Abstract
The mouse CD1d1 glycoprotein is specialized in presenting lipid antigens to a novel class of T cells called natural killer T (NKT) cells. CD1d1 is predicted to contain five potential N-linked glycosylation sites (asparagine residues at positions 25, 38, 60, 128, and 183). Glycosylation has been shown to invariably affect the molecular and functional properties of various glycoproteins, and in the current report it was found that a conservative change of the individual endogenous asparagine residues in CD1d1 to glutamine differentially affected its functional expression. Although the maturation rate of the glycosylation mutants was comparable to that of wild type, they differed in their relative levels of surface expression and in their ability to stimulate NKT cells. Mutating all five glycosylation residues resulted in the absence of detectable CD1d1 expression, with a concomitant lack of NKT cell activation. Therefore, these results demonstrate that glycosylation plays a significant role in the functional expression of CD1d1.
Keywords: antigen presentation, glycosylation, cell surface expression
Introduction
Glycoproteins undergo post-translational modifications, including the addition of oligosaccharides to asparagines within the sequence motif Asn-X-Ser/Thr, before being expressed on the cell surface.1 Glycosylation is also important in the cellular immune response with respect to the ‘immune synapse’ between antigen-presenting cells and T cells. Furthermore, the glycosylation of antigen-presenting molecules can potentially modulate various cellular immune responses.2,3 Glycosylation of leucocyte cell surface proteins (or lack thereof) has been shown to affect a variety of their biological activities, including folding,4,5 trafficking to the cell surface,4,6–8 membrane orientation,9 stability10 (e.g. protease resistance and half-life), ligand binding,11 and transport of ligand-loaded major histocompatibility complex (MHC) class I molecules to the centre of the immune synapse,9 as well as signal transduction.2,3,12
Encoded on a different chromosome from that containing the MHC region,13,14 CD1 glycoproteins are MHC class I-like molecules comprised of three immunoglobulin-like extracellular α domains (α1, α2 and α3) non-covalently associated with β2-microglobulin (β2m).15 Unlike MHC class I and class II antigens, CD1 molecules present endogenous lipids, or those derived from various micro-organisms.15–19 Humans express both group 1 (CD1a, CD1b and CD1c molecules) and group 2 (CD1d) molecules;13,20 however, mice and rats only express group 2.21,22 The ‘intermediate’cd1e gene product is not an antigen-presenting molecule; rather, it facilitates lipid loading onto group 1 CD1 molecules.23,24 CD1d molecules are related to MHC class I in structure25 and amino acid homology.15 Like CD1b, they are known to survey different acidic endocytic compartments for antigen loading.26–28 Hence, these molecules resemble MHC class II in their intracellular trafficking and antigen presentation to T cells.26,27,29 Biochemical studies have revealed that, although the CD1d heavy chain is assembled in the endoplasmic reticulum (ER), as are MHC class I molecules in the MHC pathway,30 there are distinct differences from MHC class I molecules with respect to chaperone association in the absence of β2m,31,32 and transporter associated with antigen presentation (TAP) dependence.33,34 The CD1d ligand-binding groove is occupied by an ER-resident lipid such as phosphatidylinositol,30,35 glycosylphosphatidylinositol36 or phosphatidylcholine,37 probably to maintain its conformation, until this lipid is exchanged with a natural ligand in a late endocytic compartment.27,38,39
CD1d molecules exhibit relatively little polymorphism and are found in most mammals.40 Mice have two CD1d genes, cd1d1 and cd1d2, which share about 95% nucleotide sequence homology. Mouse CD1d1 molecules are expressed primarily by cells of the haematopoietic lineage and especially on professional antigen-presenting cells.41 Although the predicted molecular mass of the mouse CD1d1 heavy chain based on its amino acid content is only 38·5 kDa, it is widely reported to be found as a 43–49-kDa protein in immunoprecipitation analyses.30 This difference in mass is attributed to glycosylation. The CD1d1 molecule contains five predicted N-linked glycosylation sites and at least three of these were clearly distinct in its crystal structure,3 and we have shown that all five glycosylation sites are indeed glycosylated.30
To date, there has been no direct evidence for the involvement of N-linked oligosaccharides in the function of CD1d1 molecules, although it has been proposed that these sugars stabilize their cell surface expression and prevent non-specific protein–protein interactions to preserve membrane orientation.3 In the current report, we examined the role of N-linked glycosylation in the functional expression of CD1d1.
Materials and methods
Generation of CD1d1 glycosylation-deficient mutants
The full-length mouse CD1d1 cDNA in pBluescript,42 kindly provided by Dr S. Balk (Beth Israel Hospital and Harvard Medical School, Boston, MA), was used as a template for site-directed mutagenesis. Glycosylation-deficient mutants were generated using the polymerase chain reaction (PCR)-based Quik-change® site-directed mutagenesis kit (Stratagene, La Jolla, CA). Individual Asn residues within the consensus sites for N-linked glycosylation were conservatively changed to glutamine according to the manufacturer's instructions to create a panel of mutant CD1d1 molecules as shown in Fig. 1. The primer pairs used were: N25Q forward, 5′-GCCCAGCAAAAGCAATACACCTTCCGC-3′; N25Q reverse, 5′-GCGGAAGGTGTATTGCTTTTGCTGGGC-3′; N38Q forward, 5′-CTTCCTTTGCACAAAGAAGCTGGTC-3′; N38Q reverse, 5′-GACCAGCTTCTTTGTGCAAAGGAAG-3′; N60Q forward, 5′-CGTTGGAGTCAAGACTCAGCC-3′; N60Q reverse, 5′-GGCTGAGTCTTGACTCCAACG-3′; N128Q forward, 5′-GTACCCTGGGCAAGCTTCGGAAAG-3′; N128Q reverse, 5′-CTTTCCGAAGCTTGCCCAGGGTAC-3′; N183 forward, 5′-GATGCTCCTGCAAGAC ACCTGCC-3′, N183 reverse, 5′-GGCAGGTGTCTTGCAGGAGCATC-3′. Bold bases are areas in the primer that have been mutated to elicit the corresponding amino acid change from asparagine to glutamine. Multiple mutations were carried out sequentially and the correct mutations were confirmed by sequence analysis following each step. Following sequence confirmation, the full-length wild-type and mutant CD1d1 cDNAs (XhoI/NotI fragments) were subcloned into the mammalian expression vector pcDNA3·1-neo (Invitrogen, Carlsbad, CA). The 1·8kb SalI/NotI fragments of wild-type or mutant CD1d1 were subcloned into the SalI/NotI sites of the vaccinia virus (VV) vector pSC11.43 The generation of recombinant VV (rVV) expressing the CD1d1 glycosylation mutants was performed as previously described.27,43
Figure 1.
