Abstract
The yeast peptide-methionine sulfoxide reductase (MsrA) was overexpressed in a Saccharomyces cerevisiae null mutant of msrA by using a high-copy plasmid harboring the msrA gene and its promoter. The resulting strain had about 25-fold higher MsrA activity than its parent strain. When exposed to either hydrogen peroxide, paraquat, or 2,2′-azobis-(2-amidinopropane) dihydrochloride treatment, the MsrA overexpressed strain grew better, had lower free and protein-bound methionine sulfoxide and had a better survival rate under these conditions than did the msrA mutant and its parent strain. Substitution of methionine with methionine sulfoxide in a medium lacking hydrogen peroxide had little effect on the growth pattern, which suggests that the oxidation of free methionine in the growth medium was not the main cause of growth inhibition of the msrA mutant. Ultraviolet A radiation did not result in obvious differences in survival rates among the three strains. An enhanced resistance to hydrogen peroxide treatment was shown in human T lymphocyte cells (Molt-4) that were stably transfected with the bovine msrA and exposed to hydrogen peroxide. The survival rate of the transfected strain was much better than its parent strain when grown in the presence of hydrogen peroxide. These results support the proposition that the msrA gene is involved in the resistance of yeast and mammalian cells to oxidative stress.
Oxidation of methionine to methionine sulfoxide can occur in the presence of various oxidants, such as hydrogen peroxide, hypochlorite, peroxynitrite, hydroxyl radicals, and ozone, as well as metal-catalyzed oxidation systems. Most, if not all, cells contain methionine sulfoxide reductases (MsrA) that catalyze the thioredoxin-dependent reduction of protein methionine sulfoxide (MetO) residues back to Met. The oxidation of methionine residues of some proteins may lead to either activation or inactivation of their biological activities (1–3), whereas the oxidation of one or more methionine residues in other proteins may have little or no effect on biological functions (4). This idea has led to the suggestion that MsrA may have multiple biological functions: (i) It may serve to repair oxidative protein damage in some proteins. (ii) It may play an important role in the regulation of enzyme (protein) activities by facilitating the interconversion of specific methionine residues of these proteins between oxidized and reduced forms [as occurs, for example, in regulating the ability of bacteria to adhere to prokaryotic cells (5)], in the regulation of various plasma proteinase activities and hormones [reviewed by Vogt (3), Brot and Weissbach (6), and Swaim and Pizzo (7)] in the calmodulin-dependent activation of plasma membrane Ca-ATPase (8), and in the modulation of potassium channel function (9). (iii) MsrA might also serve as an antioxidant enzyme to protect some enzymes from oxidative damage by various reactive oxygen species (4).
A yeast null mutant of msrA was shown to accumulate both free and protein-bound MetO, and its growth was found to be more severely inhibited by H2O2 treatment than was that of the wild-type parent strain (2). A null mutant of msrA in Escherichia coli was also more susceptible to being killed by hydrogen peroxide than its parent and msrA overproducing strains, but only when grown on plates; however, these E. coli strains were shown to be equally sensitive to methyl viologen (paraquat) treatment (10).
E. coli and S. cerevisiae both contain at least two methionine sulfoxide reductases. One (msrA) is able to reduce both free and protein-bound MetO, and the other can reduce only free MetO. The peptide-methionine sulfoxide reductase was highly expressed in macrophages (11) and neutrophils (12). The high level of msrA may reflect the need to repair oxidatively damaged Met residues of endogenous proteins during periods of enhanced oxidative stress associated with oxidative burst activity. It has been shown that preexposure to oxidative stress suppresses the ability of T lymphocytes to respond to mitogenic stimulation in terms of proliferation, interleukin 2 biosynthesis, and signal transduction and that N-acetyl cysteine protected them against the stress (13–15).
In the present study, we determine the localization of the promoter for the msrA gene in S. cerevisiae and show that overexpression of MsrA in yeast and in human T lymphocytes leads to a decrease in the cellular levels of both free and protein-bound MetO and to enhanced survival of these cells under conditions of oxidative stress.
