Abstract
Neurotrophic factors control neural cell differentiation and assembly of neural circuits. We previously showed that organophosphate pesticides differentially regulate members of the fibroblast growth factor (fgf) gene family. We administered chlorpyrifos and diazinon to neonatal rats on postnatal days 1–4 at doses devoid of systemic toxicity or growth impairment, and spanning the threshold for barely-detectable cholinesterase inhibition. We evaluated the impact on gene families for different classes of neurotrophic factors. Using microarrays, we examined the regional expression of mRNAs encoding the neurotrophins (ntfs), brain-derived neurotrophic factor (bdnf), nerve growth factor (ngf), the wnt and fzd gene families and the corresponding receptors. Chlorpyrifos and diazinon both had widespread effects on the fgf, ntf, wnt and fzd families but much less on the bdnf and ngf groups. However, the two organophosphates showed disparate effects on a number of key neurotrophic factors. To determine if the actions were mediated directly on differentiating neurons, we tested chlorpyrifos in PC12 cells, an in vitro model of neural cell development. Effects in PC12 cells mirrored many of those for members of the fgf, ntf and wnt families, as well as the receptors for the ntfs, especially during early differentiation, the stage known to be most susceptible to disruption by organophosphates. Our results suggest that actions on neurotrophic factors provide a mechanism for the developmental neurotoxicity of low doses of organophosphates, and, since effects on expression of the affected genes differed with test agent, may help explain regional disparities in effects and critical periods of vulnerability.
Keywords: Brain-derived neurotrophic factor, Brain development, Chlorpyrifos, Diazinon, Fibroblast growth factor, fzd gene family, Microarrays, Nerve growth factor, Neurotoxicity, Neurotrophic factors, Organophosphate insecticides, PC12 cells, Tyrosine kinase, wnt gene family
INTRODUCTION
The developmental neurotoxicity of organophosphate pesticides is a major concern for environmental health, given the nearly ubiquitous exposure of pregnant women and children to these agents [18,22,56,73]. Although inhibition of cholinesterase provides a common mechanism for the systemic toxicity of organophosphates in adults [56], it is increasingly evident that the developing brain is affected by other mechanisms that operate at nonsymptomatic exposures, extending below the threshold for anticholinesterase activity [18,60,72,73]. At the cellular level, divers neurodevelopmental events are targeted by organophosphates, all converging on the differentiation and assembly of neural circuits, including neuronal and glial cell replication and differentiation, specification of neurotransmitter phenotypes, axonogenesis and synaptogenesis, and synaptic function [11,39,42,60,72].
This poses a conundrum: how can such widespread disruption of brain development arise, with the vulnerability extending from the earliest stages of brain formation through late, postnatal phases of circuit formation? Recent investigations have focused on disruption of neurotrophic factors: given their vital roles in all these developmental events, perturbation of their expression or function could contribute to multiple manifestations of developmental neurotoxicity [30,31,41]. The roles of neurotrophic factors in brain development extend to cell proliferation, migration, differentiation, survival, synaptogenesis and myelination [63], with the various factors dictating regional, cellular as well as temporal specification during critical windows of development [19]. Because many of the factors have overlapping functions, it is important to note that changes in expression or function of any one neurotrophic factor may not necessarily lead to gross morphological deficits but rather to more subtle defects, akin to those seen with the organophosphate pesticides [7,79]. A number of recent studies have focused on three sets of trophic factors, nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF) and the superfamily of fibroblast growth factors (FGFs). Exposure of neonatal rats to doses of chlorpyrifos sufficient to cause cholinesterase inhibition and somatic growth impairment produced only modest reductions in NGF or BDNF [7,8]. In contrast, several key constituents of the FGF family showed deficits at much lower chlorpyrifos or diazinon exposures, specifically involving the members known to be involved in the proliferation, differentiation and survival of the neuronal phenotypes and brain regions targeted by the organophosphates [79]. These findings point to the likely involvement of a specific set of neurotrophic factors as targets underlying organophosphate developmental neurotoxicity.
In the present work, we evaluated the effects of chlorpyrifos and diazinon exposure in newborn rats on the expression of ngf and bdnf as compared to two other neurotrophins, ntf3 and ntf5, as well as with their receptors and receptor modulators, ntrk1, ntrk2, ntrk3, ngfr and ngfrap1. In addition, we assessed the expression of gene families that act in concert to control developmental patterning in the cerebral cortex and hippocampus, regions that are highly targeted by the organophosphates. The wnt family interacts with a set of receptors encoded by the fzd genes [49,86], with signals transduced by the dvl group and negatively modulated by the dkk genes [37]. Importantly for our work, the fgf and wnt families are coexpressed in adjacent locations within the developing brain and deletion of genes from either group produces parallel defects in forebrain development [9,38,66,69,70,100]. As in our earlier work [79], we utilized microarrays to examine the key members of each gene family, comparing similarities and differences in the effects of chlorpyrifos and diazinon: if the involvement of neurotrophic mechanisms is unrelated to the inhibition of cholinesterase, then there are likely to be significant disparities between the two agents. We utilized doses that straddle the threshold for barely-detectable cholinesterase inhibition, too low to elicit any signs of cholinergic hyperstimulation [81,84], and our assessments were conducted in the brainstem and forebrain, regions that differ both in anatomical attributes as well as in maturational timetables. Finally, we compared the effects of the in vivo chlorpyrifos treatment with those in PC12 cells, a standard in vitro model of mammalian neurodevelopment [89] that reproduces many of the key mechanisms and features of the adverse effects of organophosphates on neural cell replication and differentiation [6,26,42,61,72,73,85]. In response to NGF, PC12 cells exit the mitotic cycle and differentiate into cholinergic and catecholaminergic phenotypes, possessing axonal projections, electrical excitability, cholinesterase and cholinergic receptors [33,85,89]. Furthermore, the role of many of the genes examined in our study have also been confirmed in PC12 cells [86,99]. If the effects of organophosphates on neurotrophic factors are direct, then we would expect to find the same pattern of changes in the PC12 model, whereas if the effects are mediated secondarily through alterations in cell-to-cell communication or other indirect mechanisms, the effects in vivo should differ substantially from those in the in vitro model.
MATERIALS AND METHODS
Animal treatments
All experiments were carried out in accordance with federal and state guidelines and with prior approval of the Duke University Institutional Animal Care and Use Committee. All animals were treated humanely and with due care for alleviation of distress. Timed-pregnant Sprague–Dawley rats (Charles River, Raleigh, NC) were housed in breeding cages, with a 12 h light–dark cycle and free access to food and water. On the day after birth, all pups were randomized and redistributed to the dams with a litter size of 10 to maintain a standard nutritional status. Chlorpyrifos and diazinon (both from Chem Service, West Chester, PA) were dissolved in dimethylsulfoxide to provide consistent absorption [97] and were injected subcutaneously in a volume of 1 ml/kg once daily on postnatal days 1–4. Control animals received equivalent injections of the dimethylsulfoxide vehicle. For both agents, we utilized doses below the threshold for growth retardation and systemic toxicity [75,97]: 1 mg/kg of chlorpyrifos and either 1 or 2 mg/kg of diazinon. This chlorpyrifos treatment and the higher dose of diazinon produce neurotoxicity in developing rat brain while eliciting less than 20% cholinesterase inhibition, whereas the lower dose of diazinon does not produce any detectable inhibition [72,81,84,97], nor any of the symptoms of cholinergic hyperstimulation known to be characteristic of anticholinesterase activity [17]. These treatments thus resemble the nonsymptomatic exposures reported in pregnant women [28] and are pharmacodynamically comparable to expected fetal and childhood exposures after routine home application or in agricultural communities [40,58]. On postnatal day 5 (24 hr after the last dose), one male pup was selected from each of five litters in each treatment group. Animals were decapitated, the cerebellum was removed and the brainstem and forebrain were separated by a cut made rostral to the thalamus. Tissues were weighed and flash-frozen in liquid nitrogen and maintained at −45° C until analyzed. Our study design involved the analysis of 40 separate tissues: one animal from each of five litters for each of the four treatment groups, with two tissues (brainstem, forebrain) from each animal.
Cell cultures
Because of the clonal instability of the PC12 cell line [33], the experiments were performed on cells that had undergone fewer than five passages. As described previously [62,85], PC12 cells (American Type Culture Collection, 1721-CRL, obtained from the Duke Comprehensive Cancer Center, Durham, NC) were seeded onto poly-D-lysine-coated plates in RPMI-1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% inactivated horse serum (Sigma Chemical Co., St. Louis, MO), 5% fetal bovine serum (Sigma Chemical Co.), and 50 μg/ml penicillin streptomycin (Invitrogen). Incubations were carried out with 7.5% CO2 at 37 C, standard conditions for PC12 cells. For studies in the undifferentiated state, the medium was changed 24 hr after seeding to include 30 μM chlorpyrifos dissolved in dimethylsulfoxide (final concentration 0.1%), whereas control cultures received the dimethylsulfoxide only, which had no effect on the PC12 cells [61,62,85]. For studies in differentiating cells, 24 hr after seeding, the medium was changed to include 50 ng/ml of 2.5 S murine NGF (Invitrogen) and dimethylsulfoxide with or without chlorpyrifos. Cultures were examined 24 and 72 hr after commencing chlorpyrifos exposure. The 30 μM concentration was chosen because of its ability to evoke the multiple events thought to underlie the in vivo developmental neurotoxicity of chlorpyrifos without causing outright cytotoxicity [6,26,42,61,62,76,85]. Five cultures were evaluated for each treatment condition.
Microarray determinations
Total RNA was isolated from tissues or cell extracts using the Aurum total RNA Fatty and Fibrous Tissue Kit (Bio-Rad Laboratories, Hercules, CA), with RNA quality verified using the RNA 6000 LabChip Kit and the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). An aliquot of each sample used in each respective study (in vivo or in vitro) was withdrawn and combined to make the reference RNA preparation required to be included on each array to permit standardization. RNA amplification was carried out using a commercial kit (Low RNA Input Fluorescent Linear Amplification Kit, Agilent). Each RNA sample was annealed with a primer containing a polydT and a T7 polymerase promoter. Reverse transcriptase produced a first and second strand cDNA. T7 RNA polymerase then created cRNA from the double stranded cDNA by incorporating cyanine-3 (for the reference RNA) or cyanine-5 (for the sample RNA) labeled cytidine 5-triphosphate, and the quality of the labeled cRNA was again verified and the absolute concentration was measured spectrophotometrically. For each pair of reference cRNA and experimental cRNA hybridized to an array, equal amounts of cRNA (0.75 μg) were hybridized using a commercial kit (In situ Hybridization Kit-Plus, Agilent). Hybridization was performed at 60°C for 17 hr with Agilent Whole Rat Genome Arrays (G4131A). The arrays were washed with Agilent’s SSPE Wash Protocol using a solution of 6× SSPE, 0.005% N-lauroylsarcosine, a solution of 0.06× SSPE, 0.005% N-lauroylsarcosine, and Agilent’s Stabilization and Drying Solution. The arrays were scanned on an Agilent G2565BA Microarray Scanner and data from the scans were compiled with Agilent Feature Extraction Software 8.1. The steps from RNA amplification through extraction of the scanner output data were performed by a private contractor (Cogenics, Research Triangle Park, NC).
Array normalizations and error detection were carried out using Silicon Genetics’ GeneSpring GX Version 7.2 (Agilent), via the Enhanced Agilent Feature Extraction Import Preprocessor. First, values of poor quality intensity and low dependability were removed using a “filter on flags” feature, where standardized software algorithms determined which spots were “present”, “marginal”, or “absent.” Spots were considered “present” only where the output was uniform, not saturated and significant above background, whereas spots that satisfied the main requirements but were outliers relative to the typical values for the other genes were considered “marginal.” Filters were set to retain only the values that were found to be present or marginal for further analysis. However, it turned out that, of the genes that passed the filter, none was marginal.
Data were normalized in three steps, using the algorithms supplied with the Feature Extraction software. The first step divided the signal in the Cy5 channel (sample RNA) by that in the Cy3 channel (reference RNA), to give the measured ratio for each gene in the array. The second normalization adjusted the total signal of each chip to a standard value (“normalize to 50th percentile”) determined by the median of the all the reliable values on the chip. This rendered the output of each chip comparable to that of every other chip in the study. The third normalization step was applied to each gene across all the arrays in the study (“normalize to median”): the median of all the values obtained for a given gene was calculated and used as the normalization standard for that gene, so that, regardless of absolute differences in the expression of the various genes, they were placed on the same scale for comparison. After normalization, one final quality-control filter was applied, where genes showing excessive biologic variability were discarded. The criterion for retention was that more than half of the eight groupings in each study (4 treatments × 2 regions for the in vivo studies, 2 treatments × 2 states × 2 time points for the in vitro studies) had to have coefficients of variation <30%.
