Abstract
The bacterial adhesive protein, FimH, is the most common adhesin of Escherichia coli and mediates weak adhesion at low flow but strong adhesion at high flow. There is evidence that this occurs because FimH forms catch bonds, defined as bonds that are strengthened by tensile mechanical force. Here, we applied force to single isolated FimH bonds with an atomic force microscope in order to test this directly. If force was loaded slowly, most of the bonds broke up at low force (<60 piconewtons of rupture force). However, when force was loaded rapidly, all bonds survived until much higher force (140–180 piconewtons of rupture force), behavior that indicates a catch bond. Structural mutations or pretreatment with a monoclonal antibody, both of which allosterically stabilize a high affinity conformation of FimH, cause all bonds to survive until high forces regardless of the rate at which force is applied. Pretreatment of FimH bonds with intermediate force has the same strengthening effect on the bonds. This demonstrates that FimH forms catch bonds and that tensile force induces an allosteric switch to the high affinity, strong binding conformation of the adhesin. The catch bond behavior of FimH, the amount of force needed to regulate FimH, and the allosteric mechanism all provide insight into how bacteria bind and form biofilms in fluid flow. Additionally, these observations may provide a means for designing antiadhesive mechanisms.
Biological adhesion is mediated by specific noncovalent bonds between tethered ligands and receptors. When cells bind to surfaces or other cells in tissue or in fluid flow, these adhesive bonds are subjected to tensile mechanical force. Common sense, theory (1–5), and many observations (6–13) suggest that bonds should be “slip bonds” that are weakened by tensile force as the receptor and ligand are pulled apart. It is theorized, however, that at least some bonds may be “catch bonds” that are strengthened by tensile mechanical force (1, 14). Indeed, certain biological bonds have been shown to become longer lived with increased amounts of force, until a critical level, above which the bonds break more readily. One of the receptors proposed to form catch bonds is the Escherichia coli adhesin FimH (15, 16), which is the terminal adhesin on type 1 fimbriae, the most common adhesive organelles for the family Enterobacteriaceae.
Type 1 fimbriae and FimH are involved in commensal binding to the intestines (17) and the oropharynx (18) as well as pathogenic binding to lung tissue (19), urinary tract tissue (20–24), and even abiotic surfaces (25). Catch bonds allow for behavior fundamentally different from that allowed by slip bonds. Catch bonds mediate shear-enhanced adhesion in which particles bind more tightly instead of being washed off when fluid flow is increased. Catch bonds are also less susceptible to soluble inhibitors than slip bonds, since the small soluble molecules cannot apply a significant drag force, so that the bonds with inhibitors will be shorter lived than those with the surface. If FimH does form catch bonds, then understanding the mechanism by which this occurs may allow the design of alternative inhibitors that prevent activation by force. Thus, knowing whether and how FimH forms catch bonds could lead to a better understanding of the natural processes that FimH and other catch bonds mediate and may also pave the way for technological applications.
Other proposed catch bonds include the leukocyte adhesion proteins P- and L-selectin binding to endothelial sialyl-Lewis-X (26–28), the motor protein myosin binding to the cytoskeletal protein actin (29), integrins binding to various ligands, and the blood protein von Willebrand factor binding to the platelet receptor GPIb. Of these, selectin- and myosin-mediated interactions have been demonstrated directly to form catch bonds by using single molecule force spectroscopy experiments. In these experiments, conditions can be chosen in which usually only one bond forms and tensile force is applied by drawing the surfaces directly apart from each other. In contrast, the catch bond mechanism for FimH has been supported by a variety of studies showing shear-enhanced FimH-mediated adhesion of either fimbriated bacteria or functionalized beads (30–35). In these experiments, increased shear stress from fluid flow increases the time bacteria remain stationary on the surface. This occurs even when soluble mannose is added at the moment shear is increased, which should prevent the formation of new bonds between FimH and mannose on the surface (31, 36). Nevertheless, it has been suggested that shear-enhanced adhesion occurs as a result of enhanced bond formation when the sheared surfaces are pressed more closely together (37, 38) or as a result of the mechanical properties of the bacteria rather than the FimH bonds (39). Single molecule force spectroscopy studies on FimH are thus needed to demonstrate conclusively that FimH forms catch bonds.
FimH is located on the tip of a long fimbrial rod and has two domains: a lectin domain that binds the carbohydrate mannose and a pilin domain that integrates FimH into the fimbriae. Structural simulations showed that Escherichia coli FimH undergoes a force-induced conformational change that was correlated with stronger binding (15). The linker chain connecting the two domains was extended in simulations in which force was applied between the mannose-binding residues at the tip of the lectin domain and the C terminus at the base of the lectin domain that anchors it to the pilin domain. The hypothesis that this extension might somehow lead to a stronger binding state was supported by the effect of structural mutations (15). Because the predicted force-induced conformational change was far from the active site, we have hypothesized “allosteric” regulation of FimH activity, where the conformation of the active site is regulated by conformation of the interdomain region of FimH. This hypothesis was supported by a mathematical model that described an “allosteric catch bond” and explained the effect of force on the lifetime of interactions between bacteria and mannose (31). Moreover, we showed recently that FimH is indeed an allosteric protein, since disruption of the interaction between the lectin and pilin domain by a structural mutation in the interdomain region increases the affinity of the lectin domain for mannose by up to 300-fold (34). Finally, we show in a companion paper (40) that the interdomain region possesses a ligand-induced binding site (LIBS)2 epitope that is exposed in FimH only in the presence of mannose, providing a direct demonstration that FimH is an allosteric protein. Binding of monoclonal antibodies to the LIBS locks FimH in the high affinity conformation, apparently due to sustained disruption of the interaction between the lectin and pilin domains by LIBS-bound antibody wedged into the interface (40). These results suggest that mechanical force, which would facilitate separation of the two domains, would also shift FimH from low to high affinity conformation. However, this has not been tested directly at the single molecule level, so the compelling idea that mechanical force allosterically activates FimH still remains a hypothesis.
In this study, we report atomic force microscope (AFM) measurements of the strength of individual bonds between mannose and fimbrial tips-incorporated FimH. These studies show that the bond strength switches from weak to strong if force is increased, demonstrating that FimH forms catch bonds. However, the FimH-mannose bond is always strong when 1) the adhesin has a structural mutation that disrupts the interdomain interaction, 2) the fimbrial tips are pretreated with LIBS-binding monoclonal antibody, or 3) prior to testing, the FimH-mannose bond is transiently prepulled with moderate force. All of these results are quantitatively consistent with a previously proposed mathematical model for a two-state allosteric catch bond (31). Thus, this demonstrates allosteric regulation of the FimH-mannose bond under tensile force.
EXPERIMENTAL PROCEDURES
Isolation of Fimbrial Tips—Genes coding for chaperone FimC (with C-terminal His6 tag) and fimbrial tip subunits FimF, FimG, and FimH were cloned in pRSET-B plasmid, expressed in BL21(DE3) cells, and purified as described (34).
