Abstract
Among Caucasian males with normal color vision, long-wavelength sensitive (L) cones outnumber middle-wavelength sensitive (M) cones by nearly three to one, on average, and the L and M cone opsin genes are arrayed on the X-chromosome with the L opsin gene being closest to an upstream enhancer element termed the Locus Control Region (LCR). Interaction between an opsin gene promoter and the LCR is required to mediate normal opsin gene expression, and the relative proximity of the L opsin gene promoter (4,000 base pairs for L compared to 40,000 base pairs for the M opsin gene promoter) has been proposed to endow the L gene with the advantage in competing for interaction with the LCR, thereby accounting for the nearly 3:1 ratio of L:M cones. This proximal advantage hypothesis predicts that the L:M cone ratio will be similar among populations that share the same X-chromosome opsin gene array organization. Here we tested this hypothesis by examining a sample of males of African descent and found them to have a significantly different average L:M ratio compared to Caucasian males even though their L/M gene arrays were indistinguishable from arrays in males of Caucasian descent. How these observations might be reconciled is discussed.
Keywords: cone ratio, cone photopigments, human color vision, cone photoreceptors, variation in cone ratio
Introduction
The ratio of long-wavelength (L) to middle-wavelength (M) cones is widely variable among Caucasian males, averaging 2.7:1 in men with normal color vision (Carroll, Neitz, & Neitz, 2002; Hofer, Carroll, Neitz, Neitz, & Williams, 2005), and it has been hypothesized that it is determined, at least in part, by the organization of the L and M opsin genes on the X-chromosome (Smallwood, Wang, & Nathans, 2002). The most typical arrangement is an L gene followed by one or more M genes. Transcription of the X-chromosome opsin genes requires a promoter element contained within the 236 base pairs (bp) immediately 5′ to the coding sequence of each gene, and an enhancer element, also termed the Locus Control Region (LCR), contained within 600 bp of DNA that lies between 3.1 and 3.7 kilobase pairs (kb) upstream of the translational start codon of the X-chromosome opsin gene array (Nathans et al., 1989; Wang et al., 1992).
In adult human L and M cone photoreceptors only one opsin gene is expressed (Hagstrom, Neitz, & Neitz,2000), and it has been proposed that mutually-exclusive expression is mediated by the LCR (Smallwood, Wang, & Nathans, 2002; Wang et al., 1999; Nathans et al., 1989; Wang et al., 1992). From an evolutionary perspective, it appears likely that human L and M cone photoreceptors represent a single cell type, and the stochastic choice of which gene is expressed, L or M determines the cone type. The L and M cone opsin genes in humans resulted from a gene duplication that occurred in the Old World primate lineage after the divergence of Old and New World primates (Nathans, Thomas, & Hogness, 1986; Neitz, Carroll, & Neitz, 2001). Typically, New World primates have a single opsin gene on the X-chromosome, whereas Old World primates have two, an L and an M opsin gene (Boissinot et al., 1998; Jacobs & Neitz, 1985; Jacobs & Neitz, 1987; Jacobs, Neitz, & Neitz, 1993). The promoter of the ancestral gene was included in the duplication, but the LCR was not, thus the tandem L and M opsin genes in Old World primates must share a single LCR. According to the model proposed by Nathans and colleagues (Figure 1), the LCR makes a one-time, stochastic choice to form a permanent complex with the promoter of one of the X-chromosome opsin genes. The promoter of the L gene is closer to the LCR than is the promoter of the M gene (Vollrath, Nathans, & Davis, 1988), leading to the suggestion that the LCR is biased, choosing the L opsin gene more often because of its proximity (Smallwood, Wang, & Nathans, 2002), thereby accounting for the average 2.7:1 cone ratio. Indeed, proximity effects have been demonstrated in transgenic mice carrying an artificial mini opsin gene array in which the LCR was linked in tandem to two different reporter genes, the first driven by an L gene promoter, the second driven by an M gene promoter (Smallwood, Wang, & Nathans, 2002; Wang et al., 1999). The reporter genes exhibited mutually exclusive expression in a fraction of cones. The effect of distance was tested by inserting a 9 kb spacer between the reporter genes in the artificial mini-array to increase the distance between the LCR and the distal promoter. The presence of the spacer nearly abolished expression of the distal reporter gene in retinas of transgenic mice, with more than 99% of cones expressing the proximal gene in the artificial array with the 9 kb spacer versus only 65-95% expressing the proximal gene in the absence of the spacer (Smallwood, Wang, & Nathans, 2002).
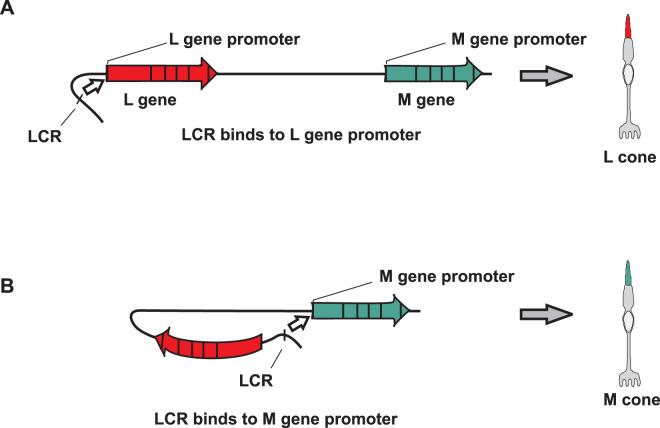
Figure 1.