For some of the genes, the arrays contain multiple probes and/or replicates of the same probe in different locations on the chip, and these were used to verify the reliability of values and the validity of the measures on the chip. In these cases, to avoid artificially inflating the number of positive findings, we limited each gene to a single set of values, selecting those obtained for the probe showing the smallest intragroup variance. The other values for that gene were used only to corroborate direction and magnitude of change. We also validated the readings on the arrays through the use of duplicate arrays for selected samples, as described previously [77,79].
Statistical procedures
Because of the requirement to normalize the data across arrays and within each gene, the absolute values for a given gene are meaningless, so only the relative differences between regions and treatments can be compared. Accordingly, results for the regional differences in gene expression in control rats or cell cultures are presented as means and standard errors of the normalized ratios for each gene, but the effects of the treatments are given as the percentage change from control to allow for visual comparison of the relative changes evoked for each gene, regardless of its control ratio. However, statistical comparisons were based on the actual ratios (log-transformed, since the data are in the form of ratios) rather than the percent change.
Our design involved planned comparisons of the organophosphate-exposed groups to the controls as well as between the two different organophosphates (in vivo studies) or between undifferentiated and differentiating PC12 cells at two time points (in vitro studies), so it was important to consider the false positive rate and to protect against type 1 errors from repeated testing of the same data base. Accordingly, before looking at effects on individual genes, we performed a global ANOVA incorporating all the variables in a single comparison: for in vivo studies, all treatments, both regions and all genes. For in vitro studies, treatment, differentiation state, time, and all genes. Lower-order ANOVAs on subdivisions of the data set were then carried out as permitted by the interactions of treatment with the other variables. Finally, differences for individual treatments for a specified gene in a single brain region (in vivo studies), or differentiation state or time point (in vitro studies) were evaluated with Fisher’s Protected Least Significant Difference. However, for a given gene where there was no treatment interaction with other variables, only the main treatment effect was reported without subtesting of effects in lower-order subdivisions. For ANOVA results, main effects were considered significant at p < 0.05 (two-tailed, since we were interested in both increases and decreases in gene expression). For interaction terms at p < 0.1, we also examined whether lower-order main effects were detectable after subdivision of the interactive variables [82]. In addition to these parametric tests of the direction and magnitude of changes in gene expression, we evaluated the incidence of significant differences as compared to the predicted false positive rate, using Fisher’s Exact Test, applying a one-tailed criterion of p < 0.05, since only an increase above the false positive rate would be predicted. Finding a significant decrease in the incidence of detected differences relative to the false positive rate would be biologically implausible and statistically meaningless.
RESULTS
For the neurotrophic factors and their receptors, 29 genes present on the microarray passed the quality control filters, encoding two of the neurotrophins (ntf3, ntf5), all three of the neurotrophic tyrosine kinase receptors (ntrk1, ntrk2, ntrk3), two of the NGF subunits (ngfb, ngfg), the low-affinity NGF receptor (ngfr) and the NGF receptor associated protein 1 (ngfrap1), brain-derived neurotrophic factor (bdnf), 15 of the FGFs and all four FGF receptors. In the entire study, 11 of these genes showed significant treatment-related changes, as opposed to a predicted false positive rate of only 1–2 genes (p < 0.005). Similarly, for wnt and fzd and their associated signaling pathways, we obtained data for 22 genes, 12 from the wnt family, 6 from the fzd family, and 2 subtypes from each of the dkk and dvl families. Of these, 20 showed treatment-related changes, as opposed to a false-positive rate of only 1 gene (p < 0.0001). Furthermore, as described below for each study (in vivo or in vitro), multivariate ANOVA combining all the variables in a single test confirmed the significant treatment differences and interactions of treatment with other variables, thus permitting appropriate subdivisions of the data for analysis of the treatment effects.
In vivo
The effects of chlorpyrifos or diazinon exposure on expression of the genes encoding the FGFs and their receptors in neonatal rat brain regions have been presented previously [79], so the present study focused on the other neurotrophic factors and their receptors, along with the wnt and fzd families and their associated signaling pathways. In control rats, values for the genes encoding the neurotrophic factors and their receptors were significantly higher overall in the brainstem than in the forebrain (main effect of region, p < 0.0001), representing 5 out of 10 genes showing preferential expression in that region, as opposed to only 1 gene more highly expressed in the forebrain (Table 1, first 10 genes). Multivariate ANOVA across all treatments, both regions and all genes indicated a significant main treatment effect (p < 0.05) and an interaction of treatment × gene (p < 0.0002) and treatment × region × gene (p< 0.05). In light of the significant interactions, the data were subdivided into the individual treatments and genes and the results were reexamined for treatment effects and treatment × region interactions.
TABLE 1.
GENE EXPRESSION IN BRAIN REGIONS OF CONTROL RATS
| Name | Gene | Genbank | Brainstem | Forebrain |
|---|---|---|---|---|
| neurotrophic factor 3* | ntf3 | NM031073 | 0.86 ± 0.06 | 1.44 ± 0.13 |
| neurotrophic factor 5* | ntf5 | NM013184 | 1.28 ± 0.08 | 0.90 ± 0.03 |
| neurotrophic tyrosine kinase receptor 1 (trkA) | ntrk1 | NM021589 | 1.04 ± 0.03 | 0.98 ± 0.01 |
| neurotrophic tyrosine kinase receptor 2 (trkB)* | ntrk2 | BG669126 | 1.13 ± 0.03 | 0.80 ± 0.05 |
| neurotrophic tyrosine kinase receptor 3 (trkC) | ntrk3 | BF398.408 | 1.07 ± 0.10 | 1.07 ± 0.16 |
| brain-derived neurotrophic factor* | bdnf | NM012513 | 1.56 ± 0.09 | 0.78 ± 0.04 |
| nerve growth factor β | ngfb | XM227525 | 1.07 ± 0.07 | 0.97 ± 0.03 |
| nerve growth factor γ* | ngfg | NM031523 | 1.15 ± 0.09 | 0.84 ± 0.05 |
| nerve growth factor receptor* | ngfr | NM012610 | 1.15 ± 0.06 | 0.88 ± 0.05 |
| nerve growth factor receptor associated protein 1 | ngfrap1 | NM053401 | 1.03 ± 0.02 | 0.96 ± 0.05 |
| wingless-type 1 | wnt1 | XM235639 | 1.02 ± 0.11 | 1.04 ± 0.03 |
| wingless-type 2 | wnt2 | XM575397 | 1.03 ± 0.03 | 1.06 ± 0.04 |
| wingless-type 2B* | wnt2b | AF204873 | 1.76 ± 0.09 | 0.58 ± 0.01 |
| wingless-type 4 | wnt4 | NM053402 | 1.27 ± 0.02 | 0.79 ± 0.03 |
| wingless-type 5A | wnt5a | NM022631 | 1.10 ± 0.09 | 0.76 ± 0.08 |
| wingless-type 7A | wnt7a | XM342723 | 0.95 ± 0.04 | 1.03 ± 0.02 |
| wingless-type 7B | wnt7b | NM001009695 | 1.01 ± 0.04 | 1.03 ± 0.08 |
| wingless-type 8A | wnt8a | XM226051 | 1.00 ± 0.07 | 1.25 ± 0.13 |
| wingless-type 9A | wnt9a | XM220556 | 1.16 ± 0.05 | 1.16 ± 0.11 |
| wingless-type 9B | wnt9b | XM221015 | 1.30 ± 0.14 | 1.25 ± 0.15 |
| wingless-type 10A | wnt10a | XM237296 | 1.20 ± 0.09 | 1.03 ± 0.05 |
| wingless-type 10B | wnt10b | XM235636 | 1.03 ± 0.06 | 0.90 ± 0.06 |
| dickkopf homolog 1* | dkk1 | XM219804 | 1.02 ± 0.01 | 0.73 ± 0.08 |
| dickkopf homolog 3* | dkk3 | NM138519 | 0.92 ± 0.02 | 1.05 ± 0.03 |
| frizzled homolog 1* | fzd1 | NM021266 | 1.44 ± 0.04 | 0.79 ± 0.01 |
| frizzled homolog 2 | fzd2 | NM172035 | 1.00 ± 0.02 | 0.99 ± 0.04 |
| frizzled homolog 3 | fzd3 | NM153474 | 1.09 ± 0.09 | 1.14 ± 0.18 |
| frizzled homolog 5* | fzd5 | NM173838 | 1.33 ± 0.13 | 0.91 ± 0.05 |
| frizzled homolog 7 | fzd7 | XM237191 | 0.92 ± 0.11 | 0.79 ± 0.06 |
| frizzled homolog 9 | fzd9 | NM153305 | 1.18 ± 0.07 | 1.08 ± 0.07 |
| dishevelled 1 | dvl1 | NM031820 | 1.00 ± 0.03 | 0.90 ± 0.04 |
| dishevelled 2* | dvl2 | XM239254 | 0.89 ± 0.02 | 1.07 ± 0.01 |
Values are given as mean±S.E. of normalized gene expression ratios to the standard mRNA (see Materials and Methods).
Significant difference between brainstem and forebrain.
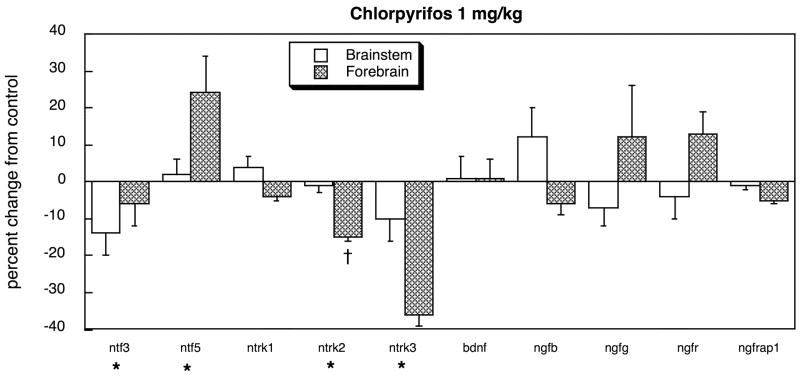
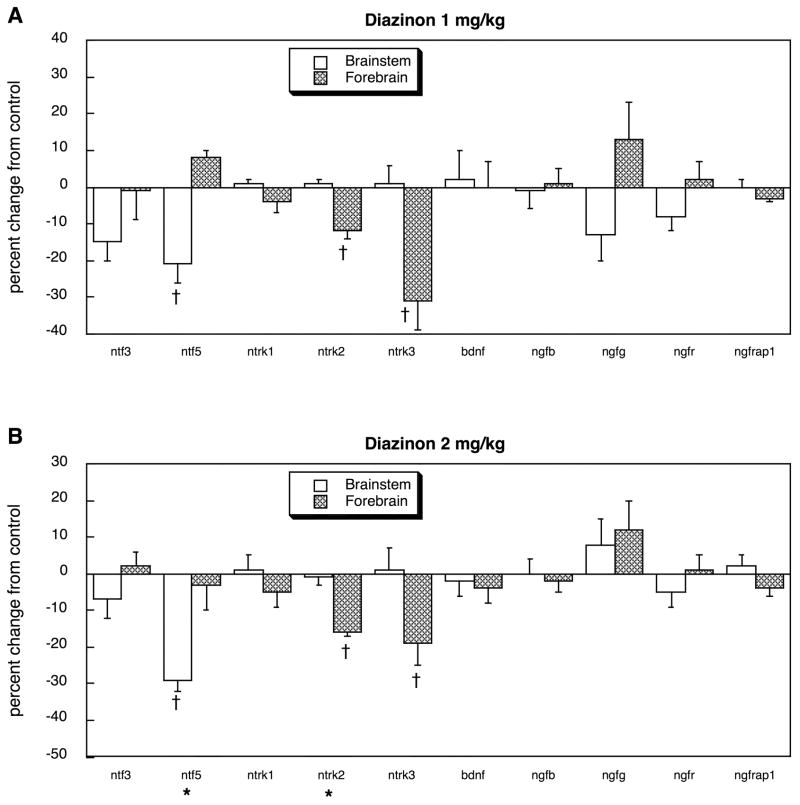
Exposure of neonatal rats to 1 mg/kg of chlorpyrifos altered the balance of neurotrophins in the developing brain, suppressing ntf3 expression while elevating ntf5 in the forebrain (Figure 1). Although ntrk1 expression was unaffected, the two other tyrosine kinase receptors (ntrk2 and ntrk3) showed significant deficits, with a pronounced preferential targeting of the forebrain (treatment × region, p < 0.04). In contrast, there were no significant effects on the other neurotrophic factors (bdnf, ngfb, ngfg) nor on the low-affinity NGF receptor (ngfr) or its adaptor protein (ngfrap1). Administration of diazinon at the same dose produced a slightly different pattern, suppressing ntf5 expression in the brainstem, while still suppressing ntrk2 and ntrk3 in the forebrain and leaving bdnf, ngfb, ngfg, ngfr and ngfrap1 unaffected (Figure 2A). At the higher dose of 2 mg/kg diazinon, we saw essentially the same pattern (Figure 2B).
