Abstract
Atrophin family proteins, including the vertebrate arginine–glutamic acid dipeptide repeats protein (RERE) and Drosophila Atrophin (Atro), constitute a new class of nuclear receptor corepressors. Both RERE and Atro share the ELM2 (EGL-27 and MTA1 homology 2) and SANT (SWI3/ADA2/N-CoR/TFIII-B) domains, which are also present in other important transcriptional cofactors. Here, we report that the SANT domain in RERE binds to the histone methyltransferase G9a, and that both the ELM2 and SANT domains orchestrate molecular events that lead to a stable methylation of histone H3-lysine 9. We establish the physiological relevance of these interactions among Atrophin, G9a, and histone deacetylases 1 and 2 in Drosophila by showing that these proteins localize to overlapping chromosomal loci, and act together to suppress wing vein and melanotic-mass formation. This study not only shows a new function of the SANT domain and establishes its connection with the ELM2 domain, but also implies that a similar strategy is used by other ELM2–SANT proteins to repress gene transcription and to exert biological effects.
Keywords: Atrophin, SANT domain, ELM2 domain, G9a, HDAC1/2
Introduction
Atrophin proteins are conserved nuclear receptor corepressors (Wang et al, 2006; Zhang et al, 2006) and include vertebrate atrophin 1 (ATN1; Koide et al, 1994; Nagafuchi et al, 1994), vertebrate arginine–glutamic acid dipeptide repeats protein (RERE, also known as atrophin 2; Yanagisawa et al, 2000) and Drosophila Atrophin (Atro, also known as Grunge; Erkner et al, 2002; Zhang et al, 2002). Human ATN1 is known to cause neurodegenerative dentatorubral–pallidoluysian atrophy when its glutamine-repeat tract is expanded (Koide et al, 1994; Nagafuchi et al, 1994). The neurotoxicity caused by glutamine-repeat-expanded ATN1 is not due to the loss of ATN1 functions, as Atn1-knockout mice show no detectable phenotype (Shen et al, 2007). By contrast, mutations of murine Rere cause embryonic lethality and severe developmental defects (Zoltewicz et al, 2004). The apparently divergent effects of RERE and ATN1 on animal development suggest that their functions are not equivalent.
The functional differences between RERE and ATN1 can be attributed to structural divergence between these two proteins. Notably, ATN1, which resembles a truncated version of RERE, lacks several conserved domains, including the BAH (bromo-adjacent homology), ELM2 (EGL-27 and MTA1 homology 2) and SANT (SWI3/ADA2/N-CoR/TFIII-B) domains, which are all located at the amino terminus of RERE. The ELM2 and SANT domains are also present in Atro and several other important transcriptional repressors such as MTA (metastasis-associated family) proteins, CoREST (REST corepressor) and MIER1 (mesoderm induction early response 1; Ding et al, 2003). The conjoined conservation of these two domains in various transcriptional repressors suggests that they have important functions.
Ours and earlier studies on the ELM2 domain have shown its direct involvement in binding to histone deacetylases 1 and 2 (HDAC1/2; Ding et al, 2003; Wang et al, 2006). In comparison, less is known about the exact function of the SANT domain of RERE/Atro, although previous studies have implicated the SANT domain in binding to histone tails (Boyer et al, 2002; Yu et al, 2003), activating the enzymatic activity of HDAC3 (Guenther et al, 2001) and stimulating the histone demethylation activity of lysine-specific histone demethylase 1 (LSD1; Shi et al, 2005). SANT is evidently a versatile domain and its properties depend not only on its coding sequences, but also on other regions of the proteins within which it resides.
Here, we expand our knowledge about the SANT domain by reporting that the SANT domain of RERE/Atro recruits the histone methyltransferase (HMTase) G9a and that it enables RERE to stably methylate lysine 9 of histone H3 (H3K9) through cross-talk with the ELM2 domain. We find a parallel relationship among Atro, dG9a and Rpd3 (the fly homologue of HDAC1/2) in Drosophila. By using the fly system, we show that Atro, dG9a and Rpd3 bind to overlapping chromosomal regions, and act together to suppress wing vein and melanotic-mass formation in adult flies. Our results indicate that recruiting HDAC1/2 and G9a is an important mechanism used by ELM2–SANT domain proteins to modify histone tails, regulate gene transcription and control animal development.
