Abstract
Cyclin E overexpression is observed in multiple human tumors and linked to poor prognosis. We have previously shown that ectopic expression of cyclin E is sufficient to induce mitogen-independent cell cycle entry in a variety of tumor/immortal cell lines. Here we have investigated the rate-limiting step leading to cell cycle entry in quiescent normal human fibroblasts (NHF) ectopically expressing cyclin E. We found that in serum-starved NHF, cyclin E forms inactive complexes with CDK2 and fails to induce DNA synthesis. Coexpression of SV40 small t antigen (st), but not other tested oncogenes, efficiently induces mitogen-independent CDK2 phosphorylation on Thr-160, CDK2 activation, and DNA synthesis. Additionally, in contact-inhibited NHF ectopically expressing cyclin E, st induces cell cycle entry, continued proliferation, and foci formation. Coexpression of cyclin E and st also bypasses G0/G1 arrests induced by CDK inhibitors. Although CDK2 is dispensable for G0/G1 cell cycle entry and normal proliferation in mammals, CDK2 activity is an essential rate-limiting step in NHF with deregulated cyclin E expression and altered PP2A activity, which endows primary cells with transformed features. Consequently, CDK2 could be targeted therapeutically in tumors that involve these alterations. These data also suggest that alterations prior to cyclin E deregulation facilitate proliferation of tumor cells by bypassing mitogenic requirements and negative regulation by adjacent cells.
Cyclin E expression is finely regulated during the cell cycle, and its expression is dramatically reduced in quiescent cells. As cells enter G1 from G0 following mitogenic stimulation, D-type cyclin-CDKs3 partially inactivate pRB/p130, which activates expression of E2F-dependent genes, including the cyclin E gene. Cyclin E binds and activates CDK2 and phosphorylates several substrates, including members of the pRB family, which further facilitates cyclin E accumulation and CDK2 activation. Cyclin E-CDK2 also phosphorylates replication factors, centrosomal proteins, and NPAT/p220, a transcription factor that controls histone synthesis. Cyclin E is subsequently degraded by the proteasome, a process that is tightly regulated and involves two separate ubiquitin ligases (reviewed in Ref. 1). The activity of the cyclin E-CDK2 complex is positively regulated by CAK, which opens the activating T loop in CDK2 via phosphorylation of threonine 160, and negatively regulated by phosphorylation of threonine 14/tyrosine 15 and binding to CDK inhibitors (CKIs), p21 and p27, of the KIP family (reviewed in Ref. 2).
Cyclin E is overexpressed in many tumors as a result of gene amplification, disrupted proteolysis, and/or alterations in the pRB/E2F pathway. Cyclin E overexpression has been linked to poor prognosis in breast cancer, non-small cell lung carcinoma, larynx squamous cell carcinoma, and adrenocortical tumors (reviewed in Ref. 3). Two main mechanisms for cyclin E-associated tumorigenesis have been considered to date: induction of genomic instability and facilitation of cell cycle progression via deregulation of the pRB pathway and other G1/S events (reviewed in Ref. 3). Previous studies have shown that ectopic expression of cyclin E in mammalian diploid fibroblasts shortens the G1 phase of the cell cycle (4, 5). Cyclin E overexpression also decreased cell size and increased saturation cell density. However, cyclin E overexpression in quiescent cells was not sufficient to bypass mitogenic stimulation for passage trough the G0/G1 transition (4, 5). In contrast, we reported later, that ectopic expression of cyclin E in serum-starved human glioblastoma T98G cells was sufficient to bypass negative controls imposed by pRB family proteins and induce more than one cell division round in the absence of mitogens (6). We also found that cyclin E induced mitogen-independent cell cycle entry in other tumor cells.4 The different outcome to cyclin E deregulation in normal (4, 5) and tumor cells (6) suggests that certain tumor cells exhibit alterations that allow cyclin E to bypass mitogenic stimulation. If this is the case, cyclin E deregulation may facilitate multistep tumorigenesis by allowing tumor cells to proliferate in growth factor deprived environments. In turn, induction of DNA synthesis by deregulated cyclin E could promote cyclin E-dependent genomic instability ensuring further accumulation of transforming alterations.
Since others have shown that microinjection of active cyclin E-CDK2 complexes in normal human fibroblasts (NHF) induces DNA synthesis (7), we sought to determine what prevents cyclin E deregulation in quiescent NHF from inducing passage trough G1 and DNA synthesis. We have found that ectopically expressed cyclin E in serum-starved NHF effectively forms complexes with CDK2, but these complexes are inactive. Coexpression of simian virus 40 small t antigen (st) cooperates with cyclin E to activate CDK2 by triggering phosphorylation of Thr-160 on the T loop and induce mitogen-independent cell cycle entry. Importantly, st and cyclin E also cooperate to bypass distinct forms of quiescence induced by cell saturation density and overexpression of CKIs from the INK and KIP families. Additionally, expression of cyclin E and st induces continued proliferation of density-arrested cells and foci formation. This process requires CDK2 activity. Moreover, since st triggers alterations of cellular signaling also observed in human cancer cells (8, 9) and cyclin E is found deregulated in multiple cancers, our results provide a mechanism for oncogenic cooperativity that may play a role in human multistep tumorigenesis.
EXPERIMENTAL PROCEDURES
Cell Culture and Cell Cycle Synchronization—All cell lines were obtained from ATCC unless otherwise indicated. BJ-hTERT immortalized fibroblasts were a gift from William C. Hahn (Harvard Medical School Dana-Farber Cancer Institute). All cell lines were maintained in Dulbecco's modified Eagle's medium (Cellgro) supplemented with 10% fetal bovine serum (FBS) (Sigma) at 37 °C in a humidified atmosphere with 5% CO2. Exit into G0 by serum starvation was performed as previously described (6). Exit into G0 by density arrest was achieved by growing cells to high density until no dividing cells were visible in the culture and maintained for additional 48 h prior to any treatment. Roscovitine was used to pharmacologically inhibit CDK2 at a concentration of 25 μm as described in Ref. 6.
Recombinant Virus Production and Infection—Recombinant adenoviruses encoding cyclin D1 and cyclin E were provided by Jeffrey H. Albrecht (Hennepin County Medical Center). Adenoviruses encoding p16 were provided by Juan Fueyo (University of Texas MD Anderson Cancer Center). Adenoviruses encoding p21 were provided by Wafik El-Deiry (University of Pennsylvania). β-Galactosidase adenoviruses were from Clontech. Adenoviruses encoding enhanced green fluorescent protein were provided by Pilar Ruiz-Lozano (Burnham Institute for Medical Research). Adenoviruses encoding st were provided by Kathleen Rundell (Northwestern University). Adenoviruses encoding p27 were provided by Frank L. Graham (McMaster University). Adenoviruses encoding activated RasL61 and c-Myc were provided by Joseph R. Nevins (Duke University). Adenoviral stocks were amplified using 293 cells and purified by using CsCl density gradient centrifugation. Viral titers were determined with the Adeno-XTM rapid titer kit (BD Bioscience). T98G and Rat-1 cells were infected as previously described (6). NHF cell lines were infected with a multiplicity of infection (MOI) of 150 plaque-forming units/cell when synchronized by serum starvation or with an MOI of 40 when synchronized by growth to high density.
pBabe-puro-cyclin E was obtained from Bruce Clurman (Fred Hutchinson Cancer Research Center). pBabe-zeo-st, pBabe-zeo-stΔ110, and pBabe-zeo-stC97S/E102Q were provided by William C. Hahn. 10 μg of retroviral plasmids were cotransfected with 5 μg of pCL-Ampho packaging vector (Imgenex) into 293T cells following the calcium phosphate precipitation method (10). Retroviral particles were harvested at 24 and 48 h post-transfection and used to superinfect exponentially growing BJ-hTERT cells. When indicated, clones were selected in the presence of 1 μg/ml puromycin.
Western Blot Analysis and Immunoprecipitations—Whole cell lysates were obtained in buffer containing 50 mm Tris-HCl (pH 7.4), 5 mm EDTA, 250 mm NaCl, 50 mm NaF, 0.1% Triton X-100, 0.1 mm Na3VO4, 2 mm phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 4 μg/ml aprotinin, and 4 μg/ml pepstatin, as previously described (6). Immunoprecipitations were performed incubating 150–250 μg of protein. Whole cell lysates or immunocomplexes were resolved by SDS-PAGE and transferred to polyvinylidene difluoride membranes for Western blot analysis as previously described (6). The following antibodies were used for Western blot analysis: cyclin A (C19), cyclin D1 (A12), cyclin D2 (M20), cyclin D3 (C16), cyclin E (HE12), cyclin H (C18), CDK2 (M20), CDK4 (C22), CDK6 (C21), CDK7 (C19), p21 (C19), p27 (C19), p107 (C19), PP2A/Aβ (C20), Ras (C20), and c-Myc (9E10) antibodies were from Santa Cruz Biotechnology. p16 INK4 and PP2A/C antibodies were from BD. pRB phosphospecific and phospho-CDK2-Thr-160 antibodies were from Cell Signaling. st (419) monoclonal antibodies were a gift from Elizabeth Moran (Temple University). For immunoprecipitations, monoclonal antibodies to p21 (Cell Signaling), p27 (BD Bioscience), and st (Abcam) were used. Anti-phosphohistone-3 (Cell Signaling) and anti-bromodeoxyuridine (BrdUrd) (Caltag) were used for immunofluorescence assays and detected with donkey anti-mouse-rhodamine RedX or -fluorescein isothiocyanate and donkey anti-rabbit-fluorescein isothiocyanate secondary antibodies (Jackson Immunoresearch).
