is no other serine or threonine residue in this fragment), in-frame with the GST tag epitope (GST-OREBP-(146-167)). In vitro kinase assays were carried out by incubating this substrate, in the presence of [γ-32P]ATP, with protein extracts obtained from HeLa cells maintained under isotonic conditions or induced with hypotonic or hypertonic conditions, respectively. We found that GST-OREBP-(146-167) was phosphorylated to the highest level in whole cell extracts obtained from cells treated with hypotonic medium as determined by 32P incorporation (Fig. 3A, W), whereas the level of phosphorylation was reduced when extracts were obtained from cells maintained in isotonic medium or hypertonic medium. The observed phosphorylation was specific to the expressed fragment, because phosphorylation signals were not detected when GST was used as substrate (Fig. 3A). Collectively, our data suggested that GST-OREBP-(146-167) is phosphorylated, presumably at Ser-155 and Ser-158, in response to hypotonic conditions.
FIGURE 3.
In vitro and in vivo phosphorylation of OREBP Ser-155 and Ser-158. A, phosphorylation of GST-OREBP-(146-167) fusion protein. HeLa cells were maintained in isotonic (Iso), hypotonic (Hypo), and hypertonic (Hyper) medium, respectively, for 30 min. Subsequently, whole cell (W), cytoplasmic (C), and nuclear (N) extracts were prepared. Fusion proteins were incubated in vitro with [γ-32P]ATP and different cellular extracts (W, C, and N) for 20 min at 30 °C. The proteins were then separated by electrophoresis, stained with Coomassie Blue (lower panel), and submitted to autoradiography (upper panel). The purity of cytoplasmic and nuclear extracts was confirmed by Western blots using antibodies against α-tubulin and TFIIB, respectively. B, treatment with hypotonic conditions induces a time-dependent increase in GST-OREBP-(146-167) phosphorylation. GST-OREBP-(146-167) fusion protein was incubated as in A with nuclear extracts prepared from HeLa cells that were treated with hypotonicity for the indicated time. C, mutant GST-OREBP-(146-167) proteins containing the alanine substitution at the indicated residues were incubated as in A with nuclear extracts prepared from HeLa cells treated with hypotonicity for 30 min. D, Western blotting analysis of FLAG-tagged recombinant OREBP and its mutants. Cells expressing the indicated recombinant proteins were treated with hypertonic (hyper) or hypotonic (hypo) medium, respectively, for 90 min. Proteins were resolved in 7.5% SDS gel and analyzed by Western blotting using anti-FLAG antibody.
To determine the subcellular location of the putative kinases, cytoplasmic and nuclear extracts were prepared and were subjected to phosphorylation assays. The majority of the kinase activity was detected in the nuclear extracts; however, kinase activity could also be detected using cytoplasmic extracts, albeit at a lower level (Fig. 3A, N and C). In addition, the putative kinase(s) appeared to be activated rapidly in response to hypotonicity, as the highest level of phosphorylation was detected when nuclear extracts obtained from cells subjected to hypotonic conditions for 20 min were used (Fig. 3B). Furthermore, to confirm whether only Ser-155 and Ser-158 were phosphorylated in vitro, and to explore the possible interaction of the two sites, we repeated the phosphorylation assay with substrates where either one or both serine residues were substituted by alanine. As shown in Fig. 3C, compared with the WT substrate, phosphorylation levels were reduced by approximately one-half when Ser-158 was mutated to alanine (S158A), suggesting that this amino acid was phosphorylated in vitro. On the other hand, mutation of Ser-155 to alanine (S155A) completely abolished phosphorylation of the substrate, suggesting that phosphorylation of Ser-158 was dependent on prior phosphorylation at Ser-155. To confirm this, we generated a phosphomimetic mutant in which Ser-155 was substituted to aspartate (S155D). The S155D mutant was highly phosphorylated (Fig. 3C), suggesting that the phosphorylation of Ser-155 and Ser-158 proceeds in a hierarchical manner, where Ser-155 phosphorylation primes phosphorylation at Ser-158. No phosphorylation signals were detected when both Ser-155 and Ser-158 were mutated to alanine (S155A/S158A) (Fig. 3C), suggesting that phosphorylation is specific to these two amino acids.
We further determined whether Ser-155 and Ser-158 were phosphorylated by hypotonicity in vivo. HeLa cells expressing FLAG-OREBP-(1-581)-Δ1-131 was treated with hypertonic and hypotonic medium, respectively. Western blots of protein extracts from these cells were examined. Under hypertonic conditions, a single band corresponding to the size of FLAG-OREBP-(1-581)-Δ1-131 was observed, whereas an additional species of recombinant protein with retarded electrophoretic mobility was detected from hypotonicity-treated cells (Fig. 3D), consistent with our hypothesis that hypotonicity induces phosphorylation of OREBP. Similarly, a band with retarded mobility was observed when mutant S158A was expressed and induced with hypotonicity. Nevertheless, only one protein species was detected in cells expressing mutants S155A/S158A or S155A (Fig. 3D). Taken together, these findings agreed with our findings from in vitro phosphorylation assays and suggested that hypotonicity induces Ser-155 and Ser-158 phosphorylation in vivo.