Nathans and colleagues proposed that the LCR forms a stable and irreversible complex with one gene in the array, activating its expression and determining the fate of the photoreceptor as L or M. (A) If the LCR forms a complex with the L gene promoter, the cone becomes an L cone. (B) If the LCR forms a complex with the M gene promoter, the cell becomes an M cone.
In experiments reported here, we tested the hypothesis that human retinas have more L than M cones because incipient L/M photoreceptors are inherently more likely to become L cones as a consequence of the L opsin gene being relatively more proximal to the LCR than the M gene. The hypothesis predicts that a preponderance of L cones would be a consistent feature of populations that share a similar array structure. We used the flicker photometric electroretinogram (FP-ERG) to estimate the L:M cone ratios and we used genetic analyses to characterize the X-chromosome opsin gene arrays in 27 males of African descent. The mean L:M ratio in the males of African descent was significantly lower than that reported for males of Caucasian descent, despite the fact that the two groups share a common gene array structure.
Methods
Subjects
Male subjects were recruited from the Medical College of Wisconsin and the surrounding Milwaukee area. They ranged in age from 15 to 43 years old, with an average age of 26 years. All subjects had normal color vision as demonstrated by color matching on a Nagel anomaloscope, and by performance on the American Optical Hardy Rand and Rittler, the Ishihara (38 plates), the Dvorine pseudoisochromatic plate tests, and the Neitz Test of Color vision (Neitz & Neitz, 2001). Informed consent was obtained after explanation of the nature and possible consequences of the study. Research on human subjects followed the tenets of the Declaration of Helsinki and was approved by the Human Research IRB at the Medical College of Wisconsin.
Measurement of L:M cone ratio
Relative spectral sensitivity functions were obtained for each subject using ERG flicker photometry, as described previously (Neitz & Jacobs, 1984; Jacobs, Neitz, & Krogh, 1996; Carroll, McMahon, Neitz, & Neitz, 2000). The instrument used here is the one described by Carroll et al., 2000. The light sources were tungsten halogen bulbs run by calibrated power supplies at 11.00 volts, stable to 0.02% thereby avoiding significant intensity changes with voltage fluctuations. For each subject, at the time of the ERG experiment, quantal intensities were obtained from direct radiometric measurements of the light at each wavelength to insure that instrumental variation did not contribute to the result. The long-term test-retest reliability of the instrument was reported by Carroll et al., 2002 as <2.2%, much less than the differences measured between the groups in this study. Subjects were fully dilated by topical administration of Tropicamide (0.5%), and images of the filaments were brought to focus at the pupil plane. The dimensions of the filament image were approximately 1.5 mm × 0.75 mm, which fit easily within the confines of the ca. 7.0 mm diameter dilated pupil. The L:M cone ratio for each subject was estimated by determining the weighted sum of an L- and an M-cone spectral sensitivity function required to best fit the spectral sensitivity data as described by Carroll et al. (2002). Spectral sensitivity functions were first corrected for lens absorption with an age-dependent correction (Pokorny, Smith, & Lutze, 1987), and for variation in the wavelength of peak sensitivity of the L pigment. Recently, estimates of L:M cone ratio from ERG flicker photometry have been compared to direct visualization of the cone mosaic using adaptive optics (AO) imaging (Hofer, Carroll, Neitz, Neitz, & Williams, 2005). Those comparisons reveal that the ERG estimates are highly correlated with results from AO imaging, however, there is a systematic difference between the two that can be explained by a 1.5 times larger contribution of each M cone to the ERG signal than each L cone. Thus, the ERG can be used to accurately estimate cone ratio as determined by AO by applying a simple correction factor. This factor was applied to obtain the final L:M ratio estimate from the ERG.
Genetic analysis
Nucleic acid extraction from whole blood was done using the PureGene Genomic DNA Purification Kit (Gentra Systems). The L opsin gene was amplified using the XL PCR kit (ABI), and it was used in a second round of PCR to separately amplify exons 2, 3, and 4, which were then directly sequenced using the AmpliTaq FS sequencing kit (ABI), and an ABI Prism 310 Genetic Analyzer. The details of the PCR primers and conditions have been described in detail elsewhere (Carroll, McMahon, Neitz, & Neitz, 2000).
The identity of the first gene in each array as encoding an L or an M pigment was determined as described previously (Kainz, Neitz, & Neitz, 1998; Neitz, Neitz, & Kainz, 1996). The assay relies on the fact that L genes contain a recognition sequence for restriction endonuclease RsaI in exon 5 that is absent from M genes. Exon 5 from the first gene in each subject’s array was incubated with restriction endonuclease RsaI. Cleavage by RsaI identifies first genes that encode an L pigment; resistance to RsaI cleavage identifies first genes that encode an M pigment. The amplified first gene in each subject’s array was also assayed by cleavage with restriction endonuclease EcoRI followed by field inversion gel electrophoresis (FIGE) to determine the size of intron 1 of the first gene.