In Vitro Kinase Assays—CDK2 kinase activity was determined using CDK2 complexes immunopurified from 50 μg of whole cell lysate as previously described (6). CDK7 kinase activity was determined using CDK7 immunocomplexes from 150 μg of whole cell lysate and 150 ng of GST-CDK2-kinase-dead substrate (a generous gift from Philip Kaldis; Institute of Molecular and Cell Biology, Singapore). CDK2 phosphorylation on Thr-160 was detected by Western blot analysis using phosphospecific antibodies.
DNA content and DNA synthesis measurements—DNA content during the cell cycle was measured on a BD Calibur flow cytometer and quantified with Cell Quest software (BD Biosciences) as described earlier (6). DNA synthesis was determined by measuring BrdUrd incorporation via immunofluorescence. Cells were plated on 12-well clusters and serumstarved for 72 h. Once arrested, cells were transduced with recombinant adenoviruses or restimulated with serum, and 50 μm BrdUrd was added 10 h later and every 12 h for a continuous labeling. Cells were fixed in 1–3% formaldehyde/PBS for 15 min at room temperature and washed with PBS, and the DNA was denatured with 1.5 n HCl for 15 min at room temperature. Nuclear membranes were permeabilized with PBS containing 0.5% Triton X-100 and 1% bovine serum albumin for 20 min at room temperature. Cells were incubated with anti-BrdUrd antibody in PBS plus 1% bovine serum albumin for 1 h at room temperature. Secondary rhodamine-RedX-conjugated antibody was incubated in PBS, 1% bovine serum albumin, 0.5% Tween 20 for 1 h at room temperature. Total DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI). Cells were visualized under an inverted fluorescence microscope.
RESULTS
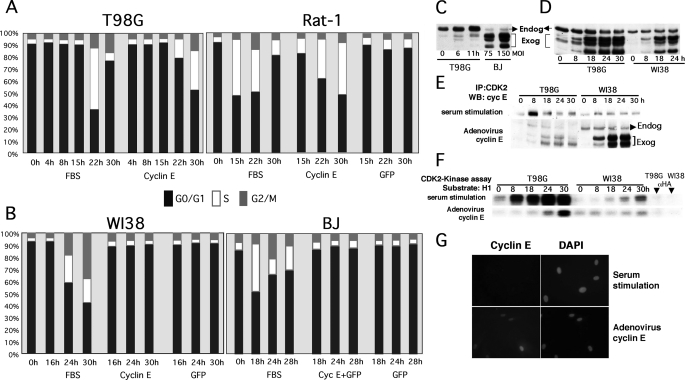
Although Cyclin E Deregulation Induces Mitogen-independent Cell Cycle Entry in Certain Tumor or Immortalized Cell Lines, It Fails to Do So in NHF—We have observed that ectopic expression of cyclin E is sufficient to induce mitogen-independent cell cycle entry in a variety of serum-starved human tumor-derived cell lines (T98G, HTB126, and MDA-MB435) and immortal nontransformed rat fibroblasts (Rat-1) (Fig. 1A) (6) (data not shown). To extend our observations to primary human cells, we compared the effects of ectopically expressing cyclin E in T98G cells, Rat-1 fibroblasts, and several primary NHF cell lines (WI38, BJ, and HLF). The cell lines indicated in Fig. 1 were serum-starved for 72 h in medium without serum, which led to accumulation of cells with a G0/G1 DNA content as determined via propidium iodide staining followed by flow-activated cell sorting (PI/FACS) analysis. As we reported earlier, T98G and Rat-1 cells entered the cell cycle following serum stimulation or transduction with cyclin E but not control adenoviruses (Fig. 1A). In contrast, although FBS stimulation induced cell cycle entry and DNA synthesis in WI38, BJ, and HLF cells (Fig. 1B) (data not shown), transduction with cyclin E and control adenoviruses failed to induce cell cycle entry. Importantly, ectopic expression of cyclin E in NHF at the MOI used throughout this study (MOI 40–150) was comparable with the levels of cyclin E expressed in serum-stimulated T98G cells (Fig. 1C).
FIGURE 1.
Ectopic expression of cyclin E induces mitogen-independent cell cycle entry in human tumor T98G cells and immortalized diploid rat fibroblasts (Rat-1) but not in NHF, where it fails to activate CDK2 despite proper complex formation and localization. T98G, Rat-1 (A), and human primary fibroblasts (WI38 and BJ) (B) were made quiescent by serum starvation (72 h). Serum-starved cells were transduced at an MOI of 50 (T98G and Rat-1) or 150 (primary fibroblasts) with the indicated recombinant adenoviruses or stimulated with FBS, harvested, and stained with propidium iodide for PI/FACS analysis at the indicated time points. The percentage of cells on each phase of the cell cycle is represented. C, T98G cells were serum-starved and restimulated with FBS and harvested at the indicated time points. Serum-starved BJ cells were transduced with cyclin E adenoviruses at the indicated MOI for 24 h. Note that ectopically expressed cyclin E is resolved as a set of bands with faster mobility than endogenous cyclin E, as previously described (see Ref. 55 and references therein). These forms, which are generated via protease cleavage, bind and activate CDK2 and form complexes with p21/p27. D, T98G and WI38 cells were serum-starved as described in A and transduced with cyclin E adenoviruses or restimulated with 10% FBS. Cells were harvested at the indicated time points. Cyclin E levels were determined by Western blot (WB) analysis. E and F, 250 μg of protein lysate were used to immunoprecipitate (IP) CDK2 complexes with specific antibodies. CDK2 complexes were resolved via SDS-PAGE and probed with cyclin E antibodies (E) or used for in vitro histone H1 kinase assays (F). G, BJ fibroblasts were seeded onto 12-well cluster plates, serum-starved for 72 h, and transduced with the cyclin E adenovirus or stimulated with FBS. After 24 h, cells were fixed, and cyclin E was detected via indirect immunofluorescence. Total DNA was visualized via 4′,6-diamidino-2-phenylindole (DAPI) staining. Cells were visualized under an inverted fluorescence microscope.
Possible reasons for the lack of effects of deregulated cyclin E expression in serum-starved NHF include that cyclin E does not form complexes with CDK2 or fails to activate CDK2. Thus, serum-starved T98G cells and WI38 NHF were transduced with cyclin E adenoviruses or stimulated with serum for the periods of time indicated in Fig. 1D. Whole cell lysates and CDK2 immunocomplexes were resolved by SDS-PAGE followed by Western blot analysis using anti-cyclin E antibodies (Fig. 1, D and E). Duplicate CDK2 immunoprecipitates were subjected to in vitro kinase assays using exogenous histone H1 as a substrate (Fig. 1F). Fig. 1D shows that exogenous cyclin E is efficiently expressed in both T98G cells and WI38 fibroblasts. Fig. 1E shows that cyclin E efficiently formed complexes with CDK2 in both T98G and WI38 cells. However, although cyclin E deregulation or FBS stimulation led to CDK2 activation in T98G cells, FBS, but not ectopic expression of cyclin E, led to CDK2 activation in WI38 fibroblasts (Fig. 1F). Thus, complexes are efficiently formed but are not active in NHF. To rule out the possibility that exogenous cyclin E is not expressed in the right cellular compartment in the absence of serum, we determined cyclin E localization. Serum-starved BJ fibroblasts were transduced with cyclin E adenoviruses or stimulated with serum. 24 h later, the cells were fixed and immunostained with antibodies directed to cyclin E. Fig. 1G shows clear nuclear labeling in serum-starved NHF transduced with adenoviral cyclin E. Under larger magnification, we also noted cyclin E staining localized as a dot within the cytoplasm of most cells, consistent with its localization to the centrosome (data not shown). Thus, lack of CDK2-associated activity is unlikely to be due to mislocalization of ectopically expressed cyclin E.
st, but Not Other Oncogenes, Efficiently Cooperates with Cyclin E to Induce Mitogen-independent Cell Cycle Entry in NHF—The observation that deregulated cyclin E bypasses mitogenic stimulation in various tumor cell lines but fails to do so in NHF indicates that G0-arrested NHF may require a serum-dependent rate-limiting event for CDK2 activation that cannot be simply bypassed by cyclin E deregulation. In contrast, cyclin E-responsive tumor/immortal cell lines have apparently lost this layer of regulation.
