Abstract
A mixture of three homologous bioactive hydroxyproline-rich glycopeptides (HypSys peptides) of 18 amino acids in length, differing only at two residues, was isolated from leaves of Ipomoea batatas, the common sweet potato. One of the peptides represented over 95% of the isolated isopeptides, which, at 2.5 nm concentration, induced the expression of sporamin, a major defense protein of I. batatas. The sequence of the major isoform was used to synthesize a primer that identified a cDNA encoding a precursor protein. The protein contained six proline-rich regions whose sequences suggested that they might be HypSys defense signals. One of the encoded peptides, called IbHypSys IV, was identical to one of two minor components of the isolated isopeptides, but neither the major isopeptide nor the other minor isoform was found within the precursor. The six peptides encoded by the precursor gene were synthesized but with hydroxyproline residues at positions found in the native isoforms and lacking carbohydrate moieties. All of the peptides were biologically active when supplied to leaves of sweet potato plants. The gene is the first ortholog of the preproHypSys gene family to be found outside of the Solanaceae family, and its encoded peptide precursor is the first example in plants of a precursor protein with six potential peptide defense signals, a scenario only found previously in animals. The data indicate that multiple copies of the HypSys peptides in a single precursor may have an important role in amplifying wound signaling in leaves in response to herbivore attacks.
Defense signaling peptides from plants include systemin (1), hydroxyproline-rich glycopeptide systemins (HypSys peptides) from solanaceous plants (2-4), and the AtPep1 peptide from Arabidopsis and its orthologs throughout the plant kingdom (5-7). The peptides are from 18 to 23 amino acids in length, are processed from wound- and jasmonate-inducible precursor proteins, and act as amplifiers of signaling for the expression of defense genes (4, 6, 7). Cell surface receptors for systemin and AtPep1 have been isolated (8-10), but a receptor for HypSys peptides has not, although the biological activities of the peptides suggest that they are mediated by cell surface receptors.
HypSys peptides are unique among plant peptide signals, being derived from polyprotein precursors: two peptides from a tobacco precursor (2), three from a tomato precursor (3), and four from a petunia precursor (4). Studies with the tomato HypSys precursor protein have demonstrated that it is synthesized through the secretory pathway and is sequestered in cell walls (11). HypSys peptides isolated from tobacco (Nicotiana tabacum) (2) and tomato (Solanum lycopersicum) (3) activate the expression of anti-herbivore protease inhibitors and polyphenol oxidase in response to wounding and MeJA, whereas HypSys peptides isolated from Petunia hybrida (4) activate defensin I, a gene associated with defense against pathogens. Tobacco plants transformed with the tobacco HypSys precursor gene driven by the 35S promoter express elevated levels of the tobacco trypsin inhibitor in leaves and exhibit increased resistance against Helicoverpa armigera larvae (12). Here we report the isolation of three 18-amino-acid HypSys isopeptides from leaves of sweet potato (Ipomoea batatas) of the Convolvulaceae family that potently induce expression of the defense gene sporamin. A cDNA was isolated that encoded the precursor of one of the isopeptides, as well as sequences of five other homologous peptides that are potential HypSys defense signals. The research is the initial demonstration of the presence of a HypSys precursor gene outside of the Solanaceae family whose multiple HypSys peptides may have signaling roles in amplifying the defense responses of sweet potato plants against attacking herbivores.
EXPERIMENTAL PROCEDURES
Alkalinization Assay—Tomato suspension cultured cells were grown with shaking at 160 rpm in Murashige and Skoog medium as described (8). Petunia, tobacco, Arabidopsis, and sweet potato suspension cells were grown similarly in Nt3 (N. tabacum) medium as described previously (2). Three ml of 7-day-old cell cultures was transferred into 35 ml of medium in 125-ml flasks and used for assay 3-6 days after transferring. One-ml aliquots of cell culture were delivered into each well of 24-well cell culture cluster plates and equilibrated on an orbital shaker at 160 rpm for 1 h. One- to 10-μl aliquots of fractions eluted from HPLC columns were added to the cells, and the pH of the medium was recorded after 30 min. Of the six peptide sequences identified in the sweet potato precursor protein, only that of IbHypSys IV (cf. Figs. 2 and 3) could be identified among the peptides isolated from sweet potato leaves using the tomato suspension culture as an assay system.
FIGURE 2.
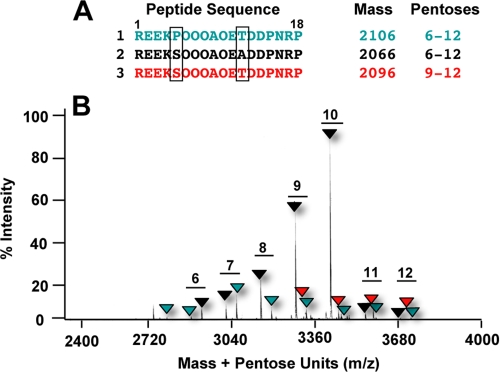
Amino acid sequences of three glycopeptides present in the purified peak from Fig. 1C, analyzed by MALDI MS. A, amino acid sequences. The boxed residues indicate the differences among the peptides. B, MALDI MS analysis of the peptide mixture from Fig. 1C, indicating the differences in masses due to the various numbers of pentose residues present in the peptides. The masses of the peptides are color-coded to match the backbone sequences shown in panel A.