Results And Discussion
RERE and Atro, but not ATN1, recruit G9a in human cells
There is increasing evidence to indicate that, in addition to histone deacetylation, histone lysine methylation (HMT) is another mechanism used by transcriptional regulators to repress gene transcription (Sims et al, 2003). For example, methylation on K9 and K27 of histone H3, and K20 of histone H4 has been shown to contribute to large-scale chromosomal repression (Lachner et al, 2003). As both the ELM2 and SANT domains are found in several transcriptional regulators known to have potent repressive activities (Ding et al, 2003), we suggested that, in addition to their known associations with HDAC1/2 (Wang et al, 2006), these two conserved domains are also connected with HMT activity.
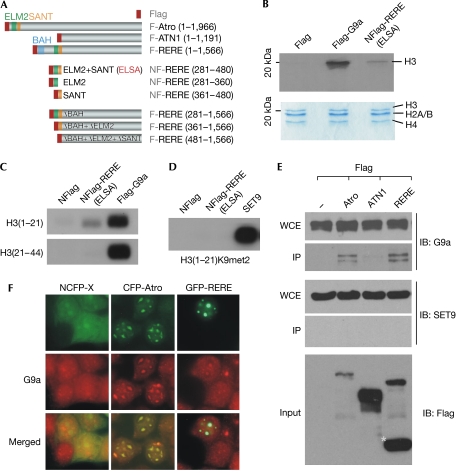
To test whether ELM2/SANT-mediated transcriptional repression involves HMT, we expressed Flag-tagged ELM2 and SANT domains of RERE (hereafter called REREELSA; Fig 1A) in human embryonic kidney cells (HEK293) and performed in vitro HMT assays on the immunoprecipitated REREELSA complex. The REREELSA complex exerts HMT activity (Fig 1B), although weaker than that of the control G9a. This result was expected because RERE is not by itself an HMTase. Therefore, we speculated that REREELSA acquires its HMT activity by associating with HMTases.
Figure 1.
Arginine–glutamic acid dipeptide repeats protein exerts histone methylation activity and associates with G9a. (A) Diagram showing the Flag-tagged Atrophin (Atro) proteins used in immunoprecipitation experiments. The conserved BAH (bromo-adjacent homology), ELM2 (EGL-27 and MTA1 homology 2) and SANT (SWI3/ADA2/N-CoR/TFIII-B) domains in each protein are highlighted in different colours. (B) Histone methylation (HMT) assays were performed on recombinant histone octamers to determine whether the ELM2–SANT domains of arginine–glutamic acid dipeptide repeats protein (RERE) mediate HMT activity. The identity of each histone is determined by its size in the Coomassie blue-stained gel; G9a, a known histone H3-specific histone methyltransferase (HMTase), was used as a control. (C,D) HMT assays were performed on the indicated peptides to determine which specific lysine residue is a target of the REREELSA complex. Recombinant SET9, a known H3K4 HMTase, was used as a control. (E) Western blot experiments were performed on the indicated immunoprecipitation products to test whether G9a is associated with Atro proteins. SET9 was used as a negative control. A cleaved product of RERE is marked with an asterisk. (F) Immunostaining experiments were performed on cells expressing green fluorescent protein (GFP)-tagged RERE or cyan fluorescent protein (CFP)-tagged Atro to determine whether endogenous G9a is recruited to the RERE/Atro-mediated nuclear foci. Nuclear localization signal-tagged CFP (NCFP) was used as a control. ELSA, ELM2+SANT domains; F, Flag; H3K9met2, dimethylated H3K9; IB, immunoblot; IP, immunoprecipitation; NF, nuclear localization signal-tagged Flag; WCE, whole-cell extract.
Our HMT assays also showed that the REREELSA complex preferentially methylates histone H3. We came to this conclusion on the basis of the following evidence: (i) the size of the methylated histone matches that of histone H3 (Fig 1B); (ii) the methylated histone migrates with G9a-methylated histone H3 (Fig 1B); and (iii) the REREELSA complex also methylates a synthetic peptide that encompasses only the first 21 amino acids of histone H3, H3(1–21) (Fig 1C). By contrast, the REREELSA immunoprecipitation complex fails to methylate H3(21–44), suggesting that the two lysine residues (K4 and K9) located within H3(1–21) are potential targets for REREELSA. Therefore, we tested an H3 (1–21)K9met2 peptide in further HMT assays. If H3K9 is a target of the REREELSA complex, prior methylation should prevent it from being methylated by the REREELSA complex. As we predicted, H3(1–21)K9met2 cannot be methylated by the REREELSA complex. In comparison, robust methylation was achieved by the control SET9, which is an H3K4 HMTase (Fig 1D). Thus our data indicate that the REREELSA complex primarily targets H3K9, but not H3K4, for methylation.
