Abstract
The bcr-abl chimeric oncoprotein exhibits deregulated protein tyrosine kinase activity and is implicated in the pathogenesis of Philadelphia chromosome (Ph)-positive human leukemias, such as chronic myelogenous leukemia (CML). Recently we have shown that the levels of the protein tyrosine phosphatase PTP1B are enhanced in p210 bcr-abl-expressing cell lines. Furthermore, PTP1B recognizes p210 bcr-abl as a substrate, disrupts the formation of a p210 bcr-abl/Grb2 complex, and inhibits signaling events initiated by this oncoprotein PTK. In this report, we have examined whether PTP1B effects transformation induced by p210 bcr-abl. We demonstrate that expression of either wild-type PTP1B or the substrate-trapping mutant form of the enzyme (PTP1B-D181A) in p210 bcr-abl-transformed Rat-1 fibroblasts diminished the ability of these cells to form colonies in soft agar, to grow in reduced serum, and to form tumors in nude mice. In contrast, TCPTP, the closest relative of PTP1B, did not effect p210 bcr-abl-induced transformation. Furthermore, neither PTP1B nor TCPTP inhibited transformation induced by v-Abl. In addition, overexpression of PTP1B or treatment with CGP57148, a small molecule inhibitor of p210 bcr-abl, induced erythroid differentiation of K562 cells, a CML cell line derived from a patient in blast crisis. These data suggest that PTP1B is a selective, endogenous inhibitor of p210 bcr-abl and is likely to be important in the pathogenesis of CML.
Chronic myelogenous leukemia (CML) is characterized by the reciprocal chromosomal translocation 9:22, which generates the Philadelphia chromosome (Ph) (1). This event occurs in the pluripotent hematopoietic stem cell and transposes the c-abl proto-oncogene on chromosome 9, encoding a protein tyrosine kinase (PTK), to a new position downstream of the second exon of the gene bcr on chromosome 22. This translocation generates a novel fusion gene, bcr-abl, that encodes a chimeric protein, p210 bcr-abl, the PTK activity of which is aberrantly regulated relative to c-Abl (2). Growth factor-dependent lymphoid and myeloid cells are transformed in culture into factor-independent and tumorigenic cells after ectopic expression of p210 bcr-abl (3, 4). Furthermore, expression of p210 bcr-abl in transgenic mice has been shown to cause a CML-like myeloproliferative disease (5–7). Therefore, p210 bcr-abl appears to play a fundamental role as the primary causative factor in CML (8).
The observation that production of p210 bcr-abl is the initiating event in CML has focused attention on the tyrosine phosphorylation-dependent signaling events triggered by this oncoprotein. Progress has been made in defining critical substrates for propagating such signals (9, 10). Nevertheless, control over the state of tyrosine phosphorylation of proteins in vivo is governed by the coordinated and competing actions of both PTKs and protein tyrosine phosphatases (PTPs) (11). PTPs have been identified in eukaryotes, prokaryotes, plants, and viruses and comprise a large family of enzymes that will rival the PTKs in structural diversity and complexity (12). PTPs may either antagonize or potentiate PTK-induced signaling in vivo and have been implicated in such fundamental physiological processes as growth and proliferation, differentiation, cytoskeletal function, as well as in the etiology and pathogenesis of certain diseases (13). Therefore, characterization of the PTPs that either potentiate or antagonize p210 bcr-abl-induced signaling and transformation would provide critical insights into the genesis and development of CML.
Recently we have shown that PTP1B is up-regulated rapidly and selectively after expression of p210 bcr-abl in a number of CML model cell lines. A substrate-trapping mutant of PTP1B (PTP1B-D181A), in which high affinity for substrate is maintained but catalytic function is impaired (14), forms a stable, direct complex with p210 bcr-abl in vitro and upon expression in COS cells. Furthermore, expression of either wild-type or substrate-trapping mutant forms of PTP1B inhibited the binding of the adaptor protein, Grb2, to p210 bcr-abl and suppressed p210 bcr-abl-induced, Ras-dependent transcriptional activation in NIH 3T3 cells. In contrast, TCPTP, the closest known relative of PTP1B, did not recognize p210 bcr-abl as a substrate nor did it effect p210 bcr-abl-induced signaling. Furthermore, neither PTP1B nor TCPTP recognized v-Abl, which shares the same catalytic domain as p210 bcr-abl, as a substrate or attenuated its signaling function. These data suggest that PTP1B may function as a specific, negative regulator of p210 bcr-abl signaling in vivo (15).
In this study, we have addressed the paramount question of whether PTP1B can influence transformation induced by p210 bcr-abl. We show that ectopic expression of either wild-type or substrate-trapping (D181A) mutant PTP1B in Rat-1 fibroblasts attenuated transformation by p210 bcr-abl, as illustrated by reduced growth in soft agar, impaired proliferation under reduced serum conditions, and decreased tumor formation in nude mice. In contrast, TCPTP did not suppress transformation by p210 bcr-abl. In addition, neither PTP1B nor TCPTP suppressed v-Abl-induced transformation. Furthermore expression of PTP1B, but not TCPTP, induced erythroid differentiation of the CML cell line, K562, similar to the effect observed after treatment of the cells with a chemical inhibitor of p210 bcr-abl. These data stress the potential importance of PTP1B as a physiological negative regulator of the p210 bcr-abl oncoprotein PTK and thus as a controlling element in the pathogenesis of CML.
MATERIALS AND METHODS
Cell Lines.