FIGURE 3.
The nucleotide sequence of a cDNA encoding a precursor protein harboring IbHypSys peptide 1 (underlined with a double line) that was purified from sweet potato leaves (cf.Fig. 1). Sequences of five homologous peptides are identified with single solid lines. The peptides are named IbHypSys I through VI from the N terminus. The 23-amino-acid leader sequence is underlined with a dotted line.
IbHypSys Peptide Isolation—Sweet potato (I. batatas cv. Georgia Jet) plants were grown under green house conditions and collected for peptide isolation 6 weeks after planting. The aerial parts of the plants were sprayed with a 1.25 mm methyl jasmonate solution (Bedoukian Research, Danbury, CT) in 0.1% Triton X-100 and incubated for 15 h, and peptide extraction from leaves was performed as previously reported (3), with the following modifications.
Stage I Purification, Crude Extract Preparation—Sweet potato leaves (1.2 kg of wet weight) were frozen in liquid N2 and homogenized in a 4-liter blender for 5 min in 2.5 liters of 1% trifluoroacetic acid. The mixture was filtered through four layers of cheesecloth and two layers of Miracloth (Calbiochem) and then centrifuged at 10,000 × g for 60 min. The supernatant was adjusted to pH 4.3 by slowly adding 10 n NaOH with stirring, and the precipitate was sedimented by centrifugation at 10,000 × g. The clear supernatant liquid was collected and readjusted to pH 2.2 with 1 n HCl and recentrifuged at 10,000 × g to clarify. The clear supernatant solution was passed through a 40-μm 3 × 25-cm C18 reversed-phase flash column (Bondesil, Varian Analytical Instruments, Walnut Creek, CA) equilibrated with 0.1% trifluoroacetic acid, H2O. The column was washed with 100 ml of 0.1% trifluoroacetic acid, H2O, and the bound material was eluted with 250 ml of 40% methanol, 0.1% trifluoroacetic acid at 8 p.s.i. compressed nitrogen gas. The eluate was dried by lyophilization. The procedure was repeated 10 times to yield a total of 12 g of dry powder.
Stage II Purification, Sephadex Chromatography—The dry powder was dissolved in 30 ml of 0.1% trifluoroacetic acid, H2O and centrifuged at 10,000 × g for 15 min to clarify. The supernatant was applied to a Sephadex G-25 column (5.5 × 35 cm) equilibrated with 0.1% trifluoroacetic acid, H2O, and the collected fractions (8 ml) were assayed for alkalinization activities with tomato suspension cells as described above using 10 μl of each fraction with 1 ml of cells. Activity was located at or near the column void, and these fractions were pooled and lyophilized, yielding about 1 gram of dry weight.
Stage III Purification, Initial HPLC Chromatography—The dry powder was dissolved in 4 ml of 0.1% trifluoroacetic acid, H2O and centrifuged at 10,000 × g for 15 min. The supernatant was filtered through a Millex-HV PVDF membrane filter (Millipore, Bedford, MA), and 1-ml aliquots were applied to a preparative reversed-phase C18-HPLC column (218TP1022, 10-μm, 2.2 cm ID × 25 cm, Vydac, Hesperia, CA) equilibrated with 0.1% trifluoroacetic acid, H2O with a flow rate of 4 ml/min. After 5 min, an elution program from 0 to 40% acetonitrile, 0.1% trifluoroacetic acid was applied over 90 min, and fractions were collected at 1-min intervals. The absorbance was monitored at 220 nm. Ten-μl aliquots from each fraction were added to 1 ml of tomato suspension cells to determine the alkalinizing activity. The active fraction at 37-39 min was lyophilized. The yield from four duplicate runs was 62 mg, dry weight.
Stage IV Purification, Cation Exchange Chromatography—One-third of the dry material was dissolved in 5 mm potassium phosphate, pH 3, in 25% acetonitrile buffer and applied to a polySULFOETHYL Aspartamide™ strong cation exchange column (5-μm, 4.6 mm ID × 200 mm, The Nest Group, Southborough, MA). The column was equilibrated with 5 mm potassium phosphate, pH 3, in 25% acetonitrile. Two min after adding the sample, a 90-min gradient from 0 to 100% elution buffer consisting of 5 mm potassium phosphate, 500 mm potassium chloride, pH 3, in 25% acetonitrile was applied to the column. Absorbance was monitored at 220 nm at a flow rate of 1 ml/min, and fractions were collected at 1-min intervals. Biological activities from each fraction were determined in the alkalinization assay using 10-μl aliquots. The activity was identified in fractions eluting at 33-35 min. The active fractions from three consecutive separations were combined and lyophilized. The dry material containing salt was not weighed.