Figure 1.
Effects of chlorpyrifos exposure in vivo (postnatal days 1–4, 1 mg/kg/day) on expression of genes encoding the neurotrophins and their receptors, shown as the percentage change from control values (Table 1). Asterisks shown below each gene denote a significant main treatment effect. Daggers denote genes for which a treatment × region interaction was detected and show the individual regions for which treatment effects were present. Multivariate ANOVA (treatment, all genes, both regions) indicates a main effect of treatment (p < 0.04) and interactions of treatment × gene (p < 0.03) and treatment × region × gene (p < 0.1).
Figure 2.
Effects of diazinon exposure in vivo (postnatal days 1–4) at 1 mg/kg/day (A) or 2 mg/kg/day (B) on expression of genes encoding the neurotrophins and their receptors, shown as the percentage change from control values (Table 1). Asterisks shown below each gene denote a significant main treatment effect. Daggers denote genes for which a treatment × region interaction was detected and show the individual regions for which treatment effects were present. Multivariate ANOVA (treatment, all genes, both regions) indicates a significant interaction of treatment × region × gene (p < 0.05).
For the wnt and fzd families and their signaling pathways, control brain regions again showed preferentially higher overall expression in the brainstem (main effect of region, p < 0.0001, Table 1, wnt, dkk, fzd and dvl genes). Multivariate ANOVA (all treatments, both regions and all genes) indicated a significant main treatment effect (p < 0.0009) and an interaction of treatment × gene (p < 0.0001) and treatment × region × gene (p< 0.04). Accordingly, the data were again subdivided into the individual treatments and genes.
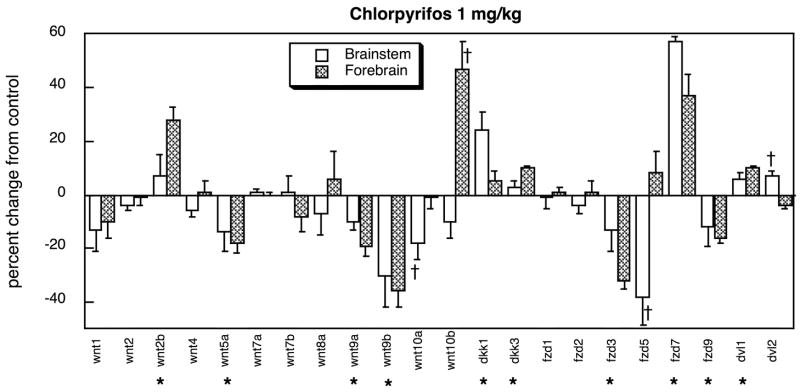
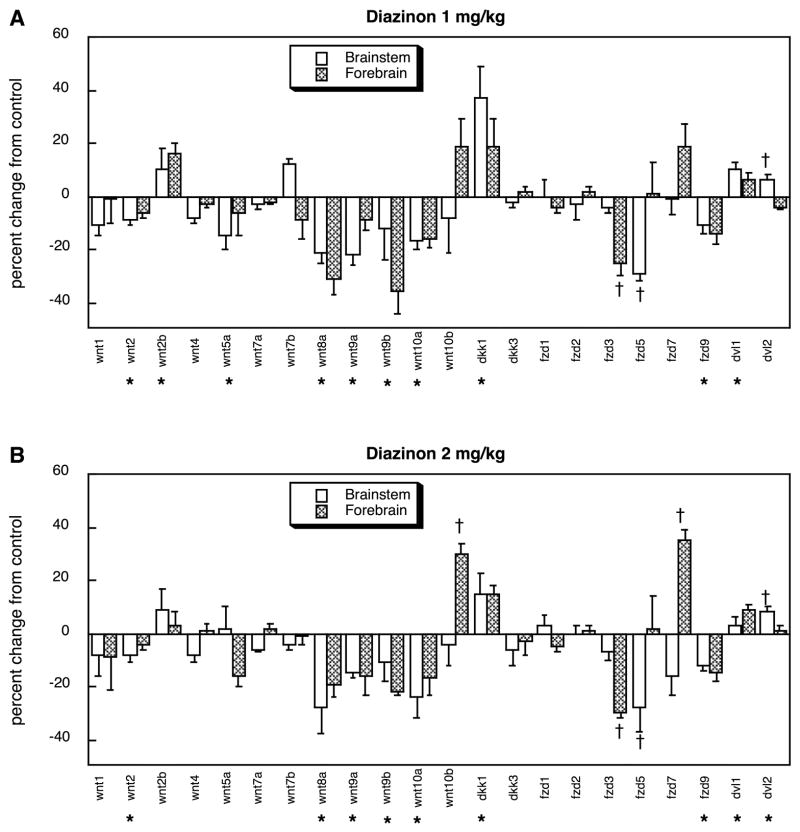
Treatment of neonatal rats with chlorpyrifos evoked robust changes in expression of genes encoding proteins in the wnt family, with suppression of wnt5a, snt9a, wnt9b and wnt10a, but stimulation of wnt2b and wnt10b (Figure 3). Smaller changes were evident for signaling molecules linked to the wnt genes, evidenced by slight but significant increases in the expression of dkk1 and dkk3. Similarly, the fzd family exhibited large reductions in two members (fzd3, fzd5), a robust increase in one (fzd7), and minor but significant alterations in one other subtype (fzd9) as well as in both downstream signaling molecules (dvl1, dvl2). Across all the genes in both families, the net effect of chlorpyrifos was a reduction in expression (main treatment effect, p < 0.04). At the lower dose, diazinon evoked an even more statistically robust overall suppression of gene expression (main treatment effect, p < 0.0006), sharing the effects of chlorpyrifos on wnt5a, wnt9a, wnt9b, wnt10a, fzd3, fzd5 and fzd9, while showing additional suppression of wnt2 and wnt8a. Stimulation of wnt2b, dkk1, dvl1 and dvl2 were seen just as for chlorpyrifos but diazinon failed to increase wnt10b, dkk3, or most notably fzd7, which had shown stimulation with chlorpyrifos treatment (figure 4A). Raising the dose of diazinon to 2 mg/kg produced a similar pattern, except that induction of fzd7 emerged in the forebrain (Figure 4B).
Figure 3.
Effects of chlorpyrifos exposure in vivo (postnatal days 1–4, 1 mg/kg/day) on expression of wnt and fzd genes and genes associated with their signaling pathways (dkk and dvl, respectively), shown as the percentage change from control values (Table 1). Asterisks shown below each gene denote a significant main treatment effect. Daggers denote genes for which a treatment × region interaction was detected and show the individual regions for which treatment effects were present. Multivariate ANOVA (treatment, all genes, both regions) indicates a main effect of treatment (p < 0.04) and interactions of treatment × gene (p < 0.0001) and treatment × region × gene (p < 0.03).
Figure 4.
Effects of diazinon exposure in vivo (postnatal days 1–4) at 1 mg/kg/day (A) or 2 mg/kg/day (B) on expression of wnt and fzd genes and genes associated with their signaling pathways (dkk and dvl, respectively), shown as the percentage change from control values (Table 1). Asterisks shown below each gene denote a significant main treatment effect. Daggers denote genes for which a treatment × region interaction was detected and show the individual regions for which treatment effects were present. Multivariate ANOVA (treatment, all genes, both regions) indicates a main effect of treatment (p < 0.0002) and interactions of treatment × gene (p < 0.0001) and treatment × region × gene (p < 0.1).
In vitro
In control PC12 cell cultures, there was a significant overall difference in expression of the neurotrophic factor genes that depended on differentiation state as well as time in culture (p < 0.05 for the main effect of state, p < 0.0001 for the interaction of state × gene, p < 0.0001 for the interaction of time × gene, p < 0.03 for the interaction of state × time × gene). The main effect of differentiation state represented higher overall values in differentiating cells than in undifferentiated cells. However, superimposed on this effect, there was a distinct dichotomy between the neurotrophic factors and their receptors (p < 0.003): of the 5 genes showing state-related differences for the factors, all of them were more highly expressed in undifferentiated cells, whereas for the 6 receptor-related genes showing such differences, every one was more highly expressed in differentiating cells (Table 2). Multivariate ANOVA across treatment, differentiation state, time and all genes indicated a strong main treatment effect (p < 0.0001) and interactions of treatment × gene (p < 0.0001) and treatment × differentiation state × gene × time (p < 0.03). Accordingly, the data were subdivided for each gene within the two states (undifferentiated and differentiating cells) and reexamined for treatment effects and treatment × time interactions.
TABLE 2.
GENE EXPRESSION IN CONTROL PC12 CELLS —NEUROTROPHIC FACTORS AND THEIR RECEPTORS
| Name | Gene | Genbank | Undifferentiated | Differentiating | ||
|---|---|---|---|---|---|---|
| 24h | 72h | 24h | 72h | |||
| neurotrophic factor 3 | ntf3 | NM031073 | 1.07 ± 0.18 | 1.46 ± 0.21 | 0.78 ± 0.18 | 1.67 ± 0.23 |
| neurotrophic factor 5 | ntf5 | NM013184 | 0.87 ± 0.06 | 0.95 ± 0.11 | 0.88 ± 0.05 | 1.16 ± 0.09 |
| neurotrophic tyrosine kinase receptor 1*† | ntrk1 | NM021589 | 0.90 ± 0.02 | 0.87 ± 0.01 | 0.99 ± 0.02 | 1.49 ± 0.03 |
| neurotrophic tyrosine kinase receptor 2* | ntrk2 | BG669126 | 0.96 ± 0.05 | 0.93 ± 0.06 | 1.10 ± 0.06 | 1.08 ± 0.07 |
| neurotrophic tyrosine kinase receptor 3 | ntrk3 | BF398.408 | 2.63 ± 0.25 | 1.20 ± 0.20 | 1.38 ± 0.23 | 0.95 ± 0.14 |
| brain-derived neurotrophic factor† | bdnf | NM012513 | 1.03 ± 0.08 | 0.98 ± 0.04 | 0.80 ± 0.03 | 1.00 ± 0.09 |
| nerve growth factor β† | ngfb | XM227525 | 0.94 ± 0.04 | 1.25 ± 0.18 | 1.17 ± 0.08 | 0.93 ± 0.05 |
| nerve growth factor γ | ngfg | NM031523 | 1.49 ± 0.25 | 2.05 ± 0.27 | 1.39 ± 0.25 | 1.73 ± 0.25 |
| nerve growth factor receptor*†§ | ngfr | NM012610 | 0.70 ± 0.02 | 0.71 ± 0.01 | 1.36 ± 0.04 | 1.71 ± 0.05 |
| nerve growth factor receptor associated protein 1*†§ | ngfrap1 | NM053401 | 0.79 ± 0.01 | 0.78 ± 0.01 | 1.16 ± 0.01 | 1.49 ± 0.03 |
| fibroblast growth factor 1 | fgf1 | NM012846 | 1.08 ± 0.20 | 0.94 ± 0.05 | 1.06 ± 0.04 | 1.34 ± 0.16 |
| fibroblast growth factor 2* | fgf2 | NM019305 | 1.19 ± 0.15 | 1.05 ± 0.08 | 0.85 ± 0.09 | 0.89 ± 0.10 |
| fibroblast growth factor 3 | fgf3 | NM130817 | 0.99 ± 0.16 | 1.50 ± 0.17 | 0.87 ± 0.04 | 1.18 ± 0.13 |
| fibroblast growth factor 9† | fgf9 | NM012952 | 0.83 ± 0.20 | 1.15 ± 0.17 | 1.46 ± 0.11 | 0.52 ± 0.11 |
| fibroblast growth factor 11 | fgf11 | NM130816 | 1.63 ± 0.25 | 1.10 ± 0.21 | 1.46 ± 0.27 | 1.42 ± 0.20 |
| fibroblast growth factor 12 | fgf12 | NM130814 | 0.94 ± 0.13 | 0.99 ± 0.13 | 1.01 ± 0.12 | 1.13 ± 0.08 |
| fibroblast growth factor 13* | fgf13 | NM053428 | 0.91 ± 0.10 | 0.60 ± 0.07 | 1.18 ± 0.22 | 1.07 ± 0.08 |
| fibroblast growth factor 14 | fgf14 | NM022223 | 1.22 ± 0.13 | 1.08 ± 0.06 | 1.22 ± 0.13 | 0.91 ± 0.02 |
| fibroblast growth factor 15 | fgf15 | NM130753 | 0.86 ± 0.07 | 0.99 ± 0.12 | 1.09 ± 0.08 | 0.99 ± 0.10 |
| fibroblast growth factor 17 | fgf17 | NM019198 | 1.06 ± 0.06 | 0.98 ± 0.03 | 0.92 ± 0.07 | 1.01 ± 0.05 |
| fibroblast growth factor 18 | fgf18 | NM019199 | 1.23 ± 0.15 | 1.11 ± 0.10 | 0.93 ± 0.11 | 1.09 ± 0.06 |
| fibroblast growth factor 20 | fgf20 | NM023961 | 3.43 ± 0.85 | 1.27 ± 0.21 | 2.22 ± 0.35 | 1.20 ± 0.20 |
| fibroblast growth factor 21 | fgf21 | NM130752 | 1.04 ± 0.09 | 0.94 ± 0.03 | 1.06 ± 0.04 | 1.02 ± 0.04 |
| fibroblast growth factor 22 | fgf22 | NM130751 | 1.01 ± 0.03 | 1.06 ± 0.06 | 1.01 ± 0.03 | 1.07 ± 0.08 |
| fibroblast growth factor 23† | fgf23 | NM130754 | 0.87 ± 0.04 | 1.28 ± 0.11 | 1.03 ± 0.06 | 1.08 ± 0.07 |
| fibroblast growth factor receptor 1*† | fgfr1 | NM024146 | 0.89 ± 0.05 | 1.04 ± 0.04 | 1.03 ± 0.03 | 1.04 ± 0.04 |
| fibroblast growth factor receptor 2* | fgfr2 | BF557.572 | 0.78 ± 0.08 | 1.00 ± 0.11 | 1.31 ± 0.11 | 1.21 ± 0.19 |
| fibroblast growth factor receptor 3 | fgfr3 | NM053429 | 0.96 ± 0.06 | 1.04 ± 0.07 | 0.93 ± 0.04 | 0.98 ± 0.03 |
| fibroblast growth factor receptor 4 | fgfr4 | XM344570 | 0.90 ± 0.05 | 1.01 ± 0.03 | 1.04 ± 0.08 | 1.01 ± 0.04 |
Values are given as mean±S.E. of normalized gene expression ratios to the standard mRNA (see Materials and Methods).