Recombinant PTP1B-, PTP1B-D181A-, and TCPTP-expressing retroviruses were constructed by inserting full-length cDNAs into the pWZL-hygro vector. Helper-free retroviral stocks were prepared by transient hyperexpression in LinX, a retroviral packaging line derived from 293 cells (L. Xie, D. Beach, and G.H., unpublished data). These retroviruses were used to infect Rat-1 fibroblasts expressing p210 bcr-abl or v-Abl. Drug-resistant populations were established by culturing the infected cells for 14 days in 100 μg/ml hygromycin, refeeding every 3 days in the presence of the drug.
Cell Culture and Preparation of Lysates.
Stable Rat-1 fibroblast lines expressing p210 bcr-abl and v-Abl were created as described (16) and cultured in DMEM with 10% fetal bovine serum (FBS). K562 cells were cultured in RPMI medium 1640 with 10% FBS. Cells were lysed in 50 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, 1% (vol/vol) Triton X-100, 10% glycerol, 1 mM EDTA, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 1 mM benzamidine, and 1 mM phenylmethylsulfonyl fluoride in the presence or absence of 1 mM Na3VO4. Protein concentrations were determined by the Bradford assay, using BSA as standard (17).
Antibodies, Immunoblotting, and Immunoprecipitation.
The following mAbs were used: anti-PTP1B, FG6, and anti-TCPTP, CF4, were kindly provided by D. Hill (Calbiochem Oncogene Research Products, MA); anti-Abl antibodies, 8E9 and K12, were from PharMingen and Santa Cruz, respectively; anti-phosphotyrosine, 4G10 was from Upstate Biotechnology; anti-Grb2 was from Transduction Laboratories (Lexington, KY); and the mAb to α-globin was kindly provided by Somatagen (Boulder, CO). For immunoprecipitation, lysates were precleared with IgG Sorb (The Enzyme Center, Malden, MA) reconstituted in lysis buffer, and then added to Protein A Sepharose-coupled antibody. After rocking at 4°C for 90 min, immune complexes were collected by centrifugation for 10 s at 1,000 × g, the supernatant was discarded, and the beads were washed five times in 20 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, 0.1% (vol/vol) Triton X-100, 10% glycerol, 1 mM EDTA, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 1 mM benzamidine, 1 mM phenylmethylsulfonyl fluoride, and 1 mM Na3VO4. Samples were subjected to SDS/PAGE and transferred to nitrocellulose membranes. The membranes were blocked with 5% nonfat dry milk in TBS-Tween, incubated with the appropriate antibody for immunoblotting, and visualized by ECL (Amersham).
Transformation Assays.
(i) Anchorage-independent growth: Equal numbers of viable cells were plated in 0.35% noble agar in 10% FBS-containing medium at a density of 104 cells per 6 cm2 plate. The number of colonies formed in agar was recorded 2 weeks after plating the cells (18). (ii) Growth in reduced serum: At day 0, p210 bcr-abl-transformed Rat-1 fibroblasts, ectopically expressing the PTPs or control vector, were seeded at a density of 104 cells per 10 cm2 plate in DMEM supplemented with 1% FBS. Subsequently, every 2 days, the culture medium was changed and cells were counted. (iii) Tumor formation: Individual clones of p210 bcr-abl-transformed Rat-1 fibroblasts expressing PTPs or control vector (1 × 106 cells) were injected into each of the flanks of BALB/c nude mice (Taconic Farms). Two separate clones were evaluated for each condition. Mice were monitored daily, and 3 weeks after inoculation they were killed and the size of the tumors recorded in grams wet weight. The data are expressed as mean ± SEM for each condition (four mice in total, two mice per cell clone, inoculated at two sites each, i.e., eight tumor masses per condition).
Erythroid Differentiation Assay.
K562 cells (2 × 106) were washed, resuspended in 0.5 ml of RPMI medium 1640 supplemented with 10% FBS, placed in a 0.4-cm gap electroporation cuvette (GIBCO/BRL), and 25 μg of plasmid DNA (pcDNA3, Invitrogen) expressing either PTP1B or TCPTP, or control vector alone, was added. After a 10-min incubation at room temperature, the cells were transiently transfected by electroporation by using a single electric pulse (960 μF, 300V) in a Bio-Rad Gene Pulser electroporator and then replated into growth medium. At 24 h after transfection, cells were resuspended in fresh media in the presence of 1 mg/ml G418. As a positive control, cells were transfected with control vector and, 24 h after transfection, resuspended in fresh media and treated with 10 μM CGP57418, a selective inhibitor of Abl PTKs (19, 20). After 48 h, cells were harvested and lysates analyzed for α-globin expression by immunoblotting.
