Abstract
Differences in neutral diversity at different loci are predicted to arise due to differences in mutation rates and from the “hitchhiking” effects of natural selection. Consistent with hitchhiking models, Drosophila melanogaster chromosome regions with very low recombination have unusually low nucleotide diversity. We compared levels of diversity from five pericentromeric regions with regions of normal recombination in Arabidopsis lyrata, an outcrossing close relative of the highly selfing A. thaliana. In contrast with the accepted theoretical prediction, and the pattern in Drosophila, we found generally high diversity in pericentromeric genes, which is consistent with the observation in A. thaliana. Our data rule out balancing selection in the pericentromeric regions, suggesting that hitchhiking is more strongly reducing diversity in the chromosome arms than the pericentromere regions.
NUCLEOTIDE diversity (the mean amount of difference between the DNA sequences of the same locus) at neutral sites in a genomic region depends on many factors, including the mutation rate in the region and the effective population size (which is influenced by population subdivision). In addition, natural selection can affect neutral diversity levels of silent and weakly selected variants that are closely linked to selected sites. For instance, selective sweeps can reduce diversity of silent and weakly selected variants (see Maynard Smith and Haigh 1974; Kaplan et al. 1989), weak Hill-Robertson effects can reduce polymorphism in genome regions with low recombination (as well as reducing levels of adaptation, see McVean and Charlesworth 2000), and balancing selection increases coalescence times of regions of a chromosome surrounding the selected site(s) (Hudson et al. 1987). Other things being equal, the size of the genome regions in which neutral sites will be affected by either of these “hitchhiking” processes will be larger, the lower the region's recombination rate. Selected variants may also impede selection acting on closely linked selected sites, and local recombination rates thus determine the ability of selection to eliminate or fix selected mutations with moderate and weak fitness effects, reducing the efficacy of selection (Charlesworth 1994).
It is well established from studies of the Drosophila melanogaster X chromosome that nucleotide diversity in chromosomal regions with very low recombination close to the centromeres is lower than in regions with higher recombination (Aguadé et al. 1989; Begun and Aquadro 1991, 1992), in contrast with the initial expectation that balancing selection would cause high diversity in these low recombination regions (Berry et al. 1991). Diversity estimates are also consistently low for D. melanogaster's fourth chromosome, also with very low recombination rates (Jensen et al. 2002; Wang et al. 2002). A mutagenic effect of recombination is excluded, since loci in low recombination regions do not have unusually low divergence from closely related Drosophila species (Aguadé et al. 1989; Begun and Aquadro 1992; Sheldahl et al. 2003; Presgraves 2005). Thus, these results suggest the action of selectively driven hitchhiking, either through selective sweeps or by “background selection” as deleterious mutations are eliminated (Charlesworth et al. 1993).
It is thus widely accepted that low diversity in centromere regions is a general phenomenon. Apart from work in Drosophila, however, DNA sequence diversity has rarely been studied for loci in such regions. Many species have chromosomal regions with low crossing over surrounding the centromeres (Choo 1997, 1998), including the plants maize and Solanum/Lycopersicon (Sherman and Stack 1995; Anderson et al. 2003). Previous studies of the relationship between diversity and recombination rates in the plants Lycopersicon (Baudry et al. 2001) and maize (Tenaillon et al. 2002) did not include loci in very low recombination regions near centromeres (and did not find clear evidence for a correlation between nucleotide diversity and estimated recombination rates). Similarly, in mammals, it has been established that diversity is higher in high recombination regions (Nachman 2001; Hellmann et al. 2003), but regions with very different recombination rates have not yet been compared, and the slight effect observed may be due to mutation rate differences, since synonymous divergence from chimpanzees and baboons also increases with increased recombination rate (Hellmann et al. 2003). More studies of the relationship between diversity and recombination rates are therefore needed.
In Arabidopsis thaliana, however, diversity in pericentromeric regions of A. thaliana (38 loci) is somewhat higher than the average for a set of 297 genes in chromosome arm regions (the difference was significant when all site types were analyzed, but not when only silent sites were compared, in the study of Schmid et al. 2005), despite rates of crossing over near the centromere regions of all five chromosomes estimated to be at least 10-fold lower than in the chromosome arms (Copenhaver et al. 1998, 1999; Haupt et al. 2001). Overall, a negative correlation with distance from the centromere was also found for nucleotide diversity estimated using an array-based sequencing method, for 4-fold degenerate sites and intergenic sites (Clark et al. 2007), and a similar, more pronounced, diversity difference was found for “single-feature polymorphisms,” which include insertion/deletion differences (Borevitz et al. 2007).
Two interacting factors are likely to explain the difference between Drosophila and A. thaliana. First, A. thaliana is highly self-fertilizing (Abbott and Gomes 1988; Berge et al. 1998), and the resulting high homozygosity causes low effective recombination rates, regardless of the crossing-over rates of different genome regions (Nordborg and Donnelly 1997). Thus nucleotide diversity may only weakly correlate with crossing-over rates, since hitchhiking is expected to be strong in both regions. Compared with Drosophila, any reduction in neutral diversity in the pericentromeric regions due to hitchhiking should therefore be less in A. thaliana, relative to the diversity level in the chromosome arms. Second, A. thaliana pericentromere regions have lower gene densities than chromosome arm regions (Arabidopsis Genome Initiative 2000; Wright et al. 2003a). It is the total number and density of closely linked genes that determines the extent to which genetic hitchhiking will affect the effective population size (Ne) that applies to a genome region. Chromosome arms thus experience high recombination rates, but their high gene numbers and densities may nevertheless lead to genetic hitchhiking (Nordborg et al. 2005).
Data from an outcrossing relative of A. thaliana (with similar relative gene numbers and densities in the pericentromere and chromosome arm regions) should be helpful in evaluating these possibilities and specifically can test whether A. thaliana's inbreeding is the cause of its similar diversity in the two types of regions. Outcrossing ensures high effective rates of recombination in the arm regions, reducing the opportunity for hitchhiking, predicting a diversity difference similar to that in Drosophila.
We therefore compared diversity at loci in several chromosome arms with loci in five pericentromeric regions, in the self-incompatible species A. lyrata, an outcrossing close relative of A. thaliana. A. lyrata has 8 chromosomes, whereas A. thaliana has 5, due to chromosome fusions (Kuittinen et al. 2004; Koch and Kiefer 2005; Yogeeswaran et al. 2005). Despite other karyotypic differences between these species, including some reciprocal translocations, most chromosome arms are syntenic, and the positions of the five centromeres in A. thaliana are probably unchanged from their ancestral positions (Berr et al. 2006; Kawabe et al. 2006b; Lysak et al. 2006). For regions corresponding to the pericentromeres of A. lyrata AL5 (homologous to A. thaliana CEN3) and AL7 (A. thaliana CEN5), the gene content is the same as in three other related species investigated: Capsella rubella, Olimarabidopsis pumila, and A. arenosa, and the A. arenosa BAC clone homologous to A. thaliana CEN3 also includes some centromeric repeat sequences, definitively identifying this as a pericentromere region (Hall et al. 2006). Finally, mapping of loci located near the centromeres of A. thaliana shows low crossing-over frequencies in A. lyrata. No recombinants were found among 99 or 198 chromosomes in putative pericentromere regions corresponding to at least 100 kb in A. thaliana, whereas the regions we classify as chromosome arms have 4 to 8 cM/Mb; furthermore, the short and long arm pericentromeric markers were completely linked for all chromosomes (Kawabe et al. 2006b). Crossing over in the A. lyrata putatively pericentromeric regions is therefore estimated to be at least 10-fold less than in the chromosome arms.