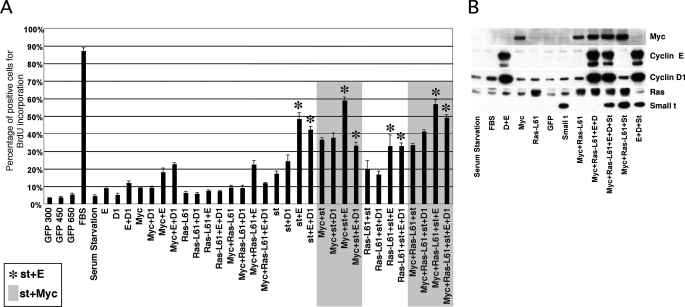
Oncogenes often cooperate to induce malignant transformation, and some of these oncogenes exhibit the ability to reduce cellular requirements for growth factors. Thus, we rationalized that one or more cellular/viral oncogenes could cooperate with cyclin E deregulation to bypass the mitogen requirement in NHF, perhaps targeting the same pathways disrupted in the tumor cell lines where cyclin E expression is sufficient to induce mitogen-independent cell cycle entry. Therefore, serum-starved BJ fibroblasts were restimulated with serum or coinfected with recombinant adenoviruses expressing cyclin E and/or c-Myc, oncogenic H-RasL61, cyclin D1, st, and control enhanced green fluorescent protein for 40 h (Fig. 2A). The proportion of cells undergoing DNA synthesis was determined by counting BrdUrd-positive nuclei following incorporation for 30 h prior to in situ cell fixation. BrdUrd incorporation was determined via indirect immunofluorescence using anti-BrdUrd-specific antibodies. As expected, serum stimulation led to labeling of ∼90% of nuclei. Fig. 2A also shows that cyclin E robustly cooperated with st and, to a lesser extent, with c-Myc, but not H-RasL61 or cyclin D1, to induce S phase entry. All of the oncogene combinations that included cyclin E and st resulted in ∼40–60% nuclei labeling (see bars marked with an asterisk). st also efficiently cooperated with c-Myc, which is consistent with the observation that st stabilizes c-Myc via PP2A inhibition (11). Since c-Myc is an upstream regulator of cyclin E (12), these results could suggest a linear st/c-Myc/cyclin E pathway. However, the results of Fig. 2A also show limited cooperation when cyclin E and c-Myc are co-expressed (only ∼20% of the nuclei were labeled), suggesting that st cooperates with cyclin E by mechanisms that are at least partially independent of c-Myc. Also, ectopic expression of st did not result in obvert up-regulation of endogenous c-Myc under these experimental conditions (Fig. 2B). Fig. 2B confirms that all oncogenes were expressed in a representative set of duplicate samples. Considering these data, st may disrupt a checkpoint pathway induced by deregulated cyclin E that prevented CDK2 activation or CDK2-independent cyclin E functions in NHF. Alternatively, st may trigger a mitogenically regulated event that is required for CDK2 activation or CDK2-independent cyclin E functions.
FIGURE 2.
st, but not other oncogenes, cooperates with cyclin E or c-Myc to induce cell cycle entry in the absence of mitogens. BJ fibroblasts were arrested in G0 as described in Fig. 1 and restimulated with FBS or transduced with the indicated recombinant adenoviruses expressing cyclin E (E), cyclin D1 (D1), Ras (Ras-L61), c-Myc (Myc), st, and/or green fluorescent protein (GFP) for 40 h. A, 50 μm BrdUrd was added to the culture plates every 12 h after transduction. BrdUrd incorporation (DNA synthesis) was determined via indirect immunofluorescence (see “Experimental Procedures”). Total DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI). One thousand nuclei were counted using Image-J software (National Institutes of Health, Bethesda, MD). The percentage of cells positive for BrdUrd incorporation is represented. Highlighted or labeled with asterisks are cells transduced with combinations of adenovirus that contain st and c-Myc or contain st and cyclin E, respectively. B, representative duplicate samples were analyzed by Western blot to monitor transgene expression.
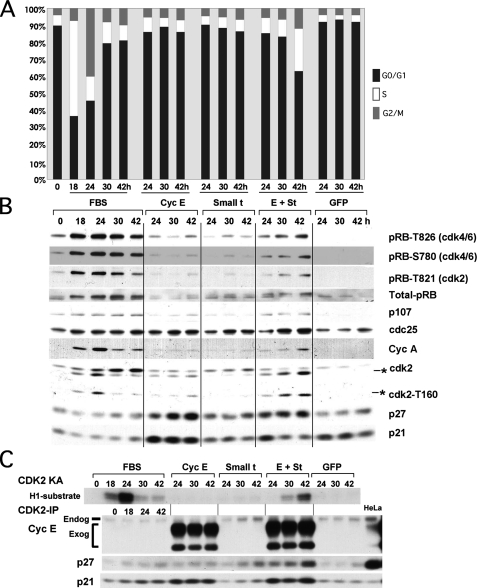
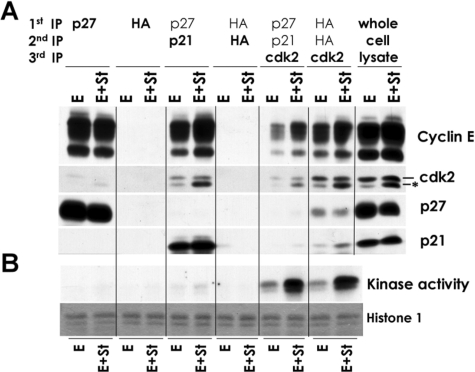
We next performed a time course experiment to determine the effects of coexpression of cyclin E and st in cell cycle entry and changes in the expression and phosphorylation of cell cycle markers. The PI/FACS analysis in Fig. 3A shows that coexpression of cyclin E and st, but not their expression alone, induces cell cycle entry and S phase progression in BJ fibroblasts, confirming the results of the BrdUrd incorporation assay (Fig. 2A). Similar results were obtained using serum-starved WI38 cells (supplemental Fig. 1). The results of Fig. 3B show that coexpression of cyclin E and st in BJ fibroblasts in the absence of mitogens, but not their independent expression, leads to pRB phosphorylation on both CDK4 (Thr-826) and CDK2 (Thr-821) preferred residues and expression of E2F-dependent gene products (p107 and cyclin A). Interestingly, examination of CDK2 via Western blot analysis revealed that coexpression of cyclin E and st induced dramatic accumulation of a faster migrating isoform (Fig. 3B), which corresponds to CDK2 phosphorylated on residue Thr-160 (13). This result was confirmed with anti-Thr-160 phosphospecific antibodies (Fig. 3B). Phosphorylation of CDK2 on Thr-160 and equivalent conserved sites in other CDKs induces a conformational change in the activating T loop of CDKs that facilitates substrate binding and stabilizes the interaction between the cyclin and CDK (reviewed in Ref. 14).
FIGURE 3.
st and cyclin E cooperate to induce phosphorylation of CDK2 on Thr-160, CDK2 activation, pocket protein hyperphosphorylation, E2F-dependent gene expression, and mitogen-independent cell cycle entry without altering the pool of cyclin E-CDK2 complexes bound to p21/p27. BJ fibroblasts were serum-starved as described in the legend to Fig. 1 and transduced with 150 MOI of the indicated adenoviruses or restimulated with FBS. Cells were harvested at the indicated time points and prepared for PI/FACS (A) or Western blot analyses (B). The Thr-160-phosphorylated form of CDK2 is marked with an asterisk. C, protein lysates from the same samples used in B were immunoprecipitated with CDK2 antibodies to determine the composition of CDK2 complexes and measure CDK2-associated kinase activity (KA).
Subsequently, we performed CDK2 immunokinase assays that demonstrate that coexpression of cyclin E and st cooperate to activate CDK2 (Fig. 3C), and this activation correlates with Thr-160 phosphorylation and cell cycle entry (Fig. 3, A and B).
CDK activation requires cyclin binding, phosphorylation by CAK, dephosphorylation by CDC25, and an absence of interactions with CKIs (reviewed in Ref. 2). Of these mechanisms, CAK is unique, since its activity is thought to be unregulated during the cell cycle. Thus, we determined whether ectopic expression of cyclin E had any effect on the expression of CKIs and if these effects were modulated by st. Western blot analysis showed that ectopic expression of cyclin E leads to accumulation of both p21 and p27 and that coexpression of st noticeably reduced the steady state levels of these two inhibitors (Fig. 3B and data not shown). However, examination of CKI association with CDK2 complexes via immunoprecipitation with CDK2 antibodies followed by Western blot analysis showed that st does not induce broad disruption of p27/p21 interactions with CDK2. On the contrary, expression of st leads to an increase in the presence of p27 in CDK2 complexes up to 42 h following transduction with the indicated adenoviruses (Fig. 3C). These data strongly suggest that the effects of st on p21/p27 expression are unrelated to CDK2 activation. To further rule out a role for p27 down-regulation in st/cyclin E cooperation in inducing cell cycle entry, we coexpressed cyclin E and SKP2 in quiescent NHF. SKP2 is an E3 ubiquitin ligase involved in targeting p27 and p130 for proteasomal degradation (15–19) and may also target p21 (20). We have previously shown that SKP2 does not induce mitogen-independent cell cycle entry in either T98G cells or NHF, despite inducing slight p27 down-regulation (19, 21). In striking contrast with st, coexpression of SKP2 with cyclin E failed to induce cell cycle entry in NHF (data not shown). Additionally, ectopic expression of cyclin E in both wild type and p21–/– murine embryo fibroblasts (MEFs) failed to induce cell cycle entry (data not shown). Taken together, these results suggest that p27/p21 inactivation is unlikely to be the rate-limiting step triggered by st to induce mitogen-independent cell cycle entry in NHF with deregulated cyclin E.
