Abstract
G93A SOD1 transgenic mice overexpressing CCS protein develop an accelerated disease course that is associated with enhanced mitochondrial pathology and increased mitochondrial localization of mutant SOD1. Because these results suggest an effect of mutant SOD1 on mitochondrial function, we assessed the enzymatic activities of mitochondrial respiratory chain complexes in the spinal cords of CCS/G93A SOD1 and control mice. CCS/G93A SOD1 mouse spinal cord demonstrates a 55% loss of complex IV (cytochrome c oxidase) activity compared with spinal cord from age-matched non-transgenic or G93A SOD1 mice. In contrast, CCS/G93A SOD1 spinal cord shows no reduction in the activities of complex I, II, or III. Blue native gel analysis further demonstrates a marked reduction in the levels of complex IV but not of complex I, II, III, or V in spinal cords of CCS/G93A SOD1 mice compared with non-transgenic, G93A SOD1, or CCS/WT SOD1 controls. With SDS-PAGE analysis, spinal cords from CCS/G93A SOD1 mice showed significant decreases in the levels of two structural subunits of cytochrome c oxidase, COX1 and COX5b, relative to controls. In contrast, CCS/G93A SOD1 mouse spinal cord showed no reduction in levels of selected subunits from complexes I, II, III, or V. Heme A analyses of spinal cord further support the existence of cytochrome c oxidase deficiency in CCS/G93A SOD1 mice. Collectively, these results establish that CCS/G93A SOD1 mice manifest an isolated complex IV deficiency which may underlie a substantial part of mutant SOD1-induced mitochondrial cytopathy.
Mutations in the Cu,Zn superoxide dismutase (SOD1)2 gene cause one form of familial amyotrophic lateral sclerosis (ALS), a neurodegenerative disorder characterized by progressive motor neuron loss leading to death of affected individuals (1). Studies using both mutant SOD1 transgenic and SOD1 knockout mice have established a toxic gain in function as the basis for disease, although precise mechanisms remain unclear (2–4). There is increasing evidence that the mitochondrial localization of mutant SOD1 may be important in disease pathogenesis (5). Mitochondrial pathology characterized by vacuolization is a principal finding in G93A and G37R SOD1 transgenic mouse lines that develop progressive motor weakness (3, 6). Importantly, mutant SOD1, once considered a cytosolic protein, has been detected within the mitochondrial intermembrane space and matrix, raising the possibility of a direct toxic effect of mutant SOD1 on mitochondrial function (7–10). Consistent with this hypothesis, the subcellular targeting of mutant SOD1 to mitochondria in vitro leads to enhanced death of N2a cells (11). An elevation in the mitochondrial load of mutant SOD1 in transgenic mice also appears to adversely impact disease. Overexpression of wild type SOD1 in G93A SOD1 transgenic mice leads to a modest but significant reduction in lifespan (by 10%) and earlier disease onset (by 25%) that is associated with an increased mitochondrial load of SOD1 aggregates (12). Recently, we have shown that overexpression of the wild type copper chaperone for SOD1 (CCS) in G93A SOD1 mice leads to an enrichment of mutant SOD1 within mitochondria and a greatly accelerated disease course (13). Compared with their G93A SOD1 littermates, CCS/G93A SOD1 mice develop an overt neurological phenotype beginning at age 10 days rather than at 6 months and succumb by 36 days rather than 242 days. Moreover, CCS/G93A SOD1 mice develop enhanced mitochondrial vacuolar pathology within motor neurons. These in vivo experiments provide additional evidence that mutant SOD1 may impact mitochondrial function and contribute significantly to the disease.
The specific aspect of mitochondrial function affected by mutant SOD1, which would lead to neuronal dysfunction, is not completely understood. G93A SOD1 mice do show a general reduction in mitochondrial respiration as disease becomes clinically evident (14–17). Mattiazzi et al. (15) found deficits in complex I, II, III, and IV activities in spinal cord from weak G93A SOD1 mice, whereas citrate synthase activity was unchanged. Kirkinezos et al. (16) observed losses in cytochrome c oxidase (COX) activity within the spinal cords of paralyzed G93A SOD1 mice that were matched by similar reductions in succinate dehydrogenase activity. Such findings, particularly in older and sicker G93A SOD1 mice, suggest that changes in general respiratory chain function are a consequence of mitochondrial degeneration and are indicators of overall mitochondrial failure (14–16).
We wished to re-examine the effect of mutant SOD1 on the function of individual complexes that participate in oxidative phosphorylation (OXPHOS). CCS/G93A SOD1 mice are suitable for this purpose because they show increased mitochondrial localization of mutant SOD1, whereas total SOD1 levels remain constant, a condition that is otherwise not easily mimicked with single mutant SOD1 mouse lines. In addition, because CCS/G93A SOD1 mice manifest an accelerated disease course with enhanced mitochondrial pathology, any mutant SOD1-induced change of OXPHOS function might likewise be accentuated. We, therefore, assessed OXPHOS function in CCS/G93A SOD1 mouse spinal cord by measuring the activities of complexes I–IV, by analyzing the abundance and assembly of complexes I–V with blue native (BN) gels, and by quantifying the levels of selected structural subunits from each of these holoenzymes with standard SDS-PAGE gels. Rather than finding a generalized reduction in either the activities or steady state levels of the different OXPHOS complexes, we observed deficits limited to a single complex. CCS/G93A SOD1 mice exhibit an isolated complex IV deficiency within spinal cord that is characterized by marked reduction in both total enzyme activity and content when compared with G93A SOD1 or CCS/WT SOD1 controls. Concordant reductions in both heme A content and the levels of COX structural subunits further validate the observed complex IV deficiency. Because complex IV is critical for normal mitochondrial respiratory function, mutant SOD1 induced complex IV deficiency may underlie a substantial part of the mitochondrial cytopathy in G93A SOD1 mice overexpressing CCS protein.
EXPERIMENTAL PROCEDURES
Transgenic Mouse Lines—Transgenic mice expressing the low copy number human G93A SOD1 mutation (B6SJL-TgNSOD1-G93A SOD1; 1Gurdl JR2300) were originally obtained from The Jackson Laboratory (Bar Harbor, ME). This line develops motor deficits at about 6 months and has a mean survival of about 240 days (18). Transgenic mice expressing wild type (WT) human SOD1 (line N29, B6SJL-WT-SOD1) were also obtained from The Jackson Laboratory. The generation and characterization of CCS transgenic mice (line 17) has recently been described (13). CCS mice do not manifest a neurological phenotype and do not show mitochondrial pathology. Line 17 CCS transgenic mice were crossed to either G93A SOD1 or WT SOD1 mice to obtain CCS/G93A SOD1 or CCS/WT SOD1 dual transgenic mice as previously described. Specific genotypes were identified by PCR on tail DNA. All animal protocols were approved by our university's Institutional Animal Care and Research Advisory Committee in compliance with National Institutes of Health guidelines.