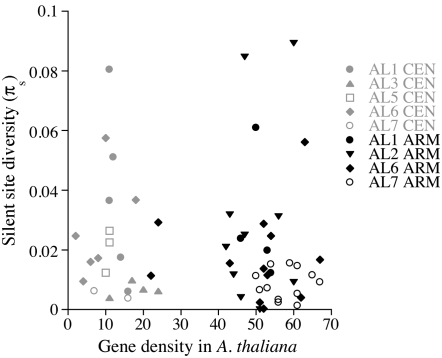
Gene densities in the A. lyrata chromosomes are not yet known (the forthcoming genome sequence assembly should reveal this information), but the A. lyrata genome probably has generally similar, or perhaps somewhat lower, densities than orthologous regions in A. thaliana, on the basis of the overall larger genome in A. lyrata. Figure 1 shows that, in A. thaliana, the pericentromeric regions studied here have roughly fourfold-lower gene densities than the chromosome arm regions we studied (see materials and methods), consistent with the overall estimated fraction of coding DNA cited above. Our comparison between chromosome arm regions and pericentromeric regions does not require detailed knowledge of gene densities in A. lyrata in the regions we compared. However, some information from related species is pertinent, since the pericentromeric regions of A. thaliana appear to be expanded relative to those of C. rubella, O. pumila and A. arenosa (Hall et al. 2006), so that pericentromeric regions of related species might not have low gene densities. The difference is, however, partly due to the large regions containing the 5S rDNA loci inserted in the A. thaliana chromosomes 3 and 5 pericentromere regions (Berr et al. 2006). The mean pericentromeric region gene densities in C. rubella and A. arenosa (0.17 and 0.12 genes per kb, respectively), are nevertheless considerably lower than in the A. thaliana chromosome arm regions (>0.25 genes/kb, see Hall et al. 2006). Gene densities are therefore probably lower than for chromosome arm regions in A. lyrata also. This is supported by our diversity results (below) showing no evidence of a reduced Ne for pericentromere region genes in A. lyrata, which would be expected if these regions had high gene densities.
Figure 1.—
Comparison of silent site diversity of 20 pericentromeric region and 38 chromosome arm loci in A. lyrata. The diversity estimates for each gene are plotted against the gene densities in A. thaliana of the regions in which they are located (estimated as number of genes in 200-kb regions centered on each gene; the counts are for functional genes only, including predicted and hypothetical genes, but excluding pseudogenes and genes related to transposons).
Our new study extends our previous analysis of the relationship between diversity and recombination rates (Wright et al. 2006), using a single A. lyrata centromere (AL1, corresponding to that of A. thaliana's chromosome 1), where higher species-wide diversity was found than for pericentromere region loci on the chromosome arms AL1 and AL2 (corresponding to the two arms of A. thaliana's chromosome 1; the AL2 centromere was not studied because it is one of the centromeres lost in a fusion in the evolution of the A. thaliana chromosome and is not yet mapped in A. lyrata). With data from just one such region, we could not definitively exclude the possibility that a single locus with long-term balancing selection could be responsible (although our previous analyses did not support this).
By extending the analysis to other chromosomes, we find that this result is general in A. lyrata, i.e., that a difference from the Drosophila pattern is found for pericentromere regions of several chromosomes (again with no evidence for balancing selection). Our additional data also allow us to conduct a novel test for the action of hitchhiking on arm loci vs. pericentromeric genes. The results suggest that the difference from the Drosophila pattern exists because hitchhiking effects have acted on A. lyrata arm loci more than on pericentromeric loci.
MATERIALS AND METHODS
Population samples:
Thirty-two plants from four natural populations of A. lyrata ssp. petraea (Mt. Esja in Iceland, Stubbsand in Sweden, Plech in Germany, and Karhumaki in Russia) and two of A. lyrata ssp. lyrata (Ontario, Canada, and Indiana) were studied (Wright et al. 2006). DNA was amplified from dried leaves from 5 to 7 plants from each population as described previously (Kawabe et al. 2006c).
Genome regions analyzed:
We analyzed a total of 58 genes. The new genes analyzed for nucleotide diversity within A. lyrata (supplemental Table 1) were the same as those used in a previous genetic mapping study (Kawabe et al. 2006b), excluding a triplicated gene found in the AL3 centromere region; the potential gene functions are given in Kawabe et al. (2006b). The genes surveyed here from putative pericentromeric regions were chosen from the known pericentromeric regions of each A. thaliana chromosome arm; one gene was as close as possible to the core centromere, and one was at least 100 kb distal to this (Kawabe et al. 2006b). All of them are within transposon-rich regions (the average density of transposon-related genes per 100 kb is 10.2, compared with a value of 0.8 for our reference chromosome arm loci).
We included the loci already analyzed previously, including 5 loci from the AL1 pericentromeric region (Wright et al. 2006) and 15 newly studied pericentromere region genes from other chromosomes. For reference loci, we used 18 chromosome arm region genes from AL6 and AL7 (Kawabe et al. 2006a) in addition to 20 loci from AL1 and AL2 previously studied (Wright et al. 2006). For the new loci studied here, regions of ∼600–800 bp, including introns, were amplified from each gene and sequenced using the DYEnamic sequence kit using the PCR products as templates. If two or more heterozygous length variants were found in a gene, the PCR product was cloned and the sequences of both alleles were determined. The same populations were used, but the samples were smaller, because cloning of sequences from plants heterozygous for length differences limited the sample size that was possible. All samples are sufficient for accurate within-population diversity estimates (since samples >5 alleles improve accuracy only slightly; see Pons and Chaouche 1995).
Data analyses:
The A. lyrata sequences for each locus were aligned manually together with those of A. thaliana and Turritis glabra. The DNAsp program version 4 (Rozas et al. 2003) was used to estimate divergence and diversity (using nucleotide diversity, π). Tajima's tests (Tajima 1989) and McDonald–Kreitman (MK) (McDonald and Kreitman 1991) tests for neutrality, estimates of Hudson's Snn statistic (Hudson 2000) and Kst (Hudson et al. 1992) were calculated using DNAsp. Multilocus HKA (Hudson et al. 1987) tests were done using the HKA test program distributed by J. Hey (http://lifesci.rutgers.edu/∼heylab/HeylabSoftware.htm#HKA) and the MLHKA program (Wright and Charlesworth 2004).