Subsequently, we investigated which HMTase lends Atrophin proteins (and REREELSA) the ability to methylate H3K9. We focused on G9a because G9a is an important HMTase known to catalyse mono- and dimethylation of H3K9 in euchromatin (Rice et al, 2003). Furthermore, both HDAC1, which binds to RERE strongly (Wang et al, 2006), and G9a have been found in the same protein complexes (Shi et al, 2003; Duan et al, 2005). A combination of co-immunoprecipitation and western blot analysis was performed on extracts derived from cells expressing Flag-tagged ATN1, RERE and Atro (Fig 1A,E). The assays confirmed that G9a is present in the immunoprecipitation complexes associated with RERE and Atro, but not with ATN1. The interaction between G9a and RERE or Atro is specific, as SET9 was not found in any of the immunoprecipitation complexes.
The physical association between RERE/Atro and G9a was further validated in HEK293 cells by using a cell biology approach. We performed co-immunostaining experiments on cells expressing green fluorescent protein (GFP)-tagged RERE or cyan fluorescent protein (CFP)-tagged Atro. These assays showed that endogenous G9a, which is known to form nuclear speckles, is recruited to the RERE/Atro-mediated nuclear foci (Fig 1F). These results provide further support to our conclusion that G9a is an associating factor of RERE and Atro. A similar interaction might occur between Atro and dG9a, the Drosophila homologue of G9a (Mis et al, 2006; Stabell et al, 2006).
The SANT domain of RERE binds to G9a directly
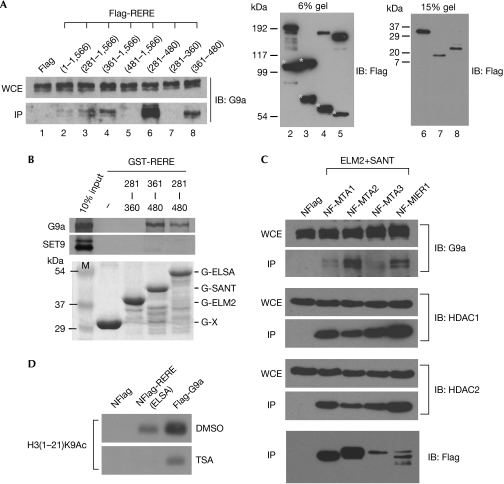
Both RERE and Atro have ELM2 and SANT domains, which are absent in ATN1. As ATN1 fails to bind to G9a (Fig 1E), we predicted that either or both of the ELM2 and SANT domains are involved in G9a association. A series of constructs expressing Flag-RERE variants with or without the ELM2 and SANT domains (Fig 1A) were generated and used for co-immunoprecipitation and western blot experiments. As shown in Fig 2A, G9a associates only with those RERE variants that contain the SANT domain (compare lanes 2–4,6,8 with lanes 5,7). The SANT domain of RERE is therefore involved in recruiting G9a.
Figure 2.
The SANT domain of arginine–glutamic acid dipeptide repeats protein binds to G9a and coordinates with the ELM2 domain to methylate H3K9. (A) Western blot experiments were performed on the indicated immunoprecipitation products to determine which region of arginine–glutamic acid dipeptide repeats protein (RERE) is responsible for G9a association. The input proteins are shown in the right panels. Cleaved products of RERE are marked with asterisks. Based on the molecular weights of the shown cleaved products of RERE, they all contain both ELM2 and SANT domains. (B) Glutathione-S-transferase (GST) pull-down experiments were used to determine whether the SANT domain of RERE binds to G9a directly. G9a and SET9 were labelled with [35S]methionine. The expression of GST-RERE proteins used in the pull-down assays is shown by Coomassie blue staining. (C) Western blot experiments were performed on the indicated immunoprecipitation products to test whether G9a is associated with additional ELM2–SANT domain proteins. Histone deacetylases 1 and 2 (HDAC1/2) were used as positive controls. (D) Histone methyltransferase assays were performed on an acetylated form of H3(1–21)K9 to determine whether REREELSA-mediated histone methylation is sensitive to trichostatin A (TSA) treatment. TSA is an HDAC inhibitor. ELM2, EGL-27 and MTA1 homology 2; ELSA, ELM2+SANT domains; G, GST; G9a, a known histone H3-specific histone methyltransferase; IB, immunoblot; IP, immunoprecipitation; MIER1, mesoderm induction early response 1; MTA1–3, metastasis-associated family proteins 1–3; NF, nuclear localization signal-tagged Flag; SANT, SWI3/ADA2/N-CoR/TFIII-B; SET9, an H3-K4 histone methyltransferase; WCE, whole-cell extract.