The Role of Ser-155 and Ser-158 Phosphorylation in Nuclear Export of OREBP/TonEBP—To determine the role of Ser-155 and Ser-158 phosphorylation in OREBP/TonEBP nuclear export, we determined the subcellular localization of FLAG-OREBP-(1-581)-Δ1-131 (WT) and its mutants (S155A, S158A, and S155A/S158A) in transfected HeLa cells in isotonic and hypotonic medium (Fig. 4A). Representative pictures of these recombinant proteins are shown in Fig. 4B. When maintained in isotonic medium, both the WT and OREBP mutants were predominantly localized to the nuclear compartment. As expected, the WT protein was exported to the cytoplasm in response to hypotonic treatment (11% nuclear and 77% cytoplasmic). In agreement with the data above (Fig. 1B), alanine substitution of Ser-155 (S155A) inhibited the hypotonic nuclear export of OREBP/TonEBP (65% nuclear and 17% cytoplasmic). Alanine substitution of Ser-158 (S158A) also inhibited nuclear export, although to a slightly lesser extent when compared with mutant S155A (54% nuclear and 24% cytoplasmic). These data suggested that both Ser-155 and Ser-158 play a role in the nuclear export process. Similarly, alanine substitution of both Ser-155 and Ser-158 (S155A/S158A) resulted in nuclear export inhibition (65% nuclear and 16% cytoplasmic). To investigate the role of phosphorylation of these amino acids in nuclear export, we determined the subcellular locations of phospho-mimetic mutants S155D, S158D, and S155D/S158D, in which Ser-155, Ser-158, or both residues were substituted with aspartate, respectively. The results are summarized in Fig. 4, C and D. Under isotonic conditions, all mutants predominantly localized to the nucleus. Under hypotonic conditions, the S155D mutant was exported to the cytoplasm (14% nuclear and 58% cytoplasmic). Conversely, the majority of the cells expressing the S158D mutant did not respond to hypotonic conditions, although cytoplasmic localization could be detected in a small percentage of cells (44% nuclear and 24% cytoplasmic). Unexpectedly, hypotonicity-induced nuclear export of the S155D/S158D double mutant was completely abolished (68% nuclear and 9% cytoplasmic). Western blotting analysis showed that the expression levels of the S155D, S158D, and S155D/S158D protein mutants were similar to those of the WT protein (Fig. 4E). These results are further discussed below.
FIGURE 4.
Role of Ser-155 and Ser-158 in OREBP nuclear export. A and C, quantification of the subcellular localization of various OREBP mutants treated with hypotonic and hypertonic medium for 90 min, respectively. For each condition >100 cells were counted. The data represented are the average of three independent experiments. I, isotonic; H, hypotonic. B and D, representative fluorescence images of fixed HeLa cells expressing OREBP mutants. Cells were treated with isotonic and hypotonic medium, respectively. Recombinant protein was visualized with a FLAG antibody and a FITC-labeled secondary antibody, counterstained with DAPI, and analyzed by fluorescence microscopy. E, expression of FLAG-tagged OREBP proteins as determined by Western blot analysis. F, live cell images of OREBP-GFP and mutant fusion proteins. Cells were moved to hypotonic medium at T = 0. Images were examined with ×63 objective. Scale bar, 20 μm.
The results obtained thus far involved the use of the FLAG-OREBP-(1-581)-Δ1-131 reporter protein, where the absence of the NES restricts the protein to the nucleus under isotonic conditions (compared with pan-cellular distribution and the ability to undergo nucleocytoplasmic shuttling) (26). To further confirm the role of Ser-155 and Ser-158 in the nucleocytoplasmic trafficking of OREBP/TonEBP, we introduced the same mutations into the OREBP-(1-158)-GFP plasmid, which contains an intact NES and AED. We have previously shown that this reporter protein is responsive to tonicity-induced subcellular redistribution similar to the endogenous OREBP/TonEBP (26). The subcellular trafficking of the fusion proteins in response to hypotonic challenge was analyzed using time-lapse fluorescence photography at 30-min intervals for 90 min. As shown in Fig. 4F, as expected, OREBP-(1-158)-GFP was exported from the nucleus under hypotonic conditions, whereas S155A-GFP (Ser-155 to alanine) or S158A-GFP (Ser-158 to alanine) was not. Similar to the results using FLAG-OREBP-(1-158)-Δ1-131, hypotonic conditions induced the nuclear export of S155D-GFP (Ser-155 to aspartate), whereas S158D-GFP (Ser-158 to aspartate) and S155D/S158D-GFP (Ser-155 and Ser-158 to aspartate) were constitutively localized to the nucleus despite hypotonic challenge. Collectively, these data suggest that the phosphorylation of Ser-155 and Ser-158 plays a critical role of in the regulation of OREBP/TonEBP trafficking.