To provide an explicit test for selection on pericentromeric and arm loci, a series of models was explored using the MLHKA program (Wright and Charlesworth 2004). We first calculated the likelihood of the data under a strictly neutral model for all loci. We then compared this with a second model that allowed one of the 58 loci to show evidence of different evolution (i.e., potentially to be under some form of selection, which might include directional selection for new adaptive variants, causing selective sweeps, or balancing selection leading to higher nucleotide diversity than for other genome regions). Model two was run for each of the 58 loci, i.e., allowing each individual locus to deviate from neutrality, and, in each case, the likelihood of the second model was compared with that of the neutral model by computing twice the difference in log likelihoods. The significance of this statistic was assessed for each locus, using the χ2 distribution, with one degree of freedom. We also tested for selection on the pericentromeric genes as a class and the arm genes as a class (see text).
Linkage disequilibrium (LD) among pairs of sites was calculated using Weir's (Weir 1996) EM algorithm for estimating the squared correlation coefficient, r2, from diploid genotype data, as implemented by MacDonald et al. (2005). Only sites segregating at a frequency >5% were used in the analysis. R code for the r2 estimates was kindly provided by S. MacDonald.
Permutation tests of the silent site nucleotide diversity (π) and Kst values were performed by taking the data set of all loci (from both the pericentromeric and the chromosome arm regions of chromosomes AL6 and AL7), randomly choosing a set of 20 loci 10,000 times and asking what proportion of such samples have Kst (or πsilent) as high or higher than the observed mean value at the 20 pericentromeric loci.
RESULTS
Nucleotide diversity in A. lyrata and tests for hitchhiking effects of selection:
Overall, there is little difference in species-wide diversity in A. lyrata between genes in the pericentromeric and chromosome arm regions (Figure 1). The overall mean synonymous site π values for arm and pericentromere loci, respectively, are 0.0195 and 0.0226, a nonsignificant difference (P = 0.33, using a permutation test). Genes in the pericentromeric regions of chromosomes AL5 and AL6, like those in AL1 (Wright et al. 2006), frequently have high diversity (for AL6, the synonymous and silent site π values are 0.0292 and 0.0262, respectively, for the arm loci, vs. 0.0398 and 0.0269 for the pericentromeric loci), whereas the loci in two other centromere regions (chromosomes AL3 and AL7), mostly have low or moderate diversity). We could not compare diversity for the loci near the AL3 and AL5 centromeres, with the respective chromosome arm loci, since diversity on these chromosomes has so far been surveyed only for the pericentromeric loci.
Because we compared synonymous and silent site diversity, it seems unlikely that the unexpectedly high diversity for pericentromere region loci could be due to direct effects of lower selective constraints on these loci, but we first evaluate this possibility and then test between several other possible explanations, which are not mutually exclusive.
No accumulation of replacement substitutions:
If nonsynonymous sites in the pericentromere region experience weaker selection than such sites in genes in other genome regions, this could lead to higher diversity of sequences in this region. Nonsynonymous site divergence from the A. thaliana orthologs should then also be high, compared with the arm loci. However, this is not seen. Both sets of genes show signs of selective constraint; their mean Ks values are very similar (0.160 with standard deviation 0.020 for pericentromere region loci, vs. 0.145 ± 0.007 for the chromosome arm loci, or 91% of the former value), while the mean Ka value is much lower for the pericentromere region loci (0.0115 with standard deviation 0.0032, vs. about double, 0.0224 ± 0.0023, for the chromosome arm loci).
We did MK tests to assess the significance of any differences in fixation of either slightly deleterious or advantageous mutations, by comparing the ratios of replacement to synonymous changes between sites diverged between A. lyrata and A. thaliana, and polymorphic within A. lyrata [supplemental Table 1 shows the results for the pericentromere region loci and loci from chromosomes AL6 and AL7; the AL1 and AL2 arm locus data are published in Foxe et al. (2008) and are not shown here]. No significant differences were observed between the low- and high-diversity centromeres, and there were no significant MK test results for any individual pericentromere region loci, or in pooled data for each centromere.
We therefore conclude that these loci do not lack effective selective constraints (for example, due to their having functions that permit nonsynonymous changes). The low Ka value for the pericentromere regions also rules out the possibility that hitchhiking processes (either selective sweeps or background selection) have caused lower effective population sizes for the pericentromere regions, reducing the efficacy of selection in the region. There is therefore no evidence that the A. lyrata pericentromere regions have lower effective population sizes than the chromosome arm regions.
HKA tests for selection:
The main alternative possibilities to explain the finding that diversity for multiple pericentromere region genes is at least as high as for the chromosome arm loci are (i) balancing selection affecting loci in several centromere regions and leading to high diversity at synonymous and intron sites or (ii) reduced diversity of arm loci through some form of hitchhiking. We therefore tested these possibilities and conclude that the second is the more likely in A. lyrata.
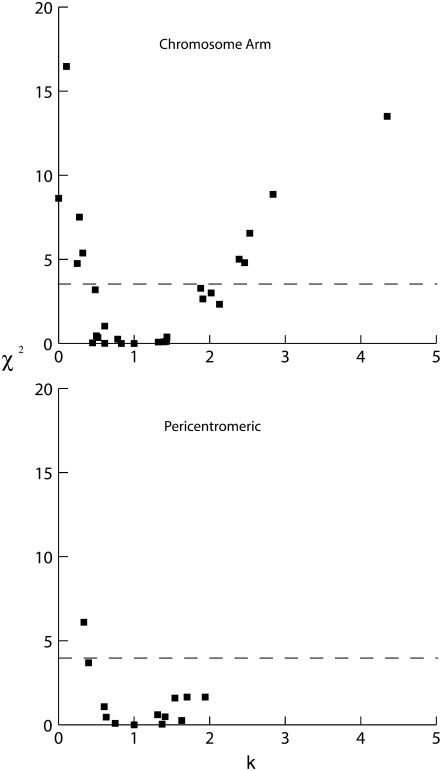
To assess whether selection affects either set of loci (possibilities i and ii above), we tested the diversity differences by multilocus HKA tests. Figure 2 displays the values of k (the estimated diversity values within A. lyrata for each locus, scaled by their divergence from A. thaliana; under neutrality, the expected value of k is 1) and the χ2 values for the loci from tests of the null hypothesis, neutrality (see materials and methods). The pericentromere region loci generally fall around the neutral expectation value of k = 1, whereas the chromosome arm loci include more extreme k values. Using likelihood ratio tests, 10 of the 38 chromosome-arm loci (26%) show evidence for selection significant at the 5% level, while only 1 of 20 (5%) pericentromere region loci is significant (Figure 2). Approximately 5% of our tests should be significant by chance, so we conclude from these results that at least some of the arm loci are subject to the effects of hitchhiking (rather than the alternative, that selection has acted to increase diversity in the pericentromere region). Several loci from the chromosome arms have reduced k values, consistent with selective sweeps and/or background selection, and some have unexpectedly high diversity, suggesting possible balancing selection. Caveats to this analysis are that the pericentromere loci are linked, reducing the number of independent tests of selection, and that the number of independent tests is lower than the number of loci tested (since the same data are included in all tests). Furthermore, population subdivision and population size changes could contribute to an excess of variance in polymorphism. Nevertheless, given that centromeric and arm loci have experienced the same demographic history, the results clearly suggest the neutrality of pericentromeric genes and the action of hitchhiking on the chromosome arms.