Spinal Cord OXPHOS Enzyme Activities—Frozen spinal cord was homogenized with a Teflon glass homogenizer in 10 vol: weight homogenization buffer (320 mm sucrose, 10 mm HEPES, 1 mm EGTA, pH 7.4) (19). A portion of the homogenate was separated into aliquots, snap-frozen in liquid nitrogen, and stored at -80 °C for complex I (NADH:ubiquinone oxidoreductase) and complex III (ubiquinol:cytochrome c oxidoreductase) activity assays and for protein assays. The remainder of the homogenate was used immediately for citrate synthase (CS), complex IV (COX), and complex II (succinate dehydrogenase) assays.
NADH:ubiquinone oxidoreductase activity was measured as described previously (20). Frozen 10% homogenate was thawed, diluted 1:1 in 20 mm KH2PO4, pH 7.2, and quickly freeze/thawed 3 times. Sample was added to 500 μl of assay buffer (3 mm KCN, 5 mm MgCl2, 75 μm coenzyme Q1, 0.13 mm NADH, 4 μg/ml antimycin, 35 mm potassium phosphate buffer, 2.5 mg/ml BSA, pH 7.2) in a cuvette at 30 °C. Oxidation of NADH was followed at 340 nm before and after the addition of 4 μg/ml rotenone. Activity of NADH:ubiquinone oxidoreductase was calculated for a time period of no less than 1 min. Complex I activity was calculated by subtracting from the overall rate, the rotenone non-inhibited rate.
Succinate dehydrogenase activity was measured by the decrease in absorbance of dichlorophenol-indophenol (DCPIP) at 600 nm coupled to oxidation of succinate to fumarate by succinate dehydrogenase. Briefly, 1–2 μl of sample was incubated in a total volume of 50 μl of 50 mm potassium phosphate, pH 7.0, and 15 mm succinate, pH 7.5, at 30 °C for 20 min. Then, KCN, DCPIP, and phenazine ethosulfate were added to final concentrations of 15 mm, 100 μm, 6 mm, respectively. Reduction of DCPIP to DCPIPH2 was followed at 600 nm. Activity of succinate dehydrogenase was calculated by using the extinction coefficient for DCPIP at 600 nm (19.1 mm-1 cm-1).
Ubiquinol:cytochrome c oxidoreductase activity was assayed as described previously (20). Reduction of cytochrome c (III) is coupled to oxidation of ubiquinol-2, and the increase in absorbance at 550 nm was followed. The reaction mix consisted of 3 mm KCN, 15 μm cytochrome c, 0.6 mm n-dodecyl-β-d-maltoside, 2 μg/ml rotenone, 60 μm ubiquinol-2 with and without sample. Activity of ubiquinol:cytochrome c oxidoreductase was calculated by using the extinction coefficient of reduced cytochrome c at 550 nm (20 mm-1 cm-1).
COX activity was determined as described previously (20). Oxidation of reduced cytochrome c by COX was followed by a decrease in absorbance at 550 nm. Sample was diluted 1:20 vol:vol in 50 mm potassium phosphate, 250 mm sucrose, and 0.5% taurodeoxycholic acid, pH 7.0. The reaction mixture included 50 mm potassium phosphate, 10 μm reduced cytochrome c, and sample. An extinction coefficient of 20 mm-1 cm-1 was used to calculate the rate of cytochrome c oxidation.
CS activity was assayed by following the release of CoA-SH at 25 °C by increasing absorbance at 412 nm due to reaction of CoA-SH with 5,5′-dithiobis-2-nitrobenzoic acid (Ellman's reagent) to yield. The reaction mixture consisted of 0.1 mm 5,5′-dithiobis-2-nitrobenzoic acid, 0.3 mm acetyl-CoA, and 0.5 mm oxaloacetate, and the mitochondria sample was diluted 1:10 vol:vol in 50 mm potassium phosphate, 250 mm sucrose, 0.5% taurodeoxycholic acid, pH 7.0, to break open the mitochondrial membranes and release citrate synthase. An extinction coefficient of 13.6 mm-1 cm-1 was used to calculate the rate of 5-thio-2-nitro-benzoic acid formation and CS activity.
Blue native (BN) Gel Electrophoresis—Spinal cords were homogenized on ice with a Dounce homogenizer in phosphate-buffered saline supplemented with a protease inhibitor mixture (Roche Applied Science) and 0.5 mm phenylmethylsulfonyl fluoride. The protein concentration of homogenates was adjusted to 5 mg/ml with ice-cold phosphate-buffered saline. Homogenates were then incubated on ice for 10 min in an equal volume of digitonin (4 mg/ml) to generate mitoplasts (21). BN-PAGE was subsequently performed on mitoplasts that had been solubilized in 1% dodecyl maltoside (22). Protocols for immunoblotting of first-dimension BN-PAGE gels and second-dimension BN/SDS-PAGE gel electrophoresis were as previously described (23). Individual structural subunits of complexes I–V were detected by Western blots using the following antibodies: mouse anti-complex I 39-kDa subunit, mouse anti-complex II Fp70 subunit, mouse anti-complex III core 1 subunit, mouse-anti-complex IV COX1 subunit, and mouse-anti-complex V ATPase α subunit (Mitosciences). Multiple exposures within the dynamic range of the film were scanned for each individual OXPHOS complex and quantified densitometrically using ImageQuant software (GE Healthcare). In all cases the levels of complexes I, II, III, IV, and V were expressed as a ratio of the median non-transgenic (NTG) value. One way ANOVA was performed followed by Tukey's post hoc test for p 0.05, whereas t tests were used to compare CCS/G93A SOD1 and G93A SOD1 directly.
Subunit Analysis—Mice were overdosed with pentobarbital (200 mg/kg, intraperitoneal) and perfused transcardially with phosphate-buffered saline. Mitochondria were isolated from dissected spinal cords by using Sigma mitochondria isolation kit (Sigma), and Western blots were done as previously described (24). The experiments were performed using the following primary antibodies: mouse anti-TIM23, 1:2000 (BD Transduction Laboratories); rabbit anti-TOM20, 0.2 μg/ml (Santa Cruz Biotechnology, Inc.), rabbit anti-MnSOD 0.2 μg/ml (StressGen); mouse anti-COX1 subunit, 1 μg/ml (clone 1D6) (Molecular Probes, Inc., Eugene, OR); mouse anti-COX5b subunit, 1 μg/ml (clone 16H12) (Molecular Probes); mouse anti-complex I NDUFB8 subunit, 0.5 μg/ml (Mitosciences); goat anti-complex I ND2 subunit, 1 μg/ml (Santa Cruz Biotechnology); mouse anti-complex II Fp70 subunit, 0.1 μg/ml (Mitosciences); mouse anti-complex III core 1 subunit, 0.5 μg/ml (Mitosciences); mouse anti-complex V ATPase β subunit, 0.5 μg/ml (Mitosciences). Western blots for each series of experiments were repeated at least three times. The Western blots were quantified densitometrically with a GE Healthcare Storm 840 Scanner using ImageQuant 5.2 software. Mitochondrial loading was normalized for TIM23. Samples were run in triplicate. One way ANOVA was performed for statistical analysis.