Glutathione-S-transferase (GST) pull-down assays were then performed to investigate whether the observed interaction between the SANT domain of RERE (RERESANT) and G9a is direct or indirect. In our assays, GST-RERE (361–480) and GST-RERE (281–480), both of which contain the SANT domain, pulled down G9a (Fig 2B). By contrast, GST-RERE (281–360), which lacks the SANT domain, failed to do so. The direct interaction observed between SANT domain and G9a is specific because none of the GST-RERE variants tested pulled down the control SET9.
A SANT domain similar to that of RERE/Atro has been found in several other transcriptional regulators, many of which also contain the ELM2 domain. For example, MTA proteins, which are intrinsic components of the nucleosome remodelling and histone deacetylase (NuRD/Mi-2) complex (Tong et al, 1998; Xue et al, 1998; Zhang et al, 1998), and MIER1, which is encoded by a fibroblast growth factor-responsive gene (Paterno et al, 1997), contain both ELM2 and SANT domains (Ding et al, 2003). Our findings led us to investigate whether these other ELM2–SANT domain proteins also show an affinity towards G9a. By using both immunoprecipitation and western blot experiments, we first confirmed the known associations between MTAs or MIER1 and HDAC1/2, and subsequently showed that the four tested ELM2–SANT domain proteins also recruit G9a (Fig 2C). Association with both HDAC1/2 and G9a is therefore a shared property of several ELM2–SANT domain proteins.
The ELM2 and SANT domains modify histone H3K9
Acetylation and methylation on H3K9 are known to be mutually exclusive. In order for H3K9 to be methylated, the prior removal of the acetyl groups by HDACs is essential. Having linked the functions of the ELM2 and SANT domains to both HDAC1/2 and G9a, we hypothesized that coordinated actions by these two domains might be responsible for stable methylation of H3K9. To test this hypothesis, we treated the REREELSA complex with trichostatin A (TSA), a potent HDAC inhibitor. We reasoned that if deacetylation of histone H3K9 is a prerequisite for its methylation, then blocking the HDAC activity of the REREELSA complex would inevitably affect its ability to methylate an acetylated form of H3K9. As shown in Fig 2D, treating the REREELSA complex with TSA, but not the control DMSO, impaired its ability to methylate H3(1–21)K9Ac. Integrating both HDAC and HMT activities therefore seems to be a strategy used by RERE, and perhaps by other ELM2–SANT domain proteins, to methylate H3K9.
Atro–dG9a–Rpd3 binds overlapping chromosomal regions
Building on the physical connections that we established among RERE, HDAC1/2 and G9a in human cells, we investigated next the biological relevance of their interactions. We focused on the fly system, because Atrophin, HDAC1/2 and G9a are all conserved in Drosophila. Furthermore, by using immunoprecipitation experiments, we also succeeded in showing parallel interactions between Atro and Rpd3 (the fly homologue of HDAC1/2), and between Atro and dG9a (supplementary Fig 1 online).
As Atro, dG9a and Rpd3 are all expressed in the nucleus of salivary gland cells, we used these cells to investigate, by means of their mutual interactions, whether these three proteins localize to overlapping chromosomal regions. Immunostaining experiments were performed on squashed polytene chromosomes derived from wild-type and Hsp70∷Atro (Hsp for Heat-shock protein) third instar larvae (Hsp70-Gal4 is a salivary gland-specific driver (Tsai et al, 2004; Mizutani et al, 2005), which allows us to induce Atro expression in salivary gland cells). In keeping with its role as a chromatin-binding factor, both endogenous and overexpressed Atro bind to distinct chromosomal loci that are primarily within the inter-band regions (supplementary Fig 2 online). As Hsp70∷ Atro larvae gave a stronger staining pattern, these larvae were used for further analysis.