Figure 2.—
Locus-specific tests of neutrality using the maximum likelihood HKA test (MLHKA). The x-axis shows k, the ratio of observed to expected diversity levels at each locus, controlling for mutation rate (see text), and the y-axis shows the results of the corresponding χ2 tests for departures from the neutral model, using the likelihood ratio test (the dashed line shows the 5% significance level). The graph at the top shows the results for chromosome arm loci and the one at the bottom for pericentromere loci.
We also did MLHKA tests including all loci not previously analyzed by Wright et al. (2006); results could not be obtained for the entire set of loci, because the program ran too slowly. Likelihood ratio tests were done to compare a strictly neutral model with models allowing either all the pericentromeric loci, or all arm loci, to show evidence of different evolution (i.e., one class of genes to be under either directional selection for new adaptive variants, causing selective sweeps, or balancing selection leading to higher nucleotide diversity than for other genome regions). For the AL6 and AL7 loci, the results indicate a significant excess variance in diversity levels and show that this is not due to selection affecting the pericentromere region loci: a model that allows for selection on the class of chromosome arm loci significantly increases the likelihood (χ2 = 39.9, 18 d.f., P = 0.002), whereas allowing for selection acting on the centromere loci does not (χ2 = 9.32 with 15 d.f., P > 0.86). This supports the analyses of individual loci just described.
Multilocus HKA tests, using the HKA test program of J. Hey, also often detect heterogeneity between diversity levels of the arm-region loci on chromosomes AL6 and AL7 (bottom row of supplemental Table 2); AL6 arm loci have higher diversity than those on the AL7 arm studied (Figure 1), and the difference is consistent and significant in both subspecies. There is also significant heterogeneity among arm loci on these chromosomes, especially AL7, and also between arm vs. pericentromere loci of AL7 (although not within ssp. lyrata, see supplemental Table 2). In contrast to these many statistically significant differences, differences were only occasionally found among pericentromeric loci on any chromosome, mostly in ssp. lyrata. Loci in the short arm of AL1 have higher diversity than long-arm loci, although the effect is significant only for ssp. lyrata, which also shows a marginally significant effect for AL3, and there is significant heterogeneity among AL6 loci in the short-arm pericentromere region (again only in ssp. lyrata). However, the test may be unreliable in this subspecies, which has probably experienced a severe bottleneck, since it generally has very low nucleotide diversity (Wright et al. 2003b, 2006) and low microsatellite variability (Muller et al. 2008), so that the HKA test assumptions are not satisfied.
Balancing selection could account for diversity differences between chromosome arms if there are polymorphic inversions or translocations, causing regions of low recombination rates near the breakpoints (reviewed by Andolfatto et al. 2001). However, no such chromosome variants are known within A. lyrata, and gene orders are conserved between both A. lyrata subspecies (ssp. lyrata and ssp. petraea) and in C. rubella, the outgroup species (Koch and Kiefer 2005; Yogeeswaran et al. 2005; Lysak et al. 2006), suggesting that this is the ancestral state. Two chromosomes with high pericentromeric loci nucleotide diversity, AL1 and AL5, correspond to parts of A. thaliana chromosomes 1 and 3, respectively, and their gene order is the same in A. thaliana and A. lyrata. Our tests (below) for balancing selection at pericentromeric loci were also uniformly negative.
Population structure:
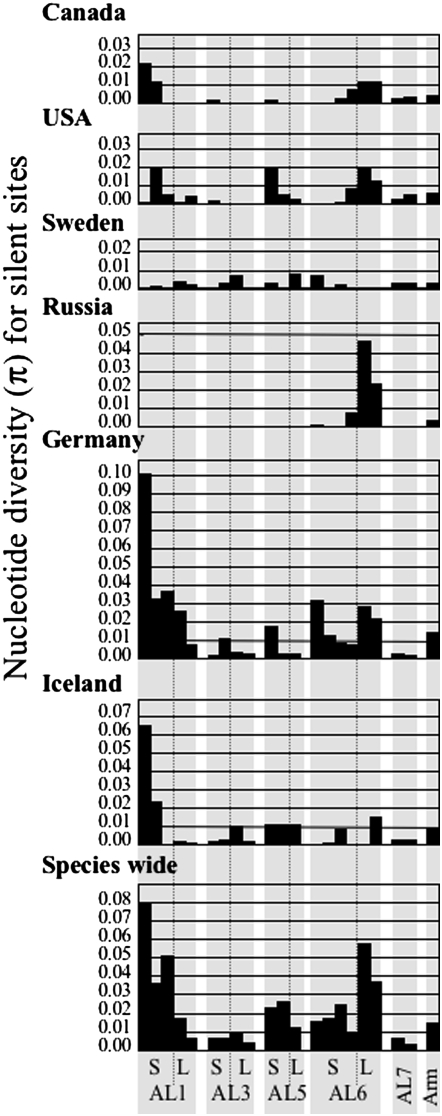
The six populations studied are from very distant geographic locations, and there is evidence of strong differentiation between these populations (Wright et al. 2003b; Ramos-Onsins et al. 2004). We therefore examined nucleotide diversity at all loci within each of the populations, to test whether the high nucleotide diversity we observed for the pericentromeric regions of chromosomes AL1, AL5, and AL6 could be due to fixed differences between populations, or to population-specific polymorphisms, and whether these regions differ in this respect from genes in chromosome arm regions. Only one population (Karhumaki, in Russia) has low centromere region diversity. No polymorphic sites were found in any of the loci from the putatively low recombination regions of AL1 (Wright et al. 2006) or in the loci from three of the four other pericentromeric regions studied here (AL3, AL5, and AL7). The AL6 pericentromeric region, however, has very high nucleotide diversity in this population, as in the other populations (Figure 3). Apart from this population, our sequence results rule out recent selective sweeps in the pericentromeric regions in the populations we surveyed.
Figure 3.—
Silent site diversity of the pericentromeric region of each chromosome and mean silent site diversity of chromosome arm loci, within six natural populations of A. lyrata. The letters S and L indicate genes in the pericentromeric regions of the short and long arms of each chromosome.
Apart from the dearth of polymorphic sites in the Karhumaki population, the nucleotide diversity within each population is similar to the species-wide estimates (Figure 3). The Plech population from Germany, which is likely to be the closest to equilibrium of the populations analyzed so far (Clauss and Mitchell-Olds 2003, 2006), has nucleotide diversity very similar to the species-wide estimates (supplemental Table 1), and Figure 3 shows that three pericentromere regions have high diversity in most populations, including the two North American A. lyrata ssp. lyrata populations, which have generally lower overall diversity than the European ssp. petraea (Wright et al. 2003b; Ramos-Onsins et al. 2004).