Heme A Analysis—Heme A analysis was conducted on 20–50 mg of spinal cord tissue extracted with 0.5 ml of acetone containing 2.5% HCl as described previously (25). The pH of the extract was adjusted to 4 by the addition of 1 μl of formic acid and titration of a KOH solution. The sample was clarified by centrifugation at 13,000 rpm for 5 min, and 1 ml was injected onto a 3.9 × 300-mm C18 Bondclone column (Phenomex). Heme A was quantified by integrating the respective peaks of the HPLC profile. Values were then normalized to wet weight.
RESULTS
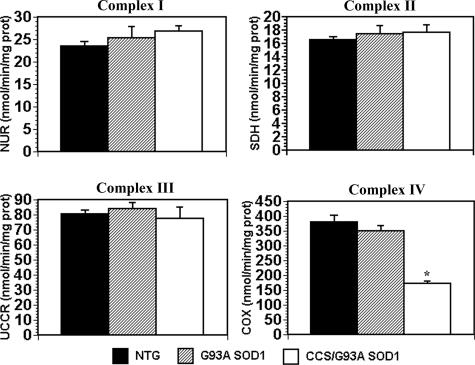
Loss of Complex IV Activity in CCS/G93A SOD1 Spinal Cord—CCS/G93A SOD1 transgenic mice show increased mitochondrial localization of G93A SOD1(13). To determine whether this change in cellular distribution of mutant SOD1 impacted mitochondrial respiratory chain function, we assayed the enzymatic activities of complexes I (NADH cytochrome c reductase), II (succinate dehydrogenase), III (ubiquinone cytochrome c reductase), and IV (cytochrome c oxidase) in spinal cord extracts from 3–4-week-old CCS/G93A SOD1 and age-matched controls (Fig. 1). COX activity is reduced by 55% in the spinal cords of CCS/G93A SOD1 mice compared with non-transgenic and by 50% when compared with G93A SOD1 mice. In contrast, the activities for complexes I, II, and III in spinal cords of CCS/G93A SOD1 are preserved and do not show any significant difference from G93A SOD1 or non-transgenic controls. CCS/G93A SOD1 mouse spinal cords show impaired cytochrome c oxidase function, whereas activities of the other mitochondrial respiratory complexes are preserved.
FIGURE 1.
Mitochondrial respiratory chain activities. Enzymatic activities of mitochondrial respiratory complexes I (NADH:ubiquinone oxidoreductase (NUR)), II (succinate dehydrogenase (SDH)), III (ubiquinone cytochrome c reductase (UCCR)), and IV (cytochrome c oxidase) in 3-week-old NTG, G93A SOD1 transgenic, and CCS/G93A SOD1 transgenic mouse spinal cords. Values are normalized to citrate synthase activity. 6–8 animals were used per genotype per complex activity. Samples were run in duplicate. CCS/G93A SOD1 mice show significant reductions in complex IV activity. *, p < 0.001 for CCS/G93A SOD1 versus NTG and G93A SOD1 by ANOVA followed by a Bonferroni t test for multiple comparisons. There is no statistical difference among the three genotypes for the activities of complex I, II, or III.
Reduction of Complex IV Levels in CCS/G93A SOD1 Spinal Cord—Because CCS/G93A SOD1 mouse spinal cord shows an isolated reduction in complex IV activity, we wished to determine whether there was a corresponding change in the amount of the fully assembled holoenzyme. We, therefore, performed one-dimension BN-PAGE gels on spinal cord tissue from 3-week-old CCS/G93A SOD1 mice and age-matched controls (Fig. 2). Consistent with the biochemical analyses, spinal cords from CCS/G93A SOD1 mice show a selective decrease in complex IV levels (Fig. 2A). Quantitative analysis reveals a 50–60% reduction in complex IV levels compared with the controls (Fig. 2B). In contrast, spinal cords from CCS/G93A SOD1 mice show no reduction in the levels of complex I, II, III, or V (ATP synthase) compared with controls but rather mild increases in complexes I and II and larger increases in complex III levels. Importantly, spinal cords from control CCS/WT SOD1 transgenic mice show no loss of complex IV content, indicating that increased expression of CCS and WT SOD1 proteins does not cause complex IV deficiency. These results indicate that CCS/G93A SOD1 mice manifest a selective reduction in the levels of mitochondrial complex IV within their spinal cords that corresponds to the deficit in complex IV enzymatic activity.
FIGURE 2.
Analysis of OXPHOS complex levels by BN-PAGE. A, mitochondrial extracts isolated from spinal cords of 3–4-week-old NTG, G93A SOD1, CCS/WT SOD1, and CCS/G93A SOD1 mice fractionated by BN-PAGE and probed with anti-39-kDa (complex I), anti-Fp70 (complex II), anti-core-1 (complex III), anti-COX1 (complex IV)- and anti-ATPase α (complex V) antibodies, 10 μg of protein/lane. Each lane represents a unique animal. B, quantification of complex levels on blue native gel. CCS/G93A SOD1 show significant a decrease in levels of complex IV. #, p < 0.04 for CCS/G93A SOD1 versus NTG by ANOVA followed by Tukey's post-hoc test. CCS/G93A SOD1 mice show significant increases in complex I, II, and III levels. p < 0.004 (*) and p < 0.001 (**) for CCS/G93A SOD1 versus NTG. n = 3–5 animals for each genotype. Levels of complexes I, II, III, IV, and V are normalized to the median NTG value.