The number and ratio of L- and M- opsin genes in each subject’s array were determined using two quantitative real-time PCR assays. One assay provided an estimate of the relative ratio of L:M genes, and the second assay provided an estimate of the relative ratio of the number of genes in the first position in the array to the number of genes in downstream positions. Details of these assays can be found elsewhere (Neitz & Neitz, 2001).
DNA from each subject was used in the PCR to separately amplify and sequence the L and M gene promoters. Hot start PCR was done using the XL PCR kit with AmpliWax Gem-100s. Each reaction contained 100 uM each of dATP, dCTP, dGTP, and dTTP, and 1.4 mM magnesium acetate. A 762 bp fragment containing the M gene promoter was amplified with 300 nM forward primer (5′cctgcaagtgggaatctaaacacaga) and 100 nM reverse primer (5′tgggtgctgtcctcatagctg). Reactions were subjected to 95°C for 5 minutes, 40 cycles of 94°C for 1 minute, 60°C for 1 minute, 72°C for 1.5 minutes and incubated at 72°C for 10 minutes. A 489 bp fragment containing the L gene promoter was amplified with 300 nM forward primer (5′cctgggctttcaagagaaccacatg) and 300 nM reverse primer (5′tgggtgctgtcctcatagctg). Reactions were subjected to 95°C for 5 minutes, 40 cycles of 94°C for 1 minute, 63°C for 1 minute, 72°C for 1 minute and incubated at 72°C for 10 minutes. Fluorescent sequencing of PCR products was done using AmpliTaq FS, and sequencing reactions were analyzed on an ABI Prism 310 Genetic Analyzer. One strand of the L and M promoter amplicons was directly sequenced using the same reverse primer used for PCR.
Results
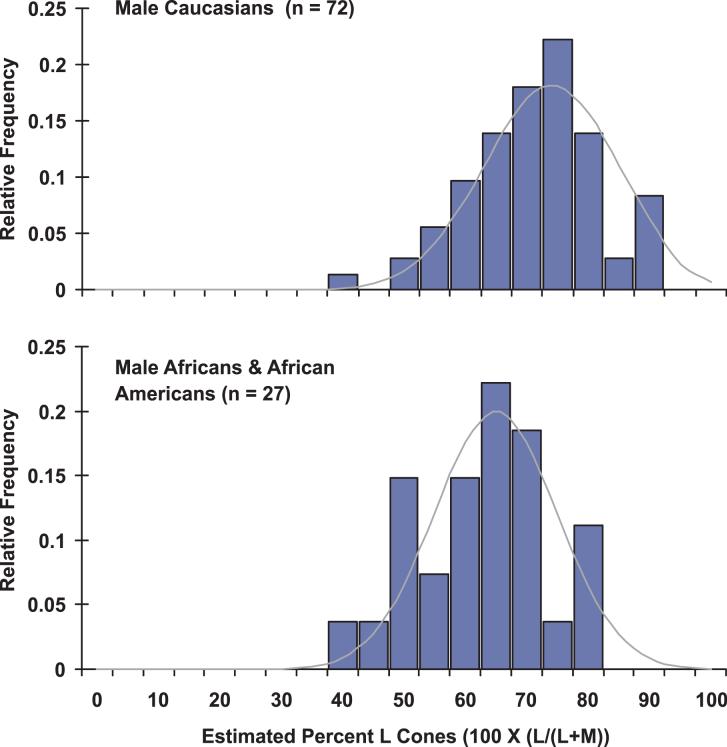
The ratio of L to M cones for each subject was estimated by determining the weighted sum of the L- and M-cone spectral sensitivity functions that best fit the FP-ERG-derived luminosity function obtained from each subject (Carroll, McMahon, Neitz, & Neitz, 2000). The L cone spectra were individualized in that each subject’s L opsin gene was sequenced, and the wavelength of maximal sensitivity (λmax) was inferred from previous studies (Carroll, McMahon, Neitz, & Neitz, 2000). Summarized in Table 1 are both the estimated L:M cone ratios and the λmax values used to calculate the ratio for each subject. The distributions of cone ratios for Africans and Caucasians are compared in Figure 2. The ratios for the males of African descent ranged from 42% to 85% L cones, with an average of 65% L cones and a standard deviation of 10.7%. Males of Caucasian descent ranged from 37% to 95% L cones, with an average of 73% L cones and a standard deviation of 11.1%. Thus the degree of variance for these two groups was similar; however, the mean ratio for the Africans was significantly lower than for the Caucasians (p = 0.0015 student t test, and p = 0.0013 Mann Whitney non-parametric test).
Table 1.