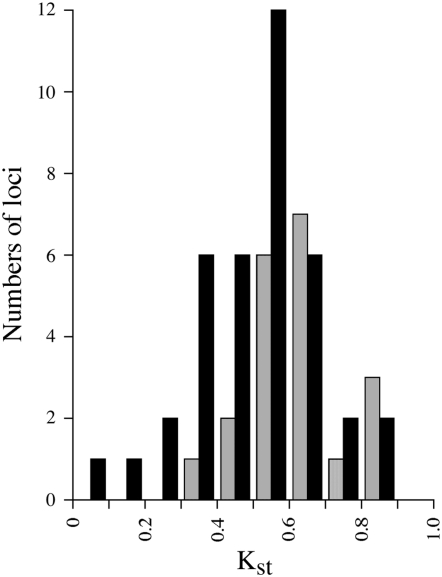
To test whether the high pericentromere locus nucleotide diversity values are due to population differentiation, we analyzed variants of all site types. The populations studied are significantly differentiated for almost all loci in both types of genome region, on the basis of analyses including all site types (P < 0.01, using the Snn test of Hudson 2000). Kst values (which estimate Fst taking sequence differences into account, Hudson et al. 1992) are also significantly >0. Values for the pericentromere loci are generally similar to those for chromosome arm loci (Figure 4; the very low value for one arm locus is from a gene with very low species-wide diversity), but a permutation test of the pericentromeric locus Kst values shows that their mean value over all loci (0.631) is significantly higher than those of chromosome-arm loci (permuted mean 0.501, P = 0.01).
Figure 4.—
Comparison of Kst values for the same pericentromeric region (shaded bars) and chromosome arm loci (solid bars) as in Figure 1.
Higher Kst values are often found for genes in low recombination regions, due to low within-population diversity in such regions (Charlesworth et al. 1997), but this is not the explanation here, as we find no significant diversity difference between the sets of loci. This result therefore suggests slightly higher between-population differences for the pericentromere loci than the arm loci. If this is a direct effect of selection, it should affect mainly nonsynonymous variants. However, the pericentromere loci have higher Kst (and also higher differences between total and within-population diversity, or Dst values) than the arm loci for silent and synonymous sites (the respective values for synonymous sites are 0.601 ± 0.060 and 0.0188 ± 0.0009 vs. 0.341 ± 0.039 and 0.0105 ± 0.0020), but lower values for replacement sites (0.313 ± 0.147 and 0.0009 ± 0.0003 vs. 0.444 ± 0.043 and 0.0023 ± 0.0006).
Hitchhiking effects involving advantageous alleles spreading across populations (selective sweeps) could reduce neutral differentiation at some arm loci, as can balancing selection maintaining the same variants within different populations (Schierup et al. 2000). AL1 arm region genes seem to be enriched in selected loci (Figure 2), and specifically include some loci that appear to be under balancing selection. These genes indeed have lower Kst than average (0.47 vs. 0.53 for the other arm loci), which lowers the Kst for the AL1 and AL2 arm loci as a set; the Kst difference between arm and pericentromeric loci is not significant if we exclude AL1 genes.
Linkage disequilibrium, variant frequencies, and further tests for selection:
We analyzed linkage disequilibrium to test whether the putatively pericentromeric-region loci studied (see materials and methods for details) indeed experience low recombination rates in A. lyrata, and also, as described below, to test for balancing selection. In our previous study of AL1 pericentromeric-region genes, LD was higher than for arm loci, although it was not extraordinarily high within loci, and decay of LD was observed between the loci studied (Wright et al. 2006). The pericentromeric loci analyzed here show generally similar patterns; pericentromeric genes have slightly elevated within-locus LD, but there is still evidence for a decline of LD with distance within individual sequenced regions (supplemental Figure 1). Furthermore, while several between-locus comparisons suggest significant LD, others show mean r2 values similar to those between the chromosome arm loci. These findings parallel those in Drosophila (Langley et al. 2000; Andolfatto and Wall 2003) and suggest the possibility of high rates of gene conversion in loci in regions of low crossing over, even in regions of generally low recombination rates.
A possible reason for the high silent site diversity of the pericentromeric-region genes is long-term balancing selection at a locus or loci within the low recombination regions (Berry et al. 1991). However, we showed above that HKA tests do not suggest selection acting on these loci. Consistent with this, the centromere region loci do not share unusually high numbers of variants between populations, and none of variants are shared with A. thaliana (data not shown). Within A. lyrata, the proportions of variants in different AL7 loci that are shared between populations increases with the diversity of the locus, as expected, but this proportion is similar for the centromere region and chromosome arm loci; for AL6 loci, this proportion is uncorrelated with diversity and does not differ significantly between the chromosome regions.
Long-term balancing selection should also lead to maintenance of distinct haplotypes, and high LD across the nonrecombining region, and positive Tajima's D (DT) values would also be expected for all site types. However, since our samples from individual populations are not large, variant frequencies are not well estimated, so Tajima's test is unlikely to be very informative; it is also problematic for the species-wide data set, because of high population structure (see previous section), which tends to cause positive DT values (Nordborg 1997). Taking all variants together, this test indeed yielded no significant deviations from neutrality at any pericentromere region loci, within populations or when all populations are pooled (supplemental Table 1). However, it is useful to compare DT values for different site types and different sets of loci studied in similar samples of individuals. Such comparisons show that, consistent with the evidence already described above for effective purifying selection in both sets of genes, DT values are lower for nonsynonymous than synonymous variants for both sets of loci. They are also lower for arm than pericentromere region loci, particularly for synonymous variants (0.053 ± 0.205 for arm loci, vs. 0.535 ± 0.257 for the pericentromere region loci), consistent with indirect effects of hitchhiking affecting the arm loci.
DISCUSSION
In both Arabidopsis species, pericentromeric loci often have nucleotide diversity similar to, or slightly higher than, loci in other genome regions with higher recombination rates. This is evidently not purely a consequence of high A. thaliana's high inbreeding, since it is also found in the outcrosser, A. lyrata. Indeed, selfing probably evolved quite recently in A. thaliana (Bechsgaard et al. 2006; Tang et al. 2007), and the common outcrossing ancestor would probably have had diversity patterns like those in A. lyrata. Thus A. thaliana's changed mating system and life history may have had little effect on current patterns of polymorphism in this species, and its higher pericentromeric region diversity may therefore not relate to its current recombination rates, but may be explained in the same way as in A. lyrata, in which three of the five pericentromeric regions analyzed showed high nucleotide diversity, and none had strongly reduced species-wide nucleotide diversity. We presented evidence above that each such region probably recombines rarely in A. lyrata. In the Karhumaki population (in which, as discussed below, selective sweeps may have occurred recently), the concordant low nucleotide diversity of the pericentromeric loci, except for the six AL6 loci, also suggests that the loci at each centromere have the same evolutionary history.