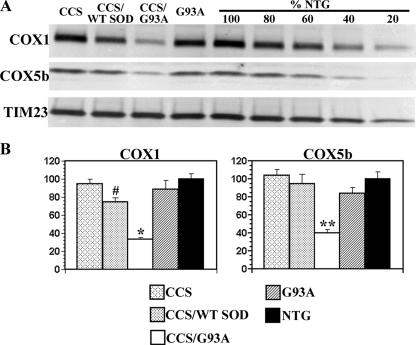
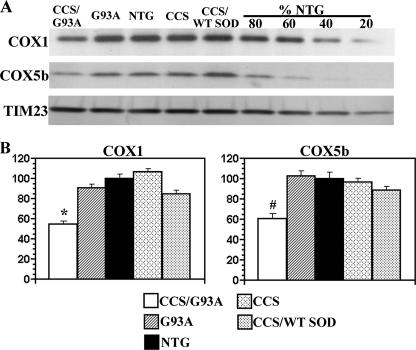
We next wished to determine whether the specific loss of complex IV holoenzyme in CCS/G93A spinal cord was accompanied by reduced levels of its individual structural subunits. Western blots were performed on mitochondrially enriched fractions from spinal cords of 3-week-old CCS/G93A SOD1 and control mice to assess levels of individual structural subunits of the differing OXPHOS complexes. Levels of COX1, a mitochondrially encoded complex IV subunit, and COX5b, a nuclear encoded complex IV subunit, were substantially reduced in the spinal cords of CCS/G93A SOD1 mice compared with G93A SOD1 and non-transgenic or CCS/WT SOD1 mice (Figs. 3, A and B). COX1 levels in CCS/G93A spinal cords were significantly reduced by >67% compared with non-transgenic and by 60% compared with G93A SOD1 controls. COX5b subunit levels were reduced by 60% in CCS/G93A SOD1 spinal cord compared with non-transgenic and by 52% compared with G93A SOD1 controls. Interestingly, we also observed a modest but significant reduction in COX1 but not in COX5b subunit levels in 3-week-old CCS/WT SOD1 compared with non-transgenic mice. In contrast, steady state levels of NDUFB8 (nuclear encoded complex I subunit), ND2 (mitochondrial encoded complex I subunit), Fp70 (nuclear encoded complex II subunit), core 1 (nuclear encoded complex III subunit), and ATPase β (nuclear encoded complex V subunit) were comparable between CCS/G93A SOD1 mice and age-matched controls (Figs. 4, A and B; supplemental Fig. S1). Furthermore, levels of manganese superoxide dismutase (SOD2), a mitochondrial matrix protein, were not different in the spinal cord mitochondrial fractions isolated from CCS/G93A SOD1 mice compared with controls. Overall, these results indicate that the loss of complex IV activity and content is associated with a reduction of its individual structural subunits within CCS/G93A SOD1 spinal cord mitochondria.
FIGURE 3.
Analysis of complex IV structural subunit levels in spinal cords from 3–4-week-old mice. A, mitochondrially enriched fractions from the spinal cords of 3–4-week NTG, CCS, G93A SOD1, CCS/WT SOD1, and CCS/G93A SOD1 mice separated on SDS-PAGE and probed with anti-COX1, anti-COX5b, and anti-TIM23 antibodies. 20 μg of protein/lane was loaded. Concentration series of 16, 12, 8, and 4 μg of NTG sample was loaded as a control for quantification. CCS/G93A SOD1 spinal cord shows marked reductions in levels of complex IV subunits, COX1, and COX5b with no appreciable change in the mitochondrial marker TIM23. B, quantification of COX1 and COX5b subunit levels. Three animals per genotype were used for analysis. Mitochondrial protein loading was normalized to TIM23 levels. Levels of COX1 and COX5b subunits are expressed as a percentage of the NTG levels. *, p < 0.001 for CCS/G93A SOD1 versus other groups by ANOVA followed by a Bonferroni t test for multiple comparisons. **, p < 0.003 for CCS/G93A SOD1 versus other groups. #, p < 0.04 CCS/WT SOD1 versus non-transgenic or CCS.
FIGURE 4.
Analysis of complex I, II, III, and V subunit levels in spinal cords from 3-week-old mice. A, mitochondrially enriched fractions from the spinal cords of 3–4-week NTG, CCS, G93A SOD1, CCS/WT SOD1, and CCS/G93A SOD1 mice separated on SDS-PAGE and probed with anti-NDUFB8 (complex I), anti-Fp70 (complex II), anti-core 1 (complex III), anti-ATPase β subunit (complex V), anti-TIM 23, and anti-MnSOD antibodies. In Western gels, 3 μg of protein/lane were loaded. B, quantification of complex I, II, III, and V subunits levels. Three animals per genotype were used. Mitochondrial loading was normalized to TIM23 levels. Levels of I, II, III, and V are expressed a percentage of the NTG values. There is no statistical difference among the differing genotypes for levels of NDUFB8, Fp70, Core 1, or ATPase β, respectively, by ANOVA. As a control for the linearity of quantification, 3, 6, 9, and 12 μg of NTG sample were loaded and quantified (data not shown).
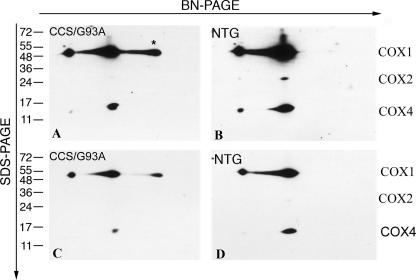
We next assessed whether complex IV deficiency in the spinal cord of CCS/G93A SOD1 mice was associated with either incomplete or abnormal holoenzyme assembly, which frequently results in the accumulation of COX subcomplexes. In mammalian cells, the mature COX holoenzyme (S4) is ultimately generated from a series of three intermediate subcomplexes termed S1–S3 by the sequential association of structural COX subunits and incorporation of relevant prosthetic groups (26). Possible changes in the abundance of complex IV assembly intermediates were evaluated by BN/SDS-PAGE using spinal cords from CCS/G93A SOD1 and control non-transgenic mice (Fig. 5). Although S2 and S3 intermediates were undetectable in the spinal cords of non-transgenics and CCS/G93A SOD1 mice, a significant accumulation of unassembled COX1 was observed in the CCS/G93A SOD1 spinal cord. Because under normal conditions nascent COX1 rapidly associates with COX4 and COX5a to form S2, these results imply that COX deficiency in CCS/G93A SOD1 spinal cord is due to mutant SOD1-induced interruption at a very early stage of holoenzyme assembly.
FIGURE 5.
Analysis of complex IV assembly in CCS/G93A SOD1 spinal cord. Mitochondrial extracts from the spinal cords of 3–4-week-old CCS/G93A SOD1 mice (A and C) and NTG controls (B and D) were analyzed by BN-PAGE and two-dimensional SDS-PAGE immunoblots. Blots were probed with antibodies against COX1, COX2, and COX4. C and D are lighter exposures of A and B, respectively. The asterisk indicates S1 subcomplex containing COX1; 20 μg of protein was loaded per lane in BN-PAGE. Experiments were performed in duplicate with two sets of animals. A representative experiment is shown.
