DNA is challenged by endogenous or environmental alkylation agents that modify heterocyclic bases and backbones.1 Many of these lesions can have mutagenic or cytotoxic consequences, and thus they must be promptly detected and repaired. Recently, it was found that Escherichia coli AlkB and its human homologues catalyze a unique oxidative repair of alkylated DNA bases. E. coli AlkB belongs to a superfamily of iron-/α-ketoglutarate-dependent dioxygenases.2 This protein and its two human homologues have been shown to oxidize the methyl groups of N1-methyladenine and N3-methylcytosine in single-stranded DNA (ssDNA), double-stranded DNA (dsDNA), and single-stranded RNA (ssRNA).3 The corresponding alcohol products spontaneously decompose in water to give the undamaged bases and formaldehyde. These proteins have also been shown to directly repair N3-methylthymine and N1-methylguanine lesions through the same proposed mechanism.3e,4 The native iron(II)-containing AlkB has been isolated from E. coli and spectroscopically characterized.5
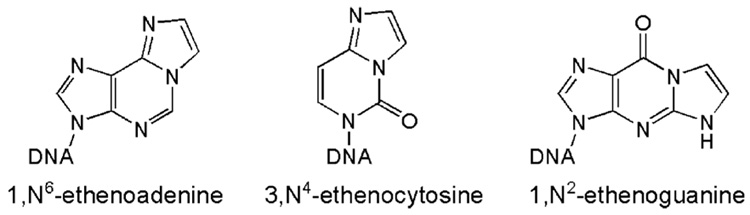
The base lesions repaired by AlkB are generated in unpaired bases in DNA.1b,c These lesions can no longer form Watson–Crick base pairs with the opposite bases and are cytotoxic. The AlkB proteins possess the ability to specifically locate the weakened base pairs containing these modifications.6 Exocyclic DNA adducts, such as 1,N6-ethenoadenine (εA), 3,N4-ethenocytosine (εC), and 1,N2-ethenoguanine (εG), also block Watson–Crick base pairing (Scheme 1).7 These adducts arise from treatment of DNA with chloroethylene oxide or chloroacetaldehyde. In particular, it has been shown that lipid peroxidation products generated under oxidative stress lead to the formation of εA, εC, and, εG,7b,d which have been linked to the cytotoxic and genotoxic effects observed for lipid peroxidation.
Scheme 1.
Exocyclic DNA Adducts
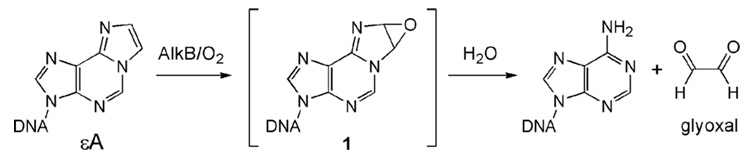
We speculated that the exocyclic lesion εA may serve as a substrate for AlkB. The putative iron(IV)–oxo intermediate formed from the reaction of Fe(II)–AlkB with dioxygen and cofactor α-ketoglutarate could oxidize the exocyclic section of εA. For instance, an epoxidation of the exocyclic double bond would afford epoxide 1, which can be hydrolyzed by water to produce the repaired base and glyoxal (Figure 1).
Figure 1.
Proposed repair of εA in DNA by the AlkB proteins.