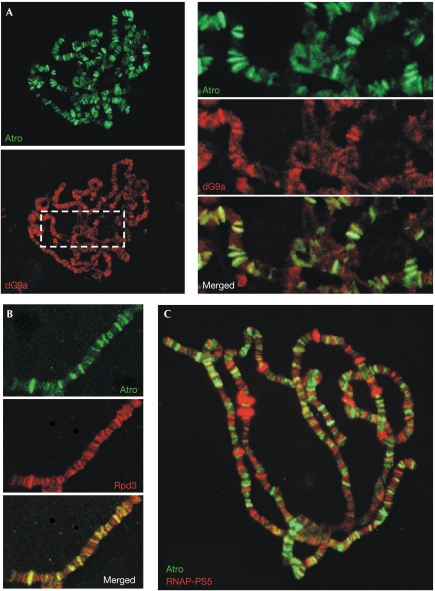
We performed co-immunostaining experiments on the polytene chromosomes from the salivary gland cells of Hsp70∷Atro larvae by using antibodies directed against Atro and against dG9a, Rpd3 or RNA polymerase II-phosphorylated-Ser5, which is a marker for transcriptional initiation. As expected, many—although not all—chromosomal regions that are enriched in Atro are also positive for dG9a or Rpd3 (Fig 3A,B). By contrast, the regions bound by Atro show little gene transcriptional initiation activity (Fig 3C). On the basis of these results, we propose that Atro, dG9a and Rpd3, by means of their mutual interactions, bind to specific chromosomal loci, where they act together to repress gene transcription.
Figure 3.
Atro, dG9a and Rpd3 bind to overlapping chromosomal regions in Drosophila. A co-immunostaining experiment was performed on salivary gland cells to determine the relative positions of Atro and dG9a, Rpd3 or a marker for transcriptional initiation (RNA polymerase II-phosphorylated-Ser5 (RNAP-p-S5)) on polytene chromosomes. Salivary gland cells derived from Hsp70∷Atro late third instar larvae were subjected to co-immunostaining using (A) Atro and dG9a antibodies, (B) Atro and Rpd3 antibodies, and (C) Atro and RNAP-p-S5 antibodies. Hsp70-Gal4 is a salivary gland-specific Gal4 driver without heat-shock treatment. The enlarged images shown in the right panels of (A) correspond to the boxed area in the lower panel. Atro, Drosophila Atrophin; dG9a, the Drosophila homologue of G9a; Hsp70, Heat-shock protein 70; Rpd3, the fly homologue of histone deacetylases 1 and 2.
Atro–dG9a–Rpd3 represses melanotic-mass formation
Next, we investigated, through genetic interactions, whether Atro, dG9a and Rpd3 participate in overlapping pathways to control Drosophila development. For this purpose, we generated two fly lines expressing Atro double-stranded (ds) RNA, Atro.IR1 and Atro.IR2, which allowed us to use the Gal4-UAS system to knock down Atro expression in a tissue-specific manner (supplementary Fig 3A,B online). Directed expression of either form of Atro dsRNA in the L3 and L4 inter-vein region, by using a dpp-Gal4 driver, causes ectopic wing vein formation (supplementary Fig 4A online). This result is consistent with the report that Atro, by antagonizing the activity of epidermal growth factor receptor (Egfr), can suppress wing vein formation (Charroux et al, 2006). The observed Atro dsRNA-mediated phenotype is specific because it can be fully rescued when both Atro dsRNA and Atro protein are simultaneously expressed in the wing (supplementary Fig 4A online).
Subsequently, we generated a recombined dpp∷Atro.IR1 line and tested it against a series of mutations, including those of Rpd3 and dG9a. Specifically, we wished to discover whether any Atro dsRNA-mediated phenotype can be modulated by the further mutation of Rpd3 or dG9a. The wing vein phenotype is enhanced when Rpd3 or dG9a is mutated, although, in comparison, Rpd3 seems to have a more prominent role than dG9a in assisting Atro to suppress wing vein formation (detailed information about these genetic experiments and results is provided in the supplementary information online).
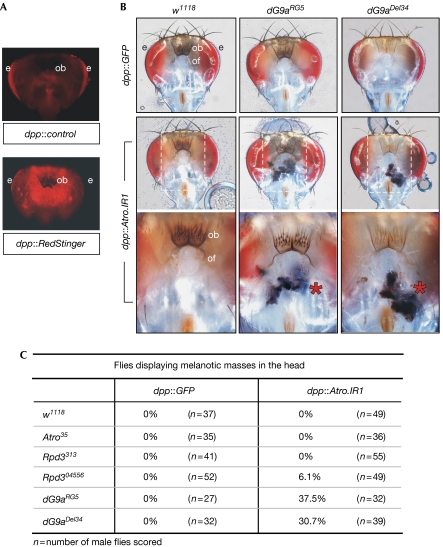
We discovered a more pronounced genetic interaction between Atro and dG9a in the adult head, where dpp-Gal4 is also active (Fig 4A). Melanotic masses, a possible consequence of aggregated haemocytes, were found in the heads of approximately 37.5% of adult dG9aRG5/Y; dpp∷Atro.IR1/+ flies and in approximately 30.7% of adult dG9aDel34/Y; dpp∷Atro.IR1/+ flies (Fig 4B,C; dG9a is an X-chromosome-linked gene, dG9aRG5 is a null allele and dG9aDel34 is a loss-of-function allele (Seum et al, 2007)). By contrast, only approximately 6.1% of dpp∷Atro.IR1/Rpd304556 (loss-of-function allele) adult flies were afflicted with melanotic masses, and none of the other fly lines tested produced melanotic lesions (Fig 4C). These genetic data for Atro, dG9a and Rpd3 observed in the Drosophila head differ from those found in the Drosophila wing (supplementary Fig 4C online), suggesting that various tissues show different sensitivity to loss of Atro, dG9a and Rpd3 in Drosophila. As no melanotic masses were detected in the heads of dG9a mutant or dpp∷Atro.IR1/Atro35 flies, we conclude that the formation of melanotic masses is due to the combined loss of Atro and dG9a or Rpd3 in the adult head.