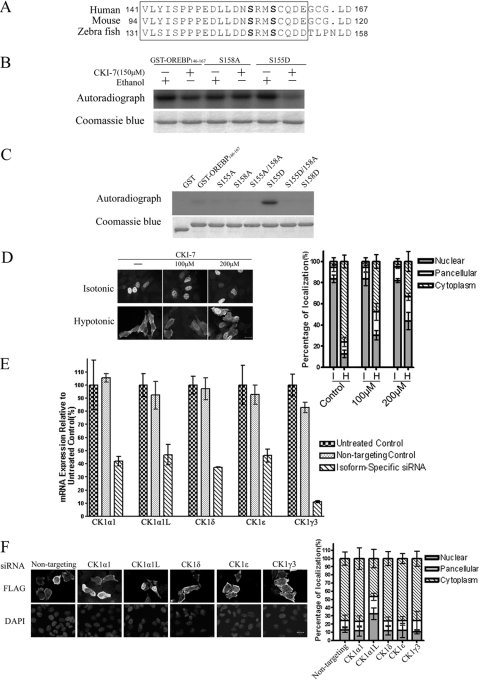
The Role of CK1α1 (CSNK1A1) and CK1α1L (CSNK1A1L) Kinases in OREBP/TonEBP Nuclear Export—Ser-155 and Ser-158 in OREBP/TonEBP, as well as their neighboring residues, are highly conserved among the human, mouse, and zebrafish homologues (Fig. 5A), suggesting that this domain may play an important regulatory role. In silico analysis of the OREBP/TonEBP sequence (PhosphoMotif Finder) (34) suggested that CK1, which phosphorylates a “primed” phosphorylation motif ((pS/T) XX(S/T)) or an (E/D)XX(S/T) motif (35, 36), was a potential Ser-158 kinase. To confirm this prediction, we first determined whether CKI-7 (37), a known CK1 inhibitor, inhibited the in vitro phosphorylation of GST-OREBP-(146-167). Consistent with our prediction, CKI-7 reduced GST-OREBP-(146-167) phosphorylation by approximately one-half in nuclear extracts, in vitro (Fig. 5B). This reduction was primarily because of the inhibition of Ser-158 phosphorylation, because the inhibitor did not reduce the phosphorylation of the GST-OREBP-(146-167) mutant where Ser-158 was replaced by alanine (GST-S158A), but potently reduced the phosphorylation of GST-S155D, where Ser-155 was replaced with aspartate, mimicking the recognition motif for CK1 (Fig. 5B). To confirm that GST-OREBP-(146-167) was the direct substrate of CK1, we performed an in vitro phosphorylation of GST-OREBP-(146-167) using recombinant CK1α1 (CSNK1A1). As shown in Fig. 5C, consistent with the results obtained above, recombinant CSNK1A1 did not phosphorylate the wild-type GST-OREBP-(146-167) substrate but efficiently phosphorylated S155D. Although CSNK1A1 efficiently phosphorylated S155D, it failed to phosphorylate S155D/S158A, suggesting that Ser-158 was specifically phosphorylated when the substrate was primed by Ser-155 phosphorylation. To further determine whether CK1 regulated the nuclear export of OREBP/TonEBP under hypotonic conditions, we examined the effects of CKI-7 on the subcellular localization of OREBP/TonEBP. HeLa cells were transfected with the FLAG-OREBP-(1-581)-Δ1-131 plasmid and then treated with CKI-7 (100 and 200 μm). As may be seen in Fig. 5D, the addition of CKI-7 did not alter the nuclear localization of the recombinant protein under isotonic conditions but significantly inhibited the translocation of the FLAG-tagged protein to the cytoplasm under hypotonic conditions. These data suggest that CK1 regulates OREBP/TonEBP nuclear export via the phosphorylation of Ser-158. On the other hand, the hypotonicity-induced cytoplasmic translocation of OREBP/TonEBP was not affected by the addition of IC261, a CK1δ- and CK1ε-specific inhibitor (38), suggesting that these two CK1 isoforms may not be responsible for the nucleocytoplasmic trafficking of this transcription factor (data not shown).
FIGURE 5.