Schematic representation of the relative N-linked glycosylation residue locations on the mouse CD1d1 glycoprotein. The asparagine residues were numbered as in Pubmed accession #P11609. N-linked glycosylation sites (N-X-S or N-X-T) are indicated by stick figures on the protein (dark line).
Cell lines and LMTK transfection
The CD1d1-specific, Vα14+ canonical NKT cell hybridomas DN32.D3,44 N38-2H445 and N38-3C3,45 and the non-canonical (but CD1d1-specific) Vα5+ hybridoma N37-1A1245, have been described elsewhere. Mouse LMTK fibroblasts (American Type Culture Collection, Manassas, VA) were cultured in Dulbecco's modified Eagle's minimal essential medium (DMEM; Cambrex, Walkersville, MD) supplemented with 10% fetal bovine serum (FBS) and 2 mm l-glutamine in the absence of antibiotics. Transfection of LMTK cells was carried out using Fugene 6 (Roche, Indianapolis, IN) as per the manufacturer's protocol. One to two weeks post-transfection, transfected cells were stained with either the mouse anti-CD1d1 monoclonal antibody (mAb) 1H6,27 or a phycoerythrin (PE)-conjugated 1B1 anti-mouse CD1d mAb (Pharmingen, San Diego, CA) and immunomagnetically sorted using either anti-mouse immunoglobulin G (IgG) or anti-PE microbeads (Miltenyi Biotec, Gladbach, Germany), respectively. The transfectants were cloned by limiting dilution (< 1 cell/well in a 96-well plate) and, after selection in 500 µg/ml Geneticin (Invitrogen), stable transfectants with comparable cell surface levels of wild-type or mutant CD1d1 [determined by fluorescence-activated cell sorter (FACS) analysis] were used for further analyses, unless otherwise noted.
NKT cell assay
Five × 105 LMTK transfectants (wild-type or mutant CD1d1) were co-cultured with 5 × 104 NKT cell hybridomas (1 × 105 for N38-3C3) in Iscove's modified Dulbecco's medium (IMDM) for 20–22 hr at 37°. The culture supernatants were harvested and assayed for interleukin (IL)-2 levels by enzyme-linked immunosorbent assay (ELISA) using anti-mouse IL-2 antibody pairs (Pharmingen). Recombinant murine IL-2 (Peprotech, Rocky Hill, NJ) was used for generating a standard curve as previously described.27 NKT cell assays with the LMTK-CD1d1 wild-type and glycosylation mutant clones pulsed with or without α-galactosylceramide were performed as we previously reported.18
Flow cytometry
Staining and analysis for flow cytometry were performed as described previously.27,46 Briefly, 5 × 105 cells were washed in ice-cold FACS buffer [Hanks' balanced salt solution (HBSS) containing 0·1% bovine serum albumin (BSA); HBSS/BSA] and incubated with the relevant mAb in FACS buffer for 30 min on ice. In indirect staining experiments following labelling with the primary mAb, the cells were washed twice in FACS buffer and then incubated with fluorescein isothiocyanate (FITC)- or PE-conjugated rabbit anti-mouse IgG (Dako, Carpinteria, CA) for 30 min on ice. Staining for CD1d1 was also performed using the 19G1147 mAb (kindly provided by A. Bendelac, University of Chicago). To assess the level of VV infection, infected cells [at a multiplicity of infection (MOI) of 5 for 6 hr] were permeabilized with FACS buffer containing 0·1% saponin (Sigma-Aldrich, St Louis, MO) and the VV E3L-specific mAb TW2·348 was diluted 1 : 5 in saponin-containing FACS buffer. The PE-conjugated anti-mouse immunoglobulin antiserum was also diluted in saponin-containing FACS buffer. Controls included staining with an isotype-matched irrelevant mAb or second antibody alone, as indicated.
Confocal microscopy
Immunofluorescent staining and analysis of CD1d1 by confocal microscopy have been previously described.27 In brief, 5 × 106 P13·9 cells were plated in sterile glass-bottom 35-mm dishes coated with poly d-lysine (MatTek, Ashland, MA). After overnight adherence, the cells were infected with rVV encoding wild-type or mutant CD1d1 molecules at a MOI of 5 for 6 hr. The cells were washed twice in ice-cold HBSS/BSA and fixed in 1% paraformaldehyde for 10 min at 4°. CD1d1 was stained using the anti-mouse CD1d mAb 1H6,27 followed by a FITC-conjugated anti-mouse immunoglobulin antiserum (Dako). Late endosomal/lysosomal compartments were identified with anti-mouse lysosome-associated membrane protein-1 (LAMP-1; Pharmingen) followed by a Texas Red-labelled anti-rat immunoglobulin antibody (Jackson Immunoresearch Laboratories, West Grove, PA). The stained cells were stored in phosphate-buffered saline (PBS) containing 0·05% azide in the dark at 4° and viewed with a Bio-Rad MRC-1024 confocal laser-scanning microscope (Bio-Rad, Hercules, CA) equipped with a krypton-argon laser. The Texas Red and FITC emissions were recorded sequentially using a 60× lens and pinhole aperture adjustment to obtain 0·3- to 0·5-µm sections.
Pulse-chase experiments
LMTK cells were starved for 30 min in methionine-free DMEM (Biowhittaker) prior to infection with rVV expressing the influenza virus nucleoprotein (NP-βGal; negative control, kindly provided by J. Yewdell and J. Bennink, National Institutes of Health) or wild-type and mutant forms of CD1d1. The cells were then labelled with 100 µCi of [35S]-methionine (Easy Tag Express –[35S] protein labeling mix; Perkin-Elmer, Wellesley, MA) for 30 min. After labelling, the cells were cultured in regular medium containing a molar excess of cold methionine for the indicated time periods. The labelled cells were lysed for 30 min on ice with 10 mm Tris-buffered saline containing 1% Triton X-100, 0·1% deoxycholate, 5 mm ethylenediaminetetraacetic acid (EDTA), and complete protease inhibitor tablets (Roche, Indianapolis, IN). CD1d1 molecules immunoprecipitated using the mouse monoclonal antibody 1H6 were separated on a non-denaturing 10% sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) gel. Dried gels were exposed on Kodak BioMax XAR film and the scanned autoradiographs were subjected to densitometric analysis. For the endoglycosidase H (EndoH) digestion experiment, the immunoprecipitated protein pellet was resuspended in 50 mm citrate buffer (pH 5·5) and divided into two equal portions. One half of the protein sample was digested with 2 mU of EndoH (Roche) at 37° overnight, whereas the other half served as the control. These samples were then loaded onto an SDS–PAGE gel for analysis as above.