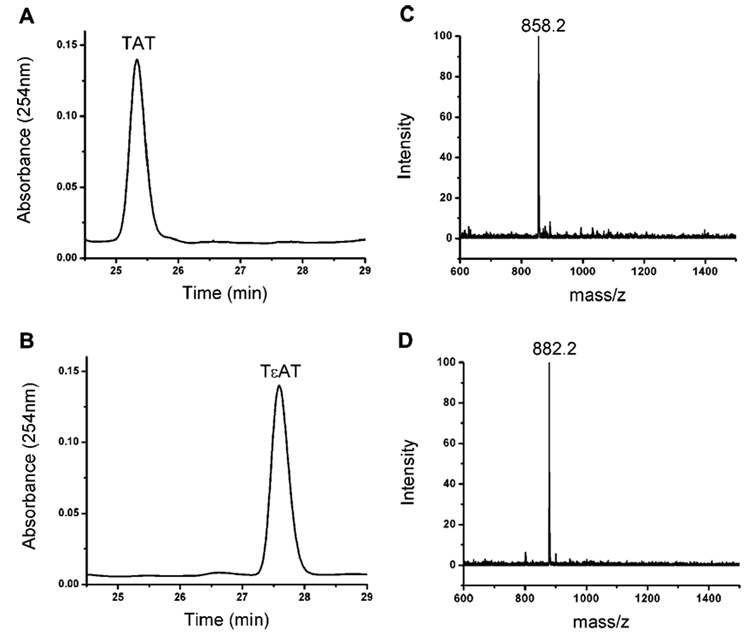
To test this hypothesis, εA was incorporated into a 3-mer DNA T(εA)T. This substrate (0.1 mM) was treated with E. coli AlkB (0.01 mM, 10 mol %) under standard conditions in 100 µL of buffer solution at pH 7.0 for 1 h.3a,d,h The HPLC analysis of the product mixture showed the complete disappearance of the T(εA)T peak and an appearance of a new peak that represents TAT (Figure 2A). TAT was added into the same mixture, which was then analyzed by HPLC. An intensity increase in the new peak confirmed that TAT was obtained as the final product. A control experiment was performed under the exact same conditions with T(εA)T, Fe(NH4)2(SO4)2, α-ketoglutarate, and ascorbate in MES or HEPES buffer while excluding AlkB. HPLC analysis of this mixture indicated that no repair reaction had occurred in the absence of AlkB (Figure 2B). The identities of TAT and TεAT in solutions were further confirmed by MALDI mass spectrometry (graphs C and D of Figure 2).
Figure 2.
Repair of T(εA)T by E. coli AlkB. (A) A reaction of T(εA)T (0.1 mM) with E. coli AlkB (0.01 mM) under the standard repair conditions (50 mM MES or HEPES, 0.2 mM Fe(NH4)2(SO4)2, 2.6 mM α-ketoglutarate, 5.2 mM ascorbate) at pH 7.0 for 1 h at 37 °C led to the formation of TAT, which was resolved by HPLC and detected at A254. (B) A control experiment under the same conditions as (A) but in the absence of AlkB gave only the starting material T(εA)T. (C) The reaction mixture of (A) was analyzed by MALDI-TOF mass spectrometry. The negative molecular peak of TAT at 858.2 was detected. (D) The mixture of the control reaction in (B) was analyzed by MALDI-TOF mass spectrometry, and the negative molecular peak of T(εA)T at 882.2 was observed.
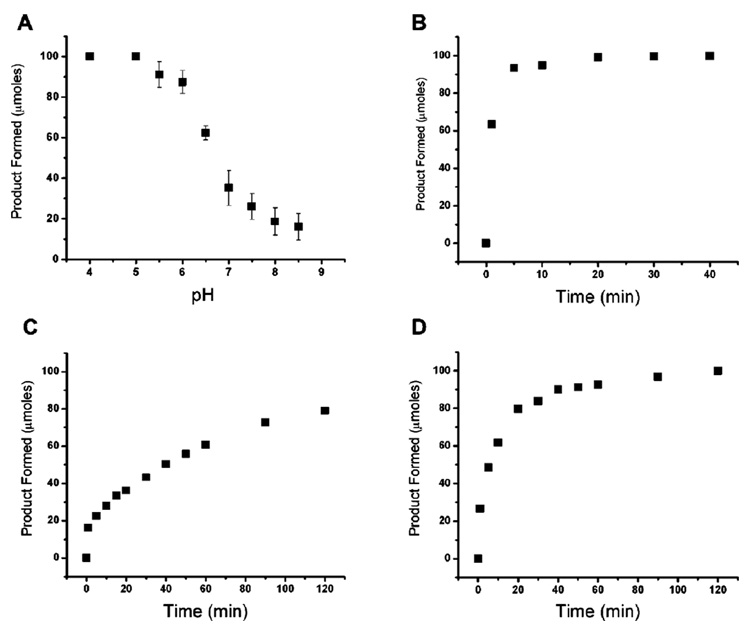
We next studied the pH dependency of this repair reaction. Quantitative conversion of T(εA)T to TAT was observed with 5 mol % of AlkB after 5 min incubation at both pH 4.0 and 5.0 (Figure 3A). Complete repair was also observed at pH 6.0 after 20 min reaction (Figure 3B). At neutral pH, the reaction proceeds catalytically as 8 turnovers were achieved after 30 min. Doubling the concentration of AlkB led to an increase of the repair rate by approximately 2-fold. The pH-rate profile of this repair reaction may suggest an acid-assisted step in the repair process. The kinetics shown here and factors that may determine the pH dependency will be carefully studied in the future.