Stage V Purification, Sequential HPLC Separations—The dried material was dissolved in 1 ml of 0.1% trifluoroacetic acid, H2O and applied to an analytical reversed-phase C18 HPLC column (218TP54, 5-μm 4.6 mm ID × 250 mm, Vydac) equilibrated with 0.1% trifluoroacetic acid, H2O at a flow rate of 1 ml/min. A gradient was performed from 0 to 40% methanol, 0.05% trifluoroacetic acid 2 min after sample injection. The absorbance was monitored at 214 nm, and 10-μl aliquots from each fraction were assayed using 1 ml of tomato suspension cells. Active fractions eluting at 44-46 min were pooled, and methanol was removed by vacuum evaporation. The combined fractions were loaded onto a narrow bore reversed-phase C18 HPLC column (218TP52, 5-μm, 2.1 mm ID × 250 mm, Vydac) equilibrated with 0.1% trifluoroacetic acid, H2O at a flow rate of 0.25 ml/min. After 2 min, an elution program from 0 to 30% acetonitrile, 0.1% trifluoroacetic acid was performed over 90 min, and 0.25-ml fractions were collected at 1-min intervals. The absorbance was monitored at 214 nm. Aliquots (2 μl) were added to 1 ml of tomato suspension cells to determine the alkalinizing activity from each fraction. Active fractions eluted at 50-53 min and were pooled (1 ml total). The pooled sample was rechromatographed on a polySULFOETHYL Aspartamide™ column as described above. The purification conditions were the same as described above, only the 90-min gradient was from 0 to 45% elution buffer. Alkalinization activities in each fraction were determined using 10-μl aliquots. Lyophilization of the active fractions (55-56 min) yielded a dry salty powder that was dissolved in 1 ml of 0.1% trifluoroacetic acid, H2O. To further purify the active fraction, a narrow bore reversed-phase C18 HPLC column was utilized as described previously but with a gradient from 0 to 30% methanol, 0.05% trifluoroacetic acid over 90 min, and fractions were collected at 1-min intervals. The absorbance was monitored at 214 nm. Two-μl aliquots were used to determine the alkalinizing activity from each fraction. The active fraction was found at 73-75 min. Methanol was removed by vacuum evaporation, and the peptide sample was loaded on the narrow bore reversed-phase C18 HPLC column again. The column was equilibrated with 10 mm potassium phosphate, pH 6, at a flow rate of 0.25 ml/min. A 90-min gradient was performed from 0 to 40% with an elution buffer consisting of 10 mm potassium phosphate in 50% acetonitrile, pH 6. The alkalinization assay revealed active fractions eluting at 45-47 min. These fractions were pooled and further purified and desalted on a narrow bore reversed-phase C18 HPLC column with a 0-40% acetonitrile elution gradient for 90 min. The purified peptides were quantified by the peak areas relative to areas measured with known quantities of synthetic NtHypSys I and II. The total yield was 125 pmol.
Peptide Sequence Analysis and Synthesis—Peptide sequence was analyzed by Edman chemistry on an Applied Biosystems (Foster City, CA) Procise model 492 protein sequencer. Peptides were synthesized by N-(9-fluorenyl) methoxycarbonyl chemistry by solid-phase techniques using an Applied Biosystems model 431 synthesizer and purified by reversed-phase HPLC. MALDI spectra were obtained on an Applied Biosystems 4800 tandem time-of-flight mass spectrometer with 200-Hz neodymium-doped yttrium aluminum garnet laser. Concentrated samples in aqueous solution were mixed 1:1 with matrix solution (α-cyano-4-hydroxycinnamic acid, 6 mg/ml in 50:50 acetonitrile:0.25% trifluoroacetic acid in water) and air-dried. Calibrated MS spectra (±0.02 Da) were obtained as the summations of 1000-2000 laser shots, whereas MS/MS spectra (±0.1 Da) were summations of 10,000-20,000 laser shots. The MS/MS spectra of the glycosylated and proline- and hydroxyproline-rich peptides show nearly complete series of b and y ions.
Cloning of IbpreproHypSys cDNA—Total RNA was prepared from sweet potato leaves by extraction with TRIzol reagent (Invitrogen). 3′-RACE and 5′-RACE reactions were performed by using a SMART™ RACE cDNA amplification kit (Clontech) according to the manufacturer's instructions. Based on the amino acid sequence of IbHypSys isopeptide 2, a degenerate primer, 26-mer oligonucleotides 5′-GA(A/G)GA(A/G)AA(A/G)(A/T)(G/C)(A/T/C/G)CC(A/T/C/G)CC(A/T/C/G)CC(A/T/C/G)GC(A/T/C/G)CC-3′ (sense) encoding EEKSPPPAP, was synthesized and used for the 3′-RACE reaction.
The 25-mer oligonucleotide antisense primer, 5′-GCTCAGGCTGATGCCAGGAAATCAC-3′, in the C-terminal was used for the 5′-RACE reaction. The cDNA was subcloned into the pCR 2.1-TOPO vector (Invitrogen) and sequenced by using the M13 primers (forward, 5′-GTAAAACGACGGCCAG-3′; reverse, 5′-CAGGAAACAGCTATGAC-3′).