Flow Chamber Experiments—3-μm diameter polystyrene beads were incubated with 200 μg/ml mannosylated bovine serum albumin (man-BSA (54), gift from Y. C. Lee (Johns Hopkins University) or from EY Laboratories) for 75 min and washed with 0.2% bovine serum albumin in phosphate-buffered saline (PBS-BSA) to reduce nonspecific binding. Fimbrial tips were immobilized on a Corning brand polystyrene tissue culture dish at 10 ng/ml total protein for 1.25 h at 37 °C and then blocked with BSA-PBS overnight. To measure the rate of accumulated binding of beads to the surface, the suspended beads were washed over the surface at the indicated shear stress levels for 5 min, and the number of adherent beads at the end of 5 min was measured. To measure the lifetime of interactions between beads and the surface, beads were accumulated on the surface for 5 min, suspended beads were washed out, and the adherent beads were detached by turning the flow off for 10 min. After this time, flow was started at the indicated level, the beads were monitored with 27 frames/s digital video microscopy, and the pause times were measured as described previously (31). Only the new pauses that started after the start of the video at the given shear stress were measured in this analysis, so that any beads that failed to detach during the 10 min without flow did not affect the data.
In the interaction lifetime experiments, 10 ng/ml fimbrial tips were incubated with a surface. The highest concentration of FimH that would be expected even if 100% of tips in solution bound to the surface and remained functional would be 123 tips/μm2, or an average of two fimbrial tips in the area that is within one tip length of a bead touching the surface. The number of functional FimH may be lower, as is the number that happen to form bonds at a given time. Since each fimbrial tip contains only one molecule of FimH, these conditions were appropriate for measuring single FimH-mannose bonds, which is validated by the lack of change in distribution of pause times with a 2-fold increase in fimbrial tip concentration.
Constant Velocity AFM Experiments—Fimbrial tips were immobilized on plates as described above except at a much higher concentration on account of the small size of the AFM cantilever tip. Olympus Biolever cantilevers were incubated with 100 μg/ml man-BSA at 37 °C for 1.25 h and blocked overnight in PBS-BSA. An Asylum MFP-3D AFM was used to probe the forces on single bonds between the cantilever and surface in PBS-BSA. The tip was pressed to the surface for 1 s with 100 pN of force, and then the tip was withdrawn at a constant velocity of 46 nm/s for a 3000-pN/s loading rate, for example. This velocity was calculated to create the desired loading rate at which force is increased, given the spring constant of the cantilever tips (calculated with the thermal method and found to vary between 4.38 and 6.36 pN/nm for different tips). The actual loading rates also reflect the spring constant of the fimbrial tip-mannose bonds, which were calculated to be 8.75 ± 1.91 pN/nm by measuring the slope of the force-separation curves for hundreds of curves. The actual loading rates stated in the figures were thus 83 ± 4% of the predicted loading rates. The force at rupture was calculated as the difference between the peak of tensile force and the average base-line force following rupture, using an automated script. Nonspecific interactions between the tip and surface were measured by adding 4% α-methyl mannose to the PBS-BSA solution to prevent specific bonds from forming.
Controlling for Spatial and Temporal Variation—To ensure that the experiments showing the effect of pull velocity were not affected by any variability between different fimbrial tips or surface locations, each location at which data were taken was probed at each pull velocity. In some cases, the experiments were repeated on multiple days in order to perform enough pulls to determine rupture distributions with statistical relevance. The order in which the pulls were performed was alternated on different days to ensure there was no time-dependent shift in the distribution.
mAb Incubation—Briefly, fimbriated plates (as above) were preincubated with 1:500 dilution of mAb 21 in the presence of 1% α-methyl-d-mannopyranoside at 37 °C for 2 h. Then plates were washed with 0.2% bovine serum albumin in phosphate-buffered saline (PBS-BSA) to wash out antibody and α-methyl-d-mannopyranoside and to reduce nonspecific binding. It was not possible to perform the mAb 21 incubation as well as test both the experimental conditions and the negative control at multiple loading rates on the same day, but the negative control experiments have been very reproducible and virtually identical at all loading rates and conditions. Thus, an average negative control curve obtained in other experiments could be subtracted from each curve in this experiment to give the specific binding shown in Fig. 5C.
FIGURE 5.
Effect of preloading FimH-mannose bonds. a, schematic for preloading K12 tips with force just before pulling (bottom), and control experiments (top). Note that the force snaps back to zero when the bond breaks regardless of the programmed force load, and this gives the measured rupture force. b, comparison of rupture force histograms for preloaded tips (254 pulls) versus standard conditions (222 pulls) at a low loading rate of 100 pN/s.
Model Fitting—The allosteric catch bond model is described by the coupled set of ordinary differential equations as described previously (31),
 |
(Eq. 1) |
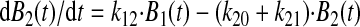 |
(Eq. 2) |
where Bi(t) is the fraction of bonds remaining in
state i, and kij(f) =
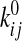 are the force-dependent rate constants shown
in Fig. 5; each depends on a
transition state distance, Δxij, and an unforced
rate constant
are the force-dependent rate constants shown
in Fig. 5; each depends on a
transition state distance, Δxij, and an unforced
rate constant 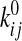 . We numerically solved the
ordinary differential equation model for the bond state over time for the
constant force and for linearly increasing force conditions for comparison
with the data. The initial conditions were determined as before by the
requirements of detailed balance given the equilibrium rate constants, so that
B1(0) = J1/J1 +
J2) =
. We numerically solved the
ordinary differential equation model for the bond state over time for the
constant force and for linearly increasing force conditions for comparison
with the data. The initial conditions were determined as before by the
requirements of detailed balance given the equilibrium rate constants, so that
B1(0) = J1/J1 +
J2) =
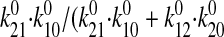 and B2(0) = 1 - B1(0), and we assumed
that the same fraction of pulls resulted in bonds in all experiments. The
numerical solution to the differential equation model was fit to the data by
minimizing the least squares of the error between the data points and the
model, after binning the probability distribution into 20-pN bins identical to
those used to make the histograms for the experimental data. The parameters of
the strong state (Δx20 and
and B2(0) = 1 - B1(0), and we assumed
that the same fraction of pulls resulted in bonds in all experiments. The
numerical solution to the differential equation model was fit to the data by
minimizing the least squares of the error between the data points and the
model, after binning the probability distribution into 20-pN bins identical to
those used to make the histograms for the experimental data. The parameters of
the strong state (Δx20 and
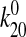 ) were used to predict the behavior of
FimH-A188D and the mAb 21-activated FimH-K12 using the assumption that they
were a simple slip bond that was identical to the strong state of FimH-K12.
That is, the differential equation model for these experiments was as follows,
) were used to predict the behavior of
FimH-A188D and the mAb 21-activated FimH-K12 using the assumption that they
were a simple slip bond that was identical to the strong state of FimH-K12.