RESULTS
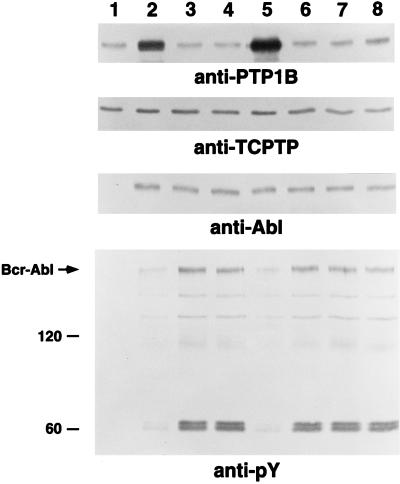
As we reported previously, Rat-1 fibroblasts responded to expression of p210 bcr-abl by increasing the levels of PTP1B protein (Fig. 1, lanes 1 and 2), (15). Strikingly, we observed that this increase was reversed in clonal, transformed lines selected in soft agar from the mass (heterogeneous) population of p210 bcr-abl-expressing Rat-1 fibroblasts (Fig. 1, compare lanes 3 and 4 with lane 2). In contrast, the levels of TCPTP and p210 bcr-abl remained unaltered after clonal isolation of transformed cells. Immunoblotting of cell lysates with antibodies to phosphotyrosine revealed that upon expression of p210 bcr-abl, the phosphorylation of tyrosyl residues in proteins in the mass population of Rat-1 fibroblasts was induced only marginally. A robust tyrosine phosphorylation response was detected only after clonal selection of transformed lines and concomitant down-regulation of PTP1B. These data are consistent with the possibility that PTP1B may function to antagonize p210 bcr-abl-induced signaling and thus transformation. To examine this issue further, we expressed PTP1B ectopically in two, distinct p210 bcr-abl-transformed Rat-1 fibroblast clones in which the level of the phosphatase had been down-regulated as a result of clonal selection. After infection of these cells with recombinant retroviruses, the levels of PTP1B reached those observed after expression of p210 bcr-abl in parental Rat-1 fibroblasts. As shown in Fig. 1 (lane 5), enforced expression of PTP1B in one of these lines attenuated tyrosine phosphorylation. Similar results were obtained in both lines. This observation prompted us to investigate in detail the effects of PTP1B on p210 bcr-abl-induced transformation.
Figure 1.
Levels of PTP1B in p210 bcr-abl-expressing Rat-1 fibroblasts. Protein expression was determined by immunoblotting equal quantities of lysate with the appropriate antibodies. Two gels were run, one to determine the levels of PTP1B (Top panel) and p210 bcr-abl (Third panel), the other to assess the state of tyrosine phosphorylation of the various proteins in the lysate (Bottom panel) and the levels of TCPTP (Second panel). Immunoblots were visualized by ECL. The lanes represent: 1, parental Rat-1 fibroblasts; 2, heterogeneous (mass) population of p210 bcr-abl-expressing Rat-1 fibroblasts; 3 and 4, soft agar-derived clones of p210 bcr-abl-transformed Rat-1 fibroblasts, lane 3 = clone 5, lane 4 = clone 10; 5, p210 bcr-abl-transformed clone 5 ectopically expressing PTP1B; 6–8, clones recovered in soft agar from p210 bcr-abl-transformed clone 5 ectopically expressing PTP1B.
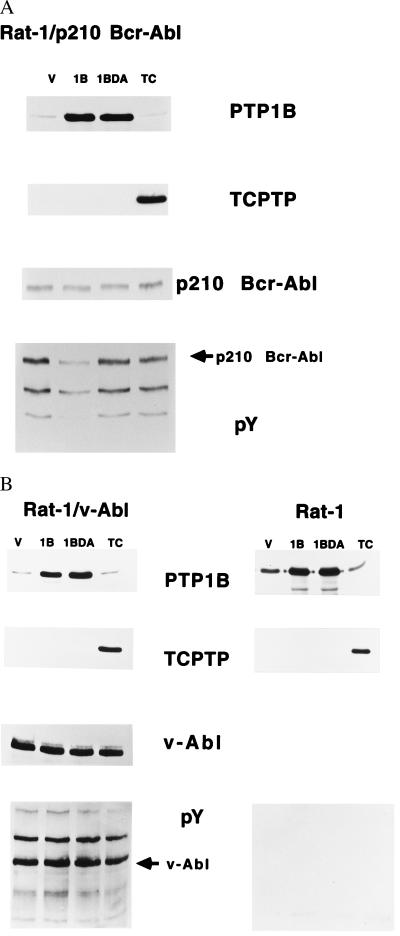
Overexpression of PTP1B, but Not TCPTP, Suppressed p210 bcr-abl-Induced Transformation of Rat-1 Fibroblasts.
Parental Rat-1 fibroblasts, Rat-1 fibroblasts expressing v-Abl, and two independent soft agar-derived clones transformed by p210 bcr-abl (clones 5 and 10) were infected with the PTP1B- or TCPTP-expressing retroviruses. After selection of infected cells, the levels of p210 bcr-abl, v-Abl, PTP1B, and TCPTP, as well as the extent of tyrosine phosphorylation, were evaluated by immunoblotting with appropriate antibodies (Fig. 2 A and B). Expression of wild-type PTP1B in the p210 bcr-abl-transformed Rat-1 fibroblast clones suppressed tyrosine phosphorylation without altering the levels of the PTK. In contrast, no effect on tyrosine phosphorylation was evident after expression of TCPTP (Fig. 2A). Tyrosine phosphorylation was enhanced in Rat-1 fibroblasts transformed by v-Abl, which bears the same catalytic domain as p210 bcr-abl (21); however, we did not detect an effect of expression of either PTP1B or TCPTP on these phosphorylation events (Fig. 2B).
Figure 2.