MATERIALS AND METHODS
Materials.
Hydrogen peroxide was purchased from Fisher. Dabsyl chloride was purchased from Pierce. Methyl viologen (paraquat) was purchased from Sigma. The compound 2,2′-azobis-(2-amidinopropane) dihydrochloride (AAPH), was purchased from Wako Chemicals USA (Richmond, VA). G418 sulfate, a generic form of Geneticin, was purchased from GIBCO/BRL.
Overexpression of Peptide-Methionine Sulfoxide Reductase in S. cerevisiae.
To get the msrA gene subcloned into the E. coli shuttle vector YEp351 (ATCC number 37672), PCR reactions were carried out by using S. cerevisiae DNA as a template, a 5′-sense primer (H1) containing a BamHI site (5′-AGCAGTGGATCCATTGAATGAGTTAACGGG) and 3′-reverse complement primer (H2) (5′-AGGGCAAAGCTTCTAAAAAAGCTACATTTC) with HindIII site. PCR was performed for one cycle of 5 min at 94°C followed by 30 cycles of 30 sec at 94°C, 60 sec at 50°C, and 60 sec at 72°C. Both the amplified product and YEp351 were digested with BamHI and HindIII, and the PCR fragment encompassing the complete yeast msrA coding region and upstream region (217 bp) was ligated into the restricted YEp351 by using T4 DNA ligase (Boehringer Mannheim). E. coli cells (DH5α) were transformed with an aliquot of the ligation mixture and grown in Luria–Bertani medium containing ampicillin (100mg/ml). One of the positive clones was used to transform msrA yeast mutant (Mata ΔmsrA∷URA3 his6 leu2) (2) according to the lithium acetate method (16). After growth on minimal medium plates without leucine and uracil, the positive yeast clones were grown on synthetic complete medium without leucine and uracil. The null msrA mutant and its parent strain (H9 strain: Mata ura3–52 his6 leu2) were transformed with the vector YEp351 alone and served as controls for the overexpression strain. To create a frame shift mutation in the msrA coding region, the same clone was digested with NdeI and then filled in with Klenow fragment and blunt ligated to itself by the T4 DNA ligase. After transformation to E. coli cells (DH5α), one of the positive clones was used to transform the msrA yeast mutant in the same way as described above.
To simplify the presentation, these three yeast strains will be referred to as follows: The parental strain (H9+YEp351) will be referred as the wild-type (WT) strain. The strain completely lacking msrA [(H9 ΔmsrA∷URA3) + YEp351] will be referred to as the null mutant (NM) strain. The NM strain harboring the cloned msrA gene will be referred to as the overproducing (OP) strain.
Determination of Growth Pattern of Yeast Strains and Their Methionine Sulfoxide Content After Exposure to Hydrogen Peroxide.
Yeast cells inoculated at late log phase were grown aerobically in synthetic complete medium at 30°C with or without H2O2 (2 mM) and also in the absence of H2O2 in a medium in which methionine was replaced by methionine sulfoxide. Their growth pattern was monitored by measuring cell density (turbidity measured in a Klett–Summerson colorimeter at 540 nm) until they reached stationary phase. When cell density reached 150 Klett units (54 filter), some of the cells in each culture were spun down and washed five times with PBS before their disruption in a French pressure cell in buffer B [6 M guanidine chloride and 500 mM K-phosphate (pH 2.5)]. After centrifugation at 20,000 × g for 20 min, the supernatant solutions were passed through microconcentrators (microcon 3, Amicon). In each case, the flow-through was collected for free amino acid analysis, whereas the retained material was kept for protein-bound amino acid analysis after extensive washing with buffer B. The protein moiety of each preparation was subjected to CNBr cleavage as previously described to quantitate the oxidized methionine (4). CNBr cleaves peptide bonds on the carboxyl side of methionine (but not such bonds involving MetO) to yield homoserine (17). Hydrogen chloride hydrolysis and amino acid analysis were carried out on samples with and without CNBr treatment, as previously described (18).