Significant main effect between undifferentiated and differentiating cells.
Interaction of NGF × time, significant difference at 24h.
Interaction of NGF × time, significant difference at 72h.
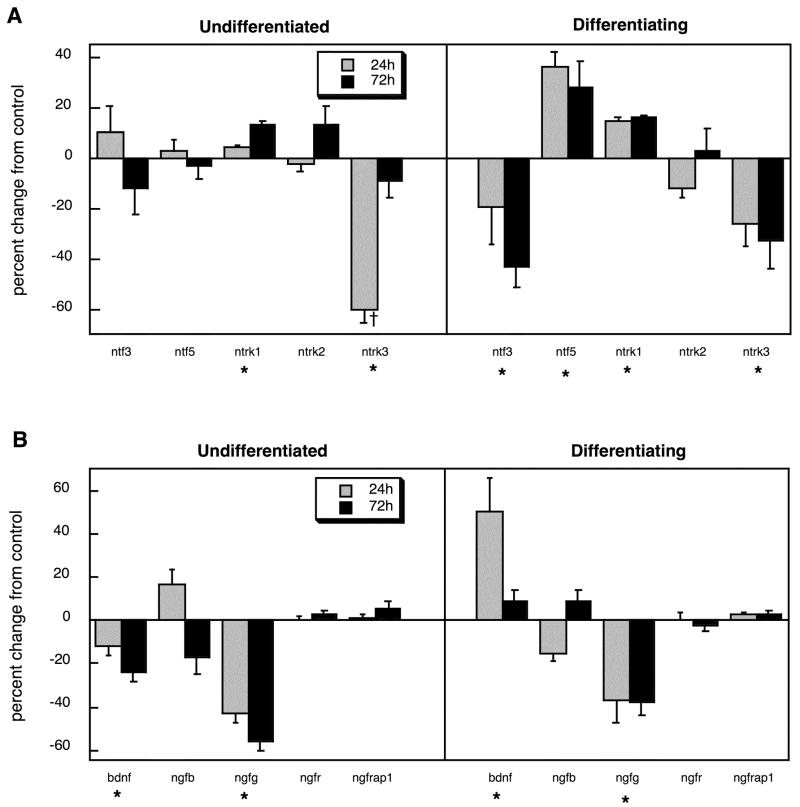
Many of the same genes related to neurotrophic factors for which we found significant effects of chlorpyrifos in vivo also showed changes in the PC12 cell model but there were also some notable disparities. In undifferentiated cells, ntrk1 was slightly but significantly increased, whereas ntrk3 showed robust suppression (Figure 5A). When chlorpyrifos exposure occurred during NGF-induced differentiation, ntf3 and ntrk3 were suppressed whereas ntf5 and ntrk1 were augmented. Examination of the other neurotrophic factors and receptors showed some major differences from the effects of chlorpyrifos in vivo (Figure 5B). Expression of bdnf was reduced in undifferentiated cells but enhanced during differentiation, whereas ngfg was suppressed under either condition.
Figure 5.
Effects of chlorpyrifos exposure in PC12 cells in vitro (30 μM) on expression of genes encoding the neurotrophins and their receptors, shown as the percentage change from control values (Table 2): (A) ntf and ntrk genes, (B) bdnf and ngf genes. Studies were conducted at two time points (24h, 72h) in undifferentiated cells and in cells undergoing NGF-induced differentiation. Asterisks shown below each gene denote a significant main treatment effect. Daggers denote genes for which a treatment × time interaction was detected and show the individual times for which treatment effects were present. Multivariate ANOVA (treatment, all genes, differentiation state, time) indicates a significant main effect of treatment (p < 0.002) and interactions of treatment × state (p < 0.05), treatment × gene (p < 0.0001) and treatment × state × gene × time (p < 0.07).
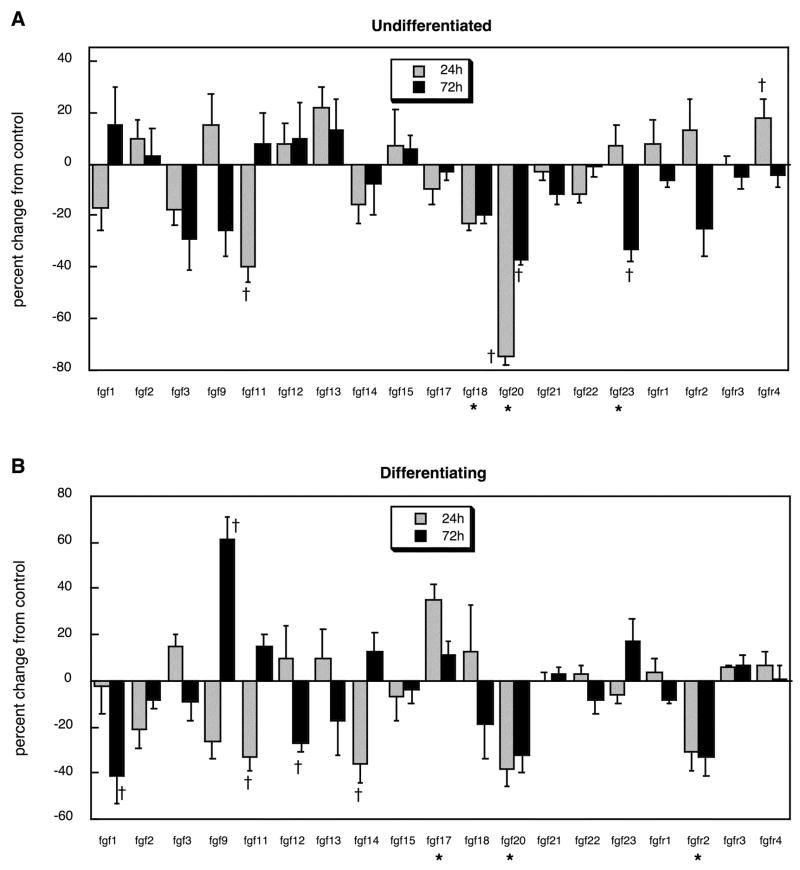
For the FGF superfamily of neurotrophic factors and their receptors, we found striking similarities of the in vitro model to the effects of chlorpyrifos in vivo as reported earlier [79]. In general, chlorpyrifos exposure evoked an overall suppression of gene expression (main effect of treatment). Superimposed on this general pattern, significant reductions were found for fgf11, fgf18, fgf20 and fgf23 in undifferentiated cells, whereas only fgfr4 showed an increase (Figure 6A). The predominant effect in differentiating cells was also to suppress gene expression, with significant decrements noted for fgf1, fgf11, fgf12, fgf14, fgf20 and fgfr2, while increases were seen only for fgf9 and fgf17 (Figure 6B).
Figure 6.
Effects of chlorpyrifos exposure in PC12 cells in vitro (30 μM) on expression of genes encoding the FGFs and their receptors, shown as the percentage change from control values (Table 2): (A) undifferentiated cells, (B) cells undergoing NGF-induced differentiation. Asterisks shown below each gene denote a significant main treatment effect. Daggers denote genes for which a treatment × time interaction was detected and show the individual times for which treatment effects were present. Multivariate ANOVA (treatment, all genes, differentiation state, time) indicates a significant main effect of treatment (p < 0.002) and interactions of treatment × gene (p < 0.02), treatment × gene × time (p < 0.1) and treatment × state × gene × time (p < 0.008).
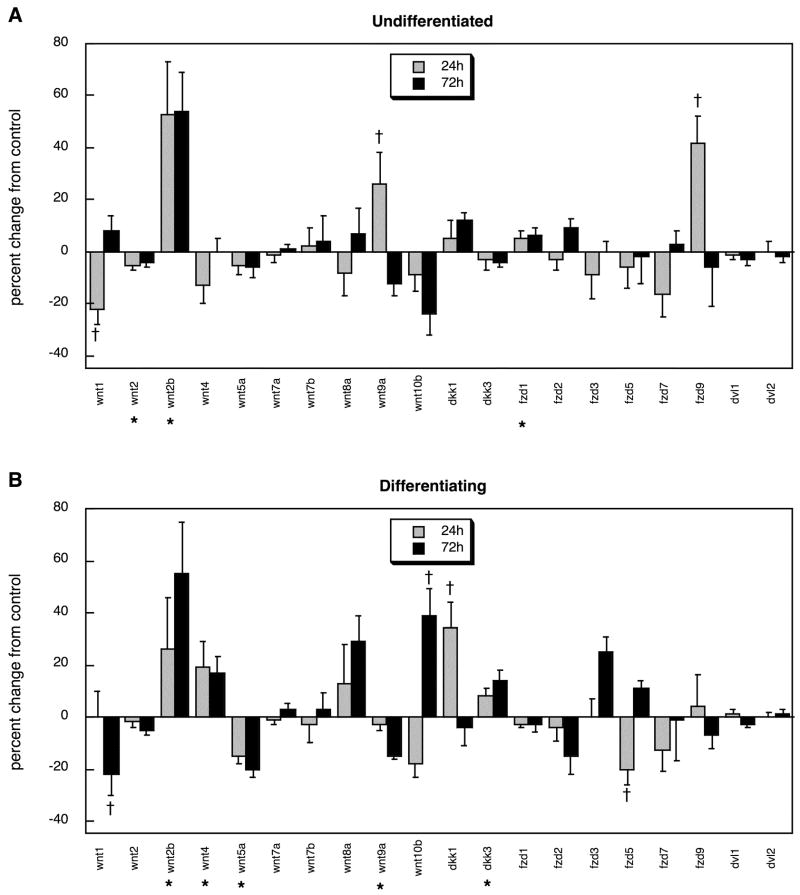
For the wnt and fzd families, control cultures again showed significant effects of differentiation state on gene expression (p < 0.0001 for the interaction of state × gene, p < 0.002 for state × gene × time), although for these families there was no main effect of state, so that both increases and decreases were seen upon differentiation (Table 3). Multivariate ANOVA (treatment, state, time, gene) indicated interactions of treatment × gene (p < 0.0001), state × gene (p < 0.0001), time × gene (p < 0.0001) and state × time × gene (p < 0.0001), necessitating subdivision of the data into the two differentiation states and lower-order examination of treatment effects and treatment × time interactions. Here, too, the PC12 cell cultures recapitulated many (but not all) of the effects seen when chlorpyrifos was given to neonatal rats. In either undifferentiated cells (Figure 7A) or differentiating cells (Figure 7B), wnt2b was augmented, and increases were seen in the differentiating cells for wnt10b, dkk1 and dkk3, all of which mirrored the in vivo effects. Similarly, differentiating PC12 cells displayed suppression of wnt5a and wnt9a, just as was true for neonatal rats given chlorpyrifos. However, the cultures also showed three changes in gene expression that were not seen in the intact rat brain, suppression of wnt1, wnt2 and wnt4.