Figure 3.
The pH dependency and time courses for the repair of T(εA)T by AlkB. (A) T(εA)T (0.1 mM) was treated with 5 µM of AlkB at different pH, and the reactions were quenched after 5 min. Time courses for reactions between 0.1 mM of T(εA)T and AlkB at 37 °C; (B) 5 µM AlkB at pH 6.0, (C) 5 µM AlkB at pH 7.0, and (D) 10 µM of AlkB with pH 8.0 are also shown. Standard repair conditions were used with MES (for pH 4.0–6.5) or HEPES (for pH 7.0–8.5) as the buffers in 100 µL of solution.
A longer DNA, TTTTT(εA)TTTTT, with εA incorporated in the middle of an 11-mer strand was prepared and tested as the substrate as well. After reaction with AlkB under standard conditions, the DNA was digested to nucleosides,9a and HPLC analysis of the digested DNA9b confirmed that treatment with 5 mol % of AlkB led to complete repair of εA to A in 30 min at both pH 6.0 and 7.0 (Figure S1). In addition, we were able to detect glyoxal, a small molecule that would be produced from the oxidative repair process proposed in Figure 1, by using a previously established procedure (Figures S2 and S3).10
ABH3 also repairs T(εA)T, but with a much lower activity compared to that of AlkB. Treatment of varying concentrations of ABH3 (10 and 50 mol %, and 1 equiv) with T(εA)T for 16 h at pH 6.0 at room temperature (to prevent aggregation of ABH3) led to a portion of the T(εA)T (16, 51, and 67%, respectively) being converted into TAT (Figures S4) as determined by HPLC analysis. It has been shown that the recombinant ABH3 overexpressed from E. coli has only 2% of AlkB’s activity in vitro when assayed with N1-methyladenine.3e The reason for the low activity observed for the human homologues of AlkB is unclear.3e,g,h The biological relevance and physiological importance of the activity reported here for ABH3 need to be carefully evaluated in the future.
The 1,N6-ethenoadenine (εA), 3,N4-ethenocytosine (εC), and 1,N2- and N2,3-ethenoguanine (εG) adducts are produced from endogenous metabolic processes, such as lipid peroxidation, in humans.8 These exocyclic adducts have miscoding potentials that can lead to mutagenic consequences. For instance, G, T, and A are readily incorporated opposite εC during DNA synthesis;11a,b formation of εA can lead to a predominant εA → G transition.11c,d It was found that εA and εC are particularly mutagenic in mammalian cells. These mutations, upon accumulation, contribute to genetic alterations that can lead to aging and disease. The repair of exocyclic DNA adducts has been shown to be mediated by DNA base glycosylases.12 We demonstrated here that one of these lesions, εA, can be directly reversed by E. coli AlkB and human ABH3 in vitro. This result suggests that the AlkB proteins may play a role in eliminating exocyclic DNA base adducts, and thus suppressing the cytotoxic and mutagenic consequences derived from the damage. Since the AlkB proteins have been shown to work on ssDNA, dsDNA, and RNA substrates,3 they may play different/complementary roles to the glycosylases to fix exocyclic DNA lesions. More thorough investigations on the mechanism and physiological importance of the repair process discovered here are in progress.