Taken together, our results suggest that the high nucleotide diversity of the centromere region genes is not due to lack of selective constraint or to balancing selection within each of these regions. It is possible that, in both A. thaliana and A. lyrata there are too few targets for purifying or positive selection in the pericentromere regions to appreciably reduce diversity below that in the chromosome arms (discussed in Wright et al. 2006). In Drosophila, current data on gene density and recombination rates in centromere regions suggest that, compared to the chromosome arms, regions of low recombination may experience higher deleterious mutation rates and/or more frequent mutations subject to weak purifying selection, perhaps through transposable-element activity driving background selection (Charlesworth 1996) and reducing Ne, and this might differ in Arabidopsis. However, the A. lyrata genome has more abundant transposable elements than in A. thaliana (Wright et al. 2001), which should make the predicted diversity difference between high and low recombination regions similar to that in Drosophila.
We also considered the possibility of balancing selection due to local selective differences, maintaining different alleles in different populations (Charlesworth et al. 1997). The Kst values for the pericentromere genes, as a set, are significantly, although slightly, higher than for the arm loci (see Figure 4 above; although high population differentiation genomewide makes it difficult to detect higher Kst values for any subset of the loci, particularly for synonymous sites, since there are few such sites in many of the loci and therefore a high variance of Kst values). If elevated Kst is caused by local selection acting on loci in the pericentromere regions, selective sweeps should have occurred within local or regional populations and might be detectable from low within-population silent site diversity (as is detected in European D. melanogaster populations Haddrill et al. 2005; Ometto et al. 2005). However, Kst mainly differs between pericentromeric and arm loci for silent sites, and for replacement variants it is somewhat lower in the pericentromeric loci than the arm loci, suggesting that locally selected differences are not the reason for high pericentromeric-region diversity. The interpretation above, that balancing selection is causing low Kst for some AL1 loci, including effects on synonymous sites in these loci, seems the most likely.
For several A. lyrata centromere regions, our results also clearly show that there have been no recent selective sweeps. This excludes yet another possible explanation for these genes' high diversity: strong selective sweeps in a subdivided population (which can, under some conditions, lead to increased diversity, even within local populations, Santiago and Caballero 2005). Assuming that, as in Drosophila, hitchhiking affects diversity at weakly selected sites in the A. lyrata pericentromeric regions with low recombination, our finding of similar diversity for genes in the chromosome arms (with higher recombination) suggests that hitchhiking effects in the pericentromeric regions are equalled, or outweighed, by those in the chromosome arm regions with higher gene density (see Figure 1).
Introgression:
The Russian population may, however, have experienced selective sweeps, which may explain its low diversity for many of the loci. For this population, there is evidence for admixture with a closely related species, possibly diploid A. arenosa (S. I. Wright, unpublished results). Spread of introgressed alleles in the population could cause selective sweeps, although it seems unlikely that such sweeps (or selective sweeps of any kind) would have been frequent enough to lower diversity for the chromosome arm loci generally.
Another possible explanation for the low chromosome arm diversity values is that introgression (or some other event in the demographic history of A. lyrata populations) has inflated the variance in diversity, such that the centromere region genes have a high mean diversity by chance. It is well established that demographic historical events, including bottlenecks and introgression, inflate the variance in diversity values (Ometto et al. 2005; Thornton and Andolfatto 2006) and that different effective sizes (Ne) of a genome region will lead to the same bottleneck affecting diversity more strongly when Ne is lower (Fay and Wu 1999), and (as discussed earlier) it is well known that genome regions with low recombination rates often have lower Ne. However, although we cannot definitively exclude this possibility, high nucleotide diversity in multiple, different pericentromere regions, such as we observe in A. lyrata, is not expected.
Conclusions:
Overall, therefore, our results suggest that the balance between hitchhiking processes of different intensities has led to similar diversity in the chromosome arms and the pericentromere regions. If selective sweeps are common in chromosome arm regions, this could lower diversity across wide regions, and, if gene density in pericentromere regions is lower, these regions would experience fewer effects of such selection, despite their low recombination. This conclusion is consistent with our HKA test results showing evidence for selection affecting the chromosome arm loci, but not the pericentromere loci.
Recently, evidence has been obtained suggesting that, in Drosophila, genome regions with highly suppressed recombination containing multiple genes may not be the only regions whose diversity is reduced by hitchhiking. Theoretical results, combined with data from Drosophila species, show that deleterious mutations may occur often enough and have selection coefficients of sufficient magnitude for background selection to affect recombining regions of genomes, at least over very short recombination distances (Loewe et al. 2006; Loewe and Charlesworth 2007). Hitchhiking, probably involving selective sweeps, may also be detectably lowering diversity in chromosome arm genes in D. melanogaster and D. simulans, since, in both species, synonymous site diversity is negatively correlated with nonsynonymous divergence between species (Andolfatto 2007; MacPherson et al. 2007). We did not apply these tests to our data, because some of our loci on the chromosome arms are probably under balancing selection (Figure 2). Unless one can distinguish these loci, and remove them from the analyses, their high diversity will mask the effects of selective sweeps lowering diversity. Even with these chromosome arm loci included, diversity is no higher than for the pericentromere region genes, suggesting that hitchhiking effects in the arm regions may have important effects on diversity in A. lyrata.
Acknowledgments
We thank Alex Berr (Institut für Pflanzengenetik und Kulturpflanzenforschung) for information about the chromosomal location of centromere satellite families and Brian Charlesworth for comments on the manuscript. This work was supported by a grant from the Natural Environment Research Council of the United Kingdom to D.C. Stephen Wright is supported by a National Sciences and Engineering Research Council discovery grant and an Alfred P. Sloan fellowship.