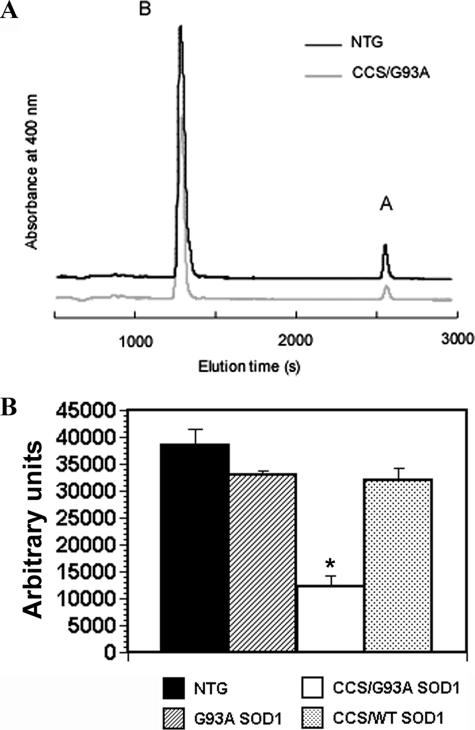
Heme A is a an important prosthetic group within complex IV that serves as a critical electron carrier (27). Because heme A is uniquely found in the COX1 subunit, levels of heme A usually reflect the abundance of the holoenzyme (28). We, therefore, measured heme A content in the spinal cords of CCS/G93A SOD1 and control mice to determine whether reductions in heme A accompanied the loss of COX 1 we observed in vivo (Fig. 6). Heme A levels are reduced by about 65% in CCS/G93A SOD1 spinal cord compared with non-transgenics, G93A SOD1, and CCS/WT SOD1 samples. Heme A values for G93A SOD1 and CCS/WT SOD1 samples are slightly decreased compared with non-transgenics, although this reduction does not reach statistical significance. Overall, reductions in heme A values correspond well to changes in COX holoenzyme and subunit levels, further supporting the existence of COX deficiency in CCS/G93A SOD1 mouse spinal cord.
FIGURE 6.
Heme A analysis in spinal cords from CCS/G93A SOD1 mice and controls. A, hemes were extracted from spinal cords and separated by reverse phase HPLC. The positions at which heme B (B) and heme A (A) elute are shown. For clarity, only two representative chromatograms are shown from 3-week-old non-transgenic and CCS/G93A SOD1 mouse spinal cords. B, the area under the curve of the heme A peak was normalized to wet weight of spinal cord tissue. The average value (n = 3) for the each genotype is shown. *, p < 0.001 for CCS/G93A SOD1 versus other groups by ANOVA followed by a Bonferroni t test for multiple comparisons.
Complex IV Deficiency in CCS/G93A SOD1 Mice Occurs Early and Is CNS-restricted—CCS/G93A SOD1 mice develop an isolated complex IV deficiency within their spinal cord by 3 weeks of age, a time when they already manifest significant neurological signs and have marked mitochondrial vacuolar pathology. To determine whether complex IV loss precedes the onset of advanced neurological deficits observed in CCS/G93A SOD1 mice, we assessed the levels of individual COX structural subunits within mitochondrially enriched fractions from the spinal cords of 7–8-day-old mice. At this age, CCS/G93A SOD1 mice are in an early stage of clinical disease with only mild mitochondrial pathology (13). Western blots show that even at this early time period, mitochondrial fractions from CCS/G93A SOD1 spinal cords show significant reductions in levels of COX1 and COX5b when compared with those derived from G93A SOD1, CCS/WT SOD1, or non-transgenic spinal cords (Figs. 7, A and B). CCS/G93A SOD1 mice show a 45 and 39% reduction in COX1 and COX5b levels, respectively, compared with age matched non-transgenic controls. In contrast, complex I, II, III, and V subunits are relatively constant among the differing genotypes (data not shown). These results indicate that complex IV deficiency is not just a marker of mitochondrial dysfunction in advanced stages of disease but instead occurs early on within CCS/G93A SOD1 spinal cord, consistent with a potentially causative role in the disease process. Of note, the degree of COX1 and COX5b reduction in 1-week-old CCS/G93A SOD1 mice is considerably less than in 3-week-old mice, indicating that the loss of complex IV subunits is progressive as the animal ages and neurological function deteriorates.
FIGURE 7.
Analysis of complex IV structural subunit levels in spinal cords from 1-week-old mice. A, mitochondrially enriched fractions from the spinal cords of 7–8-day-old NTG, CCS, G93A SOD1, CCS/WT SOD1, and CCS/G93A SOD1 mice separated on SDS-PAGE and probed with anti-COX1, anti-COX5b, and anti-TIM23 antibodies. 20 μg of protein/lane. Concentration series of 16, 12, 8, and 4 μg of non-transgenic sample was loaded as a control for quantification B, quantification of COX1 and COX5b levels. CCS/G93A SOD1 spinal cord shows significant reductions in levels of complex IV subunits, COX1 and COX5b. *, p < 0.001 for CCS/G93A SOD1 versus other groups by ANOVA followed by a Bonferroni t test for multiple comparisons. #, p < 0.001 for CCS/G93A SOD1 versus NTG, G93A SOD1, and CCS and p < 0.003 versus CCS/WT SOD1. 4–7 animals per genotype were used. Mitochondrial protein loading was normalized to TIM 23 levels.
Because clinical and pathological abnormalities are restricted to the CNS in G93A SOD1 and CCS/G93A SOD1 mice, we wished to determine whether mutant SOD1 induced complex IV deficiency in the presence of CCS overexpression was also limited to CNS tissue. We, therefore, assessed levels of individual subunit expression for the various OXPHOS complexes in kidney, which has the highest expression levels of the CCS transgene among the tissues tested outside the nervous system (13). Mitochondrially enriched fractions from the kidney of CCS/G93A SOD1 mice showed similar levels for both COX1 and COX5b when compared with fractions isolated from G93A SOD1, CCS CCS/WT SOD1, and non-transgenic controls (Fig. 8A). Expression levels for NDUFB8 (complex I), Fp70 (complex II), core 1 (complex III), and ATPase β (complex V) subunits as well as the inner membrane protein TIM23 were also comparable between the differing genotypes (Fig. 8B). These results indicate that complex IV deficiency appears to be limited to tissues that are vulnerable to mutant SOD1-induced disease such as spinal cord.
FIGURE 8.
Analysis of OXPHOS complex subunit levels in kidney from 3-week-old mice. Mitochondrially enriched fractions from kidney of 3-week-old NTG, CCS, G93A SOD1, CCS/WT SOD1, and CCS/G93A SOD1 mice were separated on SDS-PAGE and probed with following antibodies: anti-NDUFB8 (complex I), anti-Fp70 (complex II), core 1 (complex III), anti-COX1 (complex IV), anti-COX5b (complex IV), anti-ATPase β subunit (complex V), and anti-TIM 23 antibodies. A, 15 μg of protein/lane. B, 3 μg of protein/lane. CCS/G93A SOD1 mice do not show reductions in complex IV subunit levels within kidney. Experiments were done in triplicate using 2–3 animals per genotype. A representative experiment is shown.
DISCUSSION
Although mitochondrial pathology within motor neurons is characteristic of many mutant SOD1 transgenic mouse lines, its relevance for disease has been somewhat unclear. Recent work showing a severe disease phenotype in CCS/G93A SOD1 mice that is associated with increased mitochondrial vacuolation but no detectable cytosolic aggregates provides strong evidence for mutant SOD1-induced mitochondrial dysfunction and its role in disease pathogenesis (13). How mutant SOD1 might cause mitochondrial pathology and dysfunction, therefore, becomes a critical question. The present study, which finds an isolated COX deficiency in G93A SOD1 mice overexpressing CCS, may provide a potential explanation.