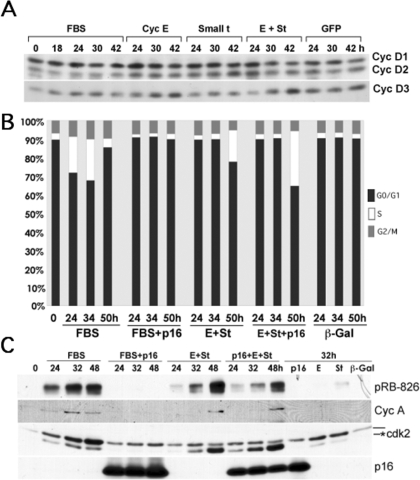
Our results also show that CDC25A expression is slightly up-regulated by serum and, to a similar extent, by ectopic expression of cyclin E, st, or both (Fig. 3B). However, coexpression of CDC25A with cyclin E did not induce cell cycle entry (data not shown). Also, an increase in CDK2-Y15 phosphorylation, rather than a decrease, was induced by coexpression of cyclin E and st (data not shown). Thus, CDC25A does not seem to be rate-limiting in cells expressing cyclin E, and CDC25A expression cannot substitute for st. On the other hand, Fig. 3B shows that pRB is phosphorylated at both D-type cyclin-CDK4(6) and cyclin E-CDK2-specific residues. Since we have previously shown that G1 cyclin-CDKs apparently maintain substrate specificity when expressed under these conditions (6, 22), these results suggest that CDK4 activity is up-regulated. However, although others have shown that st induces cyclin D1 expression (23), we observed slight down-regulation of both cyclin D1 and D2 following coexpression of cyclin E and st in BJ fibroblasts (Fig. 4A). Interestingly, cyclin D3 was clearly up-regulated. Thus, pRB phosphorylation at CDK4 preferred sites could possibly be due to an increase in cyclin D3-CDK4 activity or, alternatively, activation of D-type cyclin-CDK4 complexes (i.e. by CAK). On the other hand, we have recently shown that PP2A regulates the phosphorylation state of pocket proteins via a dynamic equilibrium with CDKs throughout the cell cycle (24). Thus, the increase in pRB phosphorylation could also be due to inhibition of PP2A activity by st. To determine whether pocket protein phosphorylation by D-type cyclin-CDKs is required for cyclin E and st to induce cell cycle entry in serum-starved NHF, we coexpressed cyclin E and st in the presence of the CDK4/6 inhibitor p16 and determined cell cycle progression and DNA synthesis by flow cytometric and BrdUrd incorporation analyses as well as determining the expression and phosphorylation of relevant proteins. Fig. 4B shows that p16 prevents cell cycle entry induced by FBS but not by cyclin E and st coexpression. In agreement with these results, FBS-stimulated cells expressing p16 do not incorporate BrdUrd, but cells expressing cyclin E/st/p16 do (data not shown). Also, Fig. 4C shows that p16 expression prevents FBS-induced CDK2 activation (Thr-160 phosphorylation), pRB phosphorylation, and cyclin A expression. In contrast, p16 expression does not block cyclin E/st-induced CDK2 activation and cyclin A expression, although it clearly diminishes pRB phosphorylation on Thr-826. Cumulatively, these data demonstrate that cyclin E and st signaling converge downstream of D-type cyclin-CDK activity and that D-type cyclin-CDK activity is not required. These results also suggest that CDK2 phosphorylates pRB on “CDK4-preferred” sites. This is not completely unexpected, since “CDK4-preferred” sites are phosphorylated in MEFs lacking expression of the three D-type cyclins, albeit at a significantly lower level (25).
FIGURE 4.
D-type cyclin-CDK activity is not required for st and cyclin E cooperation to induce CDK2 activation, pocket protein hyperphosphorylation, E2F-dependent gene expression, and mitogen-independent cell cycle entry. Levels of D-type cyclins were determined using the protein samples described in the legend to Fig. 3 via Western blot analysis (A). BJ fibroblasts were synchronized as described in the legend to Fig. 1 and transduced at an MOI of 150 with the indicated adenoviruses or restimulated with FBS. Cells were harvested at the indicated time points and processed for PI/FACS (B) or for Western blot (C) analyses. The Thr-160-phosphorylated form of CDK2 is marked with an asterisk.
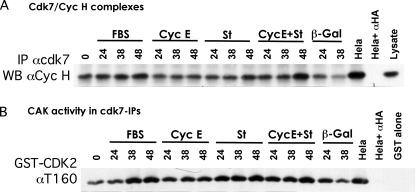
Our results show a correlation between CDK2 activation and its phosphorylation on residue Thr-160 in NHF coexpressing st and cyclin E (Fig. 3, B and C). We also observed that CDK2 Thr-160 phosphorylation correlates with pRB phosphorylation and precedes or occurs concomitantly to cyclin A accumulation, indicating that CDK2 activation may be a triggering event (Figs. 3B and 4C; also see Fig. 10B). The Thr-160 residue of CDK2 is phosphorylated by the CDK-activating enzyme designated CAK (26–32). Mammalian CAK is a trimeric CDK7-cyclin H-Mat1 complex, which activates CDKs, but also plays a role in transcriptional initiation by phosphorylating the C-terminal domain of RNA polymerase II (33, 34). Thus, we measured CDK7 assembly with cyclin H and CDK7-associated kinase activity in serum-starved hTERT-immortalized BJ (BJ-hTERT) fibroblasts stimulated with serum or transduced with the indicated adenoviruses. BJ-hTERT fibroblasts were used, since we have determined that these cells behave similarly to their primary parental cell line but are easier to grow because they do not become senescent (data not shown). CDK7 was immunoprecipitated, and cyclin H association was determined by Western blot analysis. Notably, expression of st appeared to up-regulate the cyclin H-CDK7 complex (Fig. 5A). This up-regulation was more prominent when cyclin E and st were coexpressed. CDK7-associated kinase activity was determined via CDK7 immunoprecipitation followed by in vitro kinase reactions using kinase-dead GST-CDK2 as an exogenous substrate. Fig. 5B shows that CDK7 activity is up-regulated following serum restimulation, and this correlates with formation of cyclin H-CDK7 complexes (Fig. 5A). We also detected a reproducible increase in CDK7-associated activity upon st expression to levels similar to those induced by FBS (Fig. 5B). Also, these levels of CDK7-associated kinase activity are comparable with those detected in exponentially growing HeLa cells (Fig. 5B). Similar results were obtained using WI38 fibroblasts (supplemental Fig. 2). Thus, a scenario arises where cyclin E expression and association with CDK2 could lead to partial opening of the activating T loop of CDK2, facilitating minor Thr-160 phosphorylation that is detected in NHF ectopically expressing cyclin E (Fig. 3B). Independently, expression of st up-regulates CAK activity by increasing CAK levels (Fig. 5), mediating Thr-160 phosphorylation and activation of cyclin E-CDK2 complexes. However, this increase in CDK7 activity in cells coexpressing cyclin E and st may not be the only mechanism responsible for the increase in Thr-160 phosphorylation and subsequent CDK2 activation (see below).
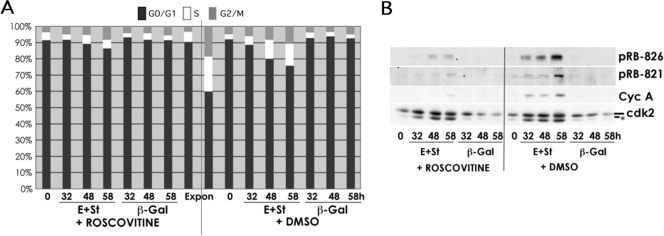
FIGURE 10.
CDK2 activity is required for the cooperating effects of cyclin E and st in density-arrested NHF. BJ-hTERT fibroblasts were grown to high density and transduced with the indicated adenoviruses. Four hours later, roscovitine (25 μm) or vehicle (Me2SO (DMSO)) was added to the medium. Cells were collected at the indicated time points and processed for PI/FACS (A) or Western blot (B) analyses.
FIGURE 5.
st and cyclin E cooperate to stabilize the cyclin H-CDK7 complex. A, formation of cyclin H-CDK7 complexes was analyzed via immunoprecipitation (IP) of lysates of BJ-hTERT cells transduced with the indicated adenoviruses or stimulated with serum followed by Western blot analysis (WB) with the indicated antibodies. B, CAK-associated kinase activity was measured in CDK7-immunoprecipitates from protein lysates obtained from the indicated treated cells. Kinase-dead GST-CDK2 was used as exogenous substrate. CAK activity was detected via Western blot analysis with a phosphospecific antibody to CDK2 phosphorylated on Thr-160.
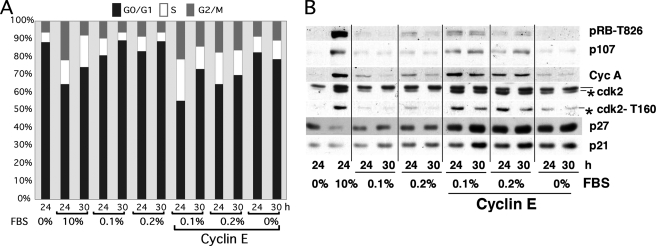
We rationalized that st is facilitating Thr-160 phosphorylation by bypassing a serum-dependent rate-limiting event for CDK2 activation. Thus, we determined whether low concentrations of serum could be sufficient to trigger this event in BJ fibroblasts with deregulated cyclin E expression. We found that concentrations as low as 0.5% FBS could trigger BrdUrd incorporation in serum-starved BJ fibroblasts (data not shown). Next, serum-starved BJ fibroblasts were transduced with cyclin E adenoviruses and subsequently stimulated with 0.1 or 0.2% FBS or left untreated. The PI/FACS analysis depicted in Fig. 6A shows dramatic cooperation between cyclin E and low serum, which also induced mitosis as well as entry into the next G1 phase by 30 h. Importantly, cyclin E expression and low serum induced phosphorylation of CDK2 on Thr-160 without apparent down-regulation of p21/p27 (Fig. 6B). Of note, these low serum concentrations are often used to maintain cells in a quiescent stage as a source of survival factors. However, it is clear from these experiments that minimal concentrations of serum supply adequate mitogenic factors to trigger signaling that suffices to induce rate-limiting steps that cannot be overcome solely by expression of cyclin E. Altogether, these results suggest that low concentrations of growth factors can be substituted by st expression to induce Thr-160 phosphorylation, CDK2 activation, and cell cycle entry.