RT-PCR Analyses of Relative Gene Expression Levels—Sweet potato leaves were either wounded with a hemostat or sprayed with a 1.25 mm methyl jasmonate solution (Bedoukian Research, Danbury, CT) in 0.1% Triton X-100 for 2, 4, and 6 h, respectively. Young excised plants were supplied with peptides in distilled water through their cut stems and incubated in light for 2 h. Leaf RNA was isolated using TRIzol reagent (Invitrogen), and 5 μg was reverse-transcribed using SuperScript™ III RNase H- reverse transcriptase (Invitrogen). Semiquantitative PCR reactions were carried out with ExTaq Hot Start polymerase (Fisher Scientific). PCR conditions were the following: an initial step of 3 min, 94 °C; 30 cycles of the following three steps, 30 s, 94 °C; 30 s, 58 °C; 1 min, 72 °C; and a terminal step of 7 min, 72 °C. The PCR products were separated on a 1.2% agarose gel and visualized on a Chemi Genius™ system using GeneSnap version 6.00.256 software (Syngene, Fredrick, MD). Analyses of gel images were performed with GeneTools software version 3.02.00 (Syngene). Relative abundance of transcripts was estimated from the expression of actin gene as the standard to give a numerical ratio.
An IbpreproHypSys forward primer (5′-TGC TAC CGG TGA TTT CCT CT-3′) and a reverse primer (5′-GTC CGC CAT TGA AAT CAC TT-3′) were employed to amplify a 676-bp product. A sporamin (U17333) forward primer (5′-CAA TCC CAT CCG CCT CCC CAC-3′) and reverse primer (5′-GTG TTA CAT TAC ACA TCG GTA GGT TTG ATG AC-3′) were used to generate a 580-bp product. Actin mRNA was used as an internal control for each PCR reaction, with a forward primer (5′-ATG GCA GAC GGT GAG GAT ATT CA-3′) and a reverse primer (5′-GCC TTT GCA ATC CAC ATC TGT TG-3′).
RESULTS
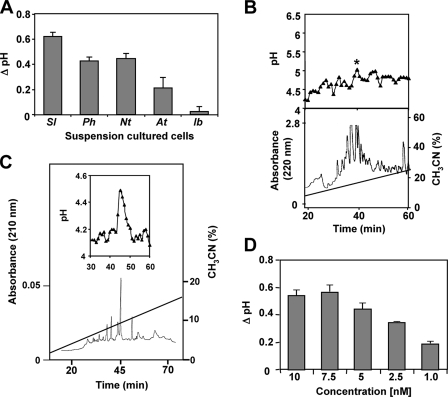
Isolation and Characterization of an Isopeptide Mixture from Sweet Potato Leaves—Systemin and HypSys peptides are defense signals that have been identified in several species of the Solanaceae family but not in species of any other plant family. A search was initiated to seek the presence of defense-signaling peptides in leaves of sweet potato (I. batatas), a species of the Convolvulaceae family, which is phytogenetically near the Solanaceae family. Alkalinization assays using plant suspension cultured cells have been the basis for detecting and isolating peptide signals originating from both plants and microorganisms (2, 13-15), and a soluble extract from sweet potato leaves enriched in peptides was assayed for its ability to cause the alkalinization of the medium of suspension cultures of sweet potato. The extract did not cause an alkalinization of sweet potato suspension cultures. In the past, we have noted that some suspension cultures do not respond to endogenous peptide signals but that peptide signals are often recognized by cultures from other species. Assaying the sweet potato extract for alkalinization of suspension cultured tomato, tobacco, and petunia cells gave a positive response, with tomato giving the strongest response (Fig. 1A). The tomato culture was therefore utilized to assay the extract components following an initial HPLC separation. Ten μl from each of the eluting fractions was added to 1 ml of tomato suspension cultured cells, and a weak but reproducible increase in alkalinizing activity was detected in some of the fractions eluting from the column. Only the fractions eluting at about 40 min, labeled in Fig. 1B with a star, when pooled and further purified using HPLC as described under “Experimental Procedures,” yielded a component with strong alkalinization activity (Fig. 1C). The active fraction was quantified by comparing the peak area with that of known synthetic peptides and found to be half-maximally active in the alkalinization assay at about 2.5 nm (Fig. 1D), the same half-maximal activity found with HypSys peptides from tobacco, tomato, and petunia (2-4). MALDI-MS/MS and amino acid sequence analyses of the peak component(s) revealed that it contained a major species having a backbone sequence of REEKSOOOAOEADDPNRP. MALDI-MS analyses also revealed two minor peptides in addition to the major peptide with substitutions at residue 5 (Ser to Pro) and residue 12 (Ala to Thr) (Fig. 2A). The major peptide (peptide 2) was identical to the peptide sequenced by Edman degradation and represented over 95% of the three peptides. All three peptides were decorated with from 6 to 12 pentose units each (Δ132 mass units/residue) (Fig. 2B), characteristic of hydroxyproline-rich glycopeptide signals isolated previously.
FIGURE 1.