Summary of data for each subject. Subjects in the African category reported that both parents and both sets of grandparents still live in Africa, and subjects in African American category have one or both parents and grandparents living in the United States. The %L cones was calculated from FP-ERG derived spectral sensitivity data as previously described (Carroll, Neitz, & Neitz, 2000) using the λmax for the L cone indicated in the table, and using a λmax of 530 nm for the M cone. In addition, a correction factor was applied to the estimate of the % L cones to account for the 1.5 times greater contribution to the ERG signal for each M cone compared to each L cone (Hofer, Carroll, Neitz, Neitz, & Williams, 2005) The λmax values used for the L cone spectra in determining the %L cones was determined based on genetic analysis of each subject’s L opsin gene, as previously described (Carroll, Neitz & Neitz, 2000). An asterisk for the λmax values for L cones indicate those subjects for whom the λmax was measured in a single gene dichromat who had the identical L cone pigment. The % L genes and % downstream (DS) genes were determined as previously described (Neitz & Neitz, 2001).
| Subject ID # | %L cones | λmax | % L Genes (No. L Genes) | % DS Genes (No. DS Genes) | Country of Origin | Size 1st Gene | |
|---|---|---|---|---|---|---|---|
| Africans | 330 | 69.51 | 559* | 46.72 (1) | 55.07 (1) | Ethiopia | L |
| 334 | 52.32 | 559 | 51.38 (1) | 52.39 (1) | Ghana | S | |
| 337 | 63.95 | 559 | 49.12 (1) | 52.44 (1) | Senegal | L | |
| 338 | 69.59 | 559 | 33.92 (1) | 67.03 (2) | Ghana | L | |
| 339 | 71.94 | 559 | 34.64 (1) | 65.73 (2) | Ghana | L | |
| 342 | 48.36 | 559* | 34.61 (1) | 69.14 (2) | Ghana | L | |
| 343 | 57.74 | 559* | 35.32 (1) | 71.43 (2) | Ghana | L | |
| 344 | 42.37 | 559* | 33.69 (1) | 70.55 (2) | Ghana | L | |
| 347 | 79.96 | 559 | 41.63 (1) | 50.88 (1) | Ghana | L | |
| 348 | 51.45 | 559 | 32.63 (1) | 66.24 (2) | Nigeria | S | |
| 351 | 64.44 | 559* | 49.25 (1) | 51.91 (1) | Kenya | L | |
| 352 | 70.14 | 559* | 32.08 (1) | 68.35 (2) | Kenya | L | |
| 353 | 57.63 | 559* | 50.19 (1) | 52.54 (1) | Kenya | L | |
| African Americans | 354 | 69.13 | 559 | 50.81 (2) | 86.27 (4) | USA | S |
| 323 | 84.52 | 555.5 | 51.29 (2) | 75.2 (3) | USA | S | |
| 324 | 81.47 | 559* | 31.71 (1) | 68.26 (2) | USA | L | |
| 325 | 81.23 | 559 | 49.75 (1) | 53.01 (1) | USA | S | |
| 328 | 63.63 | 559 | 30.62 (1) | 67.64 (2) | St Lucia | S | |
| 329 | 70.26 | 559* | 43.07 (1) | 43.15 (1) | USA | L | |
| 331 | 65.18 | 559 | 30.37 (1) | 65.82 (2) | USA | L | |
| 332 | 65.65 | 559 | 48.76 (1) | 54.52 (1) | USA | L | |
| 333 | 50.41 | 559* | 31.91 (1) | 69.36 (2) | USA | L | |
| 335 | 70.64 | 559* | 32.63 (1) | 67.74 (2) | USA | L | |
| 336 | 52.68 | 559* | 33.29 (1) | 66.99 (2) | USA | L | |
| 340 | 64.24 | 559 | 50.69 (1) | 52.66 (1) | USA | S | |
| 341 | 68.55 | 559 | 34.68 (1) | 67.11 (1) | USA | S | |
| 349 | 71.34 | 559* | 29.85 (1) | 64.92 (2) | USA | L | |
| AVERAGE | 65.12 | (1) | (1.7) | ||||
Figure 2.
L:M cone ratio in Africans, African Americans and Caucasians. L:M cone ratio is calculated as the percentage of L plus M cones that are L [%L = 100 × (L / (L+M)]. The difference between the two populations is significant (p = 0.0015 student t test and p = 0.0013 Mann Whitney non-parametric test).
We considered the possibility that differences in the structure of the X-chromosome opsin gene arrays in the African versus Caucasian subjects might account for the difference in mean cone ratio for the two groups. For example, if an M gene occupied the 5′-most position in the arrays from African subjects, this would be predicted to give rise to a lower L:M cone ratio than observed in Caucasians because the M gene would be closer to the LCR. In addition, it was reported that 35% of African Americans have a variant of the L opsin gene that is 1.9 kb shorter in intron 1 compared to L genes in Caucasians (Jørgensen, Deeb, & Motulsky, 1990). Sequence differences in the long versus short version of the L gene could potentially contribute to differences in cone ratio (Mollon, 1999). In addition, nucleotide sequence differences in the L and M gene promoters have been hypothesized to be potential candidates for influencing cone ratio (Smallwood, Wang, & Nathans, 2002). To investigate these possibilities, genetic analyses were performed on each subject.