FIGURE 6.
Deregulated cyclin E cooperates with low concentrations of FBS to induce DNA synthesis and mitosis. BJ fibroblasts were synchronized by serum starvation and transduced with 150 MOI of adenoviruses expressing cyclin E. After 15 h, cells were restimulated with the indicated concentrations of FBS and harvested at the indicated time points for PI/FACS (A) or Western blot (B) analyses. The Thr-160-phosphorylated form of CDK2 is marked with an asterisk.
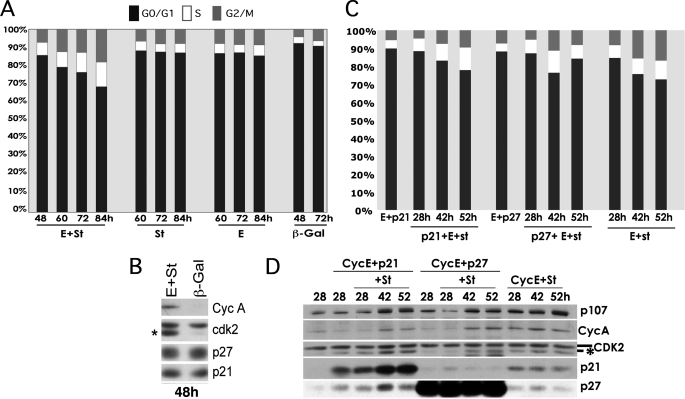
Coexpression of st and Cyclin E Also Forces Exit from Quiescence Induced by Density Arrest or CKI Overexpression—To determine whether the cooperative effects of st and cyclin E are restricted to bypass quiescence when this state is induced by serum starvation, we examined if st and cyclin E override quiescence induced by growth of cells to high density in the presence of serum, which is another physiologically relevant way to induce reversible exit into the G0 phase of the cell cycle. Thus, BJ-hTERT fibroblasts were grown 72 h past confluence and then infected with the indicated adenoviruses (Fig. 7A). As in serum-starved cells, coexpression of cyclin E and st but not their individual expression in density-arrested BJ-hTERT fibroblasts leads to cell cycle entry and progression through S phase as determined by PI/FACS analysis. Western blot analysis on whole cell lysates showed that expression of cyclin E and st, but not control β-galactosidase, led to CDK2 phosphorylation on Thr-160 without changes in the expression of p21/p27 (Fig. 7B). Considering that others have shown that the quiescent states induced by serum starvation and growth to high density are functionally distinct from the arrests induced by overexpression of p21 or p27 (35), we also sought to determine whether cyclin E and st can bypass a CKI-induced G0/G1 arrest. Exponentially growing BJ-hTERT fibroblasts were transduced with p21 or p27 adenoviruses and 8 h later with cyclin E adenoviruses. Forty hours later, the cells were arrested with a G0/G1 DNA content (Fig. 7C). These cells as well as control cells infected with cyclin E adenoviruses were transduced with st adenoviruses, and cell cycle entry was monitored by PI/FACS analysis. Fig. 7C shows that st also cooperates with cyclin E to bypass a p21- or p27-induced growth arrest, although a delay in cell cycle entry was observed. Of note, this bypass is accompanied by phosphorylation of CDK2 on Thr-160 (Fig. 7D). These data along with those shown in Figs. 3 and 4 demonstrate that st and cyclin E cooperate to bypass G0/G1 arrests, including those induced by serum starvation, growth to high density, and expression of CKIs from the INK and KIP families, and in all cases the activating T loop of CDK2 is phosphorylated.
FIGURE 7.
Coexpression of st and cyclin E forces exit from quiescence induced by density arrest or CKI overexpression. BJ-hTERT fibroblasts were grown to high density and then transduced with the indicated recombinant adenoviruses and harvested at the indicated time points for PI/FACS (A) and Western blot (B) analyses. Exponentially growing BJ-hTERT fibroblasts were transduced with recombinant adenoviruses expressing p21, p27, or control adenoviruses. Eight hours later, BJ-hTERT fibroblasts were transduced with adenoviruses expressing cyclin E. After 48 h, cells were transduced with adenoviruses expressing st. BJ-hTERT fibroblasts were harvested at the indicated time points following transduction with st adenoviruses and analyzed by PI/FACS (C) and by Western blot (D) analyses.
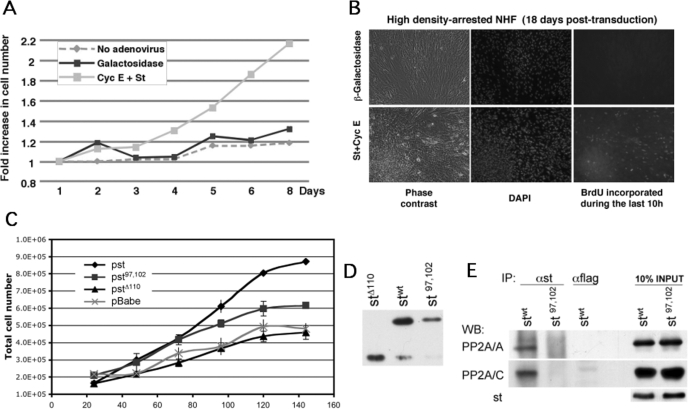
st and Cyclin E Cooperate to Induce Proliferation and Foci Formation—During the course of these experiments, we observed that although NHF ectopically expressing cyclin E and st enter and progress through the cell cycle in the absence of mitogens, they do so more slowly than cells growing in the presence of FBS. Thus, cyclin E-transduced cells are not growing optimally in the absence of serum. In contrast, density-arrested NHF expressing cyclin E and st appeared to efficiently complete normal cell cycles, as observed by the presence of mitotic figures, accumulation of cells in G2 in the presence of nocodazole, and the reaccumulation of cells in G1 after completing mitosis (data not shown). Normal cells as well as immortal/tumor cells that exhibit a normal restriction point exit the cell cycle and become quiescent when grown to high density in the presence of serum. Although the process is not fully understood, both deprivation of mitogens and inhibitory signals induced by surrounding cells are thought to be responsible for this physiological arrest. Importantly, transformed cells grow to very high densities and often can form foci. Therefore, we sought to determine whether cyclin E and st could overcome cell density-induced antiproliferative signals through multiple cycles and continue to proliferate. BJ-hTERT fibroblasts ectopically expressing cyclin E and st continued to grow past confluence for several days (Fig. 8A) and formed foci after 2–3 weeks (Fig. 8B). Of note, cyclin E/st-coexpressing cells were synthesizing DNA 18 days post-transduction. Thus, coexpression of st and cyclin E in density-arrested NHF endows these cells with transformed characteristics. However, these cells are not fully transformed, since they fail to grow in an anchorage-independent manner, even if they are immortalized by hTERT (data not shown).
FIGURE 8.
Coexpression of st and cyclin E in density-arrested cells endows NHF with unrestrained proliferation and foci formation properties, and PP2A inactivation is important for these effects. BJ-hTERT fibroblasts were grown to high density and transduced with indicated adenoviruses. A, cells were harvested and counted at the indicated time points. B, cyclin E and st induced foci formation and continued DNA synthesis 18 days post-transduction as observed by detection of BrdUrd incorporation and DNA staining with 4′,6-diamidino-2-phenylindole (DAPI). (Viral infection was repeated weekly to avoid loss of episomal exogenous DNA.) C and D, stable BJ-hTERT-Cyc E cells were transduced with the indicated retroviruses. C, 200,000 cells were seeded in 6-well plates and counted in triplicate every 24 h. Total cell number is shown. D, levels of retrovirally expressed st proteins were determined by Western blot analysis 3 days postinfection. E, wild type and mutant st proteins were immunoprecipitated from normalized protein extracts containing comparable levels of st or st97,102. Immunoprecipitations (IP) were resolved by SDS-PAGE. PP2A/A and PP2A/C were detected by Western blot analysis (WB).