Results
Individual Asn mutants of CD1d are differentially expressed on the cell surface
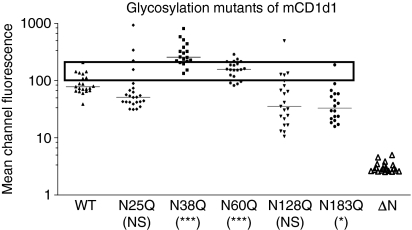
The contribution of individual glycosylation sites in the cell surface expression of CD1d1 was assessed by site-directed mutagenesis of putative Asn residues (Asn→Gln) predicted to be glycosylated.49 The mouse fibroblast cell line LMTK, which does not express detectable CD1d1 on the cell surface, was transfected with cDNA encoding either the wild type or individual glycosylation mutants of CD1d1 (Fig. 1). Preliminary experiments using G418-resistant cells selected post-transfection showed that the individual Asn→Gln mutations resulted in different levels of CD1d1 cell surface expression (data not shown). As these were heterogeneous populations, the cell surface levels could potentially be influenced by multiple factors including transfection efficiency and copy number. Therefore, in subsequent experiments, we conducted simultaneous transfections in six-well plates with equivalent amounts of plasmid DNA for the wild type and all individual CD1d1 glycosylation mutants, followed by limited dilution cloning of G418-resistant cells. Approximately 20 clones were randomly selected and analysed for their cell surface expression of CD1d1. FACS analysis of CD1d1 showed that the N38Q and N60Q CD1d1 mutants were expressed at levels two- to threefold higher than the wild type (P < 0·0001; Fig. 2). The cell surface expression of the CD1d1 glycosylation mutant N183Q was significantly lower (∼ 35%) than that of the wild type (P < 0·05). Although the difference was not statistically significant, the CD1d1 glycosylation mutants N25Q and N128Q were also expressed on the surface at lower levels than the wild type. To rule out the possibility of pleiotropic effects of drug selection on differential RNA stability, we extracted RNA from the stable transfectants and reverse-transcribed it into cDNA using CD1d1-specific primers. We could not detect any significant differences in CD1d1 mRNA levels between the different clones as analysed by semiquantitative reverse transcription (RT)–PCR (data not shown).
Figure 2.
CD1d1 cell surface expression levels in LMTK cell transfectants. Individual clones of LMTK cells transfected with cDNA encoding wild-type CD1d1 or the indicated glycosylation mutants were isolated by limiting dilution and analysed for cell surface expression using a phycoerythrin (PE)-conjugated anti-mouse CD1d antibody. The scattergram was plotted using the mean channel fluorescence of CD1d-specific staining of the individual clones normalized against cells stained with an isotype control. A second independent repeat of this cloning experiment showed a similar pattern of CD1d1 expression. The clones used for the experiments presented in Fig. 3 were selected from those within the box. *P < 0·05; ***P < 0·001; mCD1d1, murine CD1d1; NS, not significant.
Unglycosylated CD1d1 is not expressed on the cell surface
Having determined that the absence of an individual glycosylation motif does not completely block the cell surface expression of CD1d1, we sought to analyse how the lack of glycosylation might affect the cell surface expression of this molecule. Sequential PCR-based mutation steps were performed to change all five Asn residues for the analysis of unglycosylated CD1d1 (ΔNCD1). Although double (N38Q/N60Q) and triple (N25Q/N128Q/N183Q) glycosylation mutants were expressed on the cell surface (data not shown), the unglycosylated CD1d1 was undetectable. Flow cytometry analysis of the 20 randomly selected clones of wild-type and mutant CD1d1-transfected LMTK cells used in Fig. 2 was performed. Ten different antibodies50 including the 19G11 mAb47 that can potentially recognize unglycosylated CD1d were used to stain these transfectants and none of the antibodies tested could detect the expression of ΔNCD1 on the cell surface, although all were able to stain wild-type CD1d1 (data not shown). Similarly, pulsing LMTK-ΔNCD1 with α-galactosylceramide (α-GalCer) at concentrations as high as 1 µg/ml did not elicit any cytokine production from the NKT cell hybridomas, indicating a lack of functional CD1d1 on the cell surface; however, the mutation could be detected in transfected cells by RT-PCR followed by sequencing, confirming that the cDNA for ΔNCD1 was indeed expressed in these cells (data not shown).
Differential stimulation of NKT cells by individual glycosylation mutants of CD1d1
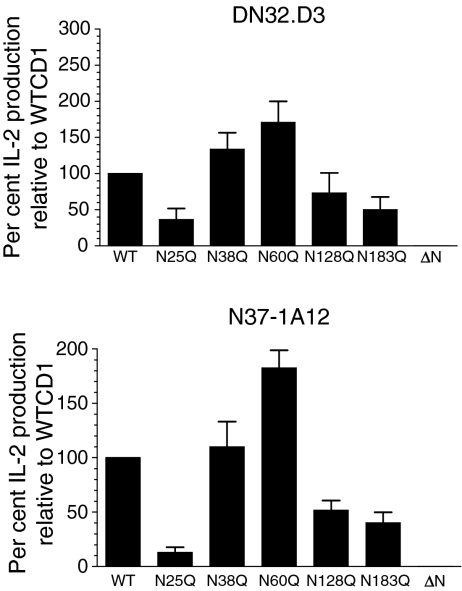
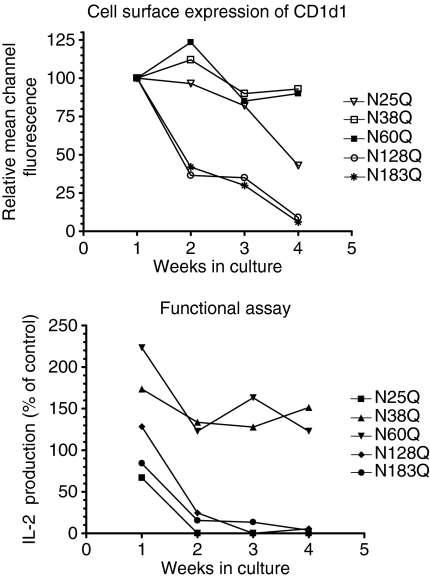
Monoclonal antibodies can vary in their capacities to bind glycosylated and unglycosylated CD1d1.47 Therefore, a functional assay (NKT cell stimulation) is the most sensitive and reliable way to determine the presence of CD1d1 on the cell surface. To compare the antigen presentation capacities of the glycosylation mutants, clones of LMTK transfectants with comparable cell surface expression of CD1d1 (except in the case of ΔNCD1) were co-cultured with two representative NKT cell hybridomas, DN32.D344 and N37-1A12.45 Although the individual Asn→Gln mutants used were selected on the basis of comparable CD1d1 cell surface expression, the antigen presentation capacity of the N25Q, N128Q and N183Q mutants was lower than that of wild-type CD1d1 (40–70% of wild type for DN32.D3 and 15–50% for N37-1A12), and endogenous antigen presentation by N38Q and N60Q was higher than that of wild-type CD1d1 (130–170% of wild-type for DN32.D3 and 110–180% for N37-1A12, respectively) (Fig. 3). Similar results were obtained with two other Vα14+ NKT cell hybridomas (N38-3C3 and N38-2H4; data not shown). As the clones tested in these NKT cell stimulation analyses (three independent experiments) expressed similar levels of CD1d1 on their cell surface, it is clear that the degree of NKT cell stimulation observed reflected their altered glycosylation status. Interestingly, when these stable transfectant clones were maintained in culture long-term, the N128Q and N183Q mutants were found to gradually lose their cell surface expression of CD1d1, and consequently their NKT cell stimulation capacity (Fig. 4). The N38Q and N60Q mutants maintained their functional activity long-term (reflecting their higher cell surface expression of CD1d1). As shown in Fig. 3, the LMTK-ΔNCD1 transfectant was consistently unable to stimulate cytokine production by NKT cells. Therefore, these results suggest that the glycosylation state of the CD1d1 molecule affects its functional cell surface expression.