COX, the terminal oxidoreductase of the mitochondrial respiratory chain, is composed of 10 nuclear-encoded subunits and three mitochondrially encoded subunits, COX1, COX2, and COX3, which form the catalytic core of the enzyme. Assembly of individual structural subunits and insertion of relevant prosthetic groups to form a functional COX holoenzyme requires a host of nuclear-encoded accessory factors (29). In human beings isolated COX deficiency is primarily associated with clinical syndromes including Leigh disease, myopathy, cardiomyopathy, encephalopathy, and hepatic failure that are characterized by neonatal/juvenile onset with early demise (29, 30). There are rare human case reports of motor neuron involvement associated with COX deficiency, although measurements of the enzymatic activities of the mitochondrial respiratory complexes (I, II, III, and IV) in spinal cord from ALS patients generally have shown no significant reductions compared with controls (31–33). However, the association between COX deficiency and human motor neuron disease has been incompletely studied in large part due to the difficulty of assessing CNS samples after long post-mortem intervals.
There are several indications that complex IV loss in the spinal cords of G93A SOD1 mice overexpressing CCS might significantly contribute to mitochondrial impairment and overall neuronal dysfunction. First, the reduction in COX subunit levels precedes the development of neurological deficits in CCS/G93A SOD1 mice. We observe reductions in COX1 and COX5b levels in the spinal cords of these mice by day 7 of life at a stage before the onset of neurological deficits and at a time when mitochondrial vacuoles are rare. Second, the reduction in COX in G93A SOD1 mice overexpressing CCS appears to be restricted to CNS tissues susceptible to mutant SOD1-induced disease. Thus, we see no reduction in COX levels in kidney, an organ resistant to mutant SOD1-induced disease which has the highest expression levels of the CCS transgene among the organs tested outside the nervous system (13). However, we cannot exclude the possibility that differential mitochondrial levels of SOD1 and CCS within spinal cord and kidney might account for the tissue specificity of complex IV deficiency. Third, the degree of COX deficiency observed in 3-week-old CCS/G93A SOD1 spinal cord likely impairs mitochondrial respiration to an extent sufficient to cause overall cellular dysfunction. Studies involving tissue from patients with isolated COX deficiencies typically have found reductions in COX activity and COX expression levels that range from 70 to 90% and 50 to 90%, respectively, in affected tissues (34–38). Spinal cords from G93A SOD1 mice overexpressing CCS show almost a 60% reduction in both COX activity and complex levels. However, these measurements likely underestimate the loss of COX in neuronal subpopulations that manifest severe mitochondrial pathology, such as motor neurons, since both COX assays and BN-PAGE analyses in CCS/G93A SOD1 spinal cord were done in whole tissue extracts, which include less affected neuronal populations, or glia (astrocytes, oligodendrocytes), which do not express the CCS transgene (13). Thus, COX deficiency occurs in the early stages of disease, in vulnerable tissues, and with sufficient severity to have a potential causative role in the dysfunction of mitochondria within motor neurons of G93A SOD1 mice overexpressing CCS.
Isolated COX deficiency represents a selective impairment of mitochondrial respiration that has been associated with only a limited number of molecular etiologies in mammalian cells and, therefore, implicates specific pathways by which mutant SOD1 may cause complex IV defects. Mutations in the mitochondrial genes encoding COX1, COX2, or COX3 subunits have been shown to cause isolated complex IV deficiency in human beings, but such an etiology is not likely in our transgenic CCS and G93A SOD1 mouse models (35, 36, 39). Although an isolated complex IV deficiency has not been associated with abnormalities in any of its nuclear-encoded structural subunits, it has been found with defects in certain nuclear-encoded accessory proteins. Defects in Surf1, a nuclear-encoded protein critical for COX assembly, have been found to cause complex IV insufficiency in both humans and mice (40–42). Similarly, an isolated COX deficiency is also caused by defects in factors that either deliver copper to COX (COX17, SCO1, SCO2) or participate in heme A biosynthesis (COX10, COX15) (38, 43–49). Mutant SOD1 may, therefore, produce a specific reduction in complex IV levels via disruption of any of these pathways or perhaps through novel interactions with specific COX translational activators, other COX assembly factors, or individual COX structural subunits. Because low heme A levels can be observed in COX deficiencies that are unrelated to defects in heme A biosynthesis, our measures of low heme A content in the CCS/G93A SOd1 spinal cord do not necessarily implicate COX10 or COX15 dysfunction (28). Moreover, affected tissues from CCS/G93A SOD1 mice have a pool of unassembled COX1 subunits detected with BN/SDS-PAGE gels that are absent in either humans or mice with genetic defects in COX10 or COX15 (47, 48, 50). Although the precise molecular pathways responsible for the observed complex IV deficiency remain unclear, understanding the mechanistic basis for mutant SOD1-induced mitochondrial dysfunction may allow for the development of effective treatment options.
Abnormalities in mitochondrial respiratory function have also been reported in other neurodegenerative diseases (51). In Parkinson disease, selective brain regions show reductions in complex I activity and subunit levels (52). Preferential decrease of complex II and III activities coupled with loss of complex II subunits has been described in Huntington disease (53, 54). In patients with Friederich ataxia, complex I, II, and III functions are compromised due to an impaired ability to synthesize iron-sulfur clusters essential for their activity (55, 56). In the brain of Alzheimer disease patients, the most consistent abnormality in mitochondrial respiratory function has been associated with reduced complex IV activity (57). For SOD1-related ALS in the context of CCS overexpression, we observe an isolated complex IV deficiency. These findings suggest that each of these particular neurodegenerative disorders has its characteristic pattern of respiratory chain deficits.
Supplementary Material
This work was supported by grants from the Muscular Dystrophy Association (to J. L. E., S. C. L., and R. G. H.), the Horace C. Cabe Foundation (to J. L. E.), the Judith and Jean Pape Adams Charitable Foundation (to J. L. E.), Veterans Affairs Merit Review (to R. G. H.), and National Institutes of Health Grants NS055315 (to J. L. E.) and GM083292 (to D. R. W.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
The abbreviations used are: SOD1, Cu,Zn-superoxide dismutase; COX, cytochrome c oxidase; CCS, copper chaperone for SOD1; OXPHOS, oxidative phosphorylation; BN, blue native; WT, wild type; CS citrate synthase; NTG, non-transgenic; HPLC, high performance liquid chromatography; ALS, amyotrophic lateral sclerosis; DCPIP, dichlorophenol-indophenol; ANOVA, analysis of variance; CNS, central nervous system.