CK1 phosphorylates GST-OREBP-(146-167) in vitro and regulates OREBP nuclear export in vivo. A, sequence alignment of relevant OREBP regions from human, mouse, and zebrafish (containing residues equivalent to Ser-155 and Ser-158 of human OREBP). Conserved residues have been boxed. B, CKI-7 inhibits in vitro phosphorylation of Ser-158. Nuclear extracts were obtained from HeLa cells treated with hypotonic growth medium for 30 min. GST-OREBP-(146-167) and its mutants were incubated with the nuclear extracts in the presence of [γ-32P]ATP and CKI-7 or ethanol (solvent) for 20 min at 30 °C. The proteins were separated by electrophoresis, stained with Coomassie Blue (lower panel), and submitted to autoradiography (upper panel). C, recombinant CK1α phosphorylates the GST-OREBP-(146-167) mutant containing a phosphomimetic mutation at Ser-155 (S155D). CK1α1 (40 ng) was incubated with GST-OREBP-(146-167) or its mutants in the presence of [γ-32P]ATP for 20 min at 30 °C. The proteins were separated by electrophoresis, stained with Coomassie Blue (lower panel), and submitted to autoradiography (upper panel). D, left, CKI-7 inhibits nuclear export of OREBP in vivo. HeLa cells transfected with FLAG-OREBP-(1-581)-Δ1-131 were pretreated with CKI-7 (100 or 200 μm) or ethanol (-) for 4 h in growth medium (Isotonic). The cells were moved to hypotonic medium supplemented with the same amount of the inhibitor or solvent for another 90 min (Hypotonic). FLAG-tagged recombinant protein was visualized after fixation with a FLAG antibody and a FITC-labeled secondary antibody; right, quantification of the subcellular localization of OREBP in the presence of CKI-7. For each condition >100 cells were counted. The data shown are the average of three independent experiments. E, real time quantitative PCR of different CK1 isoforms after siRNA knockdown. HeLa cells were transfected with the indicated siRNAs, respectively. After 72 h, mRNA was harvested from the cells and reverse-transcribed and quantified by primers specific to the isoform. Data were expressed as percentage relative to cells that were not transfected(Untreated control). Cells transfected with scramble siRNA (Nontargeting control) were also included to demonstrate the specificity of siRNA knockdown. F, left, effect of CK1 knockdown on nuclear export of OREBP. HeLa cells transfected with FLAG-OREBP-(1-581)-Δ1-131 and the indicated siRNA were treated with hypotonic medium for 90 min. FLAG-tagged recombinant protein was visualized after fixation with a FLAG antibody and a FITC-labeled secondary antibody and counterstained with DAPI; right, quantification of the subcellular localization of OREBP. For each condition >100 cells were counted. The data represented are the average of three independent experiments.
Humans contain a number of CK1 isoforms, including CK1α1, CK1-γ, CK1δ, and CK1-ε. In addition, a novel CK1 transcript, designated as CK1α1L (CSNK1A1L, GenBank™ accession number NM_145203), has been recently assigned to chromosomal location 13q13.3. Because CK1 isoforms are known to exhibit distinct biochemical properties and subcellular compartmentalization (39), we sought to determine which isoform(s) acted on the OREBP/TonEBP protein. Consequently, we used small interfering RNAs (siRNAs) to reduce the expression of the endogenous CK1 isoforms (CK1α1, CK1α1L, CK1-γ3, CK1-ε, and CK1-δ). RNA knockdown resulted in an approximately ∼60% reduction in the mRNA levels of each of the CK1 isoforms, as determined by quantitative real time PCR (Fig. 5E). Genetic “knockdown” of CK1α1, CK1-δ, CK1-ε, or CK1-γ3 did not appreciably alter the hypotonicity-induced cytoplasmic translocation of OREBP/TonEBP (Fig. 5F). However, the siRNA knockdown of CK1α1L significantly reduced the nuclear export of OREBP/TonEBP under hypotonic conditions (Fig. 5F). Taken together, these data suggest that CK1α1L is the kinase that phosphorylates Ser-158 in the regulation of OREBP/TonEBP export.
DISCUSSION
OREBP/TonEBP plays a central role in orchestrating a gene transcription program that is essential for cellular adaptation to hypertonic stress (40). The activity of OREBP/TonEBP can be regulated by its subcellular redistribution in response to changes in extracellular tonicities, where hypertonicity and hypotonicity induce its nuclear translocation and nuclear exclusion, respectively (21, 26). In addition, the transactivational activity of this transcription factor is also elevated under conditions of increased extracellular tonicity via the actions of p38, Fyn, cAMP-dependent protein kinase, and ataxia telangiectasia mutated (23-25, 41). Nevertheless, the precise regulatory mechanism underlying the subcellular localization and activation of OREBP/TonEBP remains poorly defined.