Isolation of IbHypSys. A, alkalinization of the medium of suspension cultured cells of tomato (Sl), petunia (Ph), tobacco (Nt), Arabidopsis (At), and sweet potato (Ib). Ten μl of a crude extract from sweet potato (I. batatas) leaves were added to 1 ml of cells. The change in pH was recorded after 30 min. The data represent the average of three separate experiments. B, lower panel, the crude extract was dissolved in 0.1% trifluoroacetic acid, H2O and chromatographed through a preparative reversed-phase C18-HPLC column with a 90-min gradient from 0 to 40% acetonitrile at a flow rate of 4 ml/min (Stage III of purification). Upper panel, alkalinization assays of 10 μl from each fraction when added to 1 ml of tomato suspension culture as described under “Experimental Procedures.” C, further purification of the peptide eluting in the fraction of the peak in the upper panel of B, marked (*) as described in Stages IV and V of the purification. Lower panel, the peptide mixture was separated on a narrow bore reversed-phase C18 HPLC column with 10 mm potassium phosphate, pH 6, at a flow rate of 0.25 ml/min. A 90-min gradient was performed from 0 to 40% with an elution buffer consisting of 10 mm potassium phosphate in 50% acetonitrile, pH 6. One-minute fractions were monitored at 220 nm (Stage V of purification). D, the concentration dependence of the purified isopeptides from the final purification in Stage V in the alkalinization assay. The change in pH of the suspension cell medium in response to the increasing concentration of peptides was recorded 30 min after the addition of peptide. The change in pH was compared with distilled H2O controls of equal volume (10 μl). The data represent the average of three separate experiments.
Isolation of an IbpreproHypSys cDNA—Primers constructed from the sequence of the major isopeptide in the isolated mixture of three homologous peptides from sweet potato leaves (Fig. 2A, Peptide 2) were used to isolate a cDNA (Fig. 3) from sweet potato leaves that encoded a protein of 291 amino acids containing an 18-amino-acid sequence (underlined with a double line in the figure) that was identical to peptide 1 in the peptide mixture from Fig. 2. The deduced precursor protein contained five other peptide regions of 18 amino acids each that were rich in proline residues and were suspected to be homologs of the isolated HypSys peptides. The six putative peptides were named IbHypSys I through VI in the order they occurred in the precursor protein, counting from the N terminus (Fig. 3), with peptide 1 from Fig. 2 being IbHypSys IV. The other two isolated peptides were not found among the peptide sequences within the precursor protein, suggesting that they were derived from alleles. Attempts to isolate cDNAs containing peptides 2 and 3 were unsuccessful.
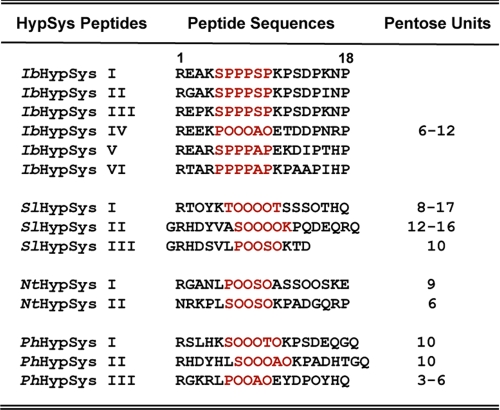
The primary amino acid sequences of the six peptides encoded in the precursor have sequences typical of HypSys peptides and were considered potential peptide signals. Post-translational hydroxylations and glycosylation of some of the prolines of the primary transcript of the precursor are reflected in the isolated HypSys peptides. In Fig. 4, the sequences of the six peptides are compared with sequences of HypSys peptides previously isolated from several Solanaceae species.
FIGURE 4.
Comparisons of the amino acid sequences of the six putative peptides encoded in the precursor protein deduced from the cDNA in Fig. 2. The peptides are aligned with the sequences of known HypSys peptides from tomato (SlHypSys I, II, and III, where Sl stands for tomato), tobacco (NtHypSys I and II), and petunia (PhHypSys I, II, and III, where Ph stands for petunia). The central cores (highlighted in red) of the deduced peptides are enriched in prolines, whereas the cores of isolated peptides contain 3-4 hydroxyproline residues each.
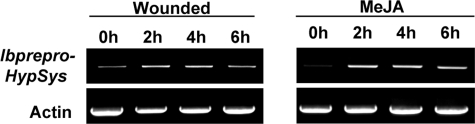
Wound and Methyl Jasmonate Induction of IbpreproHypSys Transcripts—In the Solanaceae family, the defense-signaling peptides systemin (1) and HypSys (2) and an ortholog of AtPep1 (6)4 are derived from precursor proteins that are wound-inducible and jasmonate-inducible in leaves, jasmonates being mobile signals that activate defense genes in response to both herbivore and pathogen attacks (16). To determine whether IbpreproHypSys was wound-inducible and/or methyl jasmonate-inducible, lower leaves of young sweet potato plants were either wounded using a hemostat or exposed to methyl jasmonate vapors, and the transcript levels of the gene were assayed with time using semiquantitative RT-PCR analyses. The levels of IbpreproHypSys mRNA increased over basal levels within 2-4 h after wounding and then decreased at 6 h (Fig. 5). Methyl jasmonate treatment caused the levels to increase within 2 h, but the levels remained elevated through 6 h. The time course of expression of IbpreproHypSys in response to wounding and methyl jasmonate is similar to those of HypSys precursor genes in leaves of tobacco, tomato, and petunia (2-4).
FIGURE 5.
RT-PCR analyses of the expression of IbpreproHypSys in response to wounding and methyl jasmonate in young sweet potato leaves. Total RNA was isolated from leaves at 0, 2, 4, 6 h after wounding or to exposure to methyl jasmonate vapors. mRNA levels were assayed by semiquantitative RT-PCR. Actin expression was analyzed as a control. The data are representative of three replicate experiments.