Figure 3.
Differential stimulation of natural killer T (NKT) cells by LMTK cells expressing CD1d1 glycosylation mutants. LMTK cells expressing wild-type or the indicated glycosylation mutant CD1d1 molecules were co-cultured with the indicated NKT cell hybridomas. After a 20-hr co-culture, the supernatants were harvested and assayed for interleukin (IL)-2 by enzyme-linked immunosorbent assay (ELISA). The bars represent the mean percentage of IL-2 production ± standard deviation in triplicate cultures. IL-2 production by hybridomas stimulated with LMTK cells expressing wild-type CD1d1 (WTCD1) was fixed at 100% for comparison with the glycosylation mutants.
Figure 4.
The ability of some CD1d1 glycosylation mutants to stimulate natural killer T (NKT) cells decreases over time. Clones of LMTK transfectants expressing wild-type or glycosylation mutant CD1d1 molecules were cultured for 4 weeks and monitored for their cell surface expression by staining with a phycoerythrin (PE)-conjugated anti-mouse CD1d monoclonal antibody (mAb). Concurrently, an aliquot of the same cells was used for co-culture experiments with NKT cells. Interleukin (IL)-2 secretion was measured by enzyme-linked immunosorbent assay (ELISA). (a) The relative mean fluorescence intensity of cells stained with the anti-CD1d mAb was plotted against weeks in culture, setting wild-type CD1d1 expression at 100%. (b) Co-culture of LMTK cells expressing wild-type CD1d1 or glycosylation mutants of CD1d1 with NKT cell hybridomas. For each week of analysis, IL-2 production stimulated by wild-type CD1d1-expressing cells was set at 100%.
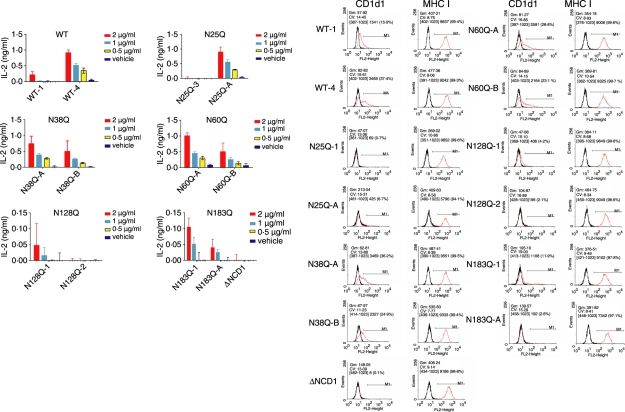
In order to determine whether the decrease in CD1d1 levels also reflected a quantitative decrease in exogenous antigen presentation, clones of LMTK-CD1d1 transfectants (four wild type and 24 mutant) were treated with vehicle [dimethyl sulphoxide (DMSO)] or α-galactosylceramide, washed, fixed and co-cultured with NKT cells as above. CD1d1 and MHC class I (as a control) surface levels were detected by FACS. As expected, the amount of CD1d1 on the cell surface was proportional to the level of NKT cell stimulation (Fig. 5) Notably, in all clones, MHC class I cell surface levels were comparable. Therefore, changes in CD1d1 glycosylation result in both qualitative and quantitative changes in the functional expression of CD1d1.
Figure 5.
α-Galactosylceramide (α-GalCer)-induced natural killer T (NKT) cell cytokine production by CD1d1 glycosylation mutants is proportional to the level of cell surface CD1d1. Representative LMTK-CD1d1 clones expressing wild-type CD1d1 or individual CD1d1 glycosylation mutants were pulsed with vehicle [dimethyl sulphoxide (DMSO)] or the indicated concentrations of α-GalCer. The cells were then fixed and co-cultured with NKT cells as above. Cell surface levels of CD1d1 and major histocompatibility complex (MHC) class I were determined by fluorescence-activated cell sorter (FACS) analysis. Black line: isotype control; red line: anti-CD1d1 (or anti-MHC class I). This experiment was performed three times.
Individual glycosylation mutants mature at approximately the same rate
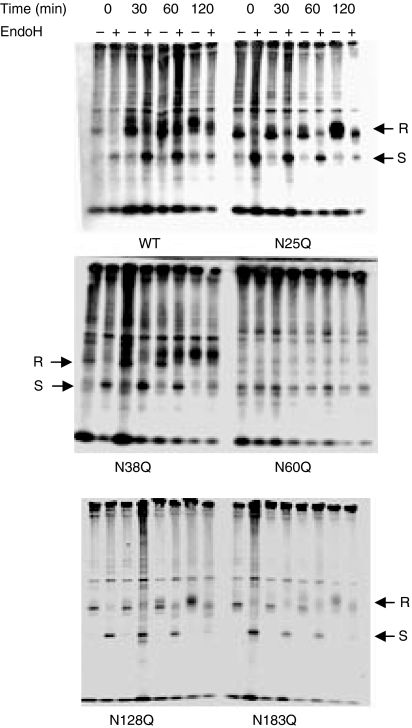
Because the observed differences in cell surface expression of the individual glycosylation mutants could simply have been a result of altered rates of biosynthesis, we performed a pulse-chase analysis on wild-type CD1d1 and the glycosylation mutants. For these experiments, LMTK cells were infected with rVV encoding the different glycosylation mutants, as rVV allows much higher levels of CD1d1 expression for use in this experimental approach.27 Following a 90-min infection, the cells were pulse-labelled with [35S]-methionine, chased for the indicated times, and immunoprecipitated with the anti-mouse CD1d1 mAb 1H6.27 The immunoprecipitates were treated with endoglycosidase H (EndoH), which removes the high-mannose oligosaccharides from ER proteins but cannot cleave the modified oligosaccharides of proteins that have traversed the Golgi (mature protein). Immature CD1d1 molecules were detected as a sharp band at the beginning of the chase and were sensitive to EndoH, as indicated by the decrease in the apparent relative mobility (Mr) (Fig. 6). The EndoH sensitivity of CD1d1 was slowly lost after 2 hr, and a diffuse heavily glycosylated band was detected as an EndoH-resistant form. The difference in molecular weight caused by the loss of a single glycosylation site is clearly distinguishable by the faster migration of these mutants in the polyacrylamide gel. There were subtle differences in the maturation rate and gain of EndoH resistance among the different mutants, but these were not significant. Repeated attempts at immunoprecipitating the N60Q and ΔNCD1 mutants using different monoclonal antibodies (and in the presence or absence of the proteasome inhibitor lactacystin) failed to yield any detectable protein. Therefore, these results suggest that the functional differences observed in the CD1d1 glycosylation mutants at the level of cell surface expression and ability to activate NKT cells were not a result of substantially altered rates of glycoprotein maturation.