Expression of PTP1B and TCPTP in p210 bcr-abl-transformed (A) and parental and v-Abl-transformed Rat-1 fibroblasts (B). (A) Equal quantities of lysate protein from Rat-1/p210 bcr-abl (clone 5) cells were analyzed by immunoblotting with an anti-PTP1B antibody (Top panel); the blot was stripped and reprobed with anti-TCPTP antibody (Second panel). Expression and activation status of p210 bcr-abl were analyzed on a second gel by immunoblotting with an anti-pTyr antibody (Bottom panel), then stripping and reblotting with anti-Abl antibody 8E9 (Third panel). (B) Equal quantities of lysate protein from parental and v-Abl-transformed Rat-1 cells were immunoblotted for expression of PTP1B (Top panel) and the blot was stripped and reprobed with anti-TCPTP antibody (Second panel). Tyrosine phosphorylation was determined on a second gel by immunoblotting with an anti-pTyr antibody (Bottom panel) and the blot was stripped and reblotted with anti-Abl antibody 8E9 (Third panel). Immunoblots were visualized by ECL.
Rat-1 fibroblasts transformed by either v-Abl or p210 bcr-abl display anchorage-independent growth, as measured by their ability to form colonies in soft agar (18, 22). Ectopic expression of wild-type or substrate-trapping mutant PTP1B in clonal, p210 bcr-abl-transformed Rat-1 cell lines decreased by ≈60% the number of colonies observed in soft agar, as compared with p210 bcr-abl-transformed Rat-1 fibroblasts expressing either TCPTP or a control vector (Table 1). Colonies derived from Rat-1/p210 cells expressing either PTP1B or PTP1B-D181A were small and acidification of the culture medium was not detected. In contrast, we detected no attenuation of the ability of v-Abl-transformed cells to form colonies in soft agar after expression of either PTP1B or TCPTP (Table 1). Thus, the effects of PTP1B appear to be selective for p210 bcr-abl.
Table 1.
PTP1B and PTP1B-D181A proteins inhibit the growth of p210 bcr-abl-transformed cells in soft agar
| Construct | No. of colonies per 104 cells
|
Acidification | No. of colonies per 104 cells
|
Acidification | |
|---|---|---|---|---|---|
| Rat-1 | Rat-1/p210 | Rat-1v-Abl | |||
| Vector | 2.0 ± 1.2 | A 676 ± 30 | ++ | 1732 ± 71 | ++ |
| B 669 ± 17 | ++ | ||||
| PTP1B | 1.5 ± 1.1 | A 265 ± 10 | − | 1826 ± 65 | ++ |
| B 260 ± 19 | − | ||||
| PTP1B-D181A | 1.8 ± 0.8 | A 325 ± 26 | − | 1782 ± 40 | ++ |
| B 309 ± 17 | − | ||||
| TCPTP | 0.5 ± 0.5 | A 608 ± 23 | ++ | 1715 ± 54 | ++ |
| B 589 ± 9 | ++ | ||||
Although expression of PTP1B markedly suppressed the ability of p210 bcr-abl to induce anchorage-independent growth of Rat-1 fibroblasts, the effect was not absolute. Therefore, we tested the colonies that escaped this suppression for expression of PTP1B. Interestingly, we noted that the level of PTP1B in these colonies was similar to that observed in parental Rat-1 fibroblasts (Fig. 1, lanes 6–8). The decline in levels of PTP1B was accompanied by enhanced tyrosine phosphorylation and occurred in the absence of an effect on the level of expression of p210 bcr-abl or endogenous TCPTP. These data suggest an inverse correlation between p210 bcr-abl-induced transformation and the levels of PTP1B. Depletion of PTP1B may be required for manifestation of the p210 bcr-abl-induced transformed phenotype.
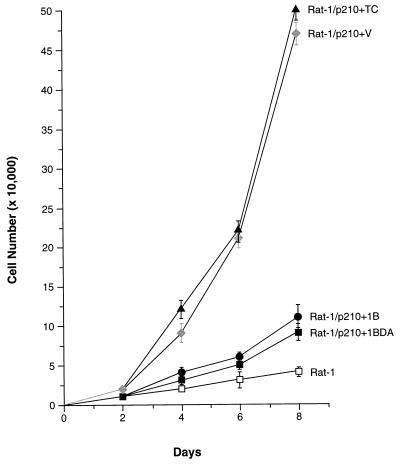
Overexpression of PTP1B Inhibited the Growth of p210 bcr-abl Transformed Rat-1 Fibroblasts in Reduced Serum and Tumor Formation in Nude Mice.
Parental Rat-1 fibroblasts typically require 10% FBS for proliferation in culture. However, transformation by p210 bcr-abl reduces this requirement, allowing growth in medium containing 1% FBS (Fig. 3). Upon transfer of either the control vector or the TCPTP expression vector, p210 bcr-abl-transformed Rat-1 cells displayed substantial growth rates in 1% FBS. In contrast, overexpression of either wild-type or substrate-trapping mutant PTP1B profoundly inhibited proliferation in 1% FBS (Fig. 3).
Figure 3.
Effect of overexpression of PTP1B or TCPTP on growth of p210 bcr-abl-transformed Rat-1 fibroblasts in reduced serum. The indicated cell lines were initially maintained in 10% FBS, then switched to 1% FBS on day 0. Cell numbers were counted and the media was changed at 2-day intervals. The graph presents the data as the mean ± SEM from three independent experiments, performed in duplicate on each of the two independent p210 bcr-abl-transformed Rat-1 clonal lines (clones 5 and 10) overexpressing the PTPs. Parental Rat-1 fibroblasts grown in 1% serum were included as a control.