Figure 4.
Atro, dG9a and Rpd3 act together to suppress melanotic-mass formation in the adult head of Drosophila. (A) The expression of RedStinger was used to determine that dpp-Gal4 is active in the adult head. The images show posterior views of the heads of dpp-Gal4 and dpp∷RedStinger flies. RedStinger is a DsRed variant. (B) Examination of adult heads to determine whether Atro interacts genetically with dG9a and Rpd3. Images show posterior views of the heads from flies with the indicated genotypes. dpp∷Atro.IR1 is an Atro double-stranded RNA-expressing line, w1118 and dpp∷GFP were used as negative control lines, dG9aRG5 is a deletion line generated by homologous recombination and dG9aDel34 is an imprecise P-element excision line. The bottom panel contains enlarged images corresponding to the boxed areas shown in the middle panel. The melanotic masses are indicated by asterisks. (C) Genetic data showing the number and percentage of flies with melanotic masses in their heads. The genotype of each examined fly line is indicated. Atro35 is a null allele, Rpd304556 is a loss-of-function allele and Rpd3313 is a hypomorphic allele. Atro, Drosophila Atrophin; dG9a, the Drosophila homologue of G9a; e, eye; ob, occipital bristles; of, occipital foramen; Rpd3, the fly homologue of histone deacetylases 1 and 2.
Prospects and implications
The results of this study suggest that RERE and Atro use their ELM2 and SANT domains to recruit HDAC1/2 and G9a, respectively. The recruited HDAC1/2 and G9a, in turn, orchestrate sequential molecular events that lead first to the deacetylation and then to the methylation of H3K9 at the chromosomal loci where RERE or Atro resides (see a model in supplementary Fig 5 online). A similar mode of action might apply to other ELM2–SANT domain proteins, such as MTA1–3 and MIER1, as these factors also show affinity towards both HDAC1/2 and G9a (Fig 2C). Although not tested in this study, another ELM2 and SANT domain protein, CoREST, has been identified in a large carboxy-terminal binding protein complex (with approximately 20 associated polypeptides), which also contains G9a and HDAC1/2 (Shi et al, 2003). Our results led us to propose that a stable CoREST–HDAC1/2–G9a protein complex forms as a result of the binding of both HDAC1/2 and G9a to the ELM2–SANT domains of CoREST.
MTA proteins and CoREST have been shown to be components of large protein complexes that show properties ranging from chromatin remodelling and histone deacetylation to transcriptional repression (Shi et al, 2003; Bowen et al, 2004). As the ELM2 and SANT domains of RERE/Atro resemble those of MTA and CoREST, we predict that RERE or Atro might also be subunits of large protein complexes. In support of this prediction, silver staining of the REREELSA immunoprecipitation complex showed that REREELSA is associated with multiple proteins (supplementary Fig 6 online). It is therefore conceivable that the ELM2–SANT domains, by acting as binding scaffolds for various transcriptional cofactors, allow proteins such as RERE, Atro, MTAs or CoREST to modify chromatin structures and to silence gene transcription efficiently.
Atrophin proteins have already been shown to interact with various transcriptional regulators, including homeobox protein (Zhang et al, 2002), nuclear receptors (Wang et al, 2006; Zhang et al, 2006) and ETO/MTG8 (which causes acute myeloid leukaemia 1; Wood et al, 2000). MTA proteins are implicated in various cancers and CoREST is involved in regulating neuronal cell fate. Our findings about the properties of the ELM2 and SANT domains suggest that the recruitment of HDAC1/2 and G9a is an important strategy used by these various transcriptional regulators to repress gene transcription; perturbation of this process might cause diseases and defects such as melanotic masses. As specific inhibitors of G9a have recently been identified (Kubicek et al, 2007), our results raise the possibility that G9a inhibitors might be useful, either alone or together with HDAC inhibitors, as therapeutic agents for treating diseases such as neurological disorders or cancers that involve Atrophin, CoREST or MTA proteins.