TABLE 3.
GENE EXPRESSION IN CONTROL PC12 CELLS — WNT AND FZD FAMILIES
| Name | Gene | Genbank | Undifferentiated | Differentiating | ||
|---|---|---|---|---|---|---|
| 24h | 72h | 24h | 72h | |||
| wingless-type 1*† | wnt1 | NM031073 | 1.07 ± 0.05 | 0.94 ± 0.06 | 0.89 ± 0.04 | 1.32 ± 0.10 |
| wingless-type 2 | wnt2 | NM013184 | 0.96 ± 0.02 | 1.05 ± 0.02 | 1.00 ± 0.01 | 1.08 ± 0.02 |
| wingless-type 2B | wnt2b | NM021589 | 1.17 ± 0.21 | 0.68 ± 0.11 | 1.03 ± 0.25 | 0.91 ± 0.14 |
| wingless-type 4* | wnt4 | BG669126 | 1.07 ± 0.06 | 1.09 ± 0.05 | 0.90 ± 0.04 | 0.82 ± 0.02 |
| wingless-type 5A*§ | wnt5a | BF398.408 | 1.11 ± 0.06 | 1.12 ± 0.03 | 1.09 ± 0.06 | 0.69 ± 0.02 |
| wingless-type 7A | wnt7a | NM012513 | 0.98 ± 0.03 | 0.96 ± 0.03 | 1.05 ± 0.03 | 1.01 ± 0.03 |
| wingless-type 7B | wnt7b | XM227525 | 0.94 ± 0.11 | 1.09 ± 0.11 | 1.14 ± 0.07 | 1.00 ± 0.10 |
| wingless-type 8A | wnt8a | NM031523 | 1.12 ± 0.13 | 0.88 ± 0.09 | 0.91 ± 0.19 | 1.12 ± 0.11 |
| wingless-type 9A | wnt9a | NM012610 | 0.84 ± 0.08 | 1.03 ± 0.05 | 1.04 ± 0.08 | 1.01 ± 0.04 |
| wingless-type 10B*§ | wnt10b | XM575397 | 0.96 ± 0.14 | 1.32 ± 0.17 | 0.92 ± 0.10 | 0.72 ± 0.05 |
| dickkopf homolog 1 | dkk1 | AF204873 | 0.96 ± 0.11 | 0.91 ± 0.04 | 0.85 ± 0.14 | 1.00 ± 0.07 |
| dickkopf homolog 3*† | dkk3 | NM053402 | 0.94 ± 0.04 | 1.08 ± 0.04 | 0.79 ± 0.02 | 1.06 ± 0.03 |
| frizzled homolog 1* | fzd1 | NM022631 | 1.17 ± 0.03 | 1.08 ± 0.02 | 0.88 ± 0.02 | 0.79 ± 0.02 |
| frizzled homolog 2 | fzd2 | XM342723 | 0.98 ± 0.06 | 1.03 ± 0.03 | 0.97 ± 0.03 | 1.06 ± 0.06 |
| frizzled homolog 3 | fzd3 | NM001009695 | 0.95 ± 0.06 | 0.91 ± 0.08 | 1.11 ± 0.10 | 0.97 ± 0.03 |
| frizzled homolog 5*§β | fzd5 | XM226051 | 1.21 ± 0.11 | 1.24 ± 0.15 | 1.08 ± 0.13 | 0.71 ± 0.07 |
| frizzled homolog 7 | fzd7 | XM220556 | 1.08 ± 0.14 | 1.02 ± 0.09 | 1.08 ± 0.11 | 0.94 ± 0.04 |
| frizzled homolog 9* | fzd9 | XM221015 | 0.52 ± 0.07 | 0.89 ± 0.09 | 1.02 ± 0.06 | 1.56 ± 0.08 |
| dishevelled 1* | dvl1 | XM237296 | 1.06 ± 0.03 | 1.09 ± 0.02 | 0.95 ± 0.03 | 0.93 ± 0.01 |
| dishevelled 2 | dvl2 | XM235636 | 0.98 ± 0.02 | 1.04 ± 0.02 | 0.98 ± 0.03 | 0.99 ± 0.04 |
Values are given as mean±S.E. of normalized gene expression ratios to the standard mRNA (see Materials and Methods).
Significant main effect between undifferentiated and differentiating cells.
Interaction of NGF × time, significant difference at 24h.
Interaction of NGF × time, significant difference at 72h..
Figure 7.
Effects of chlorpyrifos exposure in PC12 cells in vitro (30 μM) on expression of genes encoding the wnt and fzd families and their signaling partners, shown as the percentage change from control values (Table 3): (A) undifferentiated cells, (B) cells undergoing NGF-induced differentiation. Asterisks shown below each gene denote a significant main treatment effect. Daggers denote genes for which a treatment × time interaction was detected and show the individual times for which treatment effects were present. Multivariate ANOVA (treatment, all genes, differentiation state, time) indicates a significant × treatment interaction (p < 0.0001).
The most substantial differences between the in vitro and in vivo models were seen for the fzd family and the associated dvl genes. Whereas chlorpyrifos administration to developing rats evoked widespread alterations in expression of these genes, the PC12 cultures showed only sporadic effects, limited to an increase in fzd9 at one time point in undifferentiated cells (Figure 7A) and a decrease in fzd5 at one time point in differentiating cells (Figure 7B).
DISCUSSION
The results presented here reinforce the hypothesis that organophosphate exposures below the threshold for any symptoms of systemic toxicity or growth impairment, and just at or below the threshold for any discernible anticholinesterase actions, nevertheless target neurotrophic factors and their signaling pathways, effects that are highly likely to mediate many of the neurodevelopmental anomalies evoked by these agents. In our earlier work with the FGF family, we found that neonatal exposure to chlorpyrifos or diazinon both produced suppression of one particular gene, fgf20, whereas diazinon elicited much larger effects on fgf2 than did chlorpyrifos [79]. Here, in the PC12 model, we found significant decrements in fgf20 expression in both undifferentiated and differentiating cells, indicating that the actions seen in vivo are indeed a direct effect of chlorpyrifos, rather than secondary actions mediated through cell-to-cell communication in the developing brain, systemic toxicity or adverse physiological or hormonal effects on the maternal-neonatal unit. Similarly, two other members of the FGF family that were affected significantly (albeit to a lesser extent) by chlorpyrifos treatment in vivo in our earlier study [79] showed parallel changes in PC12 cells, including fgf11 and fgfr4. However, this relationship was not true for every fgf gene: fgf2 and fgf22 were significantly reduced in vivo but not in vitro, whereas the PC12 model displayed effects on fgf9, fgf12, fgf14, fgf17, fgf18, fgf23 and fgfr2 that were not observed with in vivo treatment. It is thus obvious that there are direct effects of chlorpyrifos on the expression of a few, key fgf family members, whereas other components are affected secondarily. Also, it is not surprising that the PC12 model detects changes in gene expression beyond those seen with in vivo organophosphate exposure. Within a given brain region, there are multiple types of neurons and glia, each developing with a different maturational timetable, whereas PC12 cells represent a uniform population undergoing coordinated differentiation into two specific neurotransmitter phenotypes. Thus, in the intact brain, even marked effects on a specified cell grouping may be rendered artifactually undetectable by inclusion of larger, unaffected populations. Furthermore, the concentrations used for the in vitro exposures are higher than those likely to occur in vivo. The results from PC12 cells thus provide additional information about potential changes in gene expression that may be detected with microdissection of the brain areas studied here, or alternatively may be applicable to effects seen at higher doses in vivo. In any case, these findings strengthen our suggestion that interventions aimed at restoring neurotrophic factor expression could ameliorate at least some aspects of the developmental neurotoxicity of organophosphates [79], especially those targeting factors like fgf20 that are affected directly by chlorpyrifos or diazinon. As discussed in our earlier paper on the FGF family [79], the deficits in fgf20 elicited by developmental exposure to organophosphates are likely to play a key role in the subsequent deficits that emerge in catecholaminergic systems, particularly for dopamine projections in the striatum. That paper discusses each of the affected fgf genes and their implications for mechanistic contributions to organophosphate effects, so the information will not be repeated here.
In contrast to our findings for the fgf family, earlier in vivo studies that utilized higher chlorpyrifos exposures showed only modest effects on gene expression or protein concentrations for BDNF and NGF [7,8,59], emphasizing that the effects of organophosphates on neurotrophic factors are selective for certain families and members. In the present study, we similarly found no significant effect on bdnf, ngfb, ngfg, ngfr or ngfrap1 after neonatal exposure to low doses of chlorpyrifos or diazinon. However, we cannot rule out the possibility of posttranscriptional alterations in BDNF and NGF protein, since they are secreted as precursors and processed extracellularly to the active products [57]. Notably, though, we did find changes in both bdnf and ngfg expression in the PC12 model, again suggestive of effects that may be exerted on selective cell populations or at higher exposures in vivo [7,8]. This interpretation is in keeping with a recent report of bdnf upregulation in the adult hippocampus in response to frankly neurotoxic doses of a diethyldithiocarbamate [55], especially given that these pesticides share many of the developmental neurotoxic profiles as the organophosphates [18,53,76,94]. In our study, whereas chlorpyrifos decreased bdnf expression in undifferentiated cells, it caused an increase during differentiation. In control cells, bdnf expression was inherently higher in the undifferentiated state, so that the chlorpyrifos-induced alterations tended to eliminate the normal developmental decrement in gene expression. This finding is consistent with the peak sensitivity of neural cells to chlorpyrifos at the initiation of cell differentiation [26,42,76], and may actually provide one of the seminal mechanisms by which impaired differentiation occurs. Second, there was impairment of ngfg expression in both undifferentiated and differentiating cells, again consistent with the decrements in ngf reported after in vivo exposures to higher doses of chlorpyrifos [7,8]. The selective suppression of only one out of the three ngf genes may have an important impact, since the γ subunit regulates the binding of NGFβ to its receptors [99]. Given the vital role of NGF in neuronal differentiation into the cholinergic phenotype [5,19,36], adverse effects on ngfg expression may contribute to the particular sensitivity of these neurons to organophosphates [4,42,72,73]. Similarly, BDNF plays critical roles in neuronal cell differentiation, brain development and synaptic plasticity [51], and alterations in this trophic factor have been implicated in psychiatric diseases, neurodegenerative disorders and epilepsy [34,35,44,67,68,83].
Our results for the ntf genes results clearly indicate that the direct impact of organophosphates on neurotrophic factor expression extends beyond the fgf family. Chlorpyrifos evoked parallel downregulation of ntf3 and its receptor, ntrk3, effects that were also observed in PC12 cells, particularly after the start of differentiation. Diazinon exposure in vivo also downregulated ntrk3 but failed to alter ntf3, indicating both a similarity and an important difference between the two organophosphates. Even greater disparities were evident from the other neurotrophins and related receptors: chlorpyrifos upregulated ntf5 expression both in vivo and in vitro, whereas diazinon strongly downregulated the same gene. Although both agents suppressed ntrk2, the receptor for both NTF5 and BDNF, this did not represent a direct effect of the organophosphates, since we did not observe a corresponding effect with the PC12 model. The results point to key divergences between chlorpyrifos and diazinon: just as we concluded earlier for the FGF family [79] the two organophosphates elicit differing alterations in neurotrophic factors that may ultimately contribute to disparities in the impact on neural pathways and behavior. Indeed, we already have substantial evidence for differing susceptibilities and damage to cholinergic and serotonergic pathways from neonatal chlorpyrifos exposure as compared to diazinon [75–77,81]. In turn, the fact that the two agents diverge in important ways reinforces the fact that the developmental neurotoxicity does not depend solely on the shared property of cholinesterase inhibition but rather reflects critical contributions of other mechanisms such as effects on neurotrophic factors [77,79]. Indeed, recent studies reinforce the separation of the effects observed here from those that depend on anticholinesterase effects: chronic, intermittent chlorpyrifos exposures in adults that are nonsymptomatic but well above the threshold for cholinesterase inhibition produce cognitive impairment in association with deficits in ntrk1, a subtype that was unaffected in our studies with lower exposures in the developing rat brain [90].