st Binding to PP2A Is Important for Cooperation with Cyclin E to Induce Proliferation of Density-arrested Cells—Previous work has shown that st induces transformation of human embryonic kidney cells expressing LT, hTERT, and Ras-V12 (36). In this setting, st-transforming activity can be substituted by knockdown of PP2A/B56γ3, which is consistent with the ability of st to disrupt trimeric PP2A complexes. However, st can promote cell cycle progression in reduced serum, but B56γ3 knockdown cannot (37). To determine whether the effects of st and cyclin E coexpression on cell proliferation depend on PP2A, we expressed cyclin E alone or in combination with st or st mutants that have been previously described to exhibit reduced PP2A/A binding and transformation activity. st-Δ110 lacks the C-terminal domain required for PP2A/A binding, and full-length stC97S/E102Q lacks Cys-97, which reduces binding to PP2A/A very significantly (8, 38). We generated BJ-hTERT NHF stably expressing cyclin E (BJ-hTERT-Cyc E) via retroviral infection followed by antibiotic selection. These cells proliferate slightly faster than BJ-hTERT cells in the presence of serum without loss of proliferation potential, and as expected, BJ-hTERT-Cyc E cells become quiescent following serum withdrawal (data not shown). As shown in Fig. 8C, BJ-hTERT-Cyc E cells transduced with st-Δ110 or mock retroviruses did not grow efficiently past confluence and arrested at a similar cell density by day 5. In contrast, transduction of BJ-hTERT-Cyc E cells with wild type st retroviruses led to continued proliferation past cell confluence to form a dense thick layer of cells with multiple foci that prevented accurate cell counting past day 5, since these cells clamp following trypsinization. Surprisingly, BJ-hTERT-Cyc E cells transduced with stC97S/E102Q retroviruses grew to a higher density than the cells expressing cyclin E alone or in combination with st-Δ110. This suggests that although PP2A is important for the cooperating ating effects of cyclin E and st in inducing continued proliferation past confluence, other domains of st contribute to these effects. We note, however, that wild type st is consistently expressed at higher levels than the mutants that are defective in PP2A binding (Fig. 8D) (data not shown). To rule out the possibility that stC97S/E102Q effects are due to retention of some PP2A binding activity, we performed immunoprecipitations with anti-st antibodies using extracts normalized for st expression levels, which clearly show that stC97S/E102Q does not bind PP2A/A or PP2A/C (Fig. 8E). Of note, these experiments were carried out with retroviral vectors, demonstrating that our observations are independent of the viral vector utilized to express cyclin E and st.
st Promotes CAK Access to Cyclin E-CDK2 Complexes—Because expression of st in serum-starved NHF induces CAK expression and activity consistent with Thr-160 phosphorylation (Fig. 5) (supplemental Fig. 2), we determined whether CAK is also up-regulated by st in density-arrested NHF. However, we did not detect changes in CDK7 expression or activation (data not shown). Thus, the induction of Thr-160 phosphorylation by st must occur through a different mechanism at least in density-arrested cells. One possibility is that st facilitates CAK access to CDK2, which could be otherwise prevented by CKI binding. To determine the composition of the cyclin E-CDK2 complexes harboring CDK2 phosphorylation on Thr-160 and activity, we performed sequential immunoprecipitation/depletion experiments in lysates of serum-starved BJ-hTERT cells transduced with the indicated adenoviruses for 48 h as indicated in Fig. 9. p27 antibodies immunoprecipitated p27 and a fraction of cyclin E and CDK2 (Fig. 9B). p21 antibodies immunoprecipitated p21, a fraction of cyclin E, and a larger fraction of CDK2 (Fig. 9A). Importantly, despite the presence of CDK2 phosphorylated on Thr-160, CDK2 complexes containing p21 and p27 appeared to be inactive (Fig. 9A). Subsequent parallel immunoprecipitation of p21/p27 and mock-depleted lysates with anti-CDK2 antibodies followed by a histone H1 kinase assay confirmed that only cyclin E-CDK2 complexes free of p27/p21 are active (Fig. 9B). Similar results were obtained using density-arrested BJ-hTERT cells transduced with st and/or cyclin E adenoviruses (data not shown). Considering that p27/p21 and CAK compete for the same surfaces on CDK2 (39, 40) and because a large fraction of CDK2 is phosphorylated on Thr-160 in cells expressing cyclin E and st, these results suggest that st facilitates, at least, transient CAK access to CDK2. Despite this, the majority of CDK2 ends up bound to CKIs and inactive. Thus, only a relative small fraction of CDK2 complexes is in the active state even with potent induction of Thr-160 phosphorylation by st.
FIGURE 9.
Only CKI-free CDK2 complexes are active. BJ-hTERT fibroblasts were serum-starved for 72 h and transduced with the indicated adenoviruses for 48 h. Protein extracts were obtained, and serial immunoprecipitations (IP) were performed with indicated antibodies to CKIs or hemagglutinin (HA; antibody control). The composition of the complexes (A) and their associated kinase activity (B) following each set of immunoprecipitations were determined by Western blot analysis and histone H1 kinase assays, respectively.
CDK2 Activity Is Essential for st and Cyclin E to Induce Cell Cycle Progression—The results reported here demonstrate that st and cyclin E cooperate to activate CDK2 (Fig. 3C). However, previous studies have shown that CDK2 activity is not essential for cell cycle progression in MEFs and is dispensable for mouse viability (41, 42). However, more recent work has demonstrated that in the absence of CDK2, CDK1 associates with cyclin E and compensates for its absence (43). Therefore, to determine whether the effects of cyclin E and st on cell cycle progression require CDK2 activity, we used the CDK2-CDK1 pharmacological inhibitor roscovitine. BJ-hTERT cells were density-arrested and infected with the adenoviruses indicated in Fig. 10. Roscovitine was added 4 h following transduction to prevent any possible effects on the expression of transgenes. As expected, control exponentially growing BJ-hTERT cells arrested in G1 upon roscovitine treatment (Fig. 10A). This arrest in G1 is probably due to inhibition of both CDK2 and CDK1, since CDK1 can compensate for CDK2 loss (43). Roscovitine also blocked cell cycle progression induced by coexpression of st and cyclin E, since these cells maintained a G0/G1 DNA content (Fig. 10A). Analysis of whole cell lysates via Western blot (Fig. 10B) clearly showed that CDK2 activation is required for st/cyclin E effects on pRB phosphorylation and E2F-dependent gene expression (i.e. cyclin A), indicating that roscovitine effectively inhibited CDK2 activity. However, phosphorylation of CDK2 on Thr-160 was not altered by roscovitine. This demonstrates that induction of Thr-160 phosphorylation by cyclin E and st coexpression precedes the expression of cyclin A and is not an indirect consequence of increased cyclin A expression. This result also shows that at the concentration used in these assays, roscovitine did not result in nonspecific CDK inhibition, since CAK activity was not affected.
DISCUSSION
Cyclin E deregulation in certain tumor and immortal cell lines induces mitogen-independent cell cycle entry (6). As reported earlier (5), we confirm that deregulation of cyclin E in NHF fails to induce cell cycle entry in the absence of mitogens. We show here that exogenous cyclin E effectively forms complexes with CDK2, but these complexes are inactive in serum-starved NHF. We also show that st, an oncogene known to alter pathways essential for malignant transformation in human cells (8), cooperates with deregulated cyclin E to activate CDK2 in cyclin E complexes that would otherwise be inactive. In this situation, which mimics alterations seen in tumor cells, CDK2 activity is essential for bypassing contact inhibition, inducing continuous proliferation, and endowing NHF with transformed characteristics. Since others have shown that CDK2 ablation in mice is compatible with normal development and viability (41, 42), our results predict that tumors exhibiting deregulated cyclin E expression and alterations mimicked by st expression (9) would be sensitive to CDK2 inhibition by selective pharmacologic agents, which would be expected to lack toxicity.
st Triggers CDK2 Activation by Promoting Thr-160 Phosphorylation—Our data show that st induces CAK activity by increasing the levels of cyclin H-CDK7 complexes in serum-starved NHF. st-induced CAK activity is comparable with that induced via FBS stimulation. However, st expression in density-arrested NHF does not increase CDK7 activity. Structural studies have suggested that the binding of CAK and KIPs to CDK2 is mutually exclusive (39, 40). Although CAK may only phosphorylate Thr-160 on CDK2 bound to cyclin E free of KIPs, it is clear that the result of st expression is the accumulation of CDK2 phosphorylated on Thr-160 independently of whether these complexes are bound to KIPs. Thus, our data suggest that st facilitates CAK “access” to newly synthesized CDK2 prior to its binding to KIPs and/or promotes transient dynamic exchange between KIPs and CAK that facilitate Thr-160 phosphorylation despite maintaining the levels of KIPs bound to CDK2. It is important to underscore that st can induce CDK2 phosphorylation on Thr-160 even in immortalized NHF ectopically expressing cyclin E arrested by overexpression of p21/p27 (Fig. 7D). Under these conditions, the levels of p27/p21 far exceed the levels of endogenous KIPs in quiescent NHF even when they forcibly express cyclin E. Regardless, it is clear that st shields a small portion of cyclin E-CDK2 from p27/p21 binding, and these are the complexes that exhibit CDK2 activity.
Bypassing Quiescence States Induced by Multiple Signals—Our results show that cyclin E and st cooperate to bypass quiescence induced by serum starvation, contact inhibition, and overexpression of KIPs and INKs despite the fact that these quiescent states are distinct and dependent on the initiating signal (35). Indeed, it has been reported that one characteristic of the quiescent stage induced by serum starvation and growth to high density is a pattern of gene expression that enforces the reversibility of this stage, at least in part, by suppressing terminal differentiation. These expression signatures are not recapitulated when the quiescent stage is induced by overexpression of CKIs (35). Thus, cyclin E and st are potent negative regulators of the quiescent stage when coexpressed in normal human cells.