The spatial distribution of a given protein within the cell is important for its proper function. For proteins that are shuttled in and out of the nucleus, the subcellular localization can be modulated via the phosphorylation of specific amino acid residues close to the NLS or NES (42-46). Phosphorylation may lead to a “masking” of the NLS (47, 48) or an inhibition of the interaction between the CRM1 and NES (46). Alternatively, the phosphorylation of a nuclear factor may create a novel recognition site for a cytoplasmic protein, resulting in the cytosolic sequestration of the nuclear factor (49). Previously, we have identified additional protein motifs that control the subcellular localization of OREBP/TonEBP in response to various degrees of extracellular tonicity. We have shown that the AED (amino acids 132-156) plays a critical role in the regulation of its nuclear export. Deletion of the AED led to the constitutive nuclear localization of OREBP/TonEBP despite the presence of the NES (26). Here, we demonstrate that Ser-155 within the AED, and Ser-158 located adjacent to this domain, are phosphorylated under hypotonic conditions, as evidenced by mass spectrometric analysis and in vitro phosphorylation assays. Furthermore, the phosphorylation of both Ser-155 and Ser-158 is required for the nuclear export of OREBP/TonEBP in response to hypotonic challenge, because nonphosphorylatable mutation of these two residues, either alone or in combination, inhibits the nuclear exclusion of OREBP/TonEBP. Interestingly, a reporter protein containing a Ser-155 to aspartate mutation (S155D) within the AED was not constitutively localized throughout the cytoplasm but was only translocated to the cytoplasm under hypotonic treatment. This suggests that phosphorylation of this serine alone is not sufficient to induce this protein to exit the nucleus. On the other hand, when Ser-158 was replaced by aspartate (S158D), hypotonicity-induced nuclear export of the protein was inhibited. More intriguingly, nuclear export was completely abolished when both Ser-155 and Ser-158 were replaced by the phosphomimetic aspartate residue. One possible explanation is that phosphomimetic substitutions lead to the misfolding of the nascent protein and hence disrupt the formation of the conformation required for nuclear export. If this was the case, then these proteins would be directed to the proteasome for degradation. However, all the mutant proteins were expressed to levels comparable with those of the WT, suggesting that the mutations did not lead to any major conformational changes. A more likely explanation is that phosphorylated Ser-155 and Ser-158 residues play distinct roles within different cellular compartments. The phosphorylation of Ser-155 in the nucleus may be required for the interaction with the nuclear export receptor, whereas phosphorylation at Ser-158 by CK1 may be important for its cytoplasmic retention. The phosphomimetic substitution of Ser-158 (with aspartate) may destroy the peptide motif required for efficient Ser-155 phosphorylation. Furthermore, a double phosphomimetic mutation of the two closely spaced serine residues may obliterate the recognition site for the nuclear export receptor, resulting in OREBP/TonEBP being constitutively localized throughout the cytoplasm. The development of phospho-specific antibodies against phosphorylated Ser-155 and phosphorylated Ser-155 and Ser-158 would help to clarify this disparity.
It is not clear exactly how this dual phosphorylation regulates the subcellular localization of OREBP/TonEBP. Nucleocytoplasmic trafficking of transcription factors such as FOXO1A (50), PERs (51), NFATs (52, 53), and Pho4 (54) is also regulated by phosphorylation at multiple sites. It has been suggested that multisite phosphorylation may induce conformational changes, resulting in the masking of the NLS or the exposure of the NES favoring cytoplasmic localization (50, 53, 55). In addition, the phosphorylation levels within the different domains may determine the rate of nuclear import or export (53). We speculate that NLS masking may be one of the critical steps in OREBP/TonEBP nuclear export, as Ser-155 and Ser-158 are in close proximity to the NLS, and their phosphorylation may promote interdomain interactions, resulting in the masking of the NLS. Notably, the stretch of amino acids between the AED and NLS (amino acid residues 159-197) contains a total of 11 serine and threonine residues. It is therefore important to examine whether additional phosphorylation events within this region play a role in OREBP/TonEBP nuclear export. Another salient feature of OREBP/TonEBP localization is that although it contains a canonical NES, this domain is dispensable for nuclear export, and the export does not involve CRM1 (26). We therefore postulate that phosphoserines and nearby residues may form “recognition motifs” for an as-yet unidentified export receptor. Alternatively, multiple phosphorylations may induce a conformational change within other folded motifs, which may be recognized by the receptor. Irrespective of the mechanism, the identity of the novel nuclear export receptor remains to be determined.