References
- 1.Rosen, D. R., Siddique, T., Patterson, D., Figlewicz, D. A., Sapp, P., Hentati, A., Donaldson, D., Goto, J., O'Regan, J. P., and Deng, H. X. (1993) Nature 362 59-62 [DOI] [PubMed] [Google Scholar]
- 2.Gurney, M. E., Pu, H., Chiu, A. Y., Dal Canto, M. C., Polchow, C. Y., Alexander, D. D., Caliendo, J., Hentati, A., Kwon, Y. W., and Deng, H. X. (1994) Science 264 1772-1775 [DOI] [PubMed] [Google Scholar]
- 3.Wong, P. C., Pardo, C. A., Borchelt, D. R., Lee, M. K., Copeland, N. G., Jenkins, N. A., Sisodia, S. S., Cleveland, D. W., and Price, D. L. (1995) Neuron 14 1105-1116 [DOI] [PubMed] [Google Scholar]
- 4.Reaume, A. G., Elliott, J. L., Hoffman, E. K., Kowall, N. W., Ferrante, R. J., Siwek, D. F., Wilcox, H. M., Flood, D. G., Beal, M. F., Brown, R. H., Jr., Scott, R. W., and Snider, W. D. (1996) Nat. Genet. 13 43-47 [DOI] [PubMed] [Google Scholar]
- 5.Manfredi, G., and Xu, Z. (2005) Mitochondrion 5 77-87 [DOI] [PubMed] [Google Scholar]
- 6.Sasaki, S., Warita, H., Murakami, T., Abe, K., and Iwata, M. (2004) Acta Neuropathol (Berl.) 107 461-474 [DOI] [PubMed] [Google Scholar]
- 7.Okado-Matsumoto, A., and Fridovich, I. (2001) J. Biol. Chem. 276 38388-38393 [DOI] [PubMed] [Google Scholar]
- 8.Higgins, C. M., Jung, C., Ding, H., and Xu, Z. (2002) J. Neurosci. 22 1-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu, J., Lillo, C., Jonsson, P. A., Vande Velde, C., Ward, C. M., Miller, T. M., Subramaniam, J. R., Rothstein, J. D., Marklund, S., Andersen, P. M., Brannstrom, T., Gredal, O., Wong, P. C., Williams, D. S., and Cleveland, D. W. (2004) Neuron 43 5-17 [DOI] [PubMed] [Google Scholar]
- 10.Vijayvergiya, C., Beal, M. F., Buck, J., and Manfredi, G. (2005) J. Neurosci. 25 2463-2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takeuchi, H., Kobayashi, Y., Ishigaki, S., Doyu, M., and Sobue, G. (2002) J. Biol. Chem. 277 50966-50972 [DOI] [PubMed] [Google Scholar]
- 12.Deng, H. X., Shi, Y., Furukawa, Y., Zhai, H., Fu, R., Liu, E., Gorrie, G. H., Khan, M. S., Hung, W. Y., Bigio, E. H., Lukas, T., Dal Canto, M. C., O'Halloran, T. V., and Siddique, T. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 7142-7147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Son, M., Puttaparthi, K., Kawamata, H., Rajendran, B., Boyer, P. J., Manfredi, G., and Elliott, J. L. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 6072-6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Browne, S. E., Bowling, A. C., Baik, M. J., Gurney, M., Brown, R. H., Jr., and Beal, M. F. (1998) J. Neurochem. 71 281-287 [DOI] [PubMed] [Google Scholar]
- 15.Mattiazzi, M., D'Aurelio, M., Gajewski, C. D., Martushova, K., Kiaei, M., Beal, M. F., and Manfredi, G. (2002) J. Biol. Chem. 277 29626-29633 [DOI] [PubMed] [Google Scholar]
- 16.Kirkinezos, I. G., Bacman, S. R., Hernandez, D., Oca-Cossio, J., Arias, L. J., Perez-Pinzon, M. A., Bradley, W. G., and Moraes, C. T. (2005) J. Neurosci. 25 164-172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin, L. J., Liu, Z., Chen, K., Price, A. C., Pan, Y., Swaby, J. A., and Golden, W. C. (2007) J. Comp. Neurol. 500 20-46 [DOI] [PubMed] [Google Scholar]
- 18.Puttaparthi, K., Gitomer, W. L., Krishnan, U., Son, M., Rajendran, B., and Elliott, J. L. (2002) J. Neurosci. 22 8790-8796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bertamini, M., Marzani, B., Guarneri, R., Guarneri, P., Bigini, P., Mennini, T., and Curti, D. (2002) Eur J. Neurosci. 16 2291-2296 [DOI] [PubMed] [Google Scholar]
- 20.Taivassalo, T., Shoubridge, E. A., Chen, J., Kennaway, N. G., DiMauro, S., Arnold, D. L., and Haller, R. G. (2001) Ann. Neurol. 50 133-141 [DOI] [PubMed] [Google Scholar]
- 21.Klement, P., Nijtmans, L. G., Van den Bogert, C., and Houstek, J. (1995) Anal. Biochem. 231 218-224 [DOI] [PubMed] [Google Scholar]
- 22.Schagger, H. (1995) Methods Enzymol. 260 190-202 [DOI] [PubMed] [Google Scholar]
- 23.Leary, S. C., Kaufman, B. A., Pellecchia, G., Guercin, G. H., Mattman, A., Jaksch, M., and Shoubridge, E. A. (2004) Hum. Mol. Genet. 13 1839-1848 [DOI] [PubMed] [Google Scholar]
- 24.Son, M., Cloyd, C. D., Rothstein, J. D., Rajendran, B., and Elliott, J. L. (2003) J. Biol. Chem. 278 14331-14336 [DOI] [PubMed] [Google Scholar]
- 25.Barros, M. H., Carlson, C. G., Glerum, D. M., and Tzagoloff, A. (2001) FEBS Lett. 492 133-138 [DOI] [PubMed] [Google Scholar]
- 26.Nijtmans, L. G., Taanman, J. W., Muijsers, A. O., Speijer, D., and Van den Bogert, C. (1998) Eur. J. Biochem. 254 389-394 [DOI] [PubMed] [Google Scholar]
- 27.Moraes, C. T., Diaz, F., and Barrientos, A. (2004) Biochim. Biophys. Acta 1659 153-159 [DOI] [PubMed] [Google Scholar]
- 28.Barros, M. H., and Tzagoloff, A. (2002) FEBS Lett. 516 119-123 [DOI] [PubMed] [Google Scholar]
- 29.Shoubridge, E. A. (2001) Am. J. Med. Genet. 106 46-52 [DOI] [PubMed] [Google Scholar]
- 30.Borisov, V. B. (2002) Mol. Aspects Med 23 385-412 [DOI] [PubMed] [Google Scholar]
- 31.Comi, G. P., Bordoni, A., Salani, S., Franceschina, L., Sciacco, M., Prelle, A., Fortunato, F., Zeviani, M., Napoli, L., Bresolin, N., Moggio, M., Ausenda, C. D., Taanman, J. W., and Scarlato, G. (1998) Ann. Neurol. 43 110-116 [DOI] [PubMed] [Google Scholar]