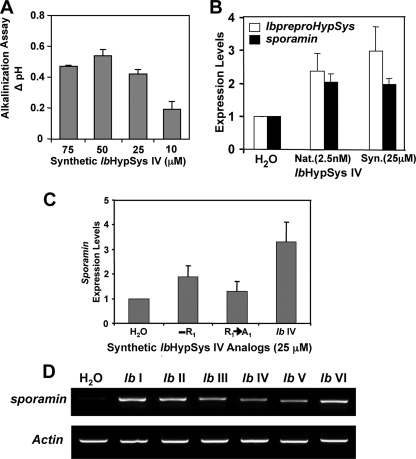
Activities of Synthetic IbHypSys Peptides—A hydroxyproline-rich peptide was synthesized based on the amino acid sequence of the isolated IbHypSys 1 isopeptide sequence (Fig. 2A) that was found in the precursor protein (Fig. 3) but lacking carbohydrate adducts. The alkalinization activity of the synthetic peptide exhibited a half-maximal alkalinizing activity at concentrations between 10 and 25 μm (Fig. 6A), which is much weaker than the activity of the isolated peptides (∼2.5 nm). The low activity of the synthetic peptide was totally abolished in a synthetic analog in which the N-terminal Arg was substituted with an Ala (data not shown).
FIGURE 6.
Biological activities of native and synthetic IbHypSys peptides. A, the change in pH of the medium of suspension cultured tomato cells in response to increasing concentrations of IbHypSys IV, when compared with H20 controls. The pH was recorded 30 min after adding 10 μl of the peptide solution to 1 ml of suspension cultured tomato cells. B, expression of IbpreproHypSys and sporamin in excised sweet potato leaves supplied for 2 h with 2.5 nm native (Nat.) IbHypSys or 25 μm synthetic (Syn.) IbHypSys IV, assayed by RT-PCR. The expression of actin was used as an internal control. Leaves supplied with water alone (H2O) were assayed as a reference. The data shown are from a representative experiment from three replications. C, sporamin expression levels induced in sweet potato leaves fed synthetic IbHypSysIV analogs. IbHypSysIV and synthetic analogs were dissolved in distilled H2Oata concentration of 25 μm and supplied to excised leaves for 2 h as described under “Experimental Procedures.” IbHypSysIV minus the N-terminal arginine (-R1), IbHypSysIV with alanine substituted for arginine at the amino terminus (R1→A1) The expression levels of sporamin were estimated using RT-PCR with actin as an internal control. Data represent three replications. Error bars indicate standard error of the mean. D, the induction of sporamin by synthetic peptides (see “Experimental Procedures”). Excised young sweet potato leaves were supplied with 25 μm of each synthetic peptide or water for 2 h, and the expression levels of sporamin were estimated using RT-PCR, with actin as an internal control.
Sporamin is the major storage protein in sweet potato tuberous roots and is a trypsin inhibitor that can also play a defensive role in leaves against herbivores (17). Sporamin is known to be wound- and methyl jasmonate-inducible in sweet potato leaves (18). Synthetic IbHypSys IV supplied to excised sweet potato leaves at 25 nmol/leaf for 2 h induced the expression of sporamin similar to levels induced by 2.5 pmol of native peptide, quantified by real-time PCR (Fig. 6B). These results paralleled the results found in the alkalinization assays.
The other five putative IbHypSys peptides in the precursor (Fig. 3, underlined) were synthesized with hydroxyproline residues substituted for prolines in the central core regions at residues where hydroxyprolines are present in the native HypSys peptides (cf. Fig. 4). All six synthetic peptides were active in inducing sporamin expression in excised leaves, assayed by RT-PCR (Fig. 6D).
DISCUSSION
This report describes for the first time the isolation of a HypSys peptide and its precursor cDNA from a species outside of the Solanaceae family, i.e. sweet potato (I. batatas) of the Convolvulaceae family. The isolation was made possible by using an alkalinization assay with tomato cell suspension cultures since we were unsuccessful in developing a sweet potato suspension culture in which the sweet potato leaf extracts would alkalinize the culture medium. In our previous studies,5 we had found that suspension cultures of some Solanaceae species could detect HypSys peptide signals derived from other Solanaceae species, whereas some cultures could not. We therefore assayed the sweet potato leaf extract with suspension cultures of tobacco, tomato, petunia, and Arabidopsis for their alkalinizing activity. The extract caused alkalinization of the medium of all of the cultures (Fig. 1A), with the tomato culture being most responsive. Using the tomato suspension culture to assay bioactive components eluting from HPLC columns, an isopeptide mixture from sweet potato leaves was obtained (Fig. 1, B and C). The half-maximal activity of the peptides in the alkalinization assay was about 2.5 nm (Fig. 1D), which was similar to the half-maximal activities of HypSys peptides that have been isolated previously from tobacco, tomato, and petunia (2-4). Amino acid sequence analysis and MALDI MS/MS indicated that the peptides contained four hydroxyproline residues in a central core consisting of -(S/P)OOOAO- and were glycosylated with varying numbers of pentose units, as evidenced by the pentose ladders (Fig. 2). These analyses revealed the presence of two minor peptides (less than 5%, Fig. 2, Peptides 1 and 3) that were homologs of the major peptide.