The targeting of ntrk2 and ntrk3 by both chlorpyrifos and diazinon, superimposed on the deficiencies in ntf3 (diazinon) or ntf5 (chlorpyrifos) is particularly noteworthy. The two tyrosine kinase receptors transduce neurotrophic signals from multiple inputs [21], and knock-downs of ntrk2 and ntrk3 produce late-emerging impairments of catecholaminergic function [95], precisely what is found with prenatal or neonatal chlorpyrifos exposure [3,80]. Other results also point to a critical role for the deficits in ntrk expression as distinct from effects on the neurotrophic factors that bind to receptors [20]. For ntf3, a deficit is likely to have devastating consequences: knockouts of this gene produce death within a few weeks of birth, whereas deletion of the ntf5 gene produces more subtle defects of neuronal structure and function [21,50]. Here, too, knockdowns of ntf3 expression gives outcomes similar to those seen with neonatal chlorpyrifos exposure, specifically involving shared effects on N-methyl-D-aspartate receptors that mediate excitatory neurotoxicity [39,77,92]. Because the organophosphates simultaneously target the neurotrophins as well as their receptors, their ultimate effects are likely to be more widespread than with knock-downs of any single gene [14,93].
Of the pathways we evaluated, the greatest changes were seen for the wnt and fzd families: chlorpyrifos or diazinon treatment in vivo elicited statistically significant changes in expression of 60–70% of the genes. For the wnt group, most of the effects in vivo were mirrored by parallel effects on PC12 cells, implying a direct effect on developing neural cells, whereas the homology was less evident for the fzd genes, indicating reactive or secondary effects for these effectors downstream from wnt. Again, the main conclusion is that organophosphates directly affect the expression of neurotrophic factors known to regulate brain development, involving direct mechanisms separate from the secondary toxicity related to cholinergic hyperstimulation consequent to cholinesterase inhibition. In contrast to the fgf and ntf families, however, chlorpyrifos and diazinon had quite similar patterns of effects on wnt and fzd gene expression, making it unlikely that these particular trophic factors contribute to disparate actions of the two agents, and rather, they would be expected to participate in those outcomes that are convergent.
The major changes in the wnt and fzd families elicited by organophosphate exposure may be particularly important. Indeed, although they are not classical neurotrophic factors, both sets of genes are critical for normal brain development [16,98] and specifically control the architectural development of the hippocampus and neocortex [32,52,54,70,100], two areas that are known to be targeted for disruption by organophosphates [1,65,72,73,91]. There is limited information linking altered expression of specific members of the wnt and fzd families to adverse neurobehavioral outcomes, but it is notable that wnt5a and fzd3, which we found to be reduced by both chlorpyrifos and diazinon, are key determinants in establishing the dopaminergic phenotype [12] and development of dopamine projections [96]. Here again, our results provide strong evidence for parallel or interactive participation of the wnt and fzd pathways with the fgf family, since we previously identified suppression of fgf members associated with the ontogeny of dopamine systems [79], which are especially sensitive to chlorpyrifos [3,25,76,78,80], and which likely underlie the association of organophosphate exposure with the subsequent development of Parkinson’s Disease [43,47]. However, the potential impact of combined or overlapping disruption of fgf, wnt and fzd pathways during critical developmental periods extends further, to include other neurodegenerative syndromes or psychiatric disorders [10,13,15,71], such as schizophrenia [23,29,45,46,96]. The same developmental mechanisms have been proposed to operate in the etiology of autism [27], a disorder in which organophosphate exposure has been implicated [24,88]. Our results thus provide some of the first mechanistic evidence to connect exposure to a specific set of environmental neurotoxicants to the subsequent emergence of major neurodevelopmental disorders [87,88].
Many of our findings also provide a mechanistic foundation for the regional selectivity seen for the effects of organophosphates on the developing brain [72,73]. In fact, both the in vivo and in vitro studies indicate important disparities that are likely to reflect the differing maturational profiles of the various brain regions. Here, we compared the brainstem, which develops earlier, and later-maturing forebrain, as well as undifferentiated vs. differentiating PC12 cells. Both chlorpyrifos and diazinon suppressed the ntrk genes specifically in the forebrain but not the brainstem, and whereas chlorpyrifos increased ntf5 in the forebrain, diazinon instead decreased the same gene in the brainstem. Again, this points to expected regional differences in the targeting of brain development by chlorpyrifos as compared to diazinon. Similarly, the PC12 model showed opposite effects on bdnf expression in undifferentiated vs. differentiating cells, implying that the maturational timetable for cells within a given brain region will determine the net outcome of organophosphate exposure. Similar examples are readily apparent from comparisons of each of the gene families between the two brain regions or differentiation states for PC12 cells, e.g. wnt10b and fzd5 for the in vivo studies, and fgf9, wnt10b and fzd9 for the in vitro studies.
There are a number of limitations to the current work. First, for the in vivo studies, we restricted our analysis to males, primarily because of limited technical and financial considerations. Certainly, the developmental neurotoxicity or organophosphates is known to have important sex-selective components [2,48,64,73,74,78,80], and it is likely that transcriptional profiles will similarly exhibit major sex disparities, a subject for future study. Similarly, the PC12 model does not permit distinction of effects that interact with sex, a clear limitation of comparing in vitro to in vivo findings. Further, we evaluated only one of the two agents with the in vitro approach, focusing on chlorpyrifos because much more is known about its developmental neurotoxicity than diazinon [60,72,73]. Nevertheless, the basic similarities between effects in neonatal rats and in the cell culture system point to mechanistic conclusions that can be garnered despite these restrictions. As a second limitation, we did not evaluate do parallel RT-PCR evaluations of the gene changes identified on the microarrays. Verification is typically required for array studies in which all the genes on the array are evaluated, a small number found to be positive, and there are a large number of false positives (e.g. the >2000 genes that would be false positives if we had considered all 42,000 probes on the array). The PCR technique is then required to ensure that the gene changes identified on the arrays are not among the false positives. For our study, we did a planned comparison of only a handful genes that would generate only 2–3 false positives, and we found alterations in over 60% of these genes, and for interpretation, relied primarily on multiple gene changes in a given pathway, as well as effects that were repeated across different treatments and/or different regions, rather than changes in any one gene. The odds of all those genes being false positives is astronomically small. However, even for individual genes, there were multiple probes and multiple spots on a given array (see Methods), so the changes cannot be “chance.” Unlike typical array studies, where a single sample derived from multiple animals might be evaluated, we evaluated individual animals and tissues, so again it is inconceivable that one could statistically produce these outcomes by accident.
In conclusion, our results suggest that organophosphate pesticides disrupt brain development through contributory actions on the expression of critical neurotrophic factors and their receptors and signaling pathways that govern neuronal cell differentiation. The impact of these agents on neurotrophic regulation provides an underlying mechanism for effects independent of cholinesterase inhibition, thus providing explanations for the targeting of brain development at otherwise subtoxic exposures, for disparate effects of the different organophosphates, for regional selectivity of the effects, and for the varied outcomes that depend upon the specific developmental period in which exposure occurs. In the long run, identification of the specific mechanisms underlying the developmental neurotoxicity of organophosphates or of related and unrelated toxicants can point toward the design of interventions that modulate neurotrophic factors so as to ameliorate or reverse damage to the developing brain.
Acknowledgments
Research was supported by NIH ES10356. The authors state that they have no competing financial interests. Theodore Slotkin and Frederic Seidler have provided expert witness testimony on behalf of government agencies, corporations and/or individuals.
Abbreviations
- ANOVA
analysis of variance
- BDNF
brain-derived neurotrophic factor
- FGF
fibroblast growth factor
- NGF
nerve growth factor
- NTF
neurotrophic factor (neurotrophin) trk, tyrosine kinase receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abdel-Rahman A, Dechkovskaia AM, Mehta-Simmons H, Guan S, Khan WA, Abou-Donia MB. Increased expression of glial fibrillary acidic protein in cerebellum and hippocampus: differential effects on neonatal brain regional acetylcholinesterase following maternal exposure to combined chlorpyrifos and nicotine. J Toxicol Environ Health A. 2003;66:2047–2066. doi: 10.1080/713853982. [DOI] [PubMed] [Google Scholar]
- 2.Aldridge JE, Levin ED, Seidler FJ, Slotkin TA. Developmental exposure of rats to chlorpyrifos leads to behavioral alterations in adulthood, involving serotonergic mechanisms and resembling animal models of depression. Environ Health Perspect. 2005;113:527–531. doi: 10.1289/ehp.7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aldridge JE, Meyer A, Seidler FJ, Slotkin TA. Alterations in central nervous system serotonergic and dopaminergic synaptic activity in adulthood after prenatal or neonatal chlorpyrifos exposure. Environ Health Perspect. 2005;113:1027–1031. doi: 10.1289/ehp.7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aluigi MG, Angelini C, Falugi C, Fossa R, Genever P, Gallus L, Layer PG, Prestipino G, Rakonczay Z, Sgro M, Thielecke H, Trombino S. Interaction between organophosphate compounds and cholinergic functions during development. Chem Biol Interact. 2005;157–158:305–316. doi: 10.1016/j.cbi.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 5.Avila AM, Davila-Garcia MI, Ascarrunz VS, Xiao Y, Kellar KJ. Differential regulation of nicotinic acetylcholine receptors in PC12 cells by nicotine and nerve growth factor. Mol Pharmacol. 2003;64:974–986. doi: 10.1124/mol.64.4.974. [DOI] [PubMed] [Google Scholar]
- 6.Bagchi D, Bagchi M, Hassoun EA, Stohs SJ. In vitro and in vivo generation of reactive oxygen species, DNA damage and lactate dehydrogenase leakage by selected pesticides. Toxicology. 1995;104:129–140. doi: 10.1016/0300-483x(95)03156-a. [DOI] [PubMed] [Google Scholar]
- 7.Betancourt AM, Burgess SC, Carr RL. Effect of developmental exposure to chlorpyrifos on the expression of neurotrophin growth factors and cell-specific markers in neonatal rat brain. Toxicol Sci. 2006;92:500–506. doi: 10.1093/toxsci/kfl004. [DOI] [PubMed] [Google Scholar]