Figure 6.
CD1d1 glycosylation mutants mature at the same rate as wild type. LMTK cells were infected with the indicated recombinant vaccinia virus (rVV) expressing wild-type CD1d1 or CD1d1 glycosylation mutants for 4 hr and pulse-labelled with [35S]-methionine. The cells were then washed and harvested immediately (time = 0) or chased for the indicated time periods before harvest. CD1d1 was immunoprecipitated from lysed cells and was treated overnight in the presence (+) or absence (–) of endoglycosidase H (EndoH). The proteins were separated on a 10% non-reducing sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) gel and detected by exposing the dried gel on film for 15–17 days. Mature EndoH-resistant (R) and immature EndoH-sensitive (S) forms of CD1d1 are indicated by the arrows. The results were confirmed in two independent experiments.
CD1d1 glycosylation mutants traffic normally to late endocytic compartments
Because CD1d1 surveys late endocytic compartments for efficient antigen loading,26,27,51 any alterations in normal trafficking might also affect the functional properties of the molecule. To determine whether changes in glycosylation status affected the intracellular localization of CD1d1, the stable transfectants were analysed by confocal microscopy. As shown in Fig. 7, all of the individual glycosylation mutants extensively co-localized with the late endosomal/lysosomal marker LAMP-1 to the same degree as wild-type CD1d1. Similar to the results obtained with cell surface immunofluorescence and immunoprecipitation, we could not detect any CD1d1 protein expression in ΔNCD1-transfected or VV-ΔNCD1-infected cells, even in the presence of the proteasome inhibitor lactacystin (Fig. 7 and data not shown). Thus, these individual N-linked glycosylation sites probably play no significant role in the intracellular trafficking of CD1d1, but a complete lack of glycosylation appears to block its functional expression.
Figure 7.
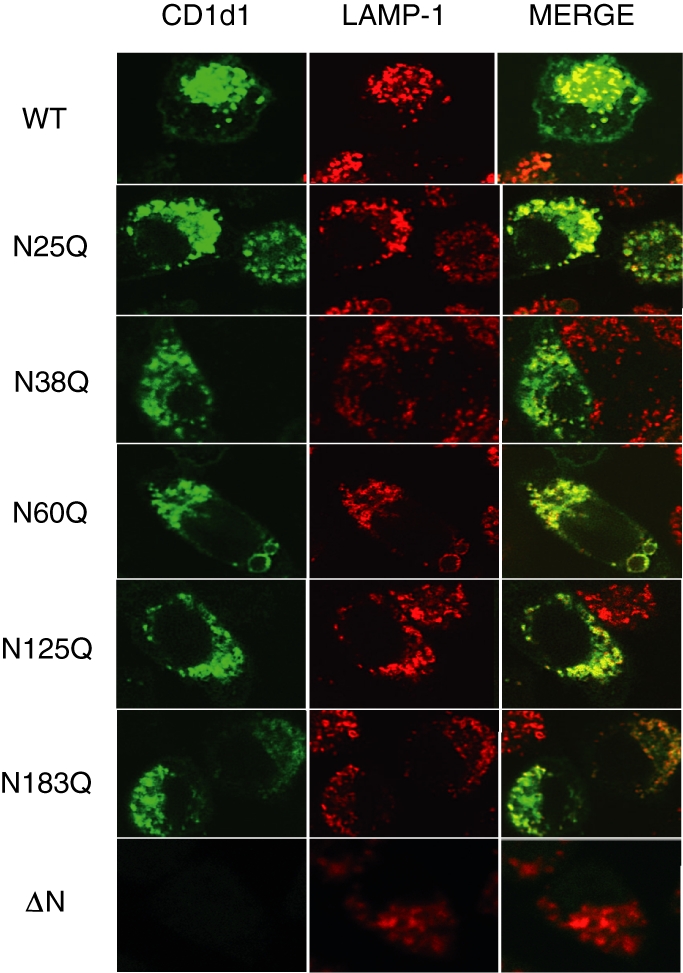
Normal trafficking of CD1d1 glycosylation mutants to late endocytic compartments. LMTK cells transfected with wild-type CD1d1 WT or the indicated CD1d1 glycosylation mutants were stained with antibodies specific for mouse CD1d (green) and lysosome-associated membrane protein-1 (LAMP-1) (late endosomes/lysosomes; red). Note that the merged image on the right shows extensive co-localization of all the individual CD1d1 glycosylation mutants with LAMP-1, comparable to that observed with wild type.
Discussion
Glycosylation is essential for the cellular stability of several integral membrane glycoproteins including CD4, CD26 and CD45,52–54 as well as the prevention of cellular protease-mediated degradation.55–58 Although the biosynthesis of CD1 molecules has been postulated to occur along the lines of MHC class I heavy chain assembly in the ER, studies characterizing this biosynthesis are relatively few in number. Huttinger et al.32 showed the involvement of calnexin and calreticulin in CD1b biogenesis and indirectly demonstrated the involvement of a terminal glucose residue in this process by using the glucosidase inhibitor castanospermine. Kang and Cresswell31 demonstrated that human CD1d also associates with calnexin, calreticulin and ERp57 to facilitate its folding and disulphide bond formation. They speculated that these chaperones may bind to different glycans of the CD1d heavy chains,31 similar to binding to the influenza virus haemagglutinin.59 Because it is well established that ER chaperones are carbohydrate-binding lectins and require the presence of a monoglucosylated N-linked glycan on the nascent polypeptide chain for association, CD1 heavy chain glycosylation is therefore expected to play an important role in its folding, conformation and surface expression.60 No differences were evident in the binding of the individual glycosylation mutants of murine CD1d1 tested with ER chaperones (S. Joyce, Vanderbilt University, Nashville, TN, personal communication). Of course, this does not preclude the possibility of other chaperones playing a role in CD1d biogenesis. The other possibility is that calnexin and calreticulin are promiscuous in their binding to CD1d as long as at least one glycosylation site is available for their interaction. Unlike MHC class I, which has only one or two glycosylation sites, the availability of five different glycosylation sites may provide redundant chaperone interactions, which may be an advantage for CD1d molecules. Given the differences in the site-specific glycosylation patterns of MHC class I alleles,60 one cannot rule out the role of microheterogeneity in the functionality of CD1d.