Survival of Yeast Strains Under Various Oxidative Stress Conditions.
Yeast strains were treated with either H2O2, methyl viologen (paraquat), or ultraviolet A radiation (UVA). The same amount of cells of each strain from late logarithmic stage of growth were diluted 1:100 into a buffer containing PBS, 2% glucose, and 0.17 mM methionine in the presence or absence of H2O2 (2 mM), paraquat (10 mM), or AAPH (10 mM). At each time point, an aliquot from each culture was taken and plated on minimal medium plates without leucine and the colonies grown were counted. In the experiment where the cells were treated with hydrogen peroxide, the consumption of hydrogen peroxide was followed by measuring the amount present by using the PeroXOquant for quantitative peroxide assay (Pierce). As for the UVA experiment, two sets of minimal medium plates without leucine were plated with the same amount of cells from each yeast strain. One set was exposed to UVA (1.2 J/min/cm2) for various time periods by using a Cube 401 Single Light High Intensity Unit with H1 filter for 100% UVA (Uvatec, Sherman Oaks, CA). The other set was not treated with UVA and served as the control experiment, and the colonies that grew were counted in each case.
Determination of MsrA Activity.
The ability of methionine sulfoxide reductase to reduce free methionine sulfoxide was assayed by using [3H]MetO as substrate, prepared as described by Brot et al. (19). The reaction mixture (30 ml) contained 15 mM DTT/25 mM Tris⋅HCl (pH 7.5)/16.7 mM [3H]MetO/cell extract. After incubation at 37°C, the reaction was stopped by adding 0.33 mM MetO, and conversion of [3H]MetO to [3H]Met was analyzed by thin-layer chromatography on a silica gel plate by using the solvent 1-butanol/acetic acid/water (60:12:25). After ninhydrin treatment of the plate, the spot that corresponded to the migration of Met was extracted by water and the radioactivity was measured. The reduction of protein-bound methionine sulfoxide by MsrA was assayed by using dabsyl-MetO as previously described (2). Briefly, reaction mixtures (100 ml) containing 20 mM Tris⋅HCl (pH 7.5)/10 mM MgCl2/30 mM KCl/20 mM DTT/dabsyl-MetO (1 mM) and an aliquot of yeast or human T lymphocyte cell (Molt-4) extract were incubated at 37°C for 30 min and were stopped by the addition of 200 ml of acetonitrile. After centrifugation, 10 ml of the clear supernatant solution was injected onto a 10-cm C18 column (Apex, Jones Chromatography, Lakewood, CO), equilibrated at 50°C with buffer [0.14 M sodium acetate, 0.5 ml/l triethylamine (pH 6.1)] containing 30% acetonitrile. By using a linear gradient from 30% to 70% acetonitrile, the dabsylmethionine was eluted at 5.0 min as monitored by peak integration at 436 nm (1 pmol of dabsylmethionine gives 340 area units). The column was washed for 5 min with the solvent containing 70% acetonitrile and reequlibrated at 30% before the next injection. For measuring MsrA activity in yeast, the cells were disrupted in 25 mM Tris⋅HCl (pH 7.5) in a French pressure cell.
Stable Transfection of Molt-4 Cells with msrAcDNA from Bovine.
The msrA from bovine cDNA was cut out and purified after digestion with HindIII and BamHI of the original clone (pJM200) (1). Then the DNA fragment harboring the msrA was subcloned in a sense orientation to the expression vector pcDNA 3.1 (Invitrogen) under the control of the cytomegalovirus enhancer–promoter for high-level expression. The resulting plasmid and pcDNA3.1 vector were linearized by digestion with BglII. Then Molt-4 cells were either transfected with this plasmid expressing msrA or with the pcDNA 3.1 vector alone, by using a liposome transfection reagent kit [N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate (DOTAP)] (Boehringer Mannheim). Stable transfectants were selected by serial dilutions in the growth medium containing RPMI1640 (GIBCO/BRL)/100 mM nonessential amino acids/4 mM l-glutamine/10 mM hepes/2 mM sodium pyruvate/50 mM β-mercaptoethanol/10% heat-inactivated fetal bovine serum/G418 sulfate (200 μg/ml) (Boehringer Mannheim).