Genomic DNA was used in long distance PCR to amplify the first gene in the array, from which exon 5 was subsequently amplified and analyzed for a polymorphism that distinguishes L from M genes. Exon 5 of the first gene was incubated with restriction endonuclease RsaI which cleaves within exon 5 of L genes but not M genes. For all of the African subjects, exon 5 from the first gene in the array was cleaved by the RsaI restriction enzyme (data not shown), indicating that all subjects had an L gene first in the array. The full-length long-distance PCR product containing the first gene in the array was also incubated with restriction enzyme EcoRI to determine whether each subject had the long or the short version of the L gene in the first position in the array. An example of this analysis is shown in Figure 3, and the data from this assay is summarized in Table 1. There was no correlation between L-gene length and the ratio of L:M cones (p > 0.999; Mann-Whitney test).
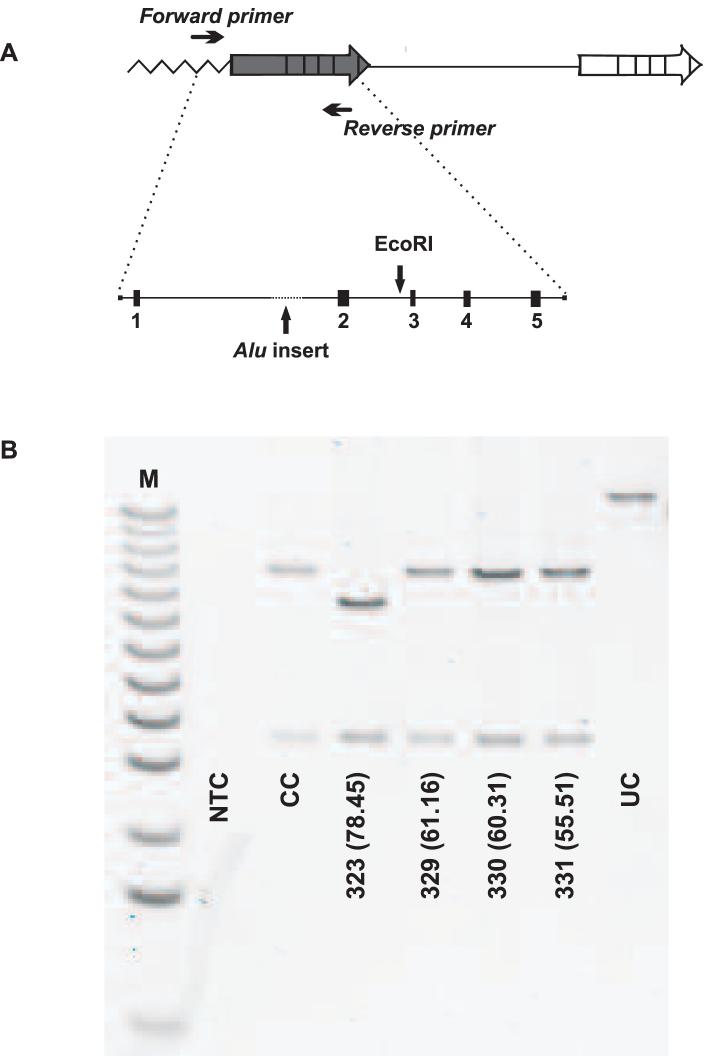
Figure 3.
FIGE analysis to determine the size of the L gene. (A) PCR strategy to amplify the first gene in the array, which encodes an L pigment. The PCR product was digested with restriction endonuclease EcoRI and the cleavage products were separated by FIGE. (B) FIGE gel comparing sizes of EcoRI cleavage products for 4 African subjects. Subjects who have the long variant of the L gene show a 3.9 kb and a 10 kb fragment. Subjects who have the short variant of the L gene show a 3.9 kb fragment and an 8.7 kb fragment. At the bottom of each lane is the subject number, followed by the % L cones in parentheses. On this gel, only subject #323 had the short L gene variant. NTC is the no template control to monitor for contamination of PCR reagents; CC is the cut control to monitor for complete digestion by EcoRI and UC is the uncut control showing that non-specific products were not generated with the PCR conditions used. M is the 1 kb ladder (Gibco Life Sciences).
Real-time quantitative PCR estimates of the ratio of L:M genes and the number of genes per array estimated from the ratio of the first:downstream genes are summarized in Table 1. Ratios of L:M genes are expressed as the percentage of L plus M genes that encode L pigment [%L genes = 100 × L/(L+M)], and the ratios of first:downstream genes are expressed as the percentage of genes in the array that are downstream of the first gene [%DS = 100 × DS/(1+DS)]. For example, if a subject was estimated to have 50% L genes and 50% downstream genes, this was interpreted as his having an array with one L and one M gene. An estimate of 33% L genes and 66 % downstream genes was interpreted as one L and two M genes, and an estimate of 50% L genes and 75% downstream genes was interpreted as two L and two M genes. There was no difference between Africans and Caucasians in the percentage of L genes (p = 0.27, Mann-Whitney test) or in the percentage of downstream genes (p = 0.61, Mann-Whitney test).
A comparison of the sequences of the 236 bp promoter-containing DNA segment upstream of the translational start codon of the L and M genes revealed the absence of nucleotide sequence variations in the L and M gene promoters among the Africans and between Africans and Caucasians; the sequences for the African males matched those previously reported for this region for the majority of Caucasian males (McMahon, Neitz, & Neitz, 2003).