Supplementary Material
Supporting Information Available: Experimental details and Figures S1–S4. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgment
We thank Dr. J. Piccirilli for the use of the DNA synthesizer, and Mr. D. Wolfgeher for assistance with the MALDI experiments. This research was supported by the G&P Foundation for Cancer Research (C.H.), the W. M. Keck Foundation Distinguished Young Scholars Program (C.H.), and the National Institutes of Health (GM071440). C.H. is also a Searle Scholar, an Arnold and Mabel Beckman Foundation Young Investigator, and a Research Corporation Cottrell Scholar.
References
- 1.a Lindahl T, Sedgewick B, Sekiguchi M, Nakabeppu Y. Annu. Rev. Biochem. 1988;57:133–157. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]; b Sedgwick B. Nat. Rev. Mol. Cell Biol. 2004;5:148–157. doi: 10.1038/nrm1312. [DOI] [PubMed] [Google Scholar]; c Drabløs F, Feyzi E, Aas PA, Vaagbø CB, Kavli B, Bratlie MS, Peña-Diaz J, Otterlei M, Slupphaug G, Krokan HE. DNA Repair. 2004;3:1389–1407. doi: 10.1016/j.dnarep.2004.05.004. [DOI] [PubMed] [Google Scholar]; d Myers LC, Terranova MP, Ferentz AE, Wagner G, Verdine GL. Science. 1993;261:1164–1167. doi: 10.1126/science.8395079. [DOI] [PubMed] [Google Scholar]
- 2.Aravind L, Koonin EV. GenomeBiology. 2001;2:007.001–007.008. [Google Scholar]
- 3.a Trewick S, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]; b Falnes PØ, Johansen RF, Seeberg E. Nature. 2002;419:178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]; c Koivisto P, Duncan T, Lindahl T, Sedgwick B. J. Biol. Chem. 2003;278:44348–44354. doi: 10.1074/jbc.M307361200. [DOI] [PubMed] [Google Scholar]; d Welford RWD, Schlemminger I, McNeill LA, Hewitson KS, Schofield CJ. J. Biol. Chem. 2003;278:10157–10161. doi: 10.1074/jbc.M211058200. [DOI] [PubMed] [Google Scholar]; e Koivisto P, Robins P, Lindahl T, Sedgwick B. J. Biol. Chem. 2004;279:40470–40474. doi: 10.1074/jbc.M407960200. [DOI] [PubMed] [Google Scholar]; f Ougland R, Zhang CM, Liiv A, Johansen RF, Seeberg E, Hou YM, Remme J, Falnes PØ. Mol. Cell. 2004;16:107–116. doi: 10.1016/j.molcel.2004.09.002. [DOI] [PubMed] [Google Scholar]; g Aas PA, Otterlei M, Falnes PØ, Vaagbo CB, Skorpen F, Akari M, Sundheim O, Bjørås M, Slupphaug G, Seeberg E, Krokan H. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]; h Duncan T, Trewick SC, Koivisto P, Bates PA, Lindahl T, Sedgwick B. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16660–16665. doi: 10.1073/pnas.262589799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a Delaney JC, Essigmann JM. Proc. Natl. Acad. Sci. U.S.A. 2004;101:14051–14056. doi: 10.1073/pnas.0403489101. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Falnes PØ. Nucleic Acids Res. 2004;32:6260–6267. doi: 10.1093/nar/gkh964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mishina Y, Chen LX, He C. J. Am. Chem. Soc. 2004;126:16930–16936. doi: 10.1021/ja045066z. [DOI] [PubMed] [Google Scholar]