Synthesis of degenerate primers based on the nucleotide sequence of the major peptide led to the isolation of a cDNA encoding a protein containing a leader sequence and an internal 18 amino acid sequence that was identical to isolated peptide 1 and not peptide 2. The cDNA that was isolated encoded a protein containing five other 18-amino-acid sequences that were proline-rich. None of the five sequences matched the sequences of the two other isolated peptides. This indicated that at least one other precursor gene is present in sweet potato plants that encode HypSys peptides. Several attempts to isolate cDNAs with coding regions for peptides 2 and 3 were unsuccessful. The reasons for not finding the cDNAs coding for peptides 2 and 3 may be a consequence of developmental or environmental factors. The isolated peptides were from large sweet potato plants consisting mainly of mature leaves, with some young developing leaves. The cDNA was isolated from small developing leaves that may have been preferentially expressing the gene from which the cDNA was isolated, a possibility that is currently under investigation.
All HypSys peptide precursor proteins identified to date, including IbpreproHypSys (cf. Fig. 2), exhibit signature motifs. One motif is within a 10-amino-acid sequence overlapping the signal peptidase splice site of -AR-. In the sweet potato precursor, the sequence at this site is -AQARRL-, and in the tomato, tobacco, and two petunia precursors, the sequences are -AQARTL-, -AEARTL-, AEARSL-, and -AEARTL-, respectively. This motif appears to be part of the substrate specificity site for recognition by the leader peptidase. Another highly conserved motif in each peptide is a -YGR- or -FGR-sequence at the N terminus of 10 of the 14 peptides in Fig. 3. These motifs appear to be part of the recognition sites for a processing enzyme(s) that accommodate large hydrophobic amino acids at the active site in which Tyr and Phe are favored, with less affinity toward other amino acids. This is the first indication in plants of a specificity site for an enzyme that processes peptides from precursor proteins and provides an opportunity to fashion synthetic peptides as substrates to seek such an enzyme. Finally, the sequence of LASA found at the C terminus of the sweet potato precursor peptide is similar to sequences in tobacco and tomato, i.e. -LASY and -QHSY, respectively, and in the two petunia HypSys precursors, -LASY and -LAST. The significance of these motifs is not known but may have a role in localizing the protein in the cell wall (11) or anchoring the precursor to cell wall components.
The six proline-rich peptide regions encoded within the cDNA were named IbHypSys I through VI, beginning from the N terminus of the encoded precursor protein (cf. Fig. 3). The IbHypSys IV sequence is identical with Fig. 2A, Peptide 1. The proline-rich regions present in all six peptides exhibit similarities to HypSys peptides previously isolated from various species of the Solanaceae family (Fig. 4), suggesting that the encoded protein might be a polyprotein precursor of six HypSys defense signaling peptides. The precursor is reminiscent of the animal polyprotein proenkephalin (19) that harbors six copies of (Met5)-enkephalin and a copy of (Leu5)-enkephalin, apparently to produce large amounts of signal in response to a variety of cues.
The six HypSys peptides encoded within the HypSys precursor cDNA were synthesized to assess their biological activities. Proline residues were replaced with hydroxyprolines at four positions where hydroxyprolines were located in the core regions of isolated IbHypSys peptides (cf. Fig. 3). However, the synthetic peptides were lacking carbohydrate adducts that are found in the native HypSys peptides. Synthetic IbHypSys IV induced the expression of its precursor, IbpreproHypSys and sporamin (Fig. 6B), but at concentrations several magnitudes higher than native IbHypSys IV, emphasizing the importance of the carbohydrate for maximal activity and similar to the activity of synthetic HypSys peptides from tobacco and tomato (2, 3). Elevated sporamin expression levels of sweet potato leaves fed synthetic IbHypSys IV were abolished by the substitution of alanine for arginine at the N terminus of the peptide (Fig. 6C), consistent with previous activity data for synthetic NtHypSys I and II (20). The other five synthetic peptides also activated the expression of sporamin when supplied at μm concentrations (Fig. 6D). The presence of pentose adducts in the HypSys peptides is clearly important for activity, and their composition, linkages, and attachment sites to the peptides are being investigated.
The biological properties of the HypSys peptides indicate that they interact with specific cell surface receptors. Peptide signals derived either from plant pathogens (e.g. flagellin fragments and elongation factor Tu fragments) or from plants (e.g. systemin and AtPep1) are known to interact with cell surface receptors and to cause alkalinization of the medium of plant cultured cells (13). The lack of an alkalinization response of the sweet potato suspension culture to IbHypSys peptides may be due to the absence of a component of the receptor system in our sweet potato suspension cultures that is present in tomato cell suspension cultures. A receptor for HypSys peptides has not been identified but is currently being sought in tomato cells. If present, a HypSys receptor would provide a useful tool to further understand the early steps of HypSys defense signaling. The tomato suspension cultures were unable to identify other IbHypSys peptides present in the precursor protein. To determine whether the sweet potato precursor produces all six of the putative hydroxyproline-rich peptides, a sweet potato suspension culture assay must be developed that can be used in an alkalinization assay to isolate additional peptides from sweet potato leaf extracts.