That is, the differential equation model for these experiments was as follows,
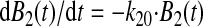 |
(Eq. 3) |
with
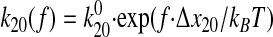 |
(Eq. 4) |
RESULTS
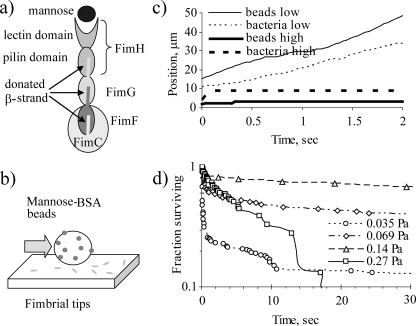
Isolated Fimbrial Tips Reproduce Force-enhanced Two-state Binding Behavior—FimH is unstable on its own, because the pilin domain lacks a β-strand (41), but when it is isolated as a complex with the β-strand-donating chaperone protein, FimC (41, 42), this FimH-FimC complex is constitutively activated (34), as is the isolated lectin domain (34, 43). Instead, we use fimbrial tips composed of FimH, FimG, FimF, and FimC polymerized via β-strand swapping (Fig. 1A). The isolated tips do not aggregate or cluster, as indicated by their elution at the expected size in a high pressure liquid chromatography sizing column (34), so they are ideal for single bond force spectroscopy. We determined whether purified FimH-containing tips of type 1 fimbriae reproduce the shear-enhanced adhesion observed in intact fully fimbriated bacteria. Recombinant fimbrial tips with FimH from E. coli K12 were purified and nonspecifically immobilized to a polystyrene surface in a parallel plate flow chamber. A solution of suspended mannose-BSA-coated polystyrene microbeads were flowed through the chamber at various levels of shear (Fig. 1B). A low concentration of receptors and ligands was used to measure the lifetime of bonds between the beads to ensure measurement of single FimH bond lifetimes. At each shear stress tested, the mannose beads alternated between pausing and moving across the FimH-coated surface (Fig. 1C). This behavior is much like the uneven rolling of live bacteria across a mannose-coated surface (Fig. 1C).
FIGURE 1.
Fimbrial tips mediate shear-enhanced adhesion. a, schematic diagram of mannose bound to a fimbrial tip with FimH, FimG, FimF, and FimC polymerized via β-strand swapping. b, schematic diagram of mannose-coated beads binding to fimbrial tip-coated surface in flow chambers. c, typical tracks showing the position as a function of time for mannose-coated beads binding to a fimbrial tip-coated surface (solid lines) or of fimbriated E. coli binding to a mannose-coated surface (dashed lines) at low flow (0.035 Pa; thin lines) or high flow (0.27 Pa for beads, 2 Pa for bacteria; thick line). d, duration of time that beads pause on surface at low receptor-ligand concentrations, expressed in number pausing for time t or longer.
Also as for intact bacteria, adhesion was enhanced at higher shear stress, where the pauses became longer. The lifetime of the beads' pauses was measured and is shown in Fig. 1D. At the lowest level of shear (0.035 Pa), a double exponential decay in pause survival was seen (i.e. the pauses had two distinct lifetimes, one very short (<<1 s) and one very long (>>1 s)). As shear increased (0.07–0.14 Pa), the fraction of long lived pauses increased until virtually all were long lived. At yet higher shear stress (0.27 Pa), the single observed lifetime became significantly shorter. Thus, shear increases the fraction of long lived pauses but then decreases the lifetime of these pauses. This again replicates the previous observations for the lifetimes of bonds formed by fully fimbriated bacteria (31). The fact that these experiments with isolated tips gave similar interaction lifetimes as the previous experiments with intact bacteria indicates that the isolation, expression, and physiosorption of fimbrial tips did not affect the activity of FimH. For example, the two distinct lifetimes cannot be due to partial inactivation or artificial activation of some fraction of the fimbrial tips, since the two lifetimes were also observed with intact bacteria (31). The two lifetimes must be due to specific FimH-mannose bonds, since they are both blocked by the addition of 1% α-methyl mannoside (aMM), a soluble competitive inhibitor for FimH binding (not shown). Both types of events are probably due to single FimH-mannose bonds, since the distribution of pause lifetimes was little changed by a 2-fold increase in the concentration of fimbrial tips used (not shown). Indeed, the two lifetimes were also observed in surface plasmon resonance experiments in which soluble fimbrial tips or fimbriae bound to mannose on a surface (40), an assay in which there is no mechanism for avidity, since there is only one FimH per fimbriae or tip.
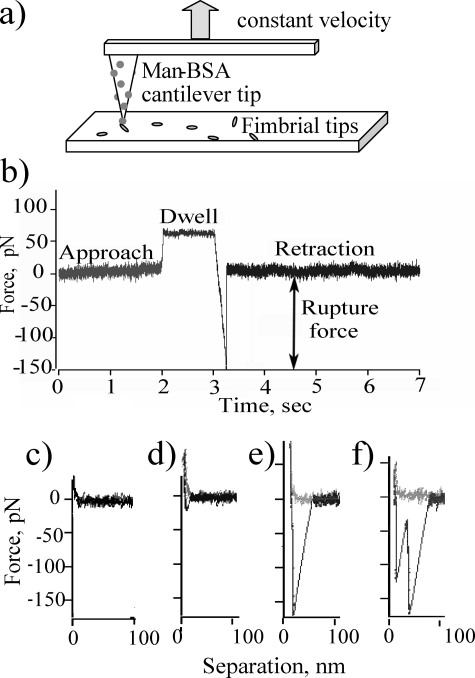
Strength of Single Bonds Probed with Atomic Force Microscopy—Single molecule AFM studies were performed with man-BSA on the AFM cantilever tip, and FimH-K12 fimbrial tips bound to a surface in essentially the same manner as for the flow chambers, except with a higher concentration of fimbrial tips (Fig. 2A) to allow for the smaller size of the cantilever tip surface in contact. The cantilever tip was pressed on the surface for 1 s to allow a bond to form and then pulled away from the surface, causing it to deflect with a constantly increasing force until the bond dissociated (Fig. 2B). The rate at which force increases is called the loading rate. Fig. 2, C–F, shows typical pulls, graphed as deflection force versus separation distance of the tip of the cantilever and the surface. In some pulls, the force returned immediately to base line during retraction, indicating that there was either no adhesion or an adhesive force of less than the base-line noise level of 10 pN (Fig. 2C). In other pulls, the force continued to ramp linearly past base line until the bond with the surface ruptured and the cantilever deflection returned to base line. This happened at a range of forces (Fig. 2, D and E). The concentration of tips was chosen such that the probability of a binding event was on average 22%, so that the probability of multiple binding events was expected to be 22% of 22%, or 4.8%. Indeed, almost exactly this fraction of pulls (5%) showed two distinct rupture peaks (e.g. see Fig. 2F) These were assumed to be caused by multiple bonds and were removed from the analysis so that very few of the events included in the histograms are caused by multiple peaks.
FIGURE 2.
Description of constant velocity AFM experiments. a, schematic of man-BSA cantilever tip and fimbrial tips on surface; b, typical curve showing force as a function of time during approach, surface dwell, and retraction of the cantilever tip; c–f, force as a function of separation for a single pull with no measurable adhesive event (c), a single weak adhesive event (d), a single strong adhesive event (e), and a rare double adhesive event (f). The separation was calculated by adding the distance the cantilever tip deflects to the position of the cantilever base.