Treating the Molt-4 Cells with Hydrogen Peroxide.
The stably transfected Molt-4 cells (either the msrA overexpressed cells or the vector transfected cells, 3 × 105 cells each) were grown with and without H2O2 at concentrations of 50, 100, and 200 μM for 24 and 48 h in the appropriate medium. Cell viability was monitored by staining the dead cells with trypan blue stain (0.4%) (GIBCO/BRL).
RESULTS
Peptide-Methionine Sulfoxide Reductase Overexpression in Yeast Cells and Its Effect on Their Growth and Methionine Sulfoxide Content.
The abilities of the three yeast strains—the null mutant lacking msrA (NM), the MsrA overproducing strain (OP), and the parental wild-type strain (WT)—to reduce free MetO and dabsyl MetO were determined. As shown in Table 1, the NM strain exhibited no detectable activity with the dabsyl MetO substrate, attesting to the fact that it is lacking the msrA gene. In contrast, the dabsyl MetO reductase activity of the OP strain was 25 times greater than that of the WT strain. In comparison, the NM strain exhibited low but significant free MetO reductase activity, and the MetO reductase activity of the OP strain was about 30 times greater than that of the WT strain (after taking into account the contribution of the MetO reductase activity of the NM strain from which it was derived). Overexpression of the msrA in the OP strain was under the control of its own promoter, which is located in the intergenic region between the msrA start codon and the upstream gene Rad2. To confirm that overexpressed MsrA activity was due to overexpression of the msrA gene, we created a frame shift mutation at Histidine 31. As shown in Table 1, this completely abolished the MsrA activity.
Table 1.
MsrA activity in yeast cells
| Enzyme | Specific activity (pmol/min/mg protein) on substrate
|
|
|---|---|---|
| Dabsyl-MetO | L-(3H) MetO | |
| Extract of yeast parental WT strain | 68 | 15 |
| Extract of yeast NM strain | 0 | 5 |
| Extract of yeast OP strain | 1700 | 300 |
| Extract of yeast NM strain + YEp351 harboring the msrA gene with a frameshift mutation | 0 | 5 |
Yeast extracts from the different strains were prepared by breaking the cells in 25 mM Tris⋅HCl (pH 7.5) in a French pressure cell. The MsrA activity was determined as described in Materials and Methods. WT, wild-type + YEp351; NM, msrA null mutant + YEp351; OP, overproducer strain (NM strain harboring YEp351 + msrA gene).
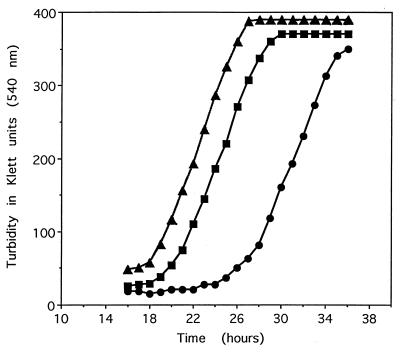
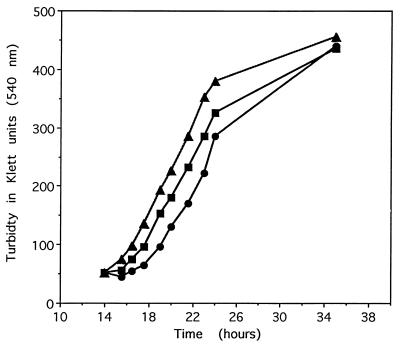
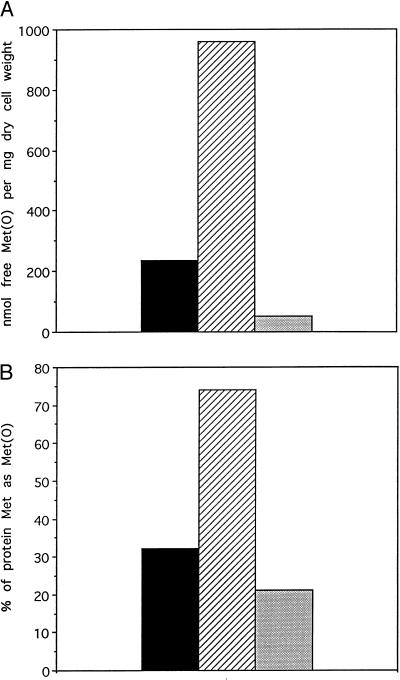
There was a little difference in the growth patterns of the three yeast strains (MN, OP, and WT) when grown under normal conditions (complete synthetic media without leucine; data not shown); however, in the presence of 2 mM H2O2, the lag phase of the NM strain was about 6 and 9 h greater than that of the WT and OP strains, respectively (Fig. 1). When MetO was substituted for Met in the growth medium, in the absence of H2O2, the lag phase of the NM strain was only 1.5 and 2.0 h longer than in WT and OP