References
- Abbott, R. J., and M. F. Gomes, 1988. Population genetic structure and outcrossing rate of Arabidopsis thaliana (L.) Heynh. Heredity 62 411–418. [Google Scholar]
- Aguadé, M., N. Miyashita and C. H. Langley, 1989. Reduced variation in the yellow-achaete-scute region in natural populations of Drosophila melanogaster. Genetics 122 607–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, L. K., G. G. Doyle, B. Brigham, J. Carter, K. D. Hooker et al., 2003. High-resolution crossover maps for each bivalent of Zea mays using recombination nodules. Genetics 165 849–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto, P., 2007. Hitchhiking effects of recurrent beneficial amino acid substitutions in the Drosophila melanogaster genome. Genome Res. 17 1755–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto, P., F. Depaulis and A. Navarro, 2001. Inversion polymorphisms and nucleotide variability in Drosophila. Genet. Res. 77 1–8. [DOI] [PubMed] [Google Scholar]
- Andolfatto, P., and J. D. Wall, 2003. Linkage disequilibrium patterns across a recombination gradient in African Drosophila melanogaster. Genetics 165 1289–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative, 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. [DOI] [PubMed] [Google Scholar]
- Baudry, E., C. Kerdelhué, H. Innan and W. Stephan, 2001. Species and recombination effects on DNA variability in the tomato genus. Genetics 158 1725–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechsgaard, J. S., V. Castric, D. Charlesworth, X. Vekemans and M. H. Schierup, 2006. The transition to self-compatibility in Arabidopsis thaliana and evolution within S-haplotypes over 10 Myr. Mol. Biol. Evol. 23 1741–1750. [DOI] [PubMed] [Google Scholar]
- Begun, D. J., and C. F. Aquadro, 1991. Molecular population genetics of the distal portion of the X chromosome in Drosophila: evidence for genetic hitchiking of the yellow-achaete region. Genetics 129 1147–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun, D. J., and C. F. Aquadro, 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356 519–520. [DOI] [PubMed] [Google Scholar]
- Berge, G., I. Nordal and G. Hestmark, 1998. The effect of breeding systems and pollination vectors on the genetic variation of small plant populations within an agricultural landscape. Oikos 81 17–29. [Google Scholar]
- Berr, A., A. Pecinka, K. G. A. Meister, A. Fuchs, J. Blattner et al. 2006. Chromosome arrangement and nuclear architecture but not centromeric sequences are conserved between Arabidopsis thaliana and Arabidopsis lyrata. Plant J. 48 771–783. [DOI] [PubMed] [Google Scholar]
- Berry, A. J., J. W. Ajioka and M. Kreitman, 1991. Lack of polymorphism on the Drosophila 4th chromosome resulting from selection. Genetics 129 1111–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz, J. O., S. P. Hazen, T. P. Michael, G. P. Morris, I. R. Baxter et al., 2007. Genome-wide patterns of single-feature polymorphism in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104 12057–12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., 1994. The effect of background selection against deleterious alleles on weakly selected, linked variants. Genet. Res. 63 213–228. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., 1996. Background selection and patterns of genetic diversity in Drosophila melanogaster. Genet. Res. 68 131–149. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., M. T. Morgan and D. Charlesworth, 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., M. Nordborg and D. Charlesworth, 1997. The effects of local selection, balanced polymorphism and background selection on equilibrium patterns of genetic diversity in subdivided inbreeding and outcrossing populations. Genet. Res. 70 155–174. [DOI] [PubMed] [Google Scholar]
- Choo, K. H. A., 1997. The Centromere. Oxford University Press, New York.
- Choo, K. H. A., 1998. Why is the centromere so cold? Genome Res. 8 81–82. [DOI] [PubMed] [Google Scholar]
- Clark, R., G. Schweikert, C. Toomajian, S. Ossowski, G. Zeller et al., 2007. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317 338–342. [DOI] [PubMed] [Google Scholar]
- Clauss, M. J., and T. Mitchell-Olds, 2003. Population genetics of tandem trypsin inhibitor genes in Arabidopsis species with contrasting ecology and life history. Mol. Ecol. 12 1287–1299. [DOI] [PubMed] [Google Scholar]
- Clauss, M. J., and T. Mitchell-Olds, 2006. Population genetic structure of Arabidopsis lyrata in Europe. Mol. Ecol. 15 2753–2766. [DOI] [PubMed] [Google Scholar]
- Copenhaver, G. P., W. E. Browne and D. Preuss, 1998. Assaying genome-wide recombination and centromere functions with Arabidopsis tetrads. Proc. Natl. Acad. Sci. USA 95 247–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver, G. P., K. Nickel, T. Kuromori, M. I. Benito, S. Kaul et al., 1999. Genetic definition and sequence analysis of Arabidopsis centromeres. Science 286 2468–2474. [DOI] [PubMed] [Google Scholar]
- Fay, J. C., and C. I. Wu, 1999. A human population bottleneck can account for the discordance between patterns of mitochondrial versus nuclear DNA variation. Mol. Biol. Evol. 16 1003–1005. [DOI] [PubMed] [Google Scholar]
- Foxe, J. P., V. Dar, H. Zheng, M. Nordborg, B. S. Gaut et al., 2008. Selection on amino acid substitutions in Arabidopsis. Mol. Biol. Evol. (in press). [DOI] [PMC free article] [PubMed]
- Haddrill, P. R., K. R. Thornton, B. Charlesworth and P. Andolfatto, 2005. Multilocus patterns of nucleotide variability and the demographic and selection history of Drosophila melanogaster populations. Genome Res. 15 790–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, A. E., G. C. Kettler and D. Preuss, 2006. Dynamic evolution at pericentromeres. Genome Res. 16 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt, W., T. Fischer, S. Winderl, P. Fransz and R. Torres-Ruiz, 2001. The CENTROMERE1 (CEN1) region of Arabidopsis thaliana: architecture and functional impact of chromatin. Plant J. 27 285–296. [DOI] [PubMed] [Google Scholar]
- Hellmann, I., I. Ebersberger, S. E. Ptak, S. Paabo and M. Przeworski, 2003. A neutral explanation for the correlation of diversity with recombination rates in humans. Am. J. Hum. Genet. 72 1527–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., 2000. A new statistic for detecting genetic differentiation. Genetics 155 2011–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., M. Kreitman and M. Aguadé, 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., D. D. Boos and N. L. Kaplan, 1992. A statistical test for detecting geographic subdivision. Mol. Biol. Evol. 9 138–151. [DOI] [PubMed] [Google Scholar]
- Jensen, M. A., B. Charlesworth and M. Kreitman, 2002. Patterns of genetic variation at a chromosome 4 locus of Drosophila melanogaster and D. simulans. Genetics 160 493–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, N. L., R. R. Hudson and C. H. Langley, 1989. The “hitchhiking effect” revisited. Genetics 123 887–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe, A., B. Hansson, A. Forrest, J. Hagenblad and D. Charlesworth, 2006. a Comparative gene mapping in Arabidopsis lyrata chromosomes 6 and 7 and A. thaliana chromosome IV: evolutionary history, rearrangements and local recombination rates. Genet. Res. 88 45–56. [DOI] [PubMed] [Google Scholar]
- Kawabe, A., B. Hansson, J. Hagenblad, A. Forrest and D. Charlesworth, 2006. b Centromere locations and associated chromosome rearrangements in A. lyrata and A. thaliana. Genetics 173 1613–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe, A., S. Nasuda and D. Charlesworth, 2006. c Duplication of centromeric histone H3 (HTR12) Gene in Arabidopsis halleri and A. lyrata, a plant species with multiple centromeric satellite sequences. Genetics 174 2021–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, M., and M. Kiefer, 2005. Genome evolution among Cruciferous plants: a lecture from the comparison of the genetic map of three diploid species —Capsella rubella, Arabidopsis lyrata subsp. petraea, and A. thaliana. Amer. J. Bot. 92 761–767. [DOI] [PubMed] [Google Scholar]