Next, we evaluated the effect of ectopic expression of PTP1B on p210 bcr-abl-induced tumorigenesis in vivo. Rat-1 fibroblasts transformed with p210 bcr-abl produced large tumors after injection into nude mice (1.10 ± 0.17 g wet tumor weight). Overexpression of PTP1B in p210 bcr-abl-transformed cells reduced, by 70–80%, the size of tumors formed at 3 weeks after injection (PTP1B 0.26 ± 0.08 g, PTP1B-D181A 0.14 ± 0.05). In contrast, the effect of expression of TCPTP was much less pronounced (0.73 ± 0.14 g). Thus, the presence of PTP1B appears to interfere with the ability of Rat-1 fibroblasts expressing p210 bcr-abl to form both colonies in soft agar and tumors in nude mice.
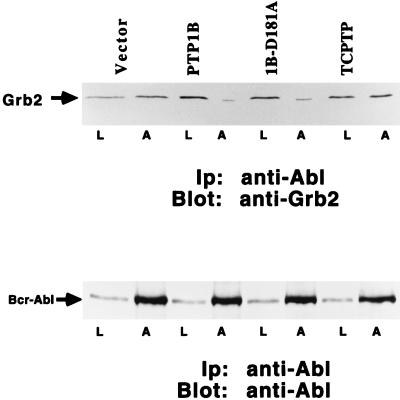
Expression of PTP1B in p210 bcr-abl-Transformed Rat-1 Cells Disrupted the Association Between Grb2 and p210 bcr-abl.
Our data illustrate that ectopic expression of PTP1B suppresses the transformed phenotype of p210 bcr-abl-expressing Rat-1 fibroblasts. Transformation of fibroblasts by p210 bcr-abl is dependent upon the adaptor protein Grb2, which links p210 bcr-abl to the Ras-signaling cascade (16, 23). We recently reported that coexpression of p210 bcr-abl with PTP1B in COS cells disrupted the association of the PTK with endogenous Grb2 (15). To address the mechanism by which PTP1B suppressed p210 bcr-abl-induced transformation, we examined the effect of the PTP on association of Grb2 with p210 bcr-abl in Rat-1 fibroblasts. Rat-1/p210 cells expressing the various PTPs were lysed, p210 bcr-abl was immunoprecipitated, and the levels of associated Grb2 were determined (Fig. 4). We observed that the association between p210 bcr-abl and Grb2 was disrupted in cells expressing either wild-type or substrate-trapping mutant PTP1B, but not in cells expressing TCPTP.
Figure 4.
Effect of overexpression of PTP1B or TCPTP on the association of Grb2 with p210 bcr-abl. Cell lysates (300 μg) from p210 bcr-abl-transformed clone 5 overexpressing PTP1B, PTP1B-D181A, TCPTP, or a control vector were prepared, and p210 bcr-abl was immunoprecipitated by using anti-Abl antibody K12 (lanes A). Immunoprecipitates were resolved by SDS/PAGE, transferred to nitrocellulose, and immunoblotted with anti-Grb2 antibody (Upper) or anti-Abl antibody 8E9 (Lower), as a control to illustrate constant levels of p210 bcr-abl. Lanes L depict 25 μg of cell lysate. Immunoblots were visualized by ECL.
PTP1B-Induced Differentiation of the Human Ph-Positive Leukemic Cell Line K562.
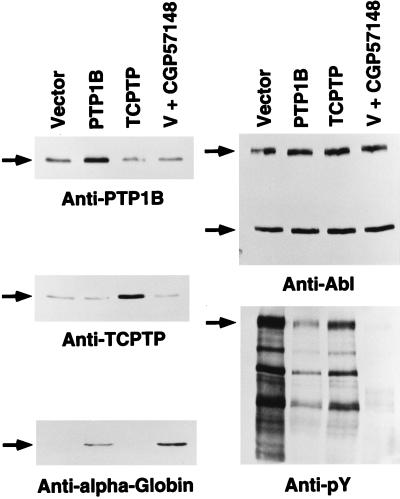
The studies described above have focused on an analysis of the effects of PTP1B on p210 bcr-abl-induced transformation of fibroblasts. However, it is important also to examine this issue in hematopoietic cells, wherein p210 bcr-abl is a causative agent in human disease. K562 cells, a p210 bcr-abl-expressing cell line established from a CML patient in blast crisis, retain their capacity to differentiate along megakaryocytic, erythroid, or monocytic lineages following appropriate stimuli (24). Previous studies have demonstrated that inhibition of the kinase activity of p210 bcr-abl induces erythroid differentiation of K562 cells (25). Recently, CGP57148, a 2-phenylaminopyrimidine derivative, was reported to inhibit the kinase activity of p210 bcr-abl with a high degree of selectivity (19, 20). Using the appearance of the α-subunit of human hemoglobin (α-globin) as a marker for the induction of erythroid differentiation, we observed that treatment of K562 cells with 10 μM CGP57148 induced differentiation (Fig. 5 Bottom Left).
Figure 5.
Effect of overexpression of PTP1B or TCPTP on differentiation of K562 cells. K562 cells were transiently transfected with pcDNA3 (control vector), pcDNA3-PTP1B, or pcDNA3-TCPTP expression plasmids. An additional aliquot of control vector-transfected K562 cells were further treated with 10 μM CGP57148 for 48 h. Lysates from each condition were analyzed by immunoblotting with anti-PTP1B antibody (Top Left), and the blot was stripped and reprobed with anti-TCPTP antibody (Middle Left). This blot also was probed with anti-α-globin antibody (Bottom Left). The expression and activation status of endogenous p210 bcr-abl were analyzed by immunoblotting with anti-pTyr antibody (Bottom Right) and the blot was stripped and reprobed with anti-Abl antibody 8E9 (Top Right). Immunoblots were visualized by ECL.