Members of the CK1 family phosphorylate a wide spectrum of substrates ranging from cytoskeletal proteins to transcription factors (36). As a result, CK1 regulates numerous cellular process such as cell cycle progression and cytokinesis (56), chromosome and microtubule dynamics (57, 58), circadian rhythm (51), and apoptosis (59, 60). CK1 is also known to regulate the subcellular localization of transcription factors. For example, the phosphorylation of Ser-322 and Ser-325 of FOXO1A by CK1 is directed by a “priming” phosphorylation at Ser-319 by another kinase, PKB (50), whereas the cytoplasmic retention of NFAT1 and NFAT4 requires CK1α1 phosphorylation at multiple sites (52, 55). Here we have demonstrated that CK1 is involved in the phosphorylation of Ser-158. Given the diverse functions of CK1, it has been proposed that the specificity of this family of kinases is because of the existence of different isoforms, each of which exhibits different substrate specificity as well as subcellular localization (39). Consistent with this notion, our data show that the CK1α1L isoform of CK1 is the major physiological kinase of OREBP/TonEBP. To our knowledge, the function of CK1α1L has not been previously characterized, and therefore our data identify the first known physiological substrate of CK1α1L. Interestingly, although CK1α1L and CK1α1 are encoded by two separate genes located on chromosome 13q13.3 and 5q32, respectively, their amino acid sequences are highly homologous to each other (85% identity) except that CK1α1 contains an extra 28 amino acid residues within the catalytic domain (Fig. 6). How such subtle differences lead to differential activity toward OREBP/TonEBP remains to be deciphered. One possibility is that the isoforms may have distinct subcellular localizations. It has been shown that CK1α1 follows a cell cycle-dependent subcellular redistribution, varying its association with cytosolic vesicles, the centrosome, and microtubules (57). Determining the precise subcellular localization of CK1α1L will greatly enhance our understanding of how this kinase is regulated by changes in tonicity.
FIGURE 6.
Amino acid sequence alignment of CK1α1 and CK1α1L. Identical amino acid residues are indicated “|”, and conservative changes are marked by “:”.
Our data also suggest that the CK1-mediated phosphorylation of OREBP/TonEBP at Ser-158 requires a priming phosphorylation at Ser-155. This implies that CK1 can only phosphorylate OREBP/TonEBP in conjunction with another, as-yet unidentified kinase. This unidentified kinase may play a major role regulating the subcellular distribution of OREBP/TonEBP through the phosphorylation of Ser-155. To the best of our knowledge, the sequence of amino acids flanking Ser-155 shares no significant homology with any previously described kinase substrate motifs. Therefore, the identification of the kinase that phosphorylates Ser-155 will be important toward a complete understanding the mechanism of nucleocytoplasmic trafficking of OREBP/TonEBP.
Acknowledgments
We thank Dr. Nathalie Wong, Dr. Anna Kashina, Dr. Stine Pedersen, and Dr. Andras Kapus for critical comments on the manuscript. We also thank Dr. Rory Watt for assistance in editing this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant P41 RR011823. This work was also supported by the Research Grant Council Grants CUHK 7419/03M and 7327/04M, by the University Grant Committee of the Hong Kong Special Administrative Region Area of Excellence Scheme Grant AoE/P-10/01, and by The Chinese University of Hong Kong Direct Grant for Research 2041316 (to B. C. B. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: OREBP, osmotic response element-binding protein; TonEBP, tonicity enhancer-binding protein; GST, glutathione S-transferase; MS/MS, tandem mass spectrometry; NLS, nuclear localization signal; NES, nuclear export sequence; AED, nuclear export sequence; siRNA, small interfering RNA; GFP, green fluorescent protein; FITC, fluorescein isothiocyanate; DAPI, 4,6-diamidino-2-phenylindole; DTT, dithiothreitol; PMSF, phenylmethylsulfonyl fluoride; NFAT, nuclear factor of activated T cells; CK1, casein kinase 1; PBS, phosphate-buffered saline; WT, wild type; MudPIT, multidimensional protein identification technology.
References
- 1.Ko, B. C., Turck, C. W., Lee, K. W., Yang, Y., and Chung, S. S. (2000) Biochem. Biophys. Res. Commun. 270 52-61 [DOI] [PubMed] [Google Scholar]