Methods
Drosophila. stocks and experiments. Stable Atro.IR1 and Atro.IR2 fly lines were generated by injecting constructs into w1118 embryos (Duke University Non-Mammalian Model Systems Flyshop). The lines w1118, dpp-Gal4, Hsp70-Gal4, ey-Gal4, UAS-GFP, UAS-RedStinger, Rpd304556, Egfrt1 and EgfrE1 were obtained from the Bloomington Stock Center. Rpd3313 was obtained from T. Grigliatti (University of British Columbia; Mottus et al, 2000); dG9aBG5 and dG9aDel34 were gifts from Dr P. Spierer (University of Geneva; Seum et al, 2007). UAS-Atro and Atro35 lines were described previously (Erkner et al, 2002; Charroux et al, 2006); dpp∷Atro.IR1 was generated by chromosomal recombination. Genetic experiments were carried out at 24.5°C.
Antibody generation. Polyclonal Atro antibodies were developed both in guinea-pigs and rabbits by using KLH-conjugated synthetic polypeptide ADTPALRQLSEYARPHVA. The polyclonal dG9a antibody was developed in guinea-pigs by using KLH-conjugated combined synthetic polypeptides AMEADRRTDDSYYFDLDN and GEEICFDYGEKFWRVEHR.
Additional materials and methods are described in the supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We thank R. Sim and B. Zhu from Danny Reinberg's laboratory for their advice on HMT assays; the Bloomington Stock Center for fly stocks; P. Spierer and C. Seum for dG9a fly lines; T.A. Grigliatti for Rpd3 fly lines; A. Lambertsson for dG9a antibody; Y. Nakatani for G9a antibody; P. Becker, A. Brehm and L. Pile for Rpd3 antibodies; R. Carthew for WIZ vector; and R. Head and R. Steward for commenting on this manuscript. C.-C.T. is a recipient of funding from the National Institutes of Health, the Leukemia Research Foundation and the New Jersey Commission on Cancer Research.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bowen NJ, Fujita N, Kajita M, Wade PA (2004) Mi-2/NuRD: multiple complexes for many purposes. Biochim Biophys Acta 1677: 52–57 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Langer MR, Crowley KA, Tan S, Denu JM, Peterson CL (2002) Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol Cell 10: 935–942 [DOI] [PubMed] [Google Scholar]
- Charroux B, Freeman M, Kerridge S, Baonza A (2006) Atrophin contributes to the negative regulation of epidermal growth factor receptor signaling in Drosophila. Dev Biol 291: 278–290 [DOI] [PubMed] [Google Scholar]
- Ding Z, Gillespie LL, Paterno GD (2003) Human MI-ER1 α and β function as transcriptional repressors by recruitment of histone deacetylase 1 to their conserved ELM2 domain. Mol Cell Biol 23: 250–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Zarebski A, Montoya-Durango D, Grimes HL, Horwitz M (2005) Gfi1 coordinates epigenetic repression of p21Cip/WAF1 by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol Cell Biol 25: 10338–10351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkner A et al. (2002) Grunge, related to human Atrophin-like proteins, has multiple functions in Drosophila development. Development 129: 1119–1129 [DOI] [PubMed] [Google Scholar]
- Guenther MG, Barak O, Lazar MA (2001) The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol 21: 6091–6101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide R et al. (1994) Unstable expansion of CAG repeat in hereditary dentatorubral–pallidoluysian atrophy (DRPLA). Nat Genet 6: 9–13 [DOI] [PubMed] [Google Scholar]
- Kubicek S et al. (2007) Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell 25: 473–481 [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Sullivan RJ, Jenuwein T (2003) An epigenetic road map for histone lysine methylation. J Cell Sci 116: 2117–2124 [DOI] [PubMed] [Google Scholar]
- Mis J, Ner SS, Grigliatti TA (2006) Identification of three histone methyltransferases in Drosophila: dG9a is a suppressor of PEV and is required for gene silencing. Mol Genet Genomics 275: 513–526 [DOI] [PubMed] [Google Scholar]