The specific objective of the current study was to understand the involvement of N-linked oligosaccharides in the functional expression of the mouse CD1d1 glycoprotein. Mouse CD1d1 shares more than 60% amino acid homology with human CD1d in its extracellular α1 and α2 domains.49 Whereas N25, N38 and N60 are part of the α1 domain, N128 and N183 are within the α2 domain of mouse CD1d1. The only difference between mouse CD1d1 and human CD1d in terms of glycosylation is the absence of a potential N-linked glycosylation site equivalent to the N25 of mouse CD1d1. It is intriguing to note that the N38 equivalent of mouse CD1d1 is the only residue that is conserved in all CD1 proteins (CD1a, b, c and d), and that mutating this residue to Gln does not decrease its cell surface expression. The cell surface expression of N60Q displays a heterogeneous surface density rather than a single high- or low-density peak as observed in the case of the other mutants by flow cytometric analysis. This peculiar cell surface expression profile along with our inability to immunoprecipitate N60Q makes it an interesting glycosylation site for further investigation.
Inhibition of glycosylation processing by castanospermine (an inhibitor of ER glucosidase I and II) has been shown to direct CD1b molecules to the proteasome for degradation.32 In the presence of lactacystin, a non-reversible inhibitor of the proteasome, we were unable to detect ΔNCD1 in transfected cells. The failure to detect ΔNCD1 in the presence of lactacystin does not necessarily suggest a proteasome-independent degradation pathway for CD1d1; instead, a lack of detection could simply be a result of the inability of the CD1d1-specific antibodies used to bind the unglycosylated form of CD1d1.
It has been shown that in vitro refolded recombinant CD1d from Escherichia coli can recognize both human and mouse NKT cells when loaded with the appropriate ligand.61 One might very well argue that the ability of unglycosylated CD1d from prokaryotic cells to activate NKT cells precludes a role for glycosylation in its function. However, our results strongly suggest that the glycosylation of CD1d1 is essential for its biosynthesis and cell surface expression in eukaryotic cells. A refolded bacterially derived (and unglycosylated) CD1d1 molecule bound to plates may functionally interact with the invariant T-cell receptor (TCR) of NKT cells, but only if loaded with a strong CD1d-dependent NKT cell stimulator, such as α-GalCer. The role of glycosylation in the stability and folding of CD1d and the involvement of intracellular chaperones in this process need to be further evaluated. This is somewhat hindered by the current lack of antibodies that can react with denatured murine CD1d1.
Given that the half-life of cell surface CD1d1 is measured in days,30 it should be noted that many studies analysing the significance of glycosylation may only provide a ‘snap-shot’ view of the molecular kinetics, and our functional analysis of CD1d1 glycosylation mutants over time helps to characterize the importance of glycosylation throughout the life of the CD1d1 glycoprotein. Thus, N-linked oligosaccharides at positions N128 and N183 may act in two ways: (1) assisting in folding and maintaining the structural conformation of CD1d1, and (2) orienting and sustaining the CD1d1–TCR interaction. Insights into the role(s) played by oligosaccharides in protein stability and T-cell stimulation may have clinical relevance with respect to congential disorders of glycosylation, and hence warrant further investigation.
Acknowledgments
The authors would like to thank Drs S. Balk, A. Bendelac, K. Hayakawa, J. Yewdell and J. Bennink for kindly providing reagents used in this study. This work was supported by grants from the National Institutes of Health to RRB (R01 AI46455 and CA89026) and funds from the Walther Cancer Institute. Excellent technical support by Philip M. Spence in the generation and titration of recombinant vaccinia viruses is appreciated. RRB is a Scholar of the Leukemia and Lymphoma Society.
References
- 1.Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–64. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 2.Rudd PM, Elliott T, Cresswell P, Wilson IA, Dwek RA. Glycosylation and the immune system. Science. 2001;291:2370–6. doi: 10.1126/science.291.5512.2370. [DOI] [PubMed] [Google Scholar]
- 3.Rudd PM, Wormald MR, Stanfield RL, et al. Roles for glycosylation of cell surface receptors involved in cellular immune recognition. J Mol Biol. 1999;293:351–66. doi: 10.1006/jmbi.1999.3104. [DOI] [PubMed] [Google Scholar]
- 4.Bergeron JJ, Zapun A, Ou WJ, Hemming R, Parlati F, Cameron PH, Thomas DY. The role of the lectin calnexin in conformation independent binding to N-linked glycoproteins and quality control. Adv Exp Med Biol. 1998;435:105–16. doi: 10.1007/978-1-4615-5383-0_11. [DOI] [PubMed] [Google Scholar]
- 5.Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–9. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, Choudhury P, Cabral CM, Sifers RN. Oligosaccharide modification in the early secretory pathway directs the selection of a misfolded glycoprotein for degradation by the proteasome. J Biol Chem. 1999;274:5861–7. doi: 10.1074/jbc.274.9.5861. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Q, Salter RD. Distinct patterns of folding and interactions with calnexin and calreticulin in human class I MHC proteins with altered N-glycosylation. J Immunol. 1998;160:831–7. [PubMed] [Google Scholar]
- 8.Hammond C, Braakman I, Helenius A. Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc Natl Acad Sci USA. 1994;91:913–7. doi: 10.1073/pnas.91.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grakoui A, Bromley SK, Sumen C, Davis MM, Shaw AS, Allen PM, Dustin ML. The immunological synapse: a molecular machine controlling T cell activation. Science. 1999;285:221–7. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- 10.Karaivanova VK, Spiro RG. Effect of proteasome inhibitors on the release into the cytosol of free polymannose oligosaccharides from glycoproteins. Glycobiology. 2000;10:727–35. doi: 10.1093/glycob/10.7.727. [DOI] [PubMed] [Google Scholar]