strains, respectively (Fig. 2). This observation suggests that the oxidation of cellular methionine, either free or protein-bound, is implicated in the lag phase. Indeed, as expected (2), the levels of free and protein-bound MetO in the MN strain grown in the presence of 2 mM H2O2 were 4- and 2.3-fold higher, respectively, than those in the parental WT strain (Fig. 3A and B). Furthermore, the levels of free and protein-bound MetO in the overproducing strain (OP) were 16.7% and 66% lower, respectively, than those in the WT strain.
Figure 1.
Growth of yeast strains in the presence of H2O2. Yeast from late log phase cultures was inoculated into yeast synthetic medium lacking leucine at 1:300 dilution and grown aerobically at 30°C with H2O2 (2 mM). ■, Wild-type parental strain; •, msrA null mutant strain; ▴, overproducer strain.
Figure 2.
Growth of yeast strain in the presence of MetO instead of Met in culture medium without H2O2. The growth of the three strains was as described in Fig. 1, except Met was replaced by MetO (0.17 mM) and the medium did not contain H2O2. ■, Wild-type parental strain; •, msrA null mutant strain; ▴, overproducer strain.
Figure 3.
Methionine sulfoxide content in yeast strains that were grown in the presence of hydrogen peroxide. Each yeast strain was grown generally in a synthetic medium lacking leucine and containing H2O2 (2 mM), as described in Fig. 1, until the cell density reached 150 Klett units in each cell culture. Cells were harvested and their extracts were measured for free methionine sulfoxide (A) or protein-bound methionine sulfoxide (B), as described in Materials and Methods. Black bars represent the wild-type parental strain, hatched bars represent the msrA null mutant strain, and dotted bars represent the overproducer strain.
The Effect of MsrA Expression Levels on the Survival Rates of Yeast and Molt-4 Cells Under Oxidative Stress Conditions.
To investigate the effect of MsrA levels on cell survival under conditions of oxidative stress, we studied the viability of the three yeast strains exposed to either 2 mM H2O2, 10 mM paraquat, or 10 mM AAPH. For comparison, the time required to obtain 50% cell killing (T50) was determined from plots of the log of percent survival vs. time (hours), which were linear for all conditions examined (data not shown). From the results summarized in Table 2, it can be seen that all forms of oxidative stress led to cell killing and that the time required to kill 50% of the cells (the T50 value) for the mutant lacking MsrA was about one-half that for the WT, which was slightly shorter than that of the OP strain. The possibility that variations in the T50 values were due to differences in the rates of oxidant inactivation was discounted, at least in the case of H2O2, by the demonstration that the rate of decomposition of H2O2 was the same for all three strains (data not shown). Treatment of these yeast cells with UVA resulted in lower survival rates but no obvious differences between the different strains (data not shown).
Table 2.