It was not possible to generate stable K562 cell lines overexpressing the PTPs; therefore, PTP1B, TCPTP, or the control vector (pcDNA3) were introduced transiently into the Ph-positive K562 cells by electroporation. After transfection and 48–60 h of culture in the presence of G418, cells were harvested and the levels of various proteins were determined by immunoblotting (Fig. 5). In each experiment, the level of ectopically expressed PTP1B was ≥2-fold that of the endogenous protein (Fig. 5 Top Left) and the level of TCPTP was increased 3- to 4-fold over the endogenous protein (Fig. 5 Middle Left). The expression of p210 bcr-abl in these cells was unchanged (Fig. 5 Top Right). Under these conditions, we observed an increase in α-globin levels in cells overexpressing PTP1B but not after overexpression of TCPTP (Fig. 5 Bottom Left). Furthermore, the level of phosphotyrosine also was markedly reduced in K562 cells overexpressing PTP1B, or treated with CGP57148, but less significantly in those cells overexpressing TCPTP (Fig. 5 Bottom Right). These data suggest that p210 bcr-abl may transform hematopoietic cells in part by blocking normal differentiation and that a specific inhibitor of the PTK (CGP57148) or a natural antagonist (PTP1B) may overcome this block.
DISCUSSION
CML represents one of the best defined examples in which aberrant protein tyrosine phosphorylation is the underlying cause of a human neoplasia (8). Deregulation of the activity of the Abl PTK domain in p210 bcr-abl, relative to c-Abl, results in aberrant tyrosine phosphorylation, which engenders the pathological effects of the oncoprotein PTK. Consequently, much research effort has focused on characterizing the aberrant signaling function of p210 bcr-abl. Nevertheless, it is important to consider that the state of tyrosine phosphorylation of proteins in vivo is governed by the coordinated actions of both PTKs and PTPs (12, 13). However, the analysis of the effects of PTPs on p210 bcr-abl function has remained largely unexplored. Characterization of the PTPs that control p210 bcr-abl function, or are themselves subject to regulation by p210 bcr-abl, represents an essential step toward gaining an understanding of the physiological changes that result in CML and in offering potential means through which the disease may be countered.
Our work has focused attention on PTP1B as a potential physiological antagonist of p210 bcr-abl. PTP1B is the prototypical PTP that was the first member of the family isolated in homogeneous form (26). It comprises an N-terminal catalytic domain fused to a regulatory C-terminal segment. The extreme C terminus is characterized by a stretch of 35 residues that are predominantly hydrophobic and both necessary and sufficient for targeting PTP1B to the cytoplasmic face of membranes of the endoplasmic reticulum (27). In light of this subcellular distribution, one would anticipate that PTP1B will be exposed to a substantial array of cellular phosphotyrosyl proteins. However, at present, insights into the physiological function of PTP1B are somewhat limited. PTP1B can revert partially the transformation of 3T3 cells by v-Src (28), protect 3T3 cells from transformation by Neu (29), and inhibit IL-3-dependent signaling in 32D-cl3 cells (30). More recently, it has been suggested that overexpression of PTP1B in Rat 3Y1 cells inhibits integrin-dependent signaling (31). In addition, the observation that PTP1B interacts with the insulin receptor (32, 33), together with the application of strategies involving overexpression (34) and the use of inhibitory antibodies (35), has implicated PTP1B in negative regulation of insulin signaling. However, it is unclear at present whether these observations reflect normal physiological functions of PTP1B and whether there is specificity for PTP1B in these processes.
The basis for the present study of the effects of PTP1B on p210 bcr-abl-induced transformation is our observation that the levels of PTP1B were enhanced rapidly and specifically in response to expression of p210 bcr-abl in several CML model systems (15). The specific induction of PTP1B by p210 bcr-abl suggests a role for this PTP in modulating the signaling events initiated by the oncoprotein PTK. However, PTP1B could either serve a permissive role in mediating signaling or function as an antagonist. The use of substrate trapping mutants revealed that PTP1B recognizes p210 bcr-abl directly as a substrate, but does not recognize v-Abl (15). Furthermore, PTP1B inhibits signaling events initiated by p210 bcr-abl, but not v-Abl (15). Such observations clearly suggest that PTP1B may be a physiological antagonist of p210 bcr-abl function but do not address the fundamentally important issue of whether PTP1B can suppress the transforming potential of this oncoprotein PTK.
In this study, we show that expression of either wild-type or substrate-trapping mutant forms of PTP1B abrogated p210 bcr-abl-induced transformation. This was manifested in the ability of PTP1B to inhibit the anchorage-independent growth, proliferation under reduced serum conditions, and tumor formation in nude mice displayed by p210 bcr-abl transformed fibroblasts. Particularly striking was the observation that the elements of specificity were maintained. Although PTP1B inhibited p210 bcr-abl-induced transformation, there was no detectable effect on transformation induced by v-Abl. Furthermore, we did not observe an effect of TCPTP on transformation induced by either PTK. The profound inhibitory effect of PTP1B on p210 bcr-abl-induced transformation is highlighted in Fig. 1. After expression of p210 bcr-abl, the levels of PTP1B were enhanced and changes in tyrosine phosphorylation were minor in the mass population of Rat-1 fibroblasts. Upon selection in soft agar, the levels of PTP1B in the clonal transformed lines that were recovered had declined to those observed in the parental cells and enhanced tyrosine phosphorylation was then apparent. Ectopic overexpression of PTP1B in the transformed cells resulted in marked tyrosine dephosphorylation and abrogation of the transformed phenotype. In the small number of clones that were recovered upon culture in soft agar after ectopic expression of PTP1B, it was observed that the levels of the phosphatase were suppressed. These data reveal that expression of p210 bcr-abl and PTP1B may be coupled as a mutually antagonistic pair of activities in vivo.