- 8.Betancourt AM, Carr RL. The effect of chlorpyrifos and chlorpyrifos-oxon on brain cholinesterase, muscarinic receptor binding, and neurotrophin levels in rats following early postnatal exposure. Toxicol Sci. 2004;77:63–71. doi: 10.1093/toxsci/kfh003. [DOI] [PubMed] [Google Scholar]
- 9.Blak AA, Naserke T, Weisenhorn DMV, Prakash N, Partanen J, Wurst W. Expression of Fgf receptors 1, 2, and 3 in the developing mid- and hindbrain of the mouse. Dev Dynamics. 2005;233:1023–1030. doi: 10.1002/dvdy.20386. [DOI] [PubMed] [Google Scholar]
- 10.Blesch A. Neurotrophic factors in neurodegeneration. Brain Pathol. 2006;16:295–303. doi: 10.1111/j.1750-3639.2006.00036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casida JE, Quistad GB. Organophosphate toxicology: safety aspects of nonacetylcholinesterase secondary targets. Chem Res Toxicol. 2004;17:983–998. doi: 10.1021/tx0499259. [DOI] [PubMed] [Google Scholar]
- 12.Castelo-Branco G, Wagner J, Rodriguez FJ, Kele J, Sousa K, Rawal N, Pasolli HA, Fuchs E, Kitajewski J, Arenas E. Differential regulation of midbrain dopaminergic neuron development by Wnt-1, Wnt-3a, and Wnt-5a. Proc Natl Acad Sci. 2003;100:12747–12752. doi: 10.1073/pnas.1534900100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castren E, Voikar V, Rantamaki T. Role of neurotrophic factors in depression. Curr Opin Pharmacol. 2007;7:18–21. doi: 10.1016/j.coph.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 14.Chang JH, Vuppalanchi D, van Niekerk E, Trepel JB, Schanen NC, Twiss JL. PC12 cells regulate inducible cyclic AMP (cAMP) element repressor expression to differentially control cAMP response element-dependent transcription in response to nerve growth factor and cAMP. J Neurochem. 2006;99:1517–1530. doi: 10.1111/j.1471-4159.2006.04196.x. [DOI] [PubMed] [Google Scholar]
- 15.Chao MV, Rajagopal R. Neurotrophin signalling in health and disease. Clin Sci. 2006;110:167–173. doi: 10.1042/CS20050163. [DOI] [PubMed] [Google Scholar]
- 16.Ciani L, Salinas PC. WNTs in the vertebrate nervous system: from patterning to neuronal connectivity. Nat Rev Neurosci. 2005;6:351–362. doi: 10.1038/nrn1665. [DOI] [PubMed] [Google Scholar]
- 17.Clegg DJ, van Gemert M. Determination of the reference dose for chlorpyrifos: proceedings of an expert panel. J Toxicol Environ Health. 1999;2:211–255. doi: 10.1080/109374099281179. [DOI] [PubMed] [Google Scholar]
- 18.Colborn T. A case for revisiting the safety of pesticides: a closer look at neurodevelopment. Environ Health Perspect. 2006;114:10–17. doi: 10.1289/ehp.7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conner JM, Varon S. Developmental profile of NGF immunoreactivity in the rat brain: a possible role of NGF in the establishment of cholinergic terminal fields in the hippocampus and cortex. Dev Brain Res. 1997;101:67–79. doi: 10.1016/s0165-3806(97)00051-5. [DOI] [PubMed] [Google Scholar]
- 20.Conover JC, Erickson JT, Katz DM, Bianchi LM, Poueymirou WT, McClain J, Pan L, Helgren M, Ip NY, Boland P, Friedman B, Wiegand S, Vejsada R, Kato AC, DeChiara TM, Yancopoulos GD. Neuronal deficits, not involving motor neurons, in mice lacking BDNF and/or NT4. Nature. 1995;375:235–238. doi: 10.1038/375235a0. [DOI] [PubMed] [Google Scholar]
- 21.Conover JC, Yancopoulos GD. Neurotrophin regulation of the developing nervous system: analyses of knockout mice. Rev Neurosci. 1997;8:13–27. doi: 10.1515/revneuro.1997.8.1.13. [DOI] [PubMed] [Google Scholar]
- 22.Costa LG. Current issues in organophosphate toxicology. Clin Chim Acta. 2006;366:1–13. doi: 10.1016/j.cca.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 23.Cotter D, Kerwin R, al-Sarraji S, Brion JP, Chadwich A, Lovestone S, Anderton B, Everall I. Abnormalities of Wnt signalling in schizophrenia--evidence for neurodevelopmental abnormality. Neuroreport. 1998;9:1379–1383. doi: 10.1097/00001756-199805110-00024. [DOI] [PubMed] [Google Scholar]
- 24.D'Amelio M, Ricci I, Sacco R, Liu X, D'Agruma L, Muscarella LA, Guarnieri V, Militerni R, Bravaccio C, Elia M, Schneider C, Melmed R, Trillo S, Pascucci T, Puglisi-Allegra S, Reichelt KL, Macciardi F, Holden JJA, Persico AM. Paraoxonase gene variants are associated with autism in North America, but not in Italy: possible regional specificity in gene-environment interactions. Mol Psychiat. 2005;10:1006–1016. doi: 10.1038/sj.mp.4001714. [DOI] [PubMed] [Google Scholar]
- 25.Dam K, Garcia SJ, Seidler FJ, Slotkin TA. Neonatal chlorpyrifos exposure alters synaptic development and neuronal activity in cholinergic and catecholaminergic pathways. Dev Brain Res. 1999;116:9–20. doi: 10.1016/s0165-3806(99)00067-x. [DOI] [PubMed] [Google Scholar]
- 26.Das KP, Barone S. Neuronal differentiation in PC12 cells is inhibited by chlorpyrifos and its metabolites: is acetylcholinesterase inhibition the site of action? Toxicol. Appl Pharmacol. 1999;160:217–230. doi: 10.1006/taap.1999.8767. [DOI] [PubMed] [Google Scholar]
- 27.De Ferrari GV, Moon RT. The ups and downs of Wnt signaling in prevalent neurological disorders. Oncogene. 2006;25:7545–7553. doi: 10.1038/sj.onc.1210064. [DOI] [PubMed] [Google Scholar]
- 28.De Peyster A, Willis WO, Molgaard CA, MacKendrick TM, Walker C. Cholinesterase and self-reported pesticide exposure among pregnant women. Arch Environ Health. 1993;48:348–352. doi: 10.1080/00039896.1993.9936724. [DOI] [PubMed] [Google Scholar]
- 29.Diwadkar VA, DeBellis MD, Sweeney JA, Pettegrew JW, Keshavan MS. Abnormalities in MRI-measured signal intensity in the corpus callosum in schizophrenia. Schizopr Res. 2004;67:277–282. doi: 10.1016/S0920-9964(03)00098-7. [DOI] [PubMed] [Google Scholar]
- 30.Dono R. Fibroblast growth factors as regulators of central nervous system development and function. Am J Physiol. 2003;284:R867–R881. doi: 10.1152/ajpregu.00533.2002. [DOI] [PubMed] [Google Scholar]
- 31.Dreyfus CF. Neurotransmitters and neurotrophins collaborate to influence brain development. Perspect Dev Neurobiol. 1998;5:389–399. [PubMed] [Google Scholar]
- 32.Fischer T, Guimera J, Wurst W, Prakash N. Distinct but redundant expression of the Frizzled Wnt receptor genes at signaling centers of the developing mouse brain. Neuroscience. 2007;147:693–711. doi: 10.1016/j.neuroscience.2007.04.060. [DOI] [PubMed] [Google Scholar]
- 33.Fujita K, Lazarovici P, Guroff G. Regulation of the differentiation of PC12 pheochromocytoma cells. Environ Health Perspect. 1989;80:127–142. doi: 10.1289/ehp.8980127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fumagalli F, Racagni G, Riva MA. Shedding light into the role of BDNF in the pharmacotherapy of Parkinson's disease. Pharmacogenomics J. 2006;6:95–104. doi: 10.1038/sj.tpj.6500360. [DOI] [PubMed] [Google Scholar]
- 35.Fumagalli F, Racagni G, Riva MA. The expanding role of BDNF: a therapeutic target for Alzheimer's disease? Pharmacogenomics J. 2006;6:8–15. doi: 10.1038/sj.tpj.6500337. [DOI] [PubMed] [Google Scholar]
- 36.Garofalo L, Ribeiro-da-Silva A, Cuello AC. Nerve growth factor-induced synaptogenesis and hypertrophy of cortical cholinergic terminals. Proc Natl Acad Sci. 1992;89:2639–2643. doi: 10.1073/pnas.89.7.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glinka A, Wu W, Delius H, Monaghan AP, Blumenstock C, Niehrs C. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature. 1998;391:357–362. doi: 10.1038/34848. [DOI] [PubMed] [Google Scholar]
- 38.Gunhaga L, Marklund M, Sjodal M, Hsieh JC, Jessell TM, Edlund T. Specification of dorsal telencephalic character by sequential Wnt and FGF signaling. Nature Neurosci. 2003;6:701–707. doi: 10.1038/nn1068. [DOI] [PubMed] [Google Scholar]
- 39.Gupta RC. Brain regional heterogeneity and toxicological mechanisms of organophosphates and carbamates. Toxicol Mech Meth. 2004;14:103–143. doi: 10.1080/15376520490429175. [DOI] [PubMed] [Google Scholar]
- 40.Gurunathan S, Robson M, Freeman N, Buckley B, Roy A, Meyer R, Bukowski J, Lioy PJ. Accumulation of chlorpyrifos on residential surfaces and toys accessible to children. Environ Health Perspect. 1998;106:9–16. doi: 10.1289/ehp.981069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henderson CE. Role of neurotrophic factors in neuronal development. Curr Opinion Neurobiol. 1996;6:64–70. doi: 10.1016/s0959-4388(96)80010-9. [DOI] [PubMed] [Google Scholar]
- 42.Jameson RR, Seidler FJ, Qiao D, Slotkin TA. Chlorpyrifos affects phenotypic outcomes in a model of mammalian neurodevelopment: critical stages targeting differentiation in PC12 cells. Environ Health Perspect. 2006;114:667–672. doi: 10.1289/ehp.8750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamel F, Hoppin JA. Association of pesticide exposure with neurologic dysfunction and disease. Environ Health Perspect. 2004;112:950–958. doi: 10.1289/ehp.7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kato T. Molecular genetics of bipolar disorder and depression. Psychiat Clin Neurosci. 2007;61:3–19. doi: 10.1111/j.1440-1819.2007.01604.x. [DOI] [PubMed] [Google Scholar]
- 45.Katsu T, Ujike H, Nakano T, Tanaka Y, Nomura A, Nakata K, Takaki M, Sakai A, Uchida N, Imamura T, Kuroda S. The human frizzled-3 (FZD3) gene on chromosome 8p21, a receptor gene for Wnt ligands, is associated with the susceptibility to schizophrenia. Neurosci Lett. 2003;353:53–56. doi: 10.1016/j.neulet.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 46.Kozlovsky N, Belmaker RH, Agam G. GSK-3 and the neurodevelopmental hypothesis of schizophrenia. Eur Neuropsychopharmacol. 2002;12:13–25. doi: 10.1016/s0924-977x(01)00131-6. [DOI] [PubMed] [Google Scholar]
- 47.Landrigan PJ, Sonawane B, Butler RN, Trasande L, Callan R, Droller D. Early environmental origins of neurodegenerative disease in later life. Environ Health Perspect. 2005;113:1230–1233. doi: 10.1289/ehp.7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levin ED, Addy N, Christopher NC, Seidler FJ, Slotkin TA. Persistent behavioral consequences of neonatal chlorpyrifos exposure in rats. Dev Brain Res. 2001;130:83–89. doi: 10.1016/s0165-3806(01)00215-2. [DOI] [PubMed] [Google Scholar]
- 49.Li F, Chong ZZ, Maiese K. Vital elements of the Wnt-Frizzled signaling pathway in the nervous system. Curr Neurovasc Res. 2005;2:331–340. doi: 10.2174/156720205774322557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu X, Ernfors P, Wu H, Jaenisch R. Sensory but not motor neuron deficits in mice lacking NT4 and BDNF. Nature. 1995;375:238–241. doi: 10.1038/375238a0. [DOI] [PubMed] [Google Scholar]
- 51.Lykissas MG, Batistatou AK, Charalabopoulos KA, Beris AE. The role of neurotrophins in axonal growth, guidance, and regeneration. Curr Neurovasc Res. 2007;4:143–151. doi: 10.2174/156720207780637216. [DOI] [PubMed] [Google Scholar]
- 52.Machon O, van den Bout CJ, Backman M, Kemler R, Krauss S. Role of beta-catenin in the developing cortical and hippocampal neuroepithelium. Neuroscience. 2003;122:129–143. doi: 10.1016/s0306-4522(03)00519-0. [DOI] [PubMed] [Google Scholar]
- 53.Makris S. Regulatory considerations in developmental neurotoxicity of organophosphorus and carbamate pesticides. In: Gupta RC, editor. Toxicity of Organophosphate and Carbamate Pesticides. Elsevier Academic Press; San Diego: 2006. pp. 633–641. [Google Scholar]
- 54.Malaterre J, Ramsay RG, Mantamadiotis T. Wnt-Frizzled signalling and the many paths to neural development and adult brain homeostasis. Frontiers Biosci. 2007;12:492–506. doi: 10.2741/2077. [DOI] [PubMed] [Google Scholar]
- 55.Micheli MR, Bova R, Laurenzi MA, Bazzucchi M, Grassi Zucconi G. Modulation of BDNF and TrkB expression in rat hippocampus in response to acute neurotoxicity by diethyldithiocarbamate. Neurosci Lett. 2006;410:66–70. doi: 10.1016/j.neulet.2006.09.069. [DOI] [PubMed] [Google Scholar]