A previous study has shown that coexpression of c-Myc and oncogenic Ras in rat embryo fibroblasts (REF52 cell line) made quiescent in the presence of 0.25% serum results in cell cycle entry (44). This oncogene combination did not result in potent induction of DNA synthesis in NHF (Fig. 2). Our serum starvation experiments were performed in the complete absence of serum, which depletes survival factors that might be required under certain cellular instances. In the absence of survival factors, certain combinations of oncogenes may fail to induce cell cycle progression if they also trigger cell senescence and/or cell death. We did not observe overt cell death induced by combinations of oncogenes that failed to induce DNA synthesis at the time points under study. However, we cannot rule out these effects at longer time points. It is also conceivable that st plays a survival function in the absence of serum that other oncogenes cannot perform.
Cyclin E and st in Transformation and Tumorigenesis—Cyclin E is overexpressed and has been linked to poor prognosis in a variety of tumors (reviewed in Ref. 3). Two mechanisms for cyclin E-associated tumorigenesis have been considered to date: induction of genomic instability and facilitation of cell cycle progression via deregulation of the pRB pathway and/or other G1/S events (reviewed in Ref. 3). Previous studies have shown that ectopic expression of cyclin E in NHF shortens the G1 phase of the cell cycle but is not sufficient to bypass mitogenic stimulation (5). In contrast, ectopic expression of cyclin E in quiescent human glioblastoma T98G cells is sufficient to bypass negative controls imposed by pRB family proteins and induce more than one round of cell division (6). This difference could be explained by our observation that cyclin E expression is not sufficient to activate CDK2 in NHF but does so in T98G cells (Figs. 1F and 3C). This is consistent with the finding that microinjection of purified active cyclin E-CDK2 complexes induces DNA synthesis in NHF (7). Together, these results suggest that certain tumor cell lines exhibit signaling alterations that make cyclin E protein levels the rate-limiting event required for CDK2 activation, which in turn mediates cyclin E-induced proliferating effects. In contrast, in NHF, cyclin E overexpression requires at least limited mitogenic stimulation (Fig. 6) or coexpression of st (Figs. 2A, 3A, and 4B) to bypass a rate-limiting step for CDK2 activation and progression through S phase. It is remarkable that in density-arrested NHF, st and cyclin E induced continued proliferation and foci formation, demonstrating that oncogene-induced signaling alterations render cyclin E transgene oncogenic in human cells. In this regard, transgenic mice expressing cyclin E under the control of a T-cell-specific CD2 promoter treated with N-methylnitrosourea develop clonal lymphomas with variable latency and penetrance, which indicates that additional alterations are needed (45). Importantly, the lymphoma T cells expressed high levels of CDK2 activity, whereas normal T cells expressing cyclin E did not. Thus, as in NHF, CDK2 activation rather than cyclin E expression is coupled to cell transformation. Indeed, we find that CDK2 activity is essential for escaping quiescence and proliferation when driven by cyclin E/st coexpression (Fig. 10).
Our results may seem to contrast with recent work indicating that the requirement of cyclin E for oncogene-mediated transformation in MEFs is independent of CDK2 activation (46). However, this is not the case, because st is not required for transformation of MEFs. Therefore, the CDK2 requirement for transformation described here might be restricted to alterations in human cells mimicked by st expression and cyclin E deregulation. In fact, CDK2 knockdown or pharmacologic inhibition specifically inhibits human melanoma cell proliferation and colony formation, indicating that CDK2 activity is required in certain human cancers (47), despite being dispensable for proliferation of other human tumor cells (48). Moreover, in mouse Ras-dependent skin tumorigenesis models where CDK4 expression is driven by a tissue specific promoter, ablation of CDK2 results in decreased incidence and multiplicity of skin tumors (49). In contrast, CDK2 ablation does not affect the incidence of c-Myc-induced tumors.
Alternatively to the proposition that cyclin E expression is not the only rate-limiting factor for CDK2 activation in quiescent NHF, normal cells could have mechanisms to prevent cyclin E activation upon its unscheduled up-regulation. In this regard, others have shown that cyclin E expression in proliferating NHF leads to p53/p21 induction and eventually growth arrest (50). Since T98G cells have a disrupted p53 pathway, this could explain why quiescent T98G cells enter the cycle when cyclin E is overexpressed. Moreover, our results show that cyclin E expression in quiescent NHF induces p21 expression. However, st induces CDK2 activation without altering the overall level of KIPs associated with the cyclin E-CDK2 complex (Fig. 3C), suggesting that st is not simply bypassing a p53/p21 checkpoint.
In primary human cells immortalized by expression of hTERT, expression of st, LT, and oncogenic Ras is sufficient for malignant transformation, which is defined as anchorage-independent growth in vitro and tumorigenesis in nude mice (8, 51). In the absence of hTERT, morphological transformation occurs, but the transformed clones are not immortal (52). Of note, st is not required for transformation of MEFs, which are efficiently transformed by LT and activated Ras (8). Thus, the pathways disrupted by st are critical for transformation of human cells, unveiling a key difference in tumorigenesis between rodent and human cells. In our study, cyclin E and st induced continued proliferation and foci formation in high density arrested cells. However, these two oncogenes were not sufficient to induce anchorage-independent growth even in hTERT-immortalized NHF (data not shown), indicating that cyclin E and st are not sufficient for full malignant transformation in vitro. This is consistent with the observation that malignant transformation of human cells requires hTERT, oncogenic Ras, st expression, and inactivation of the p53 and pRB pathways (51). Considering that activated Ras cooperates with cyclin E in the transformation of rodent fibroblasts (53) and that Ras-CDK4-dependent tumorigenesis in the mouse skin is susceptible to CDK2 ablation (49), it is possible that full malignant transformation by coexpression of cyclin E and st in NHF requires alterations in Ras signaling. In regard to the selectivity between cooperating oncogenes, it is also important to note that st did not cooperate with cyclin D1 to induce mitogenic independent cell cycle entry (Fig. 2) despite previous observations demonstrating that cyclin D1 deregulation, like cyclin E, induces mitogenic independent growth in certain tumor cell types (6). Moreover, although c-Myc is upstream of cyclin E and cooperates with p27 loss to increase cyclin E-CDK2 activity in murine lymphomas (54), it did not cooperate as efficiently as st with cyclin E in inducing DNA synthesis in NHF (Fig. 2). This suggests that the mechanism by which st activates CDK2 is independent of the well characterized effects of st on c-Myc stabilization (11).
Taken together, these results strongly suggest that CDK2 plays an important role in tumorigenesis in certain human cells characterized by cyclin E deregulation and alterations in pathways targeted by st. Considering that CDK2 activity appears to be redundant in normal cells, CDK2 emerges as a potential therapeutic target for a defined set of tumors.
Acknowledgments
We thank P. Kaldis, J. Albrecht, E. Moran, K. Rundell, J. Fueyo, W. El-Deiry, P. Ruiz, F. Graham, J. Nevins, B. Clurman, W. Hahn, C. Carbone, and D. Haines for reagents used in this study and M. Truongcao for technical assistance.
This work was supported in part by grant projects under National Institutes of Health (NIH) Grant CA095569 (to X. G. and J. G.) and NIH Career Development Award K02 AI01823 (to X. G.). Facilities used for this work were supported in part by NIH Shared Resources for Cancer Research Grant R24 CA88261-01. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: CDK, cyclin-dependent kinase; CKI, CDK inhibitor; NHF, normal human fibroblast(s); st, small t antigen; FBS, fetal bovine serum; MOI, multiplicity of infection; PBS, phosphate-buffered saline; PI, propidium iodide; FACS, flow-activated cell sorting; BrdUrd, bromodeoxyuridine; MEF, mouse embryo fibroblast; Cyc E, cyclin E.