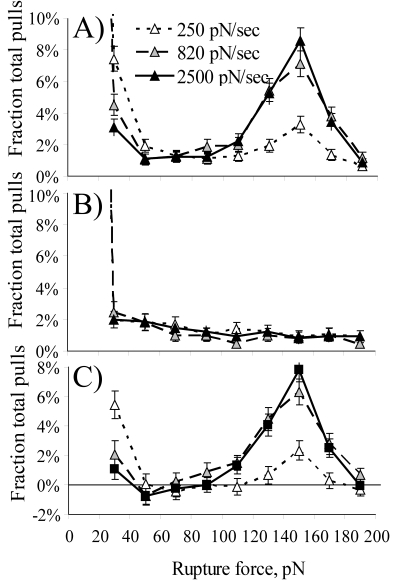
The same location was probed repeatedly at the same loading rate in order to obtain a histogram of rupture peaks (Fig. 3A). In these experiments, a bimodal distribution was observed, with many peaks at low (20–40 pN) or high (120–180 pN) force but few peaks in between. This bimodal distribution has been observed previously with catch bonds (28), since they break at low or high forces but are strongest at an intermediate range of force. The high force peaks cannot be due to simultaneous rupture of multiple low force bonds, since the ratio of force in the two peaks is 4–5-fold, and there is no probable explanation for why fimbrial tips would bind in single bonds or as part of 4–5 bonds but never as part of double or triple bonds. (Each fimbrial tip contains only one FimH with one mannose-binding site, and the tips do not cluster when analyzed with a sizing column (34).) However, a bimodal distribution could also be caused by two independent populations of slip bonds, one strong and one weak. To distinguish the two possibilities, we varied the pulling velocity in order to vary the rate at which the force was increased. A slower pulling velocity increased the number of low force peaks, whereas a faster one increased the number of high force peaks (Fig. 3A). This loading rate-dependent switch in the rupture force distribution is expected for catch bonds, since a slower loading rate would allow a higher fraction to break before force was increased enough to stabilize them. In contrast, it cannot be explained by two independent slip bonds. The FimH-mannose bonds are in series with nonspecific bonds between these proteins and surfaces. When bonds in series are subjected to tensile force, a single peak of rupture forces is observed that is below the rupture force of the weakest component (44). We describe experiments in the supplemental material that demonstrate that neither the 20–40- nor 120–180-pN force peaks reflect desorption of the proteins from the surfaces. There is thus currently no explanation other than catch bonds for a bimodal distribution that switches with loading rate. To ensure that the rupture force distribution was due to specific bonds, the experiments were also performed in the presence of aMM inhibitor. This negative control did show some rupture events, but they were distributed over the entire force range with no peaks like those observed in the absence of inhibitor (Fig. 3B). When the negative control histogram was subtracted from the experimental histogram, the bimodal distribution was not only maintained but became even clearer as the base line between the two peaks was removed (Fig. 3C). We also found that the specific and nonspecific events could be distinguished by the molecular spring constant and that filtering by the spring constant also resulted in an even clearer bimodal distribution (see supplemental material). This confirms that both low and high rupture peaks are due to specific bonds between man-BSA and fimbrial tips. A negative value means that a higher percentage of force peaks were observed in this bin when aMM inhibitor was included. A negative value is expected occasionally when the specific events are not statistically different from zero. Negative values are also expected and observed reproducibly in the lowest force bin (0–20 pN), since this bin included nonadhesive events that increased in frequency when inhibitor was added. This bin is excluded from the graphs of specific pulls, since the actual number of specific pulls in this bin cannot be determined from these data.
FIGURE 3.
Rupture force histograms for binding of FimH-K12 to man-BSA, expressed as a percentage of total pulls, for the three indicated loading rates. A, rupture force distribution in the absence of inhibitor (expressed in fraction of total pulls with 1103 pulls at 250 pN/s, 953 at 820 pN/s, and 1164 pulls at 2500 pN/s). B, rupture forces in the presence of 1% aMM for a negative control to show nonspecific binding events (652 pulls at 250 pN/s, 610 pulls at 820 pN/s, and 750 pulls at 2500 pN/s). C, the difference between experiment and control shows the distribution of specific events. The histograms are drawn with symbols and lines instead of traditional histogram bars in order to allow superposition of multiple conditions.
Effect of FimH Activation by Structural Mutation and Monoclonal Antibodies—If the high force peak in Fig. 3 is due to a high affinity FimH state, whereas the low force peak is due to a low affinity state, then converting the FimH structure to the high affinity state should eliminate the low force peak and cause FimH to behave as a simple but strong slip bond. This hypothesis was tested in AFM experiments by using FimH that is switched into the high affinity state with either a structural mutation or binding of a LIBS-specific antibody.
First, the effect of a mutation in FimH was tested in constant loading rate AFM experiments. The A188D mutation in the pilin domain enhances the affinity of FimH for mannose 10–100-fold, apparently by causing the two domains to separate (34). Even at the lowest loading rates, the low force peak was not observed, and instead only a high force peak was observed at each loading rate (Fig. 4A). The negative control with aMM looked essentially the same as for wild type FimH tips (not shown). After subtracting this negative control, the specific events still show a single force peak at each loading rate (Fig. 4B, red). Thus, using FimH that already favors the high affinity state prevented the catch bond behavior and instead produced, as predicted, behavior characteristic of a strong slip bond. Because a wider range of loading rates was used this time than in the previous figure, the experiment was repeated with the wild type FimH tips at the wider range of loading rates as well (Fig. 4B, black). In both A188D and wild type FimH tips, there is a shift in the position of the rupture peak at higher loading rates, as described previously for many other bonds. In addition, for the wild type FimH but not the A188D tips, there is also a switch from low to high force peak at higher loading rates. Although the mutation has an enormous effect at low loading rates and in static conditions, it has no significant effect at all at high loading rates, where both mutant and wild type show only the high force peak. This suggests that the A188D mutation and force induce the same high affinity state in FimH.
FIGURE 4.
Rupture force histograms of preactivated FimH. A, high affinity A188D FimH variant at the three indicated pulling speeds (239 pulls at 82 pN/s, 408 pulls at 820 pN/s, and 247 pulls at 8200 pN/s). B, comparison of a histogram of specific events (after subtracting values for nonspecific control) for A188D (red) and wild type K12 (black) at the indicated loading rates. C, distribution of rupture forces for K12 tips (778 pulls), mAb 21-preactivated K12 tips (405 pulls), and mAb 21-preactivated K12 tips with aMM at one loading rate (365 pulls). D, comparison of histograms for preactivated K12 tips (blue) to native K12 tips (black) at three loading rates.
For a second test, we utilized the monoclonal antibody (mAb 21) that is reported previously to bind to the interdomain region of FimH and increase its affinity for mannose by over 100-fold (40). The surface-bound fimbrial tips with wild-type FimH were co-incubated with soluble mAb 21 and 1% aMM, since the antibody recognizes and stabilizes a LIBS that is only induced by mannose. With aMM still in the buffer, a typical negative control curve was observed (Fig. 4C), demonstrating that the antibodies by themselves did not cause any new nonspecific binding. After the aMM inhibitor was washed out, however, specific binding peaks to the mannose on the AFM cantilever tip were again observed. However, in contrast to two peaks observed before the mAb 21 treatment, after the mAb 21 treatment, the low force peak was gone, and a single rupture force peak was observed at ∼150 pN at multiple loading rates (Fig. 4D, blue). Thus, preincubation with mAb 21 had the same effect as did introduction of the A188D point mutation in all regards, reinforcing the conclusion that the force-modulated bimodal distribution is due to the ability of FimH to switch between a weak and strong state.