It has been shown that p210 bcr-abl autophosphorylates on several sites, one of which, Y177, serves as a docking site for the adaptor protein Grb2 (16). Interaction with Grb2 initiates assembly of a multiprotein complex including SOS leading to the activation of Ras, which is known to be important for p210 bcr-abl-induced transformation (16, 23, 36–38). Mutation of Tyr-177 → Phe or expression of dominant-negative mutant forms of Ras and Grb2 has been shown to inhibit transformation by p210 bcr-abl (16, 39, 40). In this study, we observed that overexpression of either wild-type or substrate-trapping mutant PTP1B disrupted the association between endogenous Grb2 and p210 bcr-abl, which likely reflects one mechanism by which inhibition of p210 bcr-abl-induced transformation by PTP1B is achieved. However, PTP1B recognizes additional sites in p210 bcr-abl (15) and thus may abrogate other signaling events triggered by the PTK. Recently, it was shown that overexpression of PTP1B can inhibit transformation of Rat 3Y1 cells by v-Crk, v-Src, and v-Ras (41). It was suggested that the ability of PTP1B to suppress transformation in these cells correlated with the phosphorylation status of p130cas, a cytoskeletal protein that interacts with a Pro-rich sequence in PTP1B. However, we did not observe any change in the phosphorylation status of p130cas in Rat-1 fibroblasts transformed by p210 bcr-abl (data not shown). In contrast, our data suggest that PTP1B inhibits p210 bcr-abl-induced transformation directly through dephosphorylation of the PTK, thus abrogating downstream signaling events.
CML, which is initiated by the generation of the Ph+ chromosome and the expression of p210 bcr-abl, displays a predictable progression from a relatively benign chronic phase to a lethal acute leukemia, known as blast crisis. In the chronic phase of CML, the stem cells differentiate into various types of progenitor cells but in larger numbers than normal hematopoiesis (8). However, these cells retain the ability to differentiate into mature blood cells. Later, in the blast-crisis phase of CML, the progenitor cells no longer differentiate. It is interesting to speculate that perhaps in the chronic phase of the disease the function of the p210 bcr-abl PTK oncoprotein is suppressed by PTP1B and thus chronic phase CML progenitor cells are almost indistinguishable from their normal counterparts. However, after one of the many “secondary hits” that accompany progression into blast crisis, the effects of PTP1B may be lost, allowing p210 bcr-abl to exert its effects unchecked. Perhaps PTP1B offers a novel potential avenue for therapeutic intervention in CML.
Acknowledgments
We thank Drs. A. M. Pendergast (Duke University) for helpful advice and reagents and B. Clarkson (Memorial Sloan Kettering Cancer Center) for the K562 cell line and CGP57418 inhibitor. We thank L.-Y. Xie for tissue culture and L. Bianco for technical assistance in the experiments involving nude mice. This work was supported by grants from the National Institutes of Health (CA64593) and the Hansen Memorial and Lauri Strauss Leukemia Foundations.
ABBREVIATIONS
- Ph
Philadelphia chromosome
- CML
chronic myelogenous leukemia
- PTK
protein tyrosine kinase
- PTP
protein tyrosine phosphatase
- FBS
fetal bovine serum
References
- 1.Melo I V. Leukemia. 1996;10:751–756. [PubMed] [Google Scholar]
- 2.Kurzrock R, Gutterman J U, Talpaz M. N Engl J Med. 1988;319:990–998. doi: 10.1056/NEJM198810133191506. [DOI] [PubMed] [Google Scholar]
- 3.Daley G Q, Baltimore D. Proc Natl Acad Sci USA. 1988;85:9312–9316. doi: 10.1073/pnas.85.23.9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hariharan I K, Adams J M, Cory S. Oncog Res. 1988;3:387–399. [PubMed] [Google Scholar]
- 5.Daley G Q, Van Etten R A, Baltimore D. Science. 1990;247:824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- 6.Elefanty A G, Hariharan I K, Cory S. EMBO J. 1990;9:1069–1078. doi: 10.1002/j.1460-2075.1990.tb08212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelliher M A, McLaughlin J, Witte O N, Rosenberg N. Proc Natl Acad Sci USA. 1990;87:6649–6653. doi: 10.1073/pnas.87.17.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarkson B D, Strife A, Wisniewski D, Lambek C, Carpino N. Leukemia. 1997;11:1404–1428. doi: 10.1038/sj.leu.2400751. [DOI] [PubMed] [Google Scholar]
- 9.Daley G Q, Ben-Neriah Y. Adv Cancer Res. 1991;57:151–184. doi: 10.1016/s0065-230x(08)60998-7. [DOI] [PubMed] [Google Scholar]