- 11.Radcliffe CM, Diedrich G, Harvey DJ, Dwek RA, Cresswell P, Rudd PM. Identification of specific glycoforms of major histocompatibility complex class I heavy chains suggests that class I peptide loading is an adaptation of the quality control pathway involving calreticulin and ERp57. J Biol Chem. 2002;277:46415–23. doi: 10.1074/jbc.M202466200. [DOI] [PubMed] [Google Scholar]
- 12.Dwek RA. Biological importance of glycosylation. Dev Biol Stand. 1998;96:43–7. [PubMed] [Google Scholar]
- 13.Martin LH, Calabi F, Milstein C. Isolation of CD1 genes: a family of major histocompatibility complex- related differentiation antigens. Proc Natl Acad Sci USA. 1986;83:9154–8. doi: 10.1073/pnas.83.23.9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calabi F, Milstein C. A novel family of human major histocompatibility complex-related genes not mapping to chromosome 6. Nature. 1986;323:540–3. doi: 10.1038/323540a0. [DOI] [PubMed] [Google Scholar]
- 15.Brigl M, Brenner MB. CD1: antigen presentation and T cell function. Annu Rev Immunol. 2004;22:817–90. doi: 10.1146/annurev.immunol.22.012703.104608. [DOI] [PubMed] [Google Scholar]
- 16.Mattner J, Debord KL, Ismail N, et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434:525–9. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- 17.Kinjo Y, Wu D, Kim G, et al. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature. 2005;434:520–5. doi: 10.1038/nature03407. [DOI] [PubMed] [Google Scholar]
- 18.Sriram VW, Gervay-Hague J, Brutkiewicz RR. Cell wall glycosphingolipids of Sphingomonas paucimobilis are CD1d-specific ligands for NKT cells. Eur J Immunol. 2005;35:1692–701. doi: 10.1002/eji.200526157. [DOI] [PubMed] [Google Scholar]
- 19.Wu D, Xing GW, Poles MA, et al. Bacterial glycolipids and analogs as antigens for CD1d-restricted NKT cells. Proc Natl Acad Sci USA. 2005;102:1351–6. doi: 10.1073/pnas.0408696102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balk SP, Bleicher PA, Terhorst C. Isolation and characterization of a cDNA and gene coding for a fourth CD1 molecule. Proc Natl Acad Sci USA. 1989;86:252–6. doi: 10.1073/pnas.86.1.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balk SP, Bleicher PA, Terhorst C. Isolation and expression of cDNA encoding the murine homologues of CD1. J Immunol. 1991;146:768–74. [PubMed] [Google Scholar]
- 22.Katabami S, Matsuura A, Chen HZ, Imai K, Kikuchi K. Structural organization of rat CD1 typifies evolutionarily conserved CD1D class genes. Immunogenetics. 1998;48:22–31. doi: 10.1007/s002510050396. [DOI] [PubMed] [Google Scholar]
- 23.de la Salle H, Mariotti S, Angenieux C, et al. Assistance of microbial glycolipid antigen processing by CD1e. Science. 2005;310:1321–4. doi: 10.1126/science.1115301. [DOI] [PubMed] [Google Scholar]
- 24.Angenieux C, Fraisier V, Maitre B, et al. The cellular pathway of CD1e in immature and maturing dendritic cells. Traffic. 2005;6:286–302. doi: 10.1111/j.1600-0854.2005.00272.x. [DOI] [PubMed] [Google Scholar]
- 25.Zeng Z, Castano AR, Segelke BW, Stura EA, Peterson PA, Wilson IA. Crystal structure of mouse CD1: An MHC-like fold with a large hydrophobic binding groove. Science. 1997;277:339–45. doi: 10.1126/science.277.5324.339. [DOI] [PubMed] [Google Scholar]
- 26.Chiu YH, Jayawardena J, Weiss A, Lee D, Park SH, Dautry-Varsat A, Bendelac A. Distinct subsets of CD1d-restricted T cells recognize self-antigens loaded in different cellular compartments. J Exp Med. 1999;189:103–10. doi: 10.1084/jem.189.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts TJ, Sriram V, Spence PM, et al. Recycling CD1d1 molecules present endogenous antigens processed in an endocytic compartment to NKT cells. J Immunol. 2002;168:5409–14. doi: 10.4049/jimmunol.168.11.5409. [DOI] [PubMed] [Google Scholar]
- 28.Jayawardena-Wolf J, Benlagha K, Chiu YH, Mehr R, Bendelac A. CD1d endosomal trafficking is independently regulated by an intrinsic CD1d-encoded tyrosine motif and by the invariant chain. Immunity. 2001;15:897–908. doi: 10.1016/s1074-7613(01)00240-0. [DOI] [PubMed] [Google Scholar]
- 29.Sugita M, Jackman RM, van Donselaar E, Behar SM, Rogers RA, Peters PJ, Brenner MB, Porcelli SA. Cytoplasmic tail-dependent localization of CD1b antigen-presenting molecules to MIICs. Science. 1996;273:349–52. doi: 10.1126/science.273.5273.349. [DOI] [PubMed] [Google Scholar]
- 30.De Silva AD, Park JJ, Matsuki N, Stanic AK, Brutkiewicz RR, Medof ME, Joyce S. Lipid protein interactions: the assembly of CD1d1 with cellular phospholipids occurs in the endoplasmic reticulum. J Immunol. 2002;168:723–33. doi: 10.4049/jimmunol.168.2.723. [DOI] [PubMed] [Google Scholar]
- 31.Kang S-J, Cresswell P. Calnexin, calreticulin, and ERp57 cooperate in disulfide bond formation in human CD1d heavy chain. J Biol Chem. 2002;277:44838–44. doi: 10.1074/jbc.M207831200. [DOI] [PubMed] [Google Scholar]
- 32.Huttinger R, Staffler G, Majdic O, Stockinger H. Analysis of the early biogenesis of CD1b: involvement of the chaperones calnexin and calreticulin, the proteasome and β2-microglobulin. Int Immunol. 1999;11:1615–23. doi: 10.1093/intimm/11.10.1615. [DOI] [PubMed] [Google Scholar]
- 33.Brutkiewicz RR, Bennink JR, Yewdell JW, Bendelac A. TAP-independent, β2-microglobulin-dependent surface expression of functional mouse CD1.1. J Exp Med. 1995;182:1913–9. doi: 10.1084/jem.182.6.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teitell M, Holcombe HR, Brossay L, et al. Nonclassical behavior of the mouse CD1 class I-like molecules. J Immunol. 1997;158:2143–9. [PubMed] [Google Scholar]
- 35.Park JJ, Kang SJ, De Silva AD, Stanic AK, Casorati G, Hachey DL, Cresswell P, Joyce S. Lipid–protein interactions: biosynthetic assembly of CD1 with lipids in the endoplasmic reticulum is evolutionarily conserved. Proc Natl Acad Sci USA. 2004;101:1022–6. doi: 10.1073/pnas.0307847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joyce S, Woods AS, Yewdell JW, et al. Natural ligand of mouse CD1d1: cellular glycosylphosphatidylinositol. Science. 1998;279:1541–4. doi: 10.1126/science.279.5356.1541. [DOI] [PubMed] [Google Scholar]