Effect of Preactivating FimH with Force—Furthermore, we hypothesized that if FimH has two distinct states, high and low affinity, then for the mannose binding to be force-enhanced, application of force should induce the high affinity state. If this is the case, pretreatment with tensile force should have the same effect as, for example, pretreatment with antibodies. We furthermore hypothesize that the force-induced high affinity state would be stable over some period of time rather than revert instantly to low affinity upon removal of force, since we previously observed that bacteria would remain stationary for at least some time after flow was turned down. To test this, after pressing the mannose-functionalized cantilever against the surface coated with wild-type FimH tips to allow the bond to form, we pulled the cantilever quickly away at 10,000 pN/s to an intermediate force of 80 pN for 100 ms (Fig. 5a). Then the force was returned to 0 pN for 100 ms before pulling again, now slowly at 100 pN/s, until the bond ruptured. For a control, after the cantilever was pressed to the surface, we simply pulled it slowly at 100 pN/s without the pretreatment with force. As in previous slow pulling experiments (e.g. see Fig. 4c), the bonds primarily ruptured at low force in the control pulls, with the high force peak being completely lost (Fig. 5b). In contrast, when the bonds were pretreated with force, all of them ruptured at high force (Fig. 5b).
One might argue that the higher fraction of high force rupture events in the force pretreatment experiment may be the result of weak interactions breaking during the force pretreatment, resulting in preferential selection of strong interactions. However, even interactions that broke during the pretreatment were included in calculation of the total number of test pulls. Thus, the drastically increased number of high force events seen with pretreatment could not be due to selective pulls of preexisting strong bonds. Instead, before the test pull, the force apparently converted the weakly binding FimH state into the strongly binding one, and the bond remained in this state during relaxation and slow pulling.
Thus, this experiment demonstrates that pretreatment with force has the same activating effect on FimH as pretreatment with antibodies or introduction of the mutation. The fact that the bond strength depends on the history of the bond demonstrates that the bond has more than one state and that application of force induces or favors the high affinity state over the low affinity one.
Consistency with the Allosteric Catch Bond Model—We previously proposed (31) a mathematical model that calculates the probability distribution of the bond state for a bond that allosterically switches between weak and strong bound states and the unbound state (Fig. 6a). This model involves four transitions with four first order rate constants, k10, k20, k12, and k21, as diagrammed in Fig. 6A. Each of the four transition rates is exponentially affected by force according to the Bell equation,
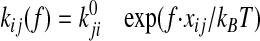 |
(Eq. 5) |
where f represents the applied force, kBT is
thermal energy, 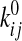 is the rate constant in the
absence of force, and xij is the characteristic distance
of the transition (5). We
simulated the model with linearly increasing force and estimated the
parameters needed to fit our AFM data. The model correctly predicts the
switch from a dominant low force shoulder or peak to a dominant high
force peak as loading rate increases in the AFM experiments
(Fig. 6b). It also
predicts the shift in peak force for the high force peak with
increased loading rate (Fig.
6b).
is the rate constant in the
absence of force, and xij is the characteristic distance
of the transition (5). We
simulated the model with linearly increasing force and estimated the
parameters needed to fit our AFM data. The model correctly predicts the
switch from a dominant low force shoulder or peak to a dominant high
force peak as loading rate increases in the AFM experiments
(Fig. 6b). It also
predicts the shift in peak force for the high force peak with
increased loading rate (Fig.
6b).
FIGURE 6.
Model for force activation of FimH through allostery. a,
schematic of a model that allows for allosteric transition between a weak
compact state and a strongly bound extended state. b, schematic of a
model for a slip bond showing a single strongly bound state. c, the
predicted rupture force probability distribution for FimH-K12 (dotted
lines) in constant loading rate experiments at three loading rates,
graphed with the K12 data from Fig.
4C (solid lines, symbols). The parameter
estimates in this fit are as follows: 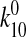 = 1.37
s-1,
= 1.37
s-1, 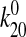 = 5.1 ×
10-6 s-1,
= 5.1 ×
10-6 s-1, 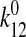 = 3.3
× 10-5 s-1,
= 3.3
× 10-5 s-1, 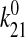 =
0.11 s-1, x10 = 2.85 Å,
x20 = 4.52 Å, x12 = 15.1
Å, x21 = -3.88 Å. d, the predicted
rupture force probability distribution for FimH that is always in the high
affinity state (dotted lines), graphed with the A188D data from
Fig. 4C (solid
lines, symbols).
=
0.11 s-1, x10 = 2.85 Å,
x20 = 4.52 Å, x12 = 15.1
Å, x21 = -3.88 Å. d, the predicted
rupture force probability distribution for FimH that is always in the high
affinity state (dotted lines), graphed with the A188D data from
Fig. 4C (solid
lines, symbols).
If the allosteric catch bond model is correct, then it should also explain
the data for FimH-A188D and mAb 21-activated FimH-K12 under the assumption
that the bond is locked into the strong state (i.e. the model is
reduced to a slip bond with a single unbinding rate,
k20(f) =
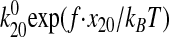 ,
as shown in Fig. 6c.
To test this, the two parameters,
,
as shown in Fig. 6c.
To test this, the two parameters, 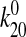 and
x20, from the fit described above were used without
further fitting to predict the behavior of FimH-A188D and mAb 21-locked
FimH-K12. The quality of this fit, shown in
Fig. 6d for A188D,
demonstrates that these two conditions do indeed behave as predicted by the
allosteric model under the assumption that they are locked at all forces into
the high affinity state. In addition, the model correctly predicts the result
of the experiment preloading the bonds with 80 pN of force, since the preload
is predicted to transition all of the bonds into the high affinity state and
not allow enough time for reversion to the low affinity state during the 100
ms for which the bond is returned to 0 pN before pulling. The correct
predictions of the model on various preactivation experiments further
strengthen the evidence that mechanical force allosterically converts FimH
from a low affinity to a high affinity state, causing it to form catch
bonds.
and
x20, from the fit described above were used without
further fitting to predict the behavior of FimH-A188D and mAb 21-locked
FimH-K12. The quality of this fit, shown in
Fig. 6d for A188D,
demonstrates that these two conditions do indeed behave as predicted by the
allosteric model under the assumption that they are locked at all forces into
the high affinity state. In addition, the model correctly predicts the result
of the experiment preloading the bonds with 80 pN of force, since the preload
is predicted to transition all of the bonds into the high affinity state and
not allow enough time for reversion to the low affinity state during the 100
ms for which the bond is returned to 0 pN before pulling. The correct
predictions of the model on various preactivation experiments further
strengthen the evidence that mechanical force allosterically converts FimH
from a low affinity to a high affinity state, causing it to form catch
bonds.