- 56.Mileson BE, Chambers JE, Chen WL, Dettbarn W, Ehrich M, Eldefrawi AT, Gaylor DW, Hamernik K, Hodgson E, Karczmar AG, Padilla S, Pope CN, Richardson RJ, Saunders DR, Sheets LP, Sultatos LG, Wallace KB. Common mechanism of toxicity: a case study of organophosphorus pesticides. Toxicol Sci. 1998;41:8–20. doi: 10.1006/toxs.1997.2431. [DOI] [PubMed] [Google Scholar]
- 57.Nomoto H, Takaiwa M, Mouri A, Furukawa S. Pro-region of neurotrophins determines the processing efficiency. Biochem Biophys Res Comm. 2007;356:919–924. doi: 10.1016/j.bbrc.2007.03.059. [DOI] [PubMed] [Google Scholar]
- 58.Ostrea EM, Morales V, Ngoumgna E, Prescilla R, Tan E, Hernandez E, Ramirez GB, Cifra HL, Manlapaz ML. Prevalence of fetal exposure to environmental toxins as determined by meconium analysis. Neurotoxicology. 2002;23:329–339. doi: 10.1016/s0161-813x(02)00077-3. [DOI] [PubMed] [Google Scholar]
- 59.Pomeroy-Black MJ, Jortner BS, Ehrich MF. Early effects of neuropathy-inducing organophosphates on in vivo concentrations of three neurotrophins. Neurotox Res. 2007;11:85–91. doi: 10.1007/BF03033387. [DOI] [PubMed] [Google Scholar]
- 60.Pope CN. Organophosphorus pesticides: do they all have the same mechanism of toxicity? J. Toxicol Environ Health. 1999;2:161–181. doi: 10.1080/109374099281205. [DOI] [PubMed] [Google Scholar]
- 61.Qiao D, Seidler FJ, Slotkin TA. Developmental neurotoxicity of chlorpyrifos modeled in vitro: comparative effects of metabolites and other cholinesterase inhibitors on DNA synthesis in PC12 and C6 cells. Environ Health Perspect. 2001;109:909–913. doi: 10.1289/ehp.01109909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qiao D, Seidler FJ, Violin JD, Slotkin TA. Nicotine is a developmental neurotoxicant and neuroprotectant: stage-selective inhibition of DNA synthesis coincident with shielding from effects of chlorpyrifos. Dev Brain Res. 2003;147:183–190. doi: 10.1016/s0165-3806(03)00222-0. [DOI] [PubMed] [Google Scholar]
- 63.Reichardt LF. Neurotrophin-regulated signalling pathways. Phil Trans Royal Soc London B. 2006;361:1545–1564. doi: 10.1098/rstb.2006.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ricceri L, Venerosi A, Capone F, Cometa MF, Lorenzini P, Fortuna S, Calamendrei G. Developmental neurotoxicity of organophosphorous pesticides: fetal and neonatal exposure to chlorpyrifos alters sex-specific behaviors at adulthood in mice. Toxicol Sci. 2006;93:105–113. doi: 10.1093/toxsci/kfl032. [DOI] [PubMed] [Google Scholar]
- 65.Roy TS, Sharma V, Seidler FJ, Slotkin TA. Quantitative morphological assessment reveals neuronal and glial deficits in hippocampus after a brief subtoxic exposure to chlorpyrifos in neonatal rats. Dev Brain Res. 2005;155:71–80. doi: 10.1016/j.devbrainres.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 66.Saarimaki-Vire J, Peltopuro P, Lahti L, Naserke T, Blak AA, Vogt Weisenhorn DM, Yu K, Ornitz DM, Wurst W, Partanen J. Fibroblast growth factor receptors cooperate to regulate neural progenitor properties in the developing midbrain and hindbrain. J Neurosci. 2007;27:8581–8592. doi: 10.1523/JNEUROSCI.0192-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scharfman HE. Brain-derived neurotrophic factor and epilepsy: a missing link? Epilepsy Curr. 2005;5:83–88. doi: 10.1111/j.1535-7511.2005.05312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shelton RC. The molecular neurobiology of depression. Psychiat Clin North Am. 2007;30:1–11. doi: 10.1016/j.psc.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shimogori T, Banuchi V, Ng HY, Strauss JB, Grove EA. Embryonic signaling centers expressing BMP, WNT and FGF proteins interact to pattern the cerebral cortex. Development. 2004;131:5639–5647. doi: 10.1242/dev.01428. [DOI] [PubMed] [Google Scholar]
- 70.Shimogori T, VanSant J, Paik E, Grove EA. Members of the Wnt, Fz, and Frp gene families expressed in postnatal mouse cerebral cortex. J Comp Neurol. 2004;473:496–510. doi: 10.1002/cne.20135. [DOI] [PubMed] [Google Scholar]
- 71.Shoval G, Weizman A. The possible role of neurotrophins in the pathogenesis and therapy of schizophrenia. Eur Neuropsychopharmacol. 2005;15:319–329. doi: 10.1016/j.euroneuro.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 72.Slotkin TA. Cholinergic systems in brain development and disruption by neurotoxicants: nicotine, environmental tobacco smoke, organophosphates. Toxicol Appl Pharmacol. 2004;198:132–151. doi: 10.1016/j.taap.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 73.Slotkin TA. Developmental neurotoxicity of organophosphates: a case study of chlorpyrifos. In: Gupta RC, editor. Toxicity of Organophosphate and Carbamate Pesticides. Elsevier Academic Press; San Diego: 2005. pp. 293–314. [Google Scholar]
- 74.Slotkin TA, Cousins MM, Tate CA, Seidler FJ. Persistent cholinergic presynaptic deficits after neonatal chlorpyrifos exposure. Brain Res. 2001;902:229–243. doi: 10.1016/s0006-8993(01)02387-3. [DOI] [PubMed] [Google Scholar]
- 75.Slotkin TA, Levin ED, Seidler FJ. Comparative developmental neurotoxicity of organophosphate insecticides: effects on brain development are separable from systemic toxicity. Environ Health Perspect. 2006;114:746–751. doi: 10.1289/ehp.8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Slotkin TA, MacKillop EA, Ryde IT, Tate CA, Seidler FJ. Screening for developmental neurotoxicity using PC12 cells: comparisons of organophosphates with a carbamate, an organochlorine and divalent nickel. Environ Health Perspect. 2007;115:93–101. doi: 10.1289/ehp.9527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Slotkin TA, Seidler FJ. Comparative developmental neurotoxicity of organophosphates in vivo: transcriptional responses of pathways for brain cell development, cell signaling, cytotoxicity and neurotransmitter systems. Brain Res Bull. 2007;72:232–274. doi: 10.1016/j.brainresbull.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Slotkin TA, Seidler FJ. Prenatal chlorpyrifos exposure elicits presynaptic serotonergic and dopaminergic hyperactivity at adolescence: critical periods for regional and sex-selective effects. Reprod Toxicol. 2007;23:421–427. doi: 10.1016/j.reprotox.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 79.Slotkin TA, Seidler FJ, Fumagalli F. Exposure to organophosphates reduces the expression of neurotrophic factors in neonatal rat brain regions: similarities and differences in the effects of chlorpyrifos and diazinon on the fibroblast growth factor superfamily. Environ Health Perspect. 2007;115:909–916. doi: 10.1289/ehp.9901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Slotkin TA, Tate CA, Cousins MM, Seidler FJ. Functional alterations in CNS catecholamine systems in adolescence and adulthood after neonatal chlorpyrifos exposure. Dev Brain Res. 2002;133:163–173. doi: 10.1016/s0165-3806(02)00284-5. [DOI] [PubMed] [Google Scholar]
- 81.Slotkin TA, Tate CA, Ryde IT, Levin ED, Seidler FJ. Organophosphate insecticides target the serotonergic system in developing rat brain regions: disparate effects of diazinon and parathion at doses spanning the threshold for cholinesterase inhibition. Environ Health Perspect. 2006;114:1542–1546. doi: 10.1289/ehp.9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Snedecor GW, Cochran WG. Statistical Methods. Iowa State University Press; Ames, Iowa: 1967. [Google Scholar]
- 83.Sohrabji F, Lewis DK. Estrogen-BDNF interactions: implications for neurodegenerative diseases. Frontiers Neuroendocrinol. 2006;27:404–414. doi: 10.1016/j.yfrne.2006.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Song X, Seidler FJ, Saleh JL, Zhang J, Padilla S, Slotkin TA. Cellular mechanisms for developmental toxicity of chlorpyrifos: targeting the adenylyl cyclase signaling cascade. Toxicol Appl Pharmacol. 1997;145:158–174. doi: 10.1006/taap.1997.8171. [DOI] [PubMed] [Google Scholar]
- 85.Song X, Violin JD, Seidler FJ, Slotkin TA. Modeling the developmental neurotoxicity of chlorpyrifos in vitro: macromolecule synthesis in PC12 cells. Toxicol Appl Pharmacol. 1998;151:182–191. doi: 10.1006/taap.1998.8424. [DOI] [PubMed] [Google Scholar]
- 86.Sutton LP, Honardoust D, Mouyal J, Rajakumar N, Rushlow WJ. Activation of the canonical Wnt pathway by the antipsychotics haloperidol and clozapine involves dishevelled-3. J Neurochem. 2007;102:153–169. doi: 10.1111/j.1471-4159.2007.04527.x. [DOI] [PubMed] [Google Scholar]
- 87.Szpir M. New thinking on neurodevelopment. Environ Health Perspect. 2006;114:A101–A107. doi: 10.1289/ehp.114-a100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Szpir M. Tracing the origins of autism: a spectrum of new studies. Environ Health Perspect. 2006;114:A412–A417. doi: 10.1289/ehp.114-a412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Teng KK, Greene LA. Cultured PC12 cells: a model for neuronal function and differentiation. In: Celis JE, editor. Cell Biology: A Laboratory Handbook. Academic Press; San Diego: 1994. pp. 218–224. [Google Scholar]
- 90.Terry AV, Gearhart DA, Beck WD, Truan JN, Middlemore ML, Williamson LN, Bartlett MG, Prendergast MA, Sickles DW, Buccafusco JJ. Chronic, intermittent exposure to chlorpyrifos in rats: protracted effects on axonal transport, neurotrophin receptors, cholinergic markers, and information processing. J Pharmacol Exp Ther. 2007;322:1117–1128. doi: 10.1124/jpet.107.125625. [DOI] [PubMed] [Google Scholar]
- 91.Terry AV, Stone JD, Buccafusco JJ, Sickles DW, Sood A, Prendergast MA. Repeated exposures to subthreshold doses of chlorpyrifos in rats: hippocampal damage, impaired axonal transport, and deficits in spatial learning. J Pharmacol Exp Ther. 2003;305:375–384. doi: 10.1124/jpet.102.041897. [DOI] [PubMed] [Google Scholar]
- 92.Torres-Peraza J, Pezzi S, Canals JM, Gavalda N, Garcia-Martinez JM, Perez-Navarro E, Alberch J. Mice heterozygous for neurotrophin-3 display enhanced vulnerability to excitotoxicity in the striatum through increased expression of N-methyl-D-aspartate receptors. Neuroscience. 2007;144:462–471. doi: 10.1016/j.neuroscience.2006.09.038. [DOI] [PubMed] [Google Scholar]
- 93.Twiss JL, Chang JH, Schanen NC. Pathophysiological mechanisms for actions of the neurotrophins. Brain Pathol. 2006;16:320–332. doi: 10.1111/j.1750-3639.2006.00039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vidair CA. Age dependence of organophosphate and carbamate neurotoxicity in the postnatal rat: extrapolation to the human. Toxicol Appl Pharmacol. 2004;196:287–302. doi: 10.1016/j.taap.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 95.von Bohlen und Halbach O, Minichiello L. Neurotrophin receptor heterozygosity causes deficits in catecholaminergic innervation of amygdala and hippocampus in aged mice. J Neural Transmission. 2006;113:1829–1836. doi: 10.1007/s00702-006-0498-2. [DOI] [PubMed] [Google Scholar]
- 96.Wang Y, Thekdi N, Smallwood PM, Macke JP, Nathans J. Frizzled-3 is required for the development of major fiber tracts in the rostral CNS. J Neurosci. 2002;22:8563–8573. doi: 10.1523/JNEUROSCI.22-19-08563.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Whitney KD, Seidler FJ, Slotkin TA. Developmental neurotoxicity of chlorpyrifos: cellular mechanisms. Toxicol Appl Pharmacol. 1995;134:53–62. doi: 10.1006/taap.1995.1168. [DOI] [PubMed] [Google Scholar]
- 98.Wilson SW, Houart C. Early steps in the development of the forebrain. Dev Cell. 2004;6:167–181. doi: 10.1016/s1534-5807(04)00027-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Woodruff NR, Neet KE. Inhibition of beta nerve growth factor binding to PC12 cells by alpha nerve growth factor and gamma nerve growth factor. Biochemistry. 1986;25:7967–7974. doi: 10.1021/bi00372a027. [DOI] [PubMed] [Google Scholar]
- 100.Zhou CJ, Borello U, Rubenstein JL, Pleasure SJ. Neuronal production and precursor proliferation defects in the neocortex of mice with loss of function in the canonical Wnt signaling pathway. Neuroscience. 2006;142:1119–1131. doi: 10.1016/j.neuroscience.2006.07.007. [DOI] [PubMed] [Google Scholar]