- 32.Rubio-Gozalbo, M. E., Smeitink, J. A., Ruitenbeek, W., Ter Laak, H., Mullaart, R. A., Schuelke, M., Mariman, E. C., Sengers, R. C., and Gabreels, F. J. (1999) Neurology 52 383-386 [DOI] [PubMed] [Google Scholar]
- 33.Wiedemann, F. R., Manfredi, G., Mawrin, C., Beal, M. F., and Schon, E. A. (2002) J. Neurochem. 80 616-625 [DOI] [PubMed] [Google Scholar]
- 34.Vesela, K., Hansikova, H., Tesarova, M., Martasek, P., Elleder, M., Houstek, J., and Zeman, J. (2004) Acta Paediatr. 93 1312-1317 [DOI] [PubMed] [Google Scholar]
- 35.Campos, Y., Garcia-Redondo, A., Fernandez-Moreno, M. A., Martinez-Pardo, M., Goda, G., Rubio, J. C., Martin, M. A., del Hoyo, P., Cabello, A., Bornstein, B., Garesse, R., and Arenas, J. (2001) Ann. Neurol. 50 409-413 [DOI] [PubMed] [Google Scholar]
- 36.Tiranti, V., Corona, P., Greco, M., Taanman, J. W., Carrara, F., Lamantea, E., Nijtmans, L., Uziel, G., and Zeviani, M. (2000) Hum. Mol. Genet. 9 2733-2742 [DOI] [PubMed] [Google Scholar]
- 37.Stiburek, L., Vesela, K., Hansikova, H., Pecina, P., Tesarova, M., Cerna, L., Houstek, J., and Zeman, J. (2005) Biochem. J. 392 625-632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Antonicka, H., Mattman, A., Carlson, C. G., Glerum, D. M., Hoffbuhr, K. C., Leary, S. C., Kennaway, N. G., and Shoubridge, E. A. (2003) Am. J. Hum. Genet. 72 101-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bruno, C., Martinuzzi, A., Tang, Y., Andreu, A. L., Pallotti, F., Bonilla, E., Shanske, S., Fu, J., Sue, C. M., Angelini, C., DiMauro, S., and Manfredi, G. (1999) Am. J. Hum. Genet. 65 611-620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tiranti, V., Hoertnagel, K., Carrozzo, R., Galimberti, C., Munaro, M., Granatiero, M., Zelante, L., Gasparini, P., Marzella, R., Rocchi, M., Bayona-Bafaluy, M. P., Enriquez, J. A., Uziel, G., Bertini, E., Dionisi-Vici, C., Franco, B., Meitinger, T., and Zeviani, M. (1998) Am. J. Hum. Genet. 63 1609-1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agostino, A., Invernizzi, F., Tiveron, C., Fagiolari, G., Prelle, A., Lamantea, E., Giavazzi, A., Battaglia, G., Tatangelo, L., Tiranti, V., and Zeviani, M. (2003) Hum. Mol. Genet. 12 399-413 [DOI] [PubMed] [Google Scholar]
- 42.Dell'agnello, C., Leo, S., Agostino, A., Szabadkai, G., Tiveron, C., Zulian, A., Prelle, A., Roubertoux, P., Rizzuto, R., and Zeviani, M. (2007) Hum. Mol. Genet. 16 431-444 [DOI] [PubMed] [Google Scholar]
- 43.Takahashi, Y., Kako, K., Kashiwabara, S., Takehara, A., Inada, Y., Arai, H., Nakada, K., Kodama, H., Hayashi, J., Baba, T., and Munekata, E. (2002) Mol. Cell. Biol. 22 7614-7621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valnot, I., Osmond, S., Gigarel, N., Mehaye, B., Amiel, J., Cormier-Daire, V., Munnich, A., Bonnefont, J. P., Rustin, P., and Rotig, A. (2000) Am. J. Hum. Genet. 67 1104-1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Papadopoulou, L. C., Sue, C. M., Davidson, M. M., Tanji, K., Nishino, I., Sadlock, J. E., Krishna, S., Walker, W., Selby, J., Glerum, D. M., Coster, R. V., Lyon, G., Scalais, E., Lebel, R., Kaplan, P., Shanske, S., De Vivo, D. C., Bonilla, E., Hirano, M., DiMauro, S., and Schon, E. A. (1999) Nat. Genet. 23 333-337 [DOI] [PubMed] [Google Scholar]
- 46.Jaksch, M., Ogilvie, I., Yao, J., Kortenhaus, G., Bresser, H. G., Gerbitz, K. D., and Shoubridge, E. A. (2000) Hum. Mol. Genet. 9 795-801 [DOI] [PubMed] [Google Scholar]
- 47.Antonicka, H., Leary, S. C., Guercin, G. H., Agar, J. N., Horvath, R., Kennaway, N. G., Harding, C. O., Jaksch, M., and Shoubridge, E. A. (2003) Hum. Mol. Genet. 12 2693-2702 [DOI] [PubMed] [Google Scholar]
- 48.Diaz, F., Thomas, C. K., Garcia, S., Hernandez, D., and Moraes, C. T. (2005) Hum. Mol. Genet. 14 2737-2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cobine, P. A., Pierrel, F., and Winge, D. R. (2006) Biochim. Biophys. Acta 1763 759-772 [DOI] [PubMed] [Google Scholar]
- 50.Bugiani, M., Tiranti, V., Farina, L., Uziel, G., and Zeviani, M. (2005) J. Med. Genet. 42 1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin, M. T., and Beal, M. F. (2006) Nature 443 787-795 [DOI] [PubMed] [Google Scholar]
- 52.Keeney, P. M., Xie, J., Capaldi, R. A., and Bennett, J. P., Jr. (2006) J. Neurosci. 26 5256-5264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tabrizi, S. J., Cleeter, M. W., Xuereb, J., Taanman, J. W., Cooper, J. M., and Schapira, A. H. (1999) Ann. Neurol. 45 25-32 [DOI] [PubMed] [Google Scholar]
- 54.Benchoua, A., Trioulier, Y., Zala, D., Gaillard, M. C., Lefort, N., Dufour, N., Saudou, F., Elalouf, J. M., Hirsch, E., Hantraye, P., Deglon, N., and Brouillet, E. (2006) Mol. Biol. Cell 17 1652-1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rotig, A., de Lonlay, P., Chretien, D., Foury, F., Koenig, M., Sidi, D., Munnich, A., and Rustin, P. (1997) Nat. Genet. 17 215-217 [DOI] [PubMed] [Google Scholar]
- 56.Thierbach, R., Schulz, T. J., Isken, F., Voigt, A., Mietzner, B., Drewes, G., von Kleist-Retzow, J. C., Wiesner, R. J., Magnuson, M. A., Puccio, H., Pfeiffer, A. F., Steinberg, P., and Ristow, M. (2005) Hum. Mol. Genet. 14 3857-3864 [DOI] [PubMed] [Google Scholar]
- 57.Bosetti, F., Brizzi, F., Barogi, S., Mancuso, M., Siciliano, G., Tendi, E. A., Murri, L., Rapoport, S. I., and Solaini, G. (2002) Neurobiol. Aging 23 371-376 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.