Discussion
The average L:M cone ratio in 27 males of African descent who have normal color vision was statistically lower than the average ratio for males of Caucasian descent with normal color vision (Figure 1). The two primary sources of error in estimates of the L:M cone ratio from FP-ERG are contributed by variation in the wavelength of peak absorption (λmax) of the L pigment and variation in the optical density of the lens (Bieber, Kraft, & Werner, 1998) The L:M cone ratios for both the Africans and Caucasians were corrected for both of these sources of error. To account for variation in the λmax of L pigments, the L cone spectrum used to determine the weighted sum of an L and an M cone spectral sensitivity that best fit the FP-ERG derived spectral sensitivity function for each subject was the one predicted for the L pigment encoded by the gene in the first position in the array for each subject. We previously catalogued the L opsin gene sequences and λmax values for dichromats who had a single X-chromosome opsin gene (Carroll, McMahon, Neitz, & Neitz, 2000). Fourteen of the subjects in the present study had an L opsin gene that encoded a pigment corresponding to one for which the spectrum had been measured in a dichromat. The remaining 13 subjects differed from catalogued pigments only by polymorphisms that have been previously demonstrated not to influence the λmax of L pigments. To account for variation in the optical density of the lens, the spectral sensitivity curves used to calculate the L:M cone ratios were determined using an age-specific correction for lens absorption (Pokorny, Smith, & Lutze, 1987).
It is possible that there is a correlation between high skin pigmentation and increased lens density (c.f. Said & Weale, 1959). We considered the possibility that the downward shift in L:M cone ratio in African subjects compared to Caucasian subjects is due to an underestimated lens density for the former. However, increasing the lens density values for a given set of spectral sensitivity data decreases the L:M ratio further. Thus we conclude that the FP-ERG-derived L:M ratio estimates reflect a real difference in the average L:M ratio between males of African versus Caucasian descent.
According to the model proposed by Nathans and colleagues, a difference in the distance between the LCR and M opsin gene promoter in Caucasians versus Africans could account for the dissimilarity in L:M cone ratio, however, molecular genetic results ruled out all of the obvious possibilities. Both Africans and Caucasians have arrays in which an L gene is closest to the LCR, so it is not due to a difference in gene order. The absence of a correlation between L:M cone ratio and the short versus long variant of the L opsin gene rules out the possibility previously suggested by Mollon (1999) that the DNA insert in the long variant has a measurable affect on L:M cone ratio. An alternative hypothesis proposed by Smallwood et al. (2002) that promoter sequence polymorphisms can account for cone ratio variation can be eliminated here by the almost complete absence of promoter sequence differences both between and among groups.
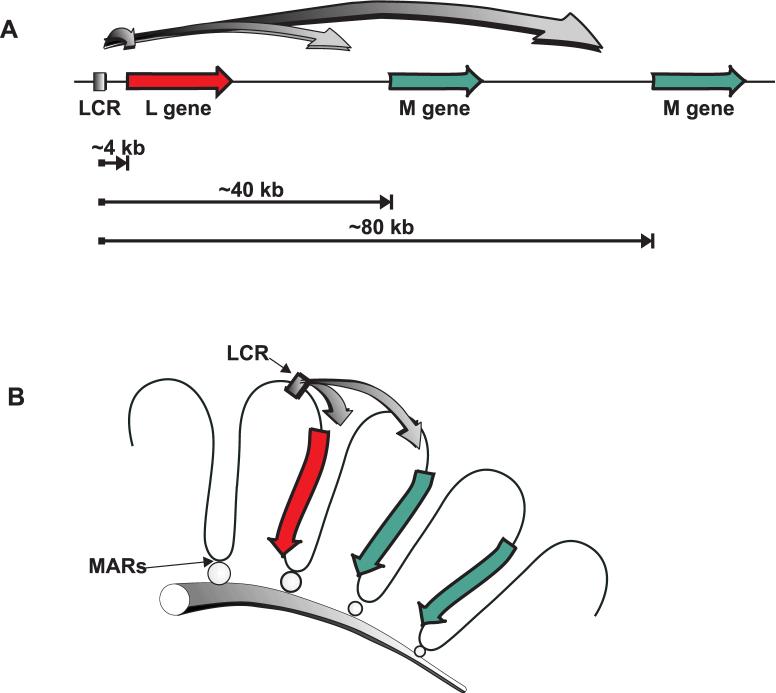
In the model illustrated in Figure 2 the DNA is envisioned to bend to allow the LCR to form a complex with the opsin gene promoter (Smallwood, Wang, & Nathans, 2002). In the absence of other constraints, the relative probability of spatial coincidence between the LCR and a promoter should decrease in proportion to the volume of the sphere, or in proportion to the cube of the distance from the LCR, so that a cone photoreceptor is 1000 times more likely to express the first gene (43) versus the second gene (403) making the expected L:M ratio approximately 1000:1 (Figure 4a). There must be constraints on the ability of the LCR to choose to complex with the L versus M gene promoter that are not accounted for in the Nathans model that prevent such a disparate ratio as predicted when only linear distance is considered.
Figure 4.