Expression of the IbHypSys peptide precursor in leaves, like other HypSys peptide precursors, is inducible by wounding and methyl jasmonate (Fig. 5). The increase in levels of the precursor protein in response to wounding (herbivore attacks) may contribute to a rapid amplification of defense signaling peptides as found with other endogenous peptide signals (7). The HypSys peptides, when supplied to young sweet potato leaves, induced sporamin, an herbivore-inducible defense gene of I. batatas (17, 21-23). The mechanism of release of HypSys peptides from cell wall-associated precursors in response to wounding is not known, but wounding causes a rapid release of linolenic acid from membranes and the subsequent production of jasmonate (24). One scenario is that jasmonate may trigger the synthesis and transport of a protease(s) to the apoplast to process the HypSys precursor and release the HypSys peptides. On the other hand, the protease(s) may already be in the cell wall as a proenzyme(s) or in an inactive state that can be activated in response to ion fluxes or other physiological changes. In any event, peptides produced in the apoplast appear to interact with a cell surface receptor to initiate HypSys defense peptide signaling.
Acknowledgments
We thank Sue Vogtman and Julia Gothard for growing plants, Guido Barona for technical assistance, and Gerhard Munske for peptide sequence analyses.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) EF621761.
This research was supported by National Science Foundation Grants IBN 0090766 and 0623029; National Institutes of Health Grant 1S10RR022538-01; the Charlotte Y. Martin Foundation; and the Washington State University College of Agriculture, Human and Natural Resources Sciences. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: Nt, N. tabacum; Ib, I. batatas; At, Arabidopsis; HPLC, high pressure liquid chromatography; MALDI, matrix-assisted laser desorption; MS, mass spectrometry; MS/MS, tandem mass spectrometry; RT-PCR, reverse transcription-PCR; RACE, rapid amplification of cDNA ends.
Y.-C. Chen and C. A. Ryan, unpublished results.
G. Pearce and C. A. Ryan, unpublished results.
References
- 1.Pearce, G., Strydom, D., Johnson, S., and Ryan, C. A. (1991) Science 253 895-898 [DOI] [PubMed] [Google Scholar]
- 2.Pearce, G., Moura, D. S., Stratmann, J., and Ryan, C. A. (2001) Nature 411 817-820 [DOI] [PubMed] [Google Scholar]
- 3.Pearce, G., and Ryan, C. A. (2003) J. Biol. Chem. 278 30044-30050 [DOI] [PubMed] [Google Scholar]
- 4.Pearce, G., Siems, W. F., Bhattacharya, R., Chen, Y.-C., and Ryan, C. A. (2007)) J. Biol. Chem. 282 17777-17784 [DOI] [PubMed] [Google Scholar]
- 5.Huffaker, A., Pearce, G., and Ryan, C. A. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 10098-10103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huffaker, A., and Ryan, C. A. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 10732-10736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ryan, C. A., Huffaker, A., and Yamaguchi, Y. (2007) Cell. Microbiol. 9 1902-1908 [DOI] [PubMed] [Google Scholar]
- 8.Scheer, J. M., and Ryan, C. A. (1999) Plant Cell 11 1525-1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamaguchi, Y., Pearce, G., and Ryan, C. A. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 10104-10109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horton, N., Cano-Delgado, A., Harrison, K., Montoya, T., Chory, J., and Bishop, G. J. (2007) Plant Cell 19 1709-1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Narváez-Vásquez, J., Pearce, G., and Ryan, C. A. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 12974-12977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren, F., and Lu, Y.-T. (2006) Plant Sci. (Shannon) 117 286-292 [Google Scholar]
- 13.Felix, G., and Boller, T. (1995) Plant J. 7 381-389 [Google Scholar]
- 14.Felix, G., Duran, J. D., Volko, S., and Boller, T. (1999) Plant J. 18 265-276 [DOI] [PubMed] [Google Scholar]
- 15.Schaller, A., and Oecking, C. (1999) Plant Cell 11 263-272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wasternack, C. (2007) Ann. Bot. (Lond.) 100 681-697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shewry, P. R. (2003) Ann. Bot. 91 755-769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang, S.-J., Lan, Y.-C., Chen, S.-F., and Yeh, K.-W. (2004) Plant Mol. Biol. 48 223-231 [DOI] [PubMed] [Google Scholar]
- 19.McLaughlin, P. J. (2006) in Handbook of Bioactive Peptides (Kasten, A. J., ed) pp, 1313-1318, Elsevier Inc., Burlington, MA
- 20.Ryan, C. A., Pearce, G., Scheer, J., and Moura, D. S. (2002) Plant Cell 14 S251-S264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai, D. G., Thurau, T., Tian, Y. Y., Lange, T., Yeh, K.-W., and Jung, C. (2003) Plant Mol. Biol. 51 (6), 839-849 [DOI] [PubMed] [Google Scholar]
- 22.Yeh, K.-W., Chen, J.-C., Lin, M.-I., Chen, Y.-M., and Lin, C.-Y. (1997) Plant Mol. Biol. 33 565-570 [DOI] [PubMed] [Google Scholar]
- 23.Yeh, K.-W., Lin, M.-I., Tuan, S.-J., Chen, Y.-M., Lin, C.-Y., and Kao, S.-S. (1997) Plant Cell Rep. 16 696-699 [DOI] [PubMed] [Google Scholar]
- 24.Conconi, A., Miquel, M., and Ryan, C. A. (1996) Plant Physiol. 111 797-803 [DOI] [PMC free article] [PubMed] [Google Scholar]