- 10.Raitano A B, Whang Y E, Sawyers C L. Biochim Biophys Acta. 1997;1333:F201–F216. doi: 10.1016/s0304-419x(97)00023-1. [DOI] [PubMed] [Google Scholar]
- 11.Hunter T. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- 12.Tonks N K, Neel B G. Cell. 1996;87:365–368. doi: 10.1016/s0092-8674(00)81357-4. [DOI] [PubMed] [Google Scholar]
- 13.Tonks N K. Adv Pharmacol. 1996;36:91–119. doi: 10.1016/s1054-3589(08)60578-5. [DOI] [PubMed] [Google Scholar]
- 14.Flint A J, Tiganis T, Barford D, Tonks N K. Proc Natl Acad Sci USA. 1997;94:1680–1685. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LaMontagne K R, Jr, Flint A J, Franza R B, Jr, Pendergast A M, Tonks N K. Mol Cell Biol. 1998;18:2965–2975. doi: 10.1128/mcb.18.5.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pendergast A M, Quilliam L A, Cripe L D, Bassing C H, Dai Z, Li N, Batzer A, Rabun K M, Der C J, Schlessinger J, et al. Cell. 1993;75:175–185. [PubMed] [Google Scholar]
- 17.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 18.Lugo T G, Witte O N. Mol Cell Biol. 1989;9:1263–1270. doi: 10.1128/mcb.9.3.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Druker B J, Tamura S, Buchdunger E, Ohno S, Segal G M, Fanning S, Zimmermann J, Lydon N B. Nat Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 20.Carroll M, Ohno-Jones S, Tamura S, Buchdunger E, Zimmermann J, Lydon N B, Gilliland D G, Druker B J. Blood. 1997;90:4947–4952. [PubMed] [Google Scholar]
- 21.Rosenberg N, Witte O N. Adv Virus Res. 1988;35:39–81. doi: 10.1016/s0065-3527(08)60708-3. [DOI] [PubMed] [Google Scholar]
- 22.Shore S K, La Cava M, Yendapalli S, Reddy E P. J Biol Chem. 1994;269:5413–5419. [PubMed] [Google Scholar]
- 23.Tauchi T, Boswell H S, Leibowitz D, Broxymeyer H E. J Exp Med. 1994;179:167–175. doi: 10.1084/jem.179.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lozzio C B, Lozzio B B. Blood. 1975;45:321–334. [PubMed] [Google Scholar]
- 25.Anafi M, Gazit A, Zehavi A, Ben-Neriah Y, Levitzki A. Blood. 1993;82:3524–3529. [PubMed] [Google Scholar]
- 26.Tonks N K, Diltz C D, Fischer E H. J Biol Chem. 1988;263:6722–6730. [PubMed] [Google Scholar]
- 27.Frangioni J V, Beahm P H, Shifrin V, Jost C A, Neel B G. Cell. 1992;68:545–560. doi: 10.1016/0092-8674(92)90190-n. [DOI] [PubMed] [Google Scholar]
- 28.Woodford-Thomas T A, Rhodes J D, Dixon J E. J Cell Biol. 1992;117:401–414. doi: 10.1083/jcb.117.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown-Shimer S, Johnson K A, Hill D E, Bruskin A M. Cancer Res. 1992;52:478–482. [PubMed] [Google Scholar]
- 30.Gelderloos J A, Anderson S M. Oncogene. 1996;13:2367–2378. [PubMed] [Google Scholar]
- 31.Liu F, Sells M A, Chernoff J. Curr Biol. 1998;8:173–176. doi: 10.1016/s0960-9822(98)70066-1. [DOI] [PubMed] [Google Scholar]
- 32.Seely B L, Staubs P A, Reichart D R, Berhanu P, Milarski K L, Saltiel A R, Kusari J, Olefsky J M. Diabetes. 1996;45:1379–1385. doi: 10.2337/diab.45.10.1379. [DOI] [PubMed] [Google Scholar]
- 33.Bandyopadhyay D, Kusari A, Kenner K A, Liu F, Chernoff J, Gustafson T A, Kusari J. J Biol Chem. 1997;272:1639–1645. doi: 10.1074/jbc.272.3.1639. [DOI] [PubMed] [Google Scholar]
- 34.Kenner K A, Anyanwu E, Olefsky J M, Kusari J. J Biol Chem. 1996;271:19810–19816. doi: 10.1074/jbc.271.33.19810. [DOI] [PubMed] [Google Scholar]
- 35.Ahmad F, Li P-M, Meyerovitch J, Goldstein B J. J Biol Chem. 1995;270:20503–20508. doi: 10.1074/jbc.270.35.20503. [DOI] [PubMed] [Google Scholar]
- 36.Puil L, Liu J, Gish G, Mbamalu G, Bowtell D, Pelicci P G, Arlinghaus R, Pawson T. EMBO J. 1994;13:764–773. doi: 10.1002/j.1460-2075.1994.tb06319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skorski T, Kanakaraj P, Ku D-H, Nieborowska-Skorska M, Canaani E, Zon G, Perussia B, Calabretta B. J Exp Med. 1994;179:1855–1865. doi: 10.1084/jem.179.6.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mandanas R A, Leibowitz D S, Gharehbaghi K, Tauchi T, Burgess G S, Miyazawa K, Jayaram H N, Boswell H S. Blood. 1993;82:1838–1847. [PubMed] [Google Scholar]
- 39.Gishizky M L, Cortez D, Pendergast A M. Proc Natl Acad Sci USA. 1995;92:10889–10893. doi: 10.1073/pnas.92.24.10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sawyers C L, McLaughlin J, Witte O N. J Exp Med. 1995;181:307–313. doi: 10.1084/jem.181.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu F, Sells M A, Chernoff J. Mol Cell Biol. 1998;18:250–259. doi: 10.1128/mcb.18.1.250. [DOI] [PMC free article] [PubMed] [Google Scholar]