E. Sotillo, J. Garriga, A. Kurimchak, and X. Graña, unpublished observations.
References
- 1.Malumbres, M., and Barbacid, M. (2005) Trends Biochem. Sci. 30 630–641 [DOI] [PubMed] [Google Scholar]
- 2.Morgan, D. O. (1995) Nature 374 131–134 [DOI] [PubMed] [Google Scholar]
- 3.Hwang, H. C., and Clurman, B. E. (2005) Oncogene 24 2776–2786 [DOI] [PubMed] [Google Scholar]
- 4.Resnitzky, D., Gossen, M., Bujard, H., and Reed, S. I. (1994) Mol. Cell. Biol. 14 1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohtsubo, M., and Roberts, J. M. (1993) Science 259 1908–1912 [DOI] [PubMed] [Google Scholar]
- 6.Calbó, J., Parreño, M., Sotillo, E., Yong, T., Mazo, A., Garriga, J., and Graña, X. (2002) J. Biol. Chem. 277 50263–50274 [DOI] [PubMed] [Google Scholar]
- 7.Connell-Crowley, L., Elledge, S. J., and Harper, J. W. (1998) Curr. Biol. 8 65–68 [DOI] [PubMed] [Google Scholar]
- 8.Hahn, W. C., Dessain, S. K., Brooks, M. W., King, J. E., Elenbaas, B., Sabatini, D. M., DeCaprio, J. A., and Weinberg, R. A. (2002) Mol. Cell. Biol. 22 2111–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deeb, K. K., Michalowska, A. M., Yoon, C. Y., Krummey, S. M., Hoenerhoff, M. J., Kavanaugh, C., Li, M. C., Demayo, F. J., Linnoila, I., Deng, C. X., Lee, E. Y., Medina, D., Shih, J. H., and Green, J. E. (2007) Cancer Res. 67 8065–8080 [DOI] [PubMed] [Google Scholar]
- 10.Ausubel, F. M., Brent, R., Kington, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., and Struhl, K. E. (1988) Current Protocols in Molecular Biology, Greene Publishing Associates and Wiley-Interscience, New York
- 11.Yeh, E., Cunningham, M., Arnold, H., Chasse, D., Monteith, T., Ivaldi, G., Hahn, W. C., Stukenberg, P. T., Shenolikar, S., Uchida, T., Counter, C. M., Nevins, J. R., Means, A. R., and Sears, R. (2004) Nat. Cell Biol. 6 308–318 [DOI] [PubMed] [Google Scholar]
- 12.Jansen, D. P., Meichle, A., Steiner, P., Pagano, M., Finke, K., Botz, J., Wessbecher, J., Draetta, G., and Eilers, M. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 3685–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu, Y., Rosenblatt, J., and Morgan, D. O. (1992) EMBO J. 11 3995–4005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morgan, D. O. (1996) Curr. Opin. Cell Biol. 8 767–772 [DOI] [PubMed] [Google Scholar]
- 15.Tsvetkov, L. M., Yeh, K. H., Lee, S. J., Sun, H., and Zhang, H. (1999) Curr. Biol. 9 661–664 [DOI] [PubMed] [Google Scholar]
- 16.Carrano, A. C., Eytan, E., Hershko, A., and Pagano, M. (1999) Nat. Cell Biol. 1 193–199 [DOI] [PubMed] [Google Scholar]
- 17.Sutterluty, H., Chatelain, E., Marti, A., Wirbelauer, C., Senften, M., Muller, U., and Krek, W. (1999) Nat. Cell Biol. 1 207–214 [DOI] [PubMed] [Google Scholar]
- 18.Tedesco, D., Lukas, J., and Reed, S. I. (2002) Genes Dev. 16 2946–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhattacharya, S., Garriga, J., Calbo, J., Yong, T., Haines, D. S., and Graña, X. (2003) Oncogene 22 2443–2451 [DOI] [PubMed] [Google Scholar]
- 20.Bornstein, G., Bloom, J., Sitry-Shevah, D., Nakayama, K., Pagano, M., and Hershko, A. (2003) J. Biol. Chem. 278 25752–25757 [DOI] [PubMed] [Google Scholar]
- 21.Garriga, J., Bhattacharya, S., Calbo, J., Marshall, R. M., Truongcao, M., Haines, D. S., and Grana, X. (2003) Mol. Cell. Biol. 23 5165–5173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.parreño, M., Garriga, J., Limon, A., Albrecht, J. H., and Graña, X. (2001) Oncogene 20 4793–4806 [DOI] [PubMed] [Google Scholar]
- 23.Watanabe, G., Howe, A., Lee, R. J., Albanese, C., Shu, I. W., Karnezis, A. N., Zon, L., Kyriakis, J., Rundell, K., and Pestell, R. G. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 12861–12866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garriga, J., Jayaraman, A. L., Limon, A., Jayadeva, G., Sotillo, E., Truongcao, M., Patsialou, A., Wadzinski, B. E., and Grana, X. (2004) Cell Cycle 3 1320–1330 [DOI] [PubMed] [Google Scholar]
- 25.Kozar, K., Ciemerych, M. A., Rebel, V. I., Shigematsu, H., Zagozdzon, A., Sicinska, E., Geng, Y., Yu, Q., Bhattacharya, S., Bronson, R. T., Akashi, K., and Sicinski, P. (2004) Cell 118 477–491 [DOI] [PubMed] [Google Scholar]
- 26.Solomon, M. J., Harper, J. W., and Shuttleworth, J. (1993) EMBO J. 12 3133–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poon, R. Y., Yamashita, K., Adamczewski, J. P., Hunt, T., and Shuttleworth, J. (1993) EMBO J. 12 3123–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fisher, R. P., and Morgan, D. O. (1994) Cell 78 713–724 [DOI] [PubMed] [Google Scholar]
- 29.Fisher, R. P., Jin, P., Chamberlin, H. M., and Morgan, D. O. (1995) Cell 83 47–57 [DOI] [PubMed] [Google Scholar]
- 30.Tassan, J. P., Schultz, S. J., Bartek, J., and Nigg, E. A. (1994) J. Cell Biol. 127 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Devault, A., Martinez, A. M., Fesquet, D., Labbe, J. C., Morin, N., Tassan, J. P., Nigg, E. A., Cavadore, J. C., and Doree, M. (1995) EMBO J. 14 5027–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, Y., Wu, C., and Galaktionov, K. (2004) J. Biol. Chem. 279 4507–4514 [DOI] [PubMed] [Google Scholar]
- 33.Serizawa, H., Maekelae, T. P., Conaway, J. W., Conaway, R. C., Weinberg, R. A., and Young, R. A. (1995) Nature 374 280–282 [DOI] [PubMed] [Google Scholar]
- 34.Shiekhattar, R., Mermelstein, F., Fisher, R. P., Drapkin, R., Dynlacht, B., Wessling, H. C., Morgan, D. O., and Reinberg, D. (1995) Nature 374 283–287 [DOI] [PubMed] [Google Scholar]
- 35.Coller, H. A., Sang, L., and Roberts, J. M. (2006) PLoS Biol. 4 e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen, W., Possemato, R., Campbell, K. T., Plattner, C. A., Pallas, D. C., and Hahn, W. C. (2004) Cancer Cell 5 127–136 [DOI] [PubMed] [Google Scholar]
- 37.Moreno, C. S., Ramachandran, S., Ashby, D. G., Laycock, N., Plattner, C. A., Chen, W., Hahn, W. C., and Pallas, D. C. (2004) Cancer Res. 64 6978–6988 [DOI] [PubMed] [Google Scholar]
- 38.Mungre, S., Enderle, K., Turk, B., Porras, A., Wu, Y. Q., Mumby, M. C., and Rundell, K. (1994) J. Virol. 68 1675–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rank, K. B., Evans, D. B., and Sharma, S. K. (2000) Biochem. Biophys. Res. Commun. 271 469–473 [DOI] [PubMed] [Google Scholar]
- 40.Aprelikova, O., Xiong, Y., and Liu, E. T. (1995) J. Biol. Chem. 270 18195–18197 [DOI] [PubMed] [Google Scholar]
- 41.Berthet, C., Aleem, E., Coppola, V., Tessarollo, L., and Kaldis, P. (2003) Curr. Biol. 13 1775–1785 [DOI] [PubMed] [Google Scholar]
- 42.Ortega, S., Prieto, I., Odajima, J., Martin, A., Dubus, P., Sotillo, R., Barbero, J. L., Malumbres, M., and Barbacid, M. (2003) Nat. Genet. 35 25–31 [DOI] [PubMed] [Google Scholar]
- 43.Aleem, E., Kiyokawa, H., and Kaldis, P. (2005) Nat. Cell Biol. 7 831–836 [DOI] [PubMed] [Google Scholar]
- 44.Leone, G., DeGregori, J., Sears, R., Jakoi, L., and Nevins, J. R. (1997) Nature 387 422–426 [DOI] [PubMed] [Google Scholar]
- 45.Karsunky, H., Geisen, C., Schmidt, T., Haas, K., Zevnik, B., Gau, E., and Moroy, T. (1999) Oncogene 18 7816–7824 [DOI] [PubMed] [Google Scholar]
- 46.Geng, Y., Lee, Y. M., Welcker, M., Swanger, J., Zagozdzon, A., Winer, J. D., Roberts, J. M., Kaldis, P., Clurman, B. E., and Sicinski, P. (2007) Mol. Cell 25 127–139 [DOI] [PubMed] [Google Scholar]
- 47.Du, J., Widlund, H. R., Horstmann, M. A., Ramaswamy, S., Ross, K., Huber, W. E., Nishimura, E. K., Golub, T. R., and Fisher, D. E. (2004) Cancer Cell 6 565–576 [DOI] [PubMed] [Google Scholar]
- 48.Tetsu, O., and McCormick, F. (2003) Cancer Cell 3 233–245 [DOI] [PubMed] [Google Scholar]
- 49.Macias, E., Kim, Y., Miliani de Marval, P. L., Klein-Szanto, A., and Rodriguez-Puebla, M. L. (2007) Cancer Res. 67 9713–9720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Minella, A. C., Swanger, J., Bryant, E., Welcker, M., Hwang, H., and Clurman, B. E. (2002) Curr. Biol. 12 1817–1827 [DOI] [PubMed] [Google Scholar]
- 51.Hahn, W. C., and Weinberg, R. A. (2002) N. Engl. J. Med. 347 1593–1603 [DOI] [PubMed] [Google Scholar]
- 52.Yu, D., Matin, A., Hinds, P. W., and Hung, M. C. (1994) Cell Growth & Differ. 5 431–438 [PubMed] [Google Scholar]
- 53.Haas, K., Johannes, C., Geisen, C., Schmidt, T., Karsunky, H., Blass-Kampmann, S., Obe, G., and Moroy, T. (1997) Oncogene 15 2615–2623 [DOI] [PubMed] [Google Scholar]
- 54.Martins, C. P., and Berns, A. (2002) EMBO J. 21 3739–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spruck, C., Sun, D., Fiegl, H., Marth, C., Mueller-Holzner, E., Goebel, G., Widschwendter, M., and Reed, S. I. (2006) Cancer Res. 66 7355–7360 [DOI] [PubMed] [Google Scholar]