Survival rates of yeast strains grown under different oxidative stress conditions
| Treatment | T50, h
|
−Δ% survival per h
|
||||
|---|---|---|---|---|---|---|
| NM | WT | OP | NM | WT | OP | |
| H2O2 | 3.5 | 6.3 | 6.9 | 14.3 | 7.9 | 7.2 |
| Paraquat | 8.1 | 16.5 | 17.6 | 6.2 | 3.0 | 2.8 |
| AAPH | 11.0 | 20.2 | 21.3 | 4.5 | 2.5 | 2.3 |
The same amount of cells of each strain from late log phase was diluted 1:100 into a buffer containing PBS, 2% glucose, and 0.17 mM methionine in the presence or absence of H2O2 (2 mM). At each time point, an aliquot from each culture was taken and plated on minimal medium plates without leucine and the colonies were counted. Each survival rate point represents the number of colonies grown when H2O2 was present in relation to the number of colonies observed when growth was performed without H2O2 in the medium. Wild-type parental strain (WT). msrA null mutant strain (NM). Overproducer strain (OP). T50 is defined as the incubation time (hours) in culture needed to achieve 50% killing.
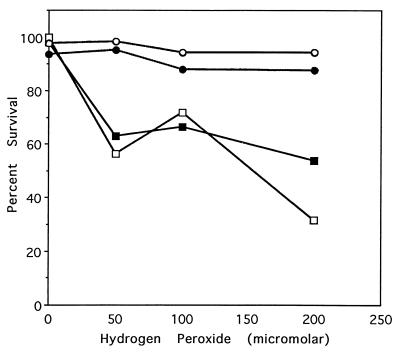
To examine the effect of MsrA overexpression in mammalian cells, we used human T lymphocyte cells (Molt-4) as a model. Stable transfection of the Molt-4 cells with bovine msrA led to a 4.8-fold increase in the MsrA activity (Table 3) and to a dramatic increase in their resistance to H2O2 toxicity, as is illustrated by the fact that 95% of the stably transfected cells survived 48 h exposure to 200 μM H2O2 as compared with only 30% survival of the parental cells (Fig. 4). Overexpression of MsrA in the Molt-4 cells is therefore more effective in resistance to induced oxidative stress than that observed in yeast (Table 2).
Table 3.
MsrA activity in Molt-4 cells
| Enzyme | Specific activity (pmol/min/mg protein) on dabsyl-MetO |
|---|---|
| Extract of Molt-4 cells transfected with pcDNA 3.1 | 11 |
| Extract of Molt-4 cells transfected with pcDNA 3.1 harboring the msrA cDNA | 53 |
Molt-4 were stably transfected with either pcDNA 3.1 or pcDNA 3.1 harboring the msrA bovine cDNA. After disruption of cells in 25 mM Tris⋅HCl (pH 7.5) by sonication, enzyme activity was measured. The details about the transfection protocol and the assay for MsrA activity are described in Materials and Methods.
Figure 4.
Effect of hydrogen peroxide on the survival rate of different Molt-4 strains. Molt-4 cells were stably transformed with either pcDNA 3.1 or pcDNA 3.1 harboring the bovine msrA cDNA, as described in Materials and Methods. Cells were grown in the presence of 50, 100, and 200 μM H2O2 for 24 and 48 h. Survival rate was represented by the percentage of living cells at each time point (as measured by using the trypan exclusion assay). Symbols are defined as follows: Molt-4 transfected only with the pcDNA 3.1 ■, at 24 h, and □, at 48 h; Molt-4 transfected with pcDNA 3.1 harboring the msrA gene •, at 24 h, and ○, at 48 h. Replicate experiments gave similar results.