- Kuittinen, H., A. A. de Haan, C. Vogl, S. Oikarinen, J. Leppälä et al., 2004. Comparing the linkage maps of the close relatives Arabidopsis lyrata and A. thaliana. Genetics 168 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley, C. H., B. P. Lazzaro, W. Phillips, E. Heikkinen and J. M. Braverman, 2000. Linkage disequilibria and the site frequency spectra in the su(s) and su(wa) regions of the Drosophila melanogaster X chromosome. Genetics 156 1837–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewe, L., and B. Charlesworth, 2007. Background selection in single genes may explain patterns of codon bias. Genetics 175 1381–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewe, L., B. Charlesworth, C. Bartolomé and V. Nöel, 2006. Estimating selection on nonsynonymous mutations. Genetics 172 1079–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak, M. A., A. Berr, A. Pecinka, R. Schmidt, K. McBreen et al., 2006. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassiceae species. Proc. Natl. Acad. Sci. USA 103 5224–5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald, S., T. Pastinen and A. Long, 2005. The effect of polymorphisms in the enhancer of split gene complex on bristle number variation in a large wild-caught cohort of Drosophila melanogaster. Genetics 171 1741–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson, J. M., G. Sella, J. C. Davis and D. A. Petrov, 2007. Genomewide spatial correspondence between nonsynonymous divergence and neutral polymorphism reveals extensive adaptation in Drosophila. Genetics 177 2083–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith, J., and J. Haigh, 1974. The hitch-hiking effect of a favorable gene. Genet. Res. 219 1114–1116. [PubMed] [Google Scholar]
- McDonald, J. H., and M. Kreitman, 1991. Accelerated protein evolution at the Adh locus in Drosophila. Nature 351 652–654. [DOI] [PubMed] [Google Scholar]
- McVean, G. A. T., and B. Charlesworth, 2000. The effects of Hill-Robertson interference between weakly selected mutations on patterns of molecular evolution and variation. Genetics 155 929–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, M.-H., J. Leppälä and O. Savolainen, 2008. Genome-wide effects of postglacial colonization in Arabidopsis lyrata. Heredity 100 47–58. [DOI] [PubMed] [Google Scholar]
- Nachman, M. W., 2001. Single nucleotide polymorphisms and recombination rate in humans. Trends Genet. 17 481–485. [DOI] [PubMed] [Google Scholar]
- Nordborg, M., 1997. Structured coalescent processes on different time scales. Genetics 146 1501–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg, M., and P. Donnelly, 1997. The coalescent process with selfing. Genetics 146 1185–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg, M., T. T. Hu, Y. Ishino, Y. Jhaveri, C. Toomajian et al., 2005. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3 e196.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ometto, L., S. Glinka, D. D. Lorenzo and W. Stephan, 2005. Inferring the effects of demography and selection on Drosophila melanogaster. Mol. Biol. Evol. 22 2119–2130. [DOI] [PubMed] [Google Scholar]
- Pons, O., and K. Chaouche, 1995. Estimation, variance and optimal sampling of gene diversity. II. Diploid locus. Theor. Appl. Genet. 91 122–130. [DOI] [PubMed] [Google Scholar]
- Presgraves, D. C., 2005. Recombination enhances protein adaptation in Drosophila melanogaster. Curr. Biol. 15 1651–1656. [DOI] [PubMed] [Google Scholar]
- Ramos-Onsins, S. E., B. E. Stranger, T. Mitchell-Olds and M. Aguadé, 2004. Multilocus analysis of variation and speciation in the closely related species Arabidopsis halleri and A. lyrata. Genetics 166 373–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., J. C. Sánchez-DelBarrio, X. Messeguer and R. Rozas, 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19 2496–2497. [DOI] [PubMed] [Google Scholar]
- Santiago, E., and A. Caballero, 2005. Variation after a selective sweep in a subdivided population. Genetics 169 475–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierup, M. H., X. Vekemans and D. Charlesworth, 2000. The effect of hitch-hiking on genes linked to a balanced polymorphism in a subdivided population. Genet. Res. 76 63–73. [DOI] [PubMed] [Google Scholar]
- Schmid, K. J., S. Ramos-Onsins, H. Ringys-Beckstein, D. Weisshaar and T. Mitchell-Olds, 2005. A multilocus sequence survey in Arabidopsis thaliana reveals a genomewide departure from a neutral model of DNA sequence polymorphism. Genetics 169 1601–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldahl, L. A., D. M. Weinreich and D. M. Rand, 2003. Recombination, dominance, and selection on amino acid polymorphism in the Drosophila genome: contrasting patterns on the X and fourth chromosomes. Genetics 165 1195–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, J. D., and S. M. Stack, 1995. Two-dimensional spreads of synaptonemal complexes from solanaceous plants. VI. High-resolution recombination map for tomato (Lycopersicon esculentum). Genetics 141 683–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis. Genetics 123 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, C., C. Toomajian, S. Sherman-Broyles, V. Plagnol, Y. Guo et al., 2007. The evolution of selfing in Arabidopsis thaliana. Science 317 1070–1072. [DOI] [PubMed] [Google Scholar]
- Tenaillon, M. I., M. C. Sawkins, L. K. Anderson, S. M. Stack, J. F. Doebley et al., 2002. Patterns of diversity and recombination along chromosome 1 of maize (Zea mays ssp. mays L.). Genetics 162 1401–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton, K., and P. Andolfatto, 2006. Approximate Bayesian inference reveals evidence for a recent, severe bottleneck in a Netherlands population of Drosophila melanogaster. Genetics 172 1607–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., K. Thornton, A. Berry and M. Long, 2002. Nucleotide variation and recombination along the Drosophila melanogaster fourth chromosome. Science 295 134–137. [DOI] [PubMed] [Google Scholar]
- Weir, B., 1996. Genetic Data Analysis II. Sinauer Associates, Sunderland, MA.
- Wright, S. I., and B. Charlesworth, 2004. The HKA test revisited: a maximum-likelihood-ratio test of the standard neutral model. Genetics 168 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S. I., N. Agrawal and T. E. Bureau, 2003. a Effects of recombination rate and gene density on transposable element distributions in Arabidopsis thaliana. Genome Res. 13 1897–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S. I., B. Lauga and D. Charlesworth, 2003. b Subdivision and haplotype structure in natural populations of Arabidopsis lyrata. Mol. Ecol. 12 1247–1263. [DOI] [PubMed] [Google Scholar]
- Wright, S. I., J. P. Foxe, L. D. Wilson, A. Kawabe, M. Looseley et al., 2006. Testing for effects of recombination rate on nucleotide diversity in natural populations of Arabidopsis lyrata. Genetics 174 1421–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S. I., Q. H. Le, D. J. Schoen and T. E. Bureau, 2001. Population dynamics of an Ac-like transposable element in self- and cross-fertilizing Arabidopsis. Genetics 158 1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogeeswaran, K., A. Frary, T. L. York, A. Amenta, A. H. Lesser et al., 2005. Comparative genome analyses of Arabidopsis spp.: inferring chromosomal rearrangement events in the evolutionary history of A. thaliana. Genome Res. 15 505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]