- 37.Giabbai B, Sidobre S, Crispin MD, Sanchez-Ruiz Y, Bachi A, Kronenberg M, Wilson IA, Degano M. Crystal structure of mouse CD1d bound to the self ligand phosphatidylcholine: a molecular basis for NKT cell activation. J Immunol. 2005;175:977–84. doi: 10.4049/jimmunol.175.2.977. [DOI] [PubMed] [Google Scholar]
- 38.Kang SJ, Cresswell P. Saposins facilitate CD1d-restricted presentation of an exogenous lipid antigen to T cells. Nat Immunol. 2004;5:175–81. doi: 10.1038/ni1034. [DOI] [PubMed] [Google Scholar]
- 39.Zhou D, Cantu C, Sagiv Y, et al. Editing of CD1d-bound lipid antigens by endosomal lipid transfer proteins. Science. 2004;303:523–7. doi: 10.1126/science.1092009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Porcelli SA, Modlin RL. The CD1 system. Antigen-presenting molecules for T cell recognition of lipids and glycolipids. Annu Rev Immunol. 1999;17:297–329. doi: 10.1146/annurev.immunol.17.1.297. [DOI] [PubMed] [Google Scholar]
- 41.Brossay L, Jullien D, Cardell S, Sydora BC, Burdin N, Modlin RL, Kronenberg M. Mouse CD1 is mainly expressed on hemopoietic-derived cells. J Immunol. 1997;159:1216–24. [PubMed] [Google Scholar]
- 42.Chen H, Paul WE. Cultured NK1.1+ CD4+ T cells produce large amounts of IL-4 and IFN-γ upon activation by anti-CD3 or CD1. J Immunol. 1997;159:2240–9. [PubMed] [Google Scholar]
- 43.Chakrabarti S, Brechling K, Moss B. Vaccinia virus expression vector: coexpression of β-galactosidase provides visual screening of recombinant virus plaques. Mol Cell Biol. 1985;5:3403–9. doi: 10.1128/mcb.5.12.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lantz O, Bendelac A. An invariant T cell receptor α chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4–8– T cells in mice and humans. J Exp Med. 1994;180:1097–106. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gui M, Li J, Wen LJ, Hardy RR, Hayakawa K. TCR β chain influences but does not solely control autoreactivity of V α 14J281T cells. J Immunol. 2001;167:6239–46. doi: 10.4049/jimmunol.167.11.6239. [DOI] [PubMed] [Google Scholar]
- 46.Sriram V, Cho S, Li P, O'Donnell PW, Dunn C, Hayakawa K, Blum JS, Brutkiewicz RR. Inhibition of glycolipid shedding rescues recognition of a CD1+ T cell lymphoma by natural killer T (NKT) cells. Proc Natl Acad Sci USA. 2002;99:8197–202. doi: 10.1073/pnas.122636199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roark JH, Park SH, Jayawardena J, Kavita U, Shannon M, Bendelac A. CD1.1 expression by mouse antigen-presenting cells and marginal zone B cells. J Immunol. 1998;160:3121–7. [PubMed] [Google Scholar]
- 48.Yuwen H, Cox JH, Yewdell JW, Bennink JR, Moss B. Nuclear localization of a double-stranded RNA-binding protein encoded by the vaccinia virus E3L gene. Virology. 1993;195:732–44. doi: 10.1006/viro.1993.1424. [DOI] [PubMed] [Google Scholar]
- 49.Porcelli SA. The CD1 family: a third lineage of antigen-presenting molecules. Adv Immunol. 1995;59:1–98. doi: 10.1016/s0065-2776(08)60629-x. [DOI] [PubMed] [Google Scholar]
- 50.Renukaradhya GJ, Sriram V, Polakova K, Russ G, Brutkiewicz RR. Development of a quantitative cell-based intracellular ELISA for the screening of B cell hybridoma supernatants: a novel rapid assay to detect positive clones. Hybridoma Hybridomics. 2004;23:373–9. doi: 10.1089/hyb.2004.23.373. [DOI] [PubMed] [Google Scholar]
- 51.Brossay L, Tangri S, Bix M, Cardell S, Locksley R, Kronenberg M. Mouse CD1-autoreactive T cells have diverse patterns of reactivity to CD1+ targets. J Immunol. 1998;160:3681–8. [PubMed] [Google Scholar]
- 52.Tifft CJ, Proia RL, Camerini-Otero RD. The folding and cell surface expression of CD4 requires glycosylation. J Biol Chem. 1992;267:3268–73. [PubMed] [Google Scholar]
- 53.Fan H, Meng W, Kilian C, Grams S, Reutter W. Domain-specific N-glycosylation of the membrane glycoprotein dipeptidylpeptidase IV (CD26) influences its subcellular trafficking, biological stability, enzyme activity and protein folding. Eur J Biochem. 1997;246:243–51. doi: 10.1111/j.1432-1033.1997.00243.x. [DOI] [PubMed] [Google Scholar]
- 54.Pulido R, Sanchez-Madrid F. Glycosylation of CD45: carbohydrate processing through Golgi apparatus is required for cell surface expression and protein stability. Eur J Immunol. 1992;22:463–8. doi: 10.1002/eji.1830220226. [DOI] [PubMed] [Google Scholar]
- 55.Rudd PM, Joao HC, Coghill E, Fiten P, Saunders MR, Opdenakker G, Dwek RA. Glycoforms modify the dynamic stability and functional activity of an enzyme. Biochemistry. 1994;33:17–22. doi: 10.1021/bi00167a003. [DOI] [PubMed] [Google Scholar]
- 56.Negroiu G, Dwek RA, Petrescu SM. The inhibition of early N-glycan processing targets TRP-2 to degradation in B16 melanoma cells. J Biol Chem. 2003;278:27035–42. doi: 10.1074/jbc.M303167200. [DOI] [PubMed] [Google Scholar]
- 57.Yoshida Y, Chiba T, Tokunaga F, et al. E3 ubiquitin ligase that recognizes sugar chains. Nature. 2002;418:438–42. doi: 10.1038/nature00890. [DOI] [PubMed] [Google Scholar]
- 58.Parodi AJ. Role of N-oligosaccharide endoplasmic reticulum processing reactions in glycoprotein folding and degradation. Biochem J. 2000;348:1–13. [PMC free article] [PubMed] [Google Scholar]
- 59.Hebert DN, Zhang J-X, Chen W, Foellmer B, Helenius A. The number and location of glycans on influenza hemagglutinin determine folding and association with calnexin and calreticulin. J Cell Biol. 1997;139:613–23. doi: 10.1083/jcb.139.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paulsson K, Wang P. Chaperones and folding of MHC class I molecules in the endoplasmic reticulum. Biochim Biophys Acta. 2003;1641:1–12. doi: 10.1016/s0167-4889(03)00048-x. [DOI] [PubMed] [Google Scholar]
- 61.Karadimitris A, Gadola S, Altamirano M, et al. Human CD1d-glycolipid tetramers generated by in vitro oxidative refolding chromatography. Proc Natl Acad Sci USA. 2001;98:3294–8. doi: 10.1073/pnas.051604498. [DOI] [PMC free article] [PubMed] [Google Scholar]