Relative accessibility of the L or M opsin gene promoters to the LCR. (A) The L gene enjoys a huge advantage compared to the M genes if linear distance is the determining factor. The L gene is only 4 kb away from the LCR whereas the distance between the LCR and an M gene is a multiple of 40 kb. (B) The first and second genes enjoy a similar degree of accessibility to the LCR if distances are determined by the packaging of DNA into chromatin loop domains. Loops are attached to the nuclear matrix via matrix attachment regions (MARS).
It has long been known that eukaryotic DNA is packaged into complex higher order structures, and there is emerging evidence that both the structure and physical position of DNA within the nucleus play key roles in regulating gene expression (Cai, Han, & Kohwi-Shigematus, 2003). Actively transcribed regions of chromosomes are organized into chromatin loop domains, and although there is considerable variability in the size of the loops, the average in humans has been estimated to be about 40 kb or about the size of the repeat unit of the L and M opsin genes (Vollrath, Nathans, & Davis, 1988). The precise organization of the chromatin loop domains for the X-chromosome opsin gene locus remains unknown; however, as illustrated in Figure 4b, packaging into chromatin loops can bring the LCR into nearly equal proximity to the promoters of both the first and second genes in the array account for the observation that the relative numbers of L and M cones is much more nearly equal than might be predicted by the extreme difference in distance between the LCR and the two promoters respectively.
In summary, the three dimensional chromatin organization of the L and M opsin gene locus must certainly play a key role in determining the probability of whether a photoreceptor cell will express an L versus an M pigment gene. The artificial L/M opsin gene array described by Wang et al. (1999) and Smallwood et al. (2002) might not mimic the normal chromatin context of the X-chromosome opsin gene locus in humans and would not provide the ideal model of the native locus in experiments aimed at understanding how variation in L:M cone ratio is regulated in humans if chromatin context is important. The challenge for the future will be to characterize the chromatin loop organization of the X-chromosome opsin gene locus, and to determine whether there are sequence differences that influence the L:M cone ratio by altering the chromatin organization.
Acknowledgements
This work was supported by NIH grants T32EY014537, EY09303, EY09620, EY01931, by Research to Prevent Blindness, the David and Ruth S. Coleman Foundation, and the Harry J. Heeb Foundation.
Footnotes
Commercial relationships: none
Contributor Information
Carrie McMahon, Department of Cell Biology, Neurobiology, & Anatomy, Medical College of Wisconsin, Milwaukee, WI, USA.
Joseph Carroll, Department Ophthalmology, Medical College of Wisconsin, Milwaukee, WI, USA.
Stella Awua, Medical College of Wisconsin, Milwaukee, WI, USA.
Jay Neitz, Department of Ophthalmology, Medical College of Wisconsin, Milwaukee, WI, USA.
Maureen Neitz, Department of Ophthalmology, Medical College of Wisconsin, Milwaukee, WI, USA.
References
- Bieber ML, Kraft JM, Werner JS. Effects of known variations in photopigments on L/M cone ratios estimated from luminous efficiency functions. Vision Research. 1998;38:1961–1966. doi: 10.1016/s0042-6989(97)00302-7. PubMed. [DOI] [PubMed] [Google Scholar]
- Boissinot S, Tan Y, Shyue S-K, Schneider H, Sampaio I, Neiswanger K, et al. Origins and antiquity of X-linked triallelic color vision systems in New World monkeys. Proceedings of the National Academy of Sciences, U.S.A. 1998;95:13749–13754. doi: 10.1073/pnas.95.23.13749. [PubMed] [Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Han H-J, Kohwi-Shigematsu T. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nature Genetics. 2003;34:42–51. doi: 10.1038/ng1146. PubMed. [DOI] [PubMed] [Google Scholar]
- Carroll J, McMahon C, Neitz M, Neitz J. Flicker-photometric electroretinogram estimates of L: M cone photoreceptor ratio in men with photopigment spectra derived from genetics. Journal of the Optical Society of America A-Optics Image Science and Vision. 2000;17:499–509. doi: 10.1364/josaa.17.000499. PubMed. [DOI] [PubMed] [Google Scholar]
- Carroll J, Neitz M, Neitz J. Estimates of L:M cone ratio from ERG flicker photometry and genetics. Journal of Vision. 2002;2:531–542. doi: 10.1167/2.8.1. [PubMed][Article] [DOI] [PubMed] [Google Scholar]
- Hagstrom SA, Neitz M, Neitz J. Cone pigment gene expression in individual photoreceptors and the chromatic topography of the retina. Journal of the Optical Society of America A-Optics Image Science and Vision. 2000;17:527–537. doi: 10.1364/josaa.17.000527. PubMed. [DOI] [PubMed] [Google Scholar]