- Mizutani A, Wang L, Rajan H, Vig PJ, Alaynick WA, Thaler JP, Tsai CC (2005) Boat, an AXH domain protein, suppresses the cytotoxicity of mutant ataxin-1. EMBO J 24: 3339–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottus R, Sobel RE, Grigliatti TA (2000) Mutational analysis of a histone deacetylase in Drosophila melanogaster: missense mutations suppress gene silencing associated with position effect variegation. Genetics 154: 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagafuchi S, Yanagisawa H, Ohsaki E, Shirayama T, Tadokoro K, Inoue T, Yamada M (1994) Structure and expression of the gene responsible for the triplet repeat disorder, dentatorubral and pallidoluysian atrophy (DRPLA). Nat Genet 8: 177–182 [DOI] [PubMed] [Google Scholar]
- Paterno GD, Li Y, Luchman HA, Ryan PJ, Gillespie LL (1997) cDNA cloning of a novel, developmentally regulated immediate early gene activated by fibroblast growth factor and encoding a nuclear protein. J Biol Chem 272: 25591–25595 [DOI] [PubMed] [Google Scholar]
- Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, Shinkai Y, Allis CD (2003) Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell 12: 1591–1598 [DOI] [PubMed] [Google Scholar]
- Seum C, Bontron S, Reo E, Delattre M, Spierer P (2007) Drosophila G9a is a nonessential gene. Genetics 177: 1955–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Lee G, Choe Y, Zoltewicz JS, Peterson AS (2007) Functional architecture of atrophins. J Biol Chem 282: 5037–5044 [DOI] [PubMed] [Google Scholar]
- Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y (2003) Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422: 735–738 [DOI] [PubMed] [Google Scholar]
- Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y (2005) Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell 19: 857–864 [DOI] [PubMed] [Google Scholar]
- Sims RJ III, Nishioka K, Reinberg D (2003) Histone lysine methylation: a signature for chromatin function. Trends Genet 19: 629–639 [DOI] [PubMed] [Google Scholar]
- Stabell M, Eskeland R, Bjorkmo M, Larsson J, Aalen RB, Imhof A, Lambertsson A (2006) The Drosophila G9a gene encodes a multi-catalytic histone methyltransferase required for normal development. Nucleic Acids Res 34: 4609–4621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL (1998) Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 395: 917–921 [DOI] [PubMed] [Google Scholar]
- Tsai CC, Kao HY, Mitzutani A, Banayo E, Rajan H, McKeown M, Evans RM (2004) Ataxin 1, a SCA1 neurodegenerative disorder protein, is functionally linked to the silencing mediator of retinoid and thyroid hormone receptors. Proc Natl Acad Sci USA 101: 4047–4052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Rajan H, Pitman JL, McKeown M, Tsai CC (2006) Histone deacetylase-associating Atrophin proteins are nuclear receptor corepressors. Genes Dev 20: 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JD, Nucifora FC Jr, Duan K, Zhang C, Wang J, Kim Y, Schilling G, Sacchi N, Liu JM, Ross CA (2000) Atrophin-1, the dentato-rubral and pallido-luysian atrophy gene product, interacts with ETO/MTG8 in the nuclear matrix and represses transcription. J Cell Biol 150: 939–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W (1998) NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell 2: 851–861 [DOI] [PubMed] [Google Scholar]
- Yanagisawa H, Bundo M, Miyashita T, Okamura-Oho Y, Tadokoro K, Tokunaga K, Yamada M (2000) Protein binding of a DRPLA family through arginine–glutamic acid dipeptide repeats is enhanced by extended polyglutamine. Hum Mol Genet 9: 1433–1442 [DOI] [PubMed] [Google Scholar]
- Yu J, Li Y, Ishizuka T, Guenther MG, Lazar MA (2003) A SANT motif in the SMRT corepressor interprets the histone code and promotes histone deacetylation. EMBO J 22: 3403–3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CL, Zou Y, Yu RT, Gage FH, Evans RM (2006) Nuclear receptor TLX prevents retinal dystrophy and recruits the corepressor atrophin1. Genes Dev 20: 1308–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Xu L, Lee J, Xu T (2002) Drosophila atrophin homolog functions as a transcriptional corepressor in multiple developmental processes. Cell 108: 45–56 [DOI] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D (1998) The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95: 279–289 [DOI] [PubMed] [Google Scholar]
- Zoltewicz JS, Stewart NJ, Leung R, Peterson AS (2004) Atrophin 2 recruits histone deacetylase and is required for the function of multiple signaling centers during mouse embryogenesis. Development 131: 3–14 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information