DISCUSSION
In this study, we describe the overexpression of MsrA in S. cerevisiae. Overexpression of MsrA in a null mutant strain lacking MsrA led to 25-fold higher MsrA activity toward protein-bound MetO and a 30-fold higher activity toward free MetO, compared with the wild-type strain (Table 1). These results with yeast are similar to those reported earlier for the overexpression of MsrA in E. coli (20). In the present study, the overexpression of MsrA was mediated by the yeast’s own promoter, which supports the assumption that the promoter for the MsrA gene is located in the intergenic area between the msrA gene and the upstream gene, Rad2. In this area, three sequences have been identified at positions −42, −72, and −114 that contain the TATA box motif and, therefore, might serve as binding sites for the RNA polymerase or other transcription factors. Overproduction of MsrA in the yeast null mutant led to increased resistance to toxic concentrations of H2O2, AAPH, and paraquat (Table 2); it led also to lower intracellular levels of both free MetO and protein-bound MetO, compared with the wild-type strain (Fig. 3 A and B).
The effects of H2O2 on the growth, toxicity, and intracellular concentrations of MetO of a mutant lacking MsrA and a strain containing highly expressed levels of MsrA are very different (Figs. 1 and 3, Table 2). However, there was only a small difference in the growth rates of these yeast strains when Met in the growth medium was replaced by MetO (Fig. 2). This unexpected result may reflect the fact that yeasts contain two different methionine sulfoxide reductases: one that is specific for free MetO and one that reduces both free MetO and protein- or peptide-bound MetO. In the studies reported here, mutants that are either lacking or overexpressing the latter form of methionine sulfoxide reductase are compared with the wild-type strain. Therefore, the failure to observe a large difference in growth rates between these various yeast strains, when Met in the growth medium was replaced by MetO, could simply reflect the possibility that the reduction of MetO by the free methionine sulfoxide reductase or the de novo synthesis of methionine, or both, could supply almost all of the methionine needed for growth in the MetO supplemented media.
It is noteworthy that although the three strains of yeast differed markedly with respect to the length of the lag phase and in their rates of cell killing during growth in the presence of H2O2, the rate of H2O2 consumption during growth of all three strains was the same (data not shown). It is therefore evident that differences in the lag time are not related to the time required to consume all of the H2O2. This result invites the speculation that differences in the lag time reflect differences in the H2O2-dependent oxidation of critical methionine residues of some proteins involved in the regulation of cell proliferation and/or cell arrest and death. This proposition is consistent with the observations: (i) that overexpression of MsrA in msrA mutant leads to reversal in the accumulation of both free and protein-bound MetO (Fig. 3 A and B), and (ii) that the survival rate of the yeast msrA mutant, when cells were exposed to H2O2, was distinctly lower than its parent and overproducer strains (Table 2).
Overexpression of the bovine MsrA in the Molt-4 cells resulted in a better resistance to H2O2 toxicity compared with the control cells (Fig. 4). This protection from H2O2-induced oxidative stress was more effective than that observed in yeast (Table 2). Perhaps this phenomenon reflects differences between the roles of MsrA in mammalian and yeast cells with respect to important regulatory functions, such as the regulation of cell proliferation. Such a possibility is highlighted by the recent demonstration that the binding of various transcription factors (NFKapaB, AP-1, and NFAT) to DNA in human T lymphocytes is abolished by exposure to H2O2 (15, 21). In addition, results of ex vivo experiments have shown that oxidative stress associated with infection of T lymphocytes with HIV participates in CD4 + T lymphocyte depletion by increasing the rate of Fas-induced apoptosis (21). These observations raise the important question of whether overexpression of MsrA in the Molt-4 cells, as shown here, will make them more resistant to HIV infection. If so, perhaps an MsrA-based therapy could be developed.
Acknowledgments
We thank Dr. Rodney L. Levine and Dr. P. Boon Chock for helpful discussions. Part of this work was funded by the RGK Foundation, Austin, TX.