DISCUSSION
Over 90% of E. coli and other enteric bacteria express the type 1 fimbrial adhesin FimH, a lectin-like protein that binds specifically to terminal mannose residues on glycoproteins on a wide range of tissues. The natural variant of FimH used in this study (from E. coli K12) is a common variant in intestinal as well as uropathogenic E. coli and is identical to the one crystallized for structural determination (41, 42, 45). Using just the type I fimbrial tip complexes containing one FimH adhesin subunit and two other minor tip subunits, FimF and FimG (complexed with FimC chaperone) but without the rod-forming major subunit FimA, we use an atomic force microscope to apply tensile force to single bonds between the fimbrial tip FimH and the ligand mannose. When force was increased at a constant loading rate, the rupture force histogram for these bonds showed two distinct force peaks, one below 40 pN and one between 140 and 160 pN. However, a slow loading rate favored the lower force peak, whereas a faster loading rate caused all of the bonds to break in the higher force peak. This behavior indicates that the bonds are catch bonds, because intermediate force (60–100 pN) protects them from rupture more than lower forces. The flow chamber data serves as additional evidence at constant force that isolated fimbrial tips form catch bonds. In contrast, other explanations, including nonspecific adhesion and the coexistence of multiple bonds in series or in parallel, cannot explain the biphasic peaks and the switch from low force to high force peaks as loading rate increases. Thus, we show here for the first time using single molecule studies that wild type FimH from E. coli forms catch bonds.
We also demonstrate here that the strongly binding A188D variant of FimH does not demonstrate catch bond behavior, at least in these assays; only a single force peak was observed at all loading rates, consistent with slip bond behavior. Although this point mutation was engineered, this result provides insight into naturally occurring point mutations. It has previously been demonstrated that the evolutionarily dominant variants of FimH bind weakly to mannose in static conditions. In contrast, point mutations in FimH often enhance adhesive strength and are observed with increased frequency in E. coli isolated from patients with urinary tract or kidney infections. The A188D point mutation, which was engineered in the laboratory, binds even more strongly under low force than any of the clinical isolates. This suggests that commensal and, to a lesser extent, pathogenic E. coli gain an advantage from the native FimH catch bonds and has raised the question of why. We demonstrate here that the commensal catch bond variant of FimH and the artificial slip bond-like A188D variant of FimH form bonds with identical strength to monomannose, as long as high levels of force are applied fast enough. Thus, the catch bond behavior in FimH provides a mechanism for down-regulation at low force but does not strengthen binding at high force (i.e. although commensal bacteria require strong adhesion at high forces, they must benefit from weak adhesion at low forces, perhaps in order to be motile in these conditions (46) or to resist binding of soluble inhibitors (15, 36)).
To better understand bacterial adhesion, it is useful to compare the quantitative response of FimH catch bonds to force with the forces involved during bacterial adhesion in flow. We show here that a commensal FimH variant rarely detaches between 60 and 100 pN, demonstrating that this is the range of force at which FimH bonds are longest lived. It has been estimated that peristalsis in intestines causes a shear stress of ∼1 Pa (47) in water, and intestinal fluid is very viscous (48), so the actual shear stress should be 2–10 Pa or even higher. E. coli binding via FimH to mannose switch to strong adhesion at 2 Pa (30), where it can be estimated that the drag force on E. coli increases at over 100,000 pN/s to 64 pN (see supplemental material). This loading rate is in huge excess of that needed to protect FimH bonds from early rupture, whereas the final force is just barely within the range where FimH is long lived, suggesting that the latter is the key determinant that controls the switch to stationary adhesion. The 60–100-pN force range where FimH is longest lived also corresponds well to the force at which the type 1 fimbriae, on which FimH is anchored, elongate dramatically by uncoiling the quaternary helix (33). We propose that fimbrial uncoiling buffers the force on bonds, allowing bacteria move along the surface, forming new bonds to share the load. This would keep the force per bond at 60–100 pN even at very high shear stress, explaining why bacteria only roll slowly at 10 Pa, where the drag force is ∼300 pN. Thus, FimH is longest lived at the same range of force that is applied by physiological shear stress and uncoiling fimbriae.
Our results here show that FimH-mannose bonds are by far the strongest receptor-ligand bonds tested to date with single molecule force spectroscopy. The catch bonds formed by P-selectin (26), L-selectin (27), and myosin (29) resist detachment in various ranges between 10 and 60 pN of force in comparison with the 60–100-pN range for FimH. When loaded at 1000 pN/s, actomyosin bonds rupture at 80 pN (29), and P-selectin-PSGL bonds rupture at 80 pN, whereas single FimH-mannose bonds rupture at 150 pN. Even biotin-streptavidin bonds, the longest lived receptor-ligand bonds known, rupture at 70 pN when loaded at 1000 pN/s (6). The unusual strength of FimH bonds is even more remarkable given the subsecond lifetime of FimH-mannose bonds when they are not activated (31). This comparison suggests that FimH catch bonds may be used as a strong yet smart adhesive for technological applications.
We have previously suggested that allosteric regulation could explain shear-enhanced bacterial adhesion (31) if force causes FimH to switch between a weak and a strong binding state. However, we have only recently presented data directly showing that FimH is allosteric (40). The monoclonal antibody mAb 21 recognizes a LIBS in the presence of mannose and enhances mannose binding (40). Thus, mannose and mAb 21 have a reciprocal activation on the binding of the other, which is a characteristic of allosteric regulation (49). Moreover, we have shown previously that the interaction of the lectin domain with the pilin domain of FimH lowers the affinity of the lectin domain for mannose (34). Here, we show that the native allosteric regulation is needed for FimH to form catch bonds, since locking FimH in the high affinity state with mAb 21 causes it to show apparent slip bond behavior (Fig. 4d). Similarly, the A188D mutation in the pilin domain, which weakens the interdomain interaction (34), also causes FimH to lose the catch bond behavior in this assay (Fig. 4b). This suggests that mechanical force applied between mannose and the pilin domain could separate the two domains (34) and extend the interdomain linker chain to activate FimH, but this remained a hypothesis. We confirm this hypothesis by showing directly that prestretching FimH transiently with force strengthens the FimH-mannose bond (Fig. 5b). Thus, we observe that all three methods of preactivating FimH (whether by mAb 21, A188D mutation, or force) result in an identical force peak, strongly suggesting that the biochemical (antibody or mutation) and mechanical (force) disruption of the interdomain interaction each results in the same high affinity state of the mannose-binding pocket. Moreover, the activation caused by prestretching was maintained when the bond was fully relaxed to zero force before testing, indicating that the state of the bond was changed (i.e. confirming that the bond has at least two distinct states). Together, this demonstrates that a force-induced allosteric switch from a low to a high affinity state is responsible for the behavior of FimH-mannose catch bonds.