- 2.Miyakawa, H., Woo, S. K., Dahl, S. C., Handler, J. S., and Kwon, H. M. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 2538-2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopez-Rodriguez, C., Aramburu, J., Rakeman, A. S., and Rao, A. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 7214-7219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hogan, P. G., Chen, L., Nardone, J., and Rao, A. (2003) Genes Dev. 17 2205-2232 [DOI] [PubMed] [Google Scholar]
- 5.Rao, A., Luo, C., and Hogan, P. G. (1997) Annu. Rev. Immunol. 15 707-747 [DOI] [PubMed] [Google Scholar]
- 6.Wu, H., Peisley, A., Graef, I. A., and Crabtree, G. R. (2007) Trends Cell Biol. 17 251-260 [DOI] [PubMed] [Google Scholar]
- 7.Handler, J. S., and Kwon, H. M. (2001) Kidney Int. 60 408-411 [DOI] [PubMed] [Google Scholar]
- 8.Ko, B. C., Ruepp, B., Bohren, K. M., Gabbay, K. H., and Chung, S. S. (1997) J. Biol. Chem. 272 16431-16437 [DOI] [PubMed] [Google Scholar]
- 9.Woo, S. K., Lee, S. D., Na, K. Y., Park, W. K., and Kwon, H. M. (2002) Mol. Cell. Biol. 22 5753-5760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takenaka, M., Preston, A. S., Kwon, H. M., and Handler, J. S. (1994) J. Biol. Chem. 269 29379-29381 [PubMed] [Google Scholar]
- 11.Rim, J. S., Atta, M. G., Dahl, S. C., Berry, G. T., Handler, J. S., and Kwon, H. M. (1998) J. Biol. Chem. 273 20615-20621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burg, M. B., Kwon, E. D., and Kultz, D. (1997) Annu. Rev. Physiol. 59 437-455 [DOI] [PubMed] [Google Scholar]
- 13.Yancey, P. H., Clark, M. E., Hand, S. C., Bowlus, R. D., and Somero, G. N. (1982) Science 217 1214-1222 [DOI] [PubMed] [Google Scholar]
- 14.Burg, M. B. (1996) Kidney Int. 49 1684-1685 [DOI] [PubMed] [Google Scholar]
- 15.Lopez-Rodriguez, C., Antos, C. L., Shelton, J. M., Richardson, J. A., Lin, F., Novobrantseva, T. I., Bronson, R. T., Igarashi, P., Rao, A., and Olson, E. N. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 2392-2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lam, A. K., Ko, B. C., Tam, S., Morris, R., Yang, J. Y., Chung, S. K., and Chung, S. S. (2004) J. Biol. Chem. 279 48048-48054 [DOI] [PubMed] [Google Scholar]
- 17.Go, W. Y., Liu, X., Roti, M. A., Liu, F., and Ho, S. N. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 10673-10678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang, Y., Ko, B. C., Yang, J. Y., Lam, T. T., Jiang, Z., Zhang, J., Chung, S. K., and Chung, S. S. (2005) J. Biol. Chem. 280 19986-19991 [DOI] [PubMed] [Google Scholar]
- 19.O'Connor, R. S., Mills, S. T., Jones, K. A., Ho, S. N., and Pavlath, G. K. (2007) J. Cell Sci. 120 149-159 [DOI] [PubMed] [Google Scholar]
- 20.Jauliac, S., Lopez-Rodriguez, C., Shaw, L. M., Brown, L. F., Rao, A., and Toker, A. (2002) Nat. Cell Biol. 4 540-544 [DOI] [PubMed] [Google Scholar]
- 21.Woo, S. K., Dahl, S. C., Handler, J. S., and Kwon, H. M. (2000) Am. J. Physiol. 278 F1006-F1012 [DOI] [PubMed] [Google Scholar]
- 22.Lee, S. D., Colla, E., Sheen, M. R., Na, K. Y., and Kwon, H. M. (2003) J. Biol. Chem. 278 47571-47577 [DOI] [PubMed] [Google Scholar]
- 23.Ferraris, J. D., Persaud, P., Williams, C. K., Chen, Y., and Burg, M. B. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 16800-16805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ko, B. C., Lam, A. K., Kapus, A., Fan, L., Chung, S. K., and Chung, S. S. (2002) J. Biol. Chem. 277 46085-46092 [DOI] [PubMed] [Google Scholar]
- 25.Irarrazabal, C. E., Liu, J. C., Burg, M. B., and Ferraris, J. D. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 8809-8814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tong, E. H., Guo, J. J., Huang, A. L., Liu, H., Hu, C. D., Chung, S. S., and Ko, B. C. (2006) J. Biol. Chem. 281 23870-23879 [DOI] [PubMed] [Google Scholar]
- 27.Cantin, G. T., Shock, T. R., Park, S. K., Madhani, H. D., and Yates, J. R., III (2007) Anal. Chem. 79 4666-4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Washburn, M. P., Wolters, D., and Yates, J. R., III (2001) Nat. Biotechnol. 19 242-247 [DOI] [PubMed] [Google Scholar]
- 29.Eng, J., McCormack, A., and Yates, J. R. (1994) J. Am. Soc. Mass Spectrom. 5 976-989 [DOI] [PubMed] [Google Scholar]
- 30.Link, A. J., Eng, J., Schieltz, D. M., Carmack, E., Mize, G. J., Morris, D. R., Garvik, B. M., and Yates, J. R., III (1999) Nat. Biotechnol. 17 676-682 [DOI] [PubMed] [Google Scholar]