- Hofer H, Carroll J, Neitz J, Neitz M, Williams DR. Organization of the human trichromatic cone mosaic. Journal of Neuroscience. 2005;25:9669–9679. doi: 10.1523/JNEUROSCI.2414-05.2005. PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs GH, Neitz J. Color vision in monkeys: sex related differences suggest the mode of inheritance. Vision Research. 1985;25:141–143. doi: 10.1016/0042-6989(85)90088-4. PubMed. [DOI] [PubMed] [Google Scholar]
- Jacobs GH, Neitz J. Inheritance of color vision in a New World monkey (Saimiri sciureus) Proceedings of the National Academy of Sciences, U.S.A. 1987;84:2545–2549. doi: 10.1073/pnas.84.8.2545. [PubMed][Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs GH, Neitz J, Krogh K. Electroretinogram flicker photometry and its applications. Journal of the Optical Society of America A. 1996;13:641–648. doi: 10.1364/josaa.13.000641. PubMed. [DOI] [PubMed] [Google Scholar]
- Jacobs GH, Neitz J, Neitz M. Genetic basis of polymorphism in the color vision of platyrrhine monkeys. Vision Research. 1993;33:269–274. doi: 10.1016/0042-6989(93)90083-9. Article. [DOI] [PubMed] [Google Scholar]
- Jørgensen AL, Deeb SS, Motulsky AG. Molecular genetics of X chromosome-linked color vision among populations of African and Japanese ancestry: High frequency of a shortened red pigment gene among Afro-Americans. Proceedings of the National Academy of Sciences, U.S.A. 1990;87:6512–6516. doi: 10.1073/pnas.87.17.6512. [PubMed][Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kainz PM, Neitz M, Neitz J. Molecular genetic detection of female carriers of protan defects. Vision Research. 1998;38:3365–3369. doi: 10.1016/s0042-6989(97)00366-0. PubMed. [DOI] [PubMed] [Google Scholar]
- McMahon C, Neitz J, Neitz M. Comparison of human and monkey pigment gene promoters to evaluate DNA sequences proposed to govern L:M cone ratio. In: Mollon JD, Knoblauch K, Pokorny J, editors. Normal and defective colour vision. Oxford University Press; Oxford, UK: 2003. pp. 51–59. [Google Scholar]
- Mollon JD. Color vision: Opsins and options. Proceedings of the National Academy of Sciences, U.S.A. 1999;96:4743–4745. doi: 10.1073/pnas.96.9.4743. [PubMed] [Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans J, Davenport CM, Maumenee IH, Lewis RA, Hejtmancik JF, Litt M, et al. Molecular genetics of blue cone monochromacy. Science. 1989;245:831–838. doi: 10.1126/science.2788922. [PubMed] [DOI] [PubMed] [Google Scholar]
- Nathans J, Thomas D, Hogness DS. Molecular genetics of human color vision: the genes encoding blue, green, and red pigments. Science. 1986;232:193–202. doi: 10.1126/science.2937147. [PubMed] [DOI] [PubMed] [Google Scholar]
- Neitz J, Carroll J, Neitz M. Color vision: Almost reason enough for having eyes. Optics & Photonics News. 2001;12:26–33. [Google Scholar]
- Neitz J, Jacobs GH. Electroretinogram measurements of cone spectral sensitivity in dichromatic monkeys. Journal of the Optical Society of America A. 1984;1:1175–1180. doi: 10.1364/josaa.1.001175. PubMed. [DOI] [PubMed] [Google Scholar]
- Neitz J, Neitz M, Kainz PM. Visual pigment gene structure and the severity of human color vision defects. Science. 1996;274:801–804. doi: 10.1126/science.274.5288.801. PubMed. [DOI] [PubMed] [Google Scholar]
- Neitz M, Neitz J. A new test for mass screening of school age children for red-green color vision defects. Color Research & Application. 2001;26(S1):S239–S249. [Google Scholar]
- Pokorny J, Smith VC, Lutze M. Aging of the human lens. Applied Optics. 1987;26(8):1437–1440. doi: 10.1364/AO.26.001437. [DOI] [PubMed] [Google Scholar]
- Said FS, Weale RA. The variation with age of the spectral transmissivity of the living human crystalline lens. Gerontologia. 1959;3:213–231. doi: 10.1159/000210900. [DOI] [PubMed] [Google Scholar]
- Smallwood PM, Wang Y, Nathans J. Role of a locus control region in the mutually exclusive expression of human red and green cone pigment genes. Proceedings of the National Academy of Sciences, U.S.A. 2002;99:1008–1011. doi: 10.1073/pnas.022629799. [PubMed][Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollrath D, Nathans J, Davis RW. Tandem array of human visual pigment genes at Xq28. Science. 1988;240:1669–1672. doi: 10.1126/science.2837827. PubMed. [DOI] [PubMed] [Google Scholar]
- Wang Y, Macke JP, Merbs SL, Zack DJ, Klaunberg B, Bennett J, et al. A locus control region adjacent to the human red and green visual pigment genes. Neuron. 1992;9:429–440. doi: 10.1016/0896-6273(92)90181-c. PubMed. [DOI] [PubMed] [Google Scholar]
- Wang Y, Smallwood PM, Cowan M, Blesh D, Lawler A, Nathans J. Mutually exclusive expression of human red and green visual pigment-reporter transgenes occurs at high frequency in murine cone photoreceptors. Proceedings of the National Academy of Sciences, U.S.A. 1999;96:5251–5256. doi: 10.1073/pnas.96.9.5251. [PubMed] [Article] [DOI] [PMC free article] [PubMed] [Google Scholar]