Proteins called selectins also form catch bonds with their ligand, called PSGL-1, and there has been considerable progress in determining the mechanism of these catch bonds. The selectin-PSGL-1 bonds show much shorter lifetimes and simpler behavior than do FimH-mannose interactions, since they display only a single rapid exponential decay at a given constant force (26). Although the two lifetimes observed for FimH (Fig. 1d) suggest two distinct states and thus allostery (31), a single lifetime is also consistent with an allosteric mechanism, since the low affinity state can simply be too short lived to be detected in a given set of experiments. Furthermore, selectins show many similarities to FimH that point toward allostery. P-selectin has been crystallized in two states that appear to be low and high affinity in the active site (50). Remarkably, the two structures also differ in the hinge angle between the lectin or the binding domain of the selectins and the neighboring epidermal growth factor domain. A mutation in this interdomain region alters the affinity of P-selectin for its ligand (51), strongly suggesting allosteric regulation. A different hinge region mutation modifies the catch bond properties of L-selectin (52). However, it remains unclear if selectin catch bonds are caused by allosteric regulation (31). Indeed, it was instead suggested that straightening the hinge angle would orient the domains to place an alternative binding site along the forced unbinding pathway, which would hold the bond together long enough for the original site to rebind (52, 53). This “sliding-rebinding” mechanism is not allosteric, since it does not require a change in the conformation of the binding site itself (52, 53), and it would not predict that affinity would be affected by the hinge angle in the absence of externally applied force (31). It should thus be noted that the sliding-rebinding model cannot apply to FimH, since interdomain disruption by structural mutation or LIBS-specific antibody changes FimH affinity for mannose in the absence of force (34, 40). The motor protein myosin has also been shown to form catch bonds with the cytoskeletal protein actin and also displays complex allosteric regulation that involves soluble nucleotides as well as a hinge angle between two domains of myosin. Thus, although selectin and myosin appear to be allosteric, it remains to be determined whether this is related to their ability to form catch bonds, as for FimH.
There is at this time no structural data that illustrate the allosteric conformational change in FimH or show the allosteric pathway by which a conformational change in the interdomain region propagates to the mannose-binding site. This is because all published crystal structures are of FimH complexed with FimC chaperone or of purified lectin domains that lack the native lectin-pilin interface and are thus in the high affinity state (34). However, the structural details of allosteric regulation are only known for a handful of the thousands of proteins that are well accepted to be allosteric. Although it would be of great interest to see the allosteric structural changes in FimH, the evidence we show here demonstrates clearly that the counterintuitive, tensile force-enhanced behavior of FimH is due to a well accepted mechanism of regulating protein activity, allostery.
To our knowledge, our observation that interdomain allosteric regulation can cause catch bonds is the first direct demonstration that allostery can result in mechanical regulation. This is significant, since many proteins are allosteric, and proteins often evolve by recombination of domains into new proteins. Thus, anchoring a protein to its allosteric regulator could provide a simple means of convergent evolution of mechanically sensitive proteins. This approach could work equally well for engineering of mechanosensors for technological applications. A second significance is that mechanical regulation that is based on allostery can be replaced by chemical regulation, as is illustrated by the ability of the monoclonal antibody to eliminate the need for mechanical force to activate FimH. This suggests that allosteric inhibitors may also be found that prevent force from activating FimH. For instance, a soluble molecule that binds in the interdomain region would not be removed by mechanical force. Allosteric inhibitors may be particularly important, since soluble inhibitors are less effective for the catch bond-forming wild type variant of FimH than for the A188D mutant that does not display catch bond behavior in our assays (34).
Acknowledgments
We heartily thank Jason Beemis of Asylum for working with us to program the MFP-3D AFM to apply user-defined variable forces.
This work was supported by National Institutes of Health Grant 1RO1 AI50940, National Science Foundation Grant CMMI-0654054, the Center for Nanotechnology at the University of Washington through an IGERT Fellowship Award (National Science Foundation Grant DGE-0504573), the University of Washington Initiatives Fund, the National Nanotechnology Infrastructure Network Research Experience for Undergraduates Program, and the Swiss Federal Institute of Technology, ETH Zurich. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
Footnotes
The abbreviations used are: LIBS, ligand-induced binding site; AFM, atomic force microscope; BSA, bovine serum albumin; man-BSA, mannosylated bovine serum albumin; PBS, phosphate-buffered saline; pN, piconewton(s); Pa, pascal(s); aMM, α-methyl mannoside.
References
- 1.Dembo, M., Torney, D. C., Saxman, K., and Hammer, D. (1988) Proc. R. Soc. Lond. B Biol. Sci. 234 55-83 [DOI] [PubMed] [Google Scholar]
- 2.Shapiro, B. E., and Qian, H. (1997) Biophys. Chem. 67 211-219 [DOI] [PubMed] [Google Scholar]
- 3.Evans, E., and Ritchie, K. (1997) Biophys. J. 72 1541-1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans, E. (2001) Annu. Rev. Biophys. Biomol. Struct. 30 105-128 [DOI] [PubMed] [Google Scholar]
- 5.Bell, G. I. (1978) Science 200 618-627 [DOI] [PubMed] [Google Scholar]
- 6.Merkel, R., Nassoy, P., Leung, A., Ritchie, K., and Evans, E. (1999) Nature 397 50-53 [DOI] [PubMed] [Google Scholar]
- 7.Moy, V. T., Florin, E. L., and Gaub, H. E. (1994) Science 266 257-259 [DOI] [PubMed] [Google Scholar]
- 8.Chilkoti, A., Boland, T., Ratner, B. D., and Stayton, P. S. (1995) Biophys. J. 69 2125-2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans, E. (1999) Biophys. Chem. 82 83-97 [DOI] [PubMed] [Google Scholar]
- 10.Hanley, W., McCarty, O., Jadhav, S., Tseng, Y., Wirtz, D., and Konstantopoulos, K. (2003) J. Biol. Chem. 278 10556-10561 [DOI] [PubMed] [Google Scholar]