- 31.Tabb, D. L., McDonald, W. H., and Yates, J. R., III (2002) J. Proteome Res. 1 21-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDonald, W. H., Ohi, R., Miyamoto, D. T., Mitchison, T. J., and Yates, J. R., III (2002) Int. J. Mass Spectrom. 219 245-251 [Google Scholar]
- 33.Lu, B., Ruse, C., Xu, T., Park, S. K., and Yates, J., III (2007) Anal. Chem. 79 1301-1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amanchy, R., Periaswamy, B., Mathivanan, S., Reddy, R., Tattikota, S. G., and Pandey, A. (2007) Nat. Biotechnol. 25 285-286 [DOI] [PubMed] [Google Scholar]
- 35.Pulgar, V., Marin, O., Meggio, F., Allende, C. C., Allende, J. E., and Pinna, L. A. (1999) Eur. J. Biochem. 260 520-526 [DOI] [PubMed] [Google Scholar]
- 36.Knippschild, U., Gocht, A., Wolff, S., Huber, N., Lohler, J., and Stoter, M. (2005) Cell. Signal. 17 675-689 [DOI] [PubMed] [Google Scholar]
- 37.Chijiwa, T., Hagiwara, M., and Hidaka, H. (1989) J. Biol. Chem. 264 4924-4927 [PubMed] [Google Scholar]
- 38.Behrend, L., Milne, D. M., Stoter, M., Deppert, W., Campbell, L. E., Meek, D. W., and Knippschild, U. (2000) Oncogene 19 5303-5313 [DOI] [PubMed] [Google Scholar]
- 39.Gross, S. D., and Anderson, R. A. (1998) Cell. Signal. 10 699-711 [DOI] [PubMed] [Google Scholar]
- 40.Burg, M. B., Ferraris, J. D., and Dmitrieva, N. I. (2007) Physiol. Rev. 87 1441-1474 [DOI] [PubMed] [Google Scholar]
- 41.Ferraris, J. D., Williams, C. K., Persaud, P., Zhang, Z., Chen, Y., and Burg, M. B. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 739-744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rihs, H. P., Jans, D. A., Fan, H., and Peters, R. (1991) EMBO J. 10 633-639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, F., White, R. L., and Neufeld, K. L. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 12577-12582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, Y., and Xiong, Y. (2001) Science 292 1910-1915 [DOI] [PubMed] [Google Scholar]
- 45.Lee, H., and Bai, W. (2002) Mol. Cell. Biol. 22 5835-5845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Blume, J., Knippschild, U., Dequiedt, F., Giamas, G., Beck, A., Auer, A., Van Lint, J., Adler, G., and Seufferlein, T. (2007) EMBO J. 26 4619-4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moll, T., Tebb, G., Surana, U., Robitsch, H., and Nasmyth, K. (1991) Cell 66 743-758 [DOI] [PubMed] [Google Scholar]
- 48.Hennekes, H., Peter, M., Weber, K., and Nigg, E. A. (1993) J. Cell Biol. 120 1293-1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grozinger, C. M., and Schreiber, S. L. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 7835-7840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rena, G., Woods, Y. L., Prescott, A. R., Peggie, M., Unterman, T. G., Williams, M. R., and Cohen, P. (2002) EMBO J. 21 2263-2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takano, A., Isojima, Y., and Nagai, K. (2004) J. Biol. Chem. 279 32578-32585 [DOI] [PubMed] [Google Scholar]
- 52.Okamura, H., Garcia-Rodriguez, C., Martinson, H., Qin, J., Virshup, D. M., and Rao, A. (2004) Mol. Cell. Biol. 24 4184-4195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okamura, H., Aramburu, J., Garcia-Rodriguez, C., Viola, J. P., Raghavan, A., Tahiliani, M., Zhang, X., Qin, J., Hogan, P. G., and Rao, A. (2000) Mol. Cell 6 539-550 [DOI] [PubMed] [Google Scholar]
- 54.Komeili, A., and O'Shea, E. K. (1999) Science 284 977-980 [DOI] [PubMed] [Google Scholar]
- 55.Zhu, J., Shibasaki, F., Price, R., Guillemot, J. C., Yano, T., Dotsch, V., Wagner, G., Ferrara, P., and McKeon, F. (1998) Cell 93 851-861 [DOI] [PubMed] [Google Scholar]
- 56.Gross, S. D., Simerly, C., Schatten, G., and Anderson, R. A. (1997) J. Cell Sci. 110 3083-3090 [DOI] [PubMed] [Google Scholar]
- 57.Brockman, J. L., Gross, S. D., Sussman, M. R., and Anderson, R. A. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 9454-9458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petronczki, M., Matos, J., Mori, S., Gregan, J., Bogdanova, A., Schwickart, M., Mechtler, K., Shirahige, K., Zachariae, W., and Nasmyth, K. (2006) Cell 126 1049-1064 [DOI] [PubMed] [Google Scholar]
- 59.Beyaert, R., Vanhaesebroeck, B., Declercq, W., Van Lint, J., Vandenabele, P., Agostinis, P., Vandenheede, J. R., and Fiers, W. (1995) J. Biol. Chem. 270 23293-23299 [DOI] [PubMed] [Google Scholar]
- 60.Zhao, Y., Qin, S., Atangan, L. I., Molina, Y., Okawa, Y., Arpawong, H. T., Ghosn, C., Xiao, J. H., Vuligonda, V., Brown, G., and Chandraratna, R. A. (2004) J. Biol. Chem. 279 30844-30849 [DOI] [PubMed] [Google Scholar]