ABBREVIATIONS
- UVA
ultraviolet A radiation
- MetO
methionine sulfoxide
- MsrA
peptide-methionine sulfoxide reductase enzyme
- AAPH
2,2′-azobis-(2-amidinopropane) dihydrochloride
- Molt-4
human T lymphocyte cells
- yeast strain ΔmsrA::URA3
yeast null mutant of peptide-methionine sulfoxide reductase gene
References
- 1.Moskovitz J, Weissbach H, Brot N. Proc Natl Acad Sci USA. 1996;93:2095–2099. doi: 10.1073/pnas.93.5.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moskovitz J, Berlett B S, Poston J M, Stadtman E R. Proc Natl Acad Sci USA. 1997;94:9585–9589. doi: 10.1073/pnas.94.18.9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vogt W. Free Radical Biol Med. 1995;18:93–105. doi: 10.1016/0891-5849(94)00158-g. [DOI] [PubMed] [Google Scholar]
- 4.Levine R L, Mosoni L, Berlett B S, Stadtman E R. Proc Natl Acad Sci USA. 1996;93:15036–15040. doi: 10.1073/pnas.93.26.15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wizemann T M, Moskovitz J, Pearce B J, Cundell D, Arvidson C G, So M, Weissbach H, Brot N, Masure R H. Proc Natl Acad Sci USA. 1996;93:7985–7990. doi: 10.1073/pnas.93.15.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brot N, Weissbach H. Arch Biochem Biophys. 1983;223:271–281. doi: 10.1016/0003-9861(83)90592-1. [DOI] [PubMed] [Google Scholar]
- 7.Swaim M W, Pizzo S V. J Leukocyte Biol. 1988;43:365–379. doi: 10.1002/jlb.43.4.365. [DOI] [PubMed] [Google Scholar]
- 8.Yao Y, Yin D, Jas G S, Kuczera K, Williams T D, Schoneich C, Squier T C. Biochemistry. 1996;35:2767–2787. doi: 10.1021/bi951712i. [DOI] [PubMed] [Google Scholar]
- 9.Ciorba M A, Heinemann S H, Weissbach H, Brot N, Hoshi T. Proc Natl Acad Sci USA. 1997;94:9932–9937. doi: 10.1073/pnas.94.18.9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moskovitz J, Rahman A M, Strassman J, Yancey S O, Kushner S R, Brot N, Weissbach H. J Bacteriol. 1995;177:502–507. doi: 10.1128/jb.177.3.502-507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moskovitz J, Jenkins N A, Gilbert D J, Copeland N G, Jursky F, Weissbach H, Brot N. Proc Natl Acad Sci USA. 1996;93:3205–3208. doi: 10.1073/pnas.93.8.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brot N, Fliss H, Coleman T, Weissbach H. Methods Enzymol. 1984;107:352–360. doi: 10.1016/0076-6879(84)07023-3. [DOI] [PubMed] [Google Scholar]
- 13.Flescher E, Bowlin T L, Talal N. J Immunol. 1989;142:907–912. [PubMed] [Google Scholar]
- 14.Flescher E, Ledbetter J A, Schieven G L, Vela-Roch N, Fossum D, Dang H, Ogawa N, Talal N. J Immunol. 1994;153:4880–4889. [PubMed] [Google Scholar]
- 15.Flescher E, Tripoli H, Salnikow K, Burns F. Clin Exp Immunol. 1998;112:242–247. doi: 10.1046/j.1365-2249.1998.00548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito H, Fukuda Y, Murata K, Kimura A. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fliss H, Weissbach H, Brot N. Proc Natl Acad Sci USA. 1983;80:7160–7164. doi: 10.1073/pnas.80.23.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reddy V Y, Desrochers P E, Pizzo S V, Gonias S L, Sahakian J A, Levine R L, Weiss S J. J Biol Chem. 1994;269:4683–4691. [PubMed] [Google Scholar]
- 19.Brot N, Werth J, Koster K, Weissbach H. Anal Biochem. 1982;122:291–294. doi: 10.1016/0003-2697(82)90283-4. [DOI] [PubMed] [Google Scholar]
- 20.Rahman M A, Nelson H, Weissbach H, Brot N. J Biol Chem. 1992;267:15549–15551. [PubMed] [Google Scholar]
- 21.Israel N, Gougerot-Pocidalo M A. Cell Mol Life Sci. 1997;53(11–12):864–870. doi: 10.1007/s000180050106. [DOI] [PMC free article] [PubMed] [Google Scholar]