- 11.Fritz, J., Katopodis, A. G., Kolbinger, F., and Anselmetti, D. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 12283-12288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doggett, T. A., Girdhar, G., Lawshe, A., Schmidtke, D. W., Laurenzi, I. J., Diamond, S. L., and Diacovo, T. G. (2002) Biophys. J. 83 194-205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arya, M., Kolomeisky, A. B., Romo, G. M., Cruz, M. A., Lopez, J. A., and Anvari, B. (2005) Biophys. J. 88 4391-4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartolo, D., Derenyi, I., and Ajdari, A. (2002) Phys. Rev. 65 051910-1-051910-4 [DOI] [PubMed] [Google Scholar]
- 15.Thomas, W. E., Trintchina, E., Forero, M., Vogel, V., and Sokurenko, E. V. (2002) Cell 109 913-923 [DOI] [PubMed] [Google Scholar]
- 16.Isberg, R. R., and Barnes, P. (2002) Cell 110 1-4 [DOI] [PubMed] [Google Scholar]
- 17.Isaacson, R. E., Fusco, P. C., Brinton, C. C., and Moon, H. W. (1978) Infect. Immun. 21 392-397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ofek, I., Mirelman, D., and Sharon, N. (1977) Nature 265 623-625 [DOI] [PubMed] [Google Scholar]
- 19.Dal Nogare, A. R. (1990) Am. J. Respir. Cell Mol. Biol. 2 433-440 [DOI] [PubMed] [Google Scholar]
- 20.Keith, B. R., Maurer, L., Spears, P. A., and Orndorff, P. E. (1986) Infect. Immun. 53 693-696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langermann, S., Mollby, R., Burlein, J. E., Palaszynski, S. R., Auguste, C. G., DeFusco, A., Strouse, R., Schenerman, M. A., Hultgren, S. J., Pinkner, J. S., Winberg, J., Guldevall, L., Soderhall, M., Ishikawa, K., Normark, S., and Koenig, S. (2000) J. Infect. Dis. 181 774-778 [DOI] [PubMed] [Google Scholar]
- 22.Langermann, S., Palaszynski, S., Barnhart, M., Auguste, G., Pinkner, J. S., Burlein, J., Barren, P., Koenig, S., Leath, S., Jones, C. H., and Hultgren, S. J. (1997) Science 276 607-611 [DOI] [PubMed] [Google Scholar]
- 23.Iwahi, T., Abe, Y., Nakao, M., Imada, A., and Tsuchiya, K. (1983) Infect. Immun. 39 1307-1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Connell, I., Agace, W., Klemm, P., Schembri, M., Marild, S., and Svanborg, C. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 9827-9832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schembri, M. A., and Klemm, P. (2001) Infect. Immun. 69 1322-1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marshall, B. T., Long, M., Piper, J. W., Yago, T., McEver, R. P., and Zhu, C. (2003) Nature 423 190-193 [DOI] [PubMed] [Google Scholar]
- 27.Sarangapani, K. K., Yago, T., Klopocki, A. G., Lawrence, M. B., Fieger, C. B., Rosen, S. D., McEver, R. P., and Zhu, C. (2004) J. Biol. Chem. 279 2291-2298 [DOI] [PubMed] [Google Scholar]
- 28.Evans, E., Leung, A., Heinrich, V., and Zhu, C. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 11281-11286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo, B., and Guilford, W. H. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 9844-9849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas, W. E., Nilsson, L., Forero, M., Sokurenko, E. V., and Vogel, V. (2004) Mol. Microbiol. 53 1545-1557 [DOI] [PubMed] [Google Scholar]
- 31.Thomas, W. E., Forero, M., Yakovenko, O., Nilsson, L., Vicini, P., Sokurenko, E. V., and Vogel, V. (2006) Biophys. J. 90 753-764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forero, M., Thomas, W. E., Bland, C., Nilsson, L., Sokurenko, E. V., and Vogel, V. (2004) Nano Lett. 4 1593-1597 [Google Scholar]
- 33.Forero, M., Yakovenko, O., Sokurenko, E. V., Thomas, W. E., and Vogel, V. (2006) PLoS Biol. 4 1509-1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aprikian, P., Tchesnokova, V., Kidd, B., Yakovenko, O., Yarov-Yarovoy, V., Trinchina, E., Vogel, V., Thomas, W., and Sokurenko, E. (2007) J. Biol. Chem. 282 23437-23446 [DOI] [PubMed] [Google Scholar]
- 35.Nilsson, L. M., Thomas, W. E., Trintchina, E., Vogel, V., and Sokurenko, E. V. (2006) J. Biol. Chem. 281 16656-16663 [DOI] [PubMed] [Google Scholar]
- 36.Nilsson, L. M., Thomas, W. E., Sokurenko, E. V., and Vogel, V. (2006) Appl. Environ. Microbiol. 72 3005-3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu, C., Lou, J., and McEver, R. P. (2005) Biorheology 42 443-462 [PubMed] [Google Scholar]
- 38.Zhu, C., and McEver, R. P. (2005) Mol. Cell Biomech. 2 91-104 [PubMed] [Google Scholar]
- 39.Miller, E., Garcia, T. I., Hultgren, S., and Oberhauser, A. (2006) Biophys. J. 91 3848-3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tchesnokova, V., Aprikian, P., Yakovenko, O., LaRock, C., Kidd, B., Vogel, V., Thomas, W., and Sokurenko, E. (2008) J. Biol. Chem. 283 7823-7833 [DOI] [PubMed] [Google Scholar]
- 41.Choudhury, D., Thompson, A., Stojanoff, V., Langermann, S., Pinkner, J., Hultgren, S. J., and Knight, S. D. (1999) Science 285 1061-1066 [DOI] [PubMed] [Google Scholar]
- 42.Hung, C. S., Bouckaert, J., Hung, D., Pinkner, J., Widberg, C., DeFusco, A., Auguste, C. G., Strouse, R., Langermann, S., Waksman, G., and Hultgren, S. J. (2002) Mol. Microbiol. 44 903-915 [DOI] [PubMed] [Google Scholar]
- 43.Bouckaert, J., Mackenzie, J., de Paz, J. L., Chipwaza, B., Choudhury, D., Zavialov, A., Mannerstedt, K., Anderson, J., Pierard, D., Wyns, L., Seeberger, P. H., Oscarson, S., De Greve, H., and Knight, S. D. (2006) Mol. Microbiol. 61 1556-1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saterbak, A., and Lauffenburger, D. A. (1996) Biotechnol. Prog. 12 682-699 [DOI] [PubMed] [Google Scholar]
- 45.Bouckaert, J., Berglund, J., Schembri, M., De Genst, E., Cools, L., Wuhrer, M., Hung, C. S., Pinkner, J., Slattegard, R., Zavialov, A., Choudhury, D., Langermann, S., Hultgren, S. J., Wyns, L., Klemm, P., Oscarson, S., Knight, S. D., and De Greve, H. (2005) Mol. Microbiol. 55 441-455 [DOI] [PubMed] [Google Scholar]
- 46.Anderson, B. N., Ding, A. M., Nilsson, L. M., Kusuma, K., Tchesnokova, V., Vogel, V., Sokurenko, E. V., and Thomas, W. E. (2007) J. Bacteriol. 189 1794-1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeffrey, B., Udaykumar, H. S., and Schulze, K. S. (2003) Am. J. Physiol. 285 G907-G918 [DOI] [PubMed] [Google Scholar]
- 48.Costerton, J. W., Rozee, K. R., and Cheng, K. J. (1983) Prog. Food Nutr. Sci. 7 91-105 [PubMed] [Google Scholar]
- 49.Jackson, M. B. (2006) Molecular and Cellular Biophysics, pp. 121-122, Cambridge University Press, New York
- 50.Somers, W. S., Tang, J., Shaw, G. D., and Camphausen, R. T. (2000) Cell 103 467-479 [DOI] [PubMed] [Google Scholar]
- 51.Phan, U. T., Waldron, T. T., and Springer, T. A. (2006) Nat. Immunol. 7 883-889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lou, J., Yago, T., Klopocki, A. G., Mehta, P., Chen, W., Zarnitsyna, V. I., Bovin, N. V., Zhu, C., and McEver, R. P. (2006) J. Cell Biol. 174 1107-1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lou, J., and Zhu, C. (2007) Biophys. J. 92 1471-1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stowell, C. P., and Lee, Y. C. (1980) Biochemistry 19 4899-4904 [DOI] [PubMed] [Google Scholar]