- 6.a Mishina Y, He C. J. Am. Chem. Soc. 2003;125:8730–8731. doi: 10.1021/ja034636c. [DOI] [PubMed] [Google Scholar]; b Mishina Y, Lee C-HJ, He C. Nucleic Acids Res. 2004;32:1548–1554. doi: 10.1093/nar/gkh319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a Choi JH, Pfeifer GP. Mutat. Res. 2004;568:245–256. doi: 10.1016/j.mrfmmm.2004.09.004. [DOI] [PubMed] [Google Scholar]; b Gros L, Ishchenko AA, Saparbaev M. Mutat. Res. 2003;531:219–229. doi: 10.1016/j.mrfmmm.2003.07.008. [DOI] [PubMed] [Google Scholar]; c Douki T, Odin F, Caillat S, Favier A, Cadet J. Free Radical Biol. Med. 2004;37:62–70. doi: 10.1016/j.freeradbiomed.2004.04.013. [DOI] [PubMed] [Google Scholar]; d Speina E, Kierzek AM, Tudek B. Mutat. Res. 2003;531:205–217. doi: 10.1016/j.mrfmmm.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 8.a Chung FL, Chen HJ, Nath RG. Carcinogenesis. 1996;17:2105–2111. doi: 10.1093/carcin/17.10.2105. [DOI] [PubMed] [Google Scholar]; b Pollack M, Oe T, Lee SH, Silva Elipe MV, Arison BH, Blair IA. Chem. Res. Toxicol. 2003;16:893–900. doi: 10.1021/tx030009p. [DOI] [PubMed] [Google Scholar]; c Nair J, Barbin A, Guichard Y, Bartsch H. Carcinogenesis. 1995;16:613–617. doi: 10.1093/carcin/16.3.613. [DOI] [PubMed] [Google Scholar]
- 9.a Crain PF. Methods Enzymol. 1990;193:782–790. doi: 10.1016/0076-6879(90)93450-y. [DOI] [PubMed] [Google Scholar]; b Pomerantz SC, McCloskey JA. Methods Enzymol. 1990;193:796–824. doi: 10.1016/0076-6879(90)93452-q. [DOI] [PubMed] [Google Scholar]
- 10.Lapolla A, Flamini R, Tonus T, Fedele D, Senesi A, Reitano R, Marrotta E, Pace G, Seraglia R, Traldi P. Rapid Commun. Mass Spectrom. 2003;17:876–878. doi: 10.1002/rcm.992. [DOI] [PubMed] [Google Scholar]
- 11.a Moriya M, Zhang W, Johnson F, Grollman AP. Proc. Natl. Acad. Sci. U.S.A. 1994;91:11899–11903. doi: 10.1073/pnas.91.25.11899. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Basu AK, Wood ML, Niedernhofer LJ, Ramos LA, Essigmann JM. Biochemistry. 1993;32:12793–12801. doi: 10.1021/bi00210a031. [DOI] [PubMed] [Google Scholar]; c Pandya GA, Moriya M. Biochemistry. 1996;35:11487–11492. doi: 10.1021/bi960170h. [DOI] [PubMed] [Google Scholar]; d Levine RL, Yang IY, Hossain M, Pandya GA, Grollman AP, Moriya M. Cancer Res. 2000;60:4098–4104. [PubMed] [Google Scholar]
- 12.a Asaeda A, Ide H, Asagoshi K, Matsuyama S, Tano K, Murakami A, Takamori Y, Kubo K. Biochemistry. 2000;39:1959–1965. doi: 10.1021/bi9917075. [DOI] [PubMed] [Google Scholar]; b O’Connor TR, Laval J. Biochem. Biophys. Res. Commun. 1991;176:1170–1177. doi: 10.1016/0006-291x(91)90408-y. [DOI] [PubMed] [Google Scholar]; c Lau AY, Wyatt MD, Glassner BJ, Samson LD, Ellenberger T. Proc. Natl. Acad. Sci. U.S.A. 2000;97:13573–13578. doi: 10.1073/pnas.97.25.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Dosanjh MK, Roy R, Mitra S, Singer B. Biochemistry. 1994;33:1624–1628. doi: 10.1021/bi00173a002. [DOI] [PubMed] [Google Scholar]; e Saparbaev M, Laval J. Proc. Natl. Acad. Sci. U.S.A. 1998;95:8508–8513. doi: 10.1073/pnas.95.15.8508. [DOI] [PMC free article] [PubMed] [Google Scholar]; f Roy R, Kennel SJ, Mitra S. Carcinogenesis. 1996;17:2177–2182. doi: 10.1093/carcin/17.10.2177. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Available: Experimental details and Figures S1–S4. This material is available free of charge via the Internet at http://pubs.acs.org.