Abstract
The Alu elements are conserved ∼300-nucleotide-long repeat sequences that belong to the SINE family of retrotransposons found abundantly in primate genomes. Pairs of inverted Alu repeats in RNA can form duplex structures that lead to hyperediting by the ADAR enzymes, and at least 333 human genes contain such repeats in their 3′-UTRs. Here, we show that a pair of inverted Alus placed within the 3′-UTR of egfp reporter mRNA strongly represses EGFP expression, whereas a single Alu has little or no effect. Importantly, the observed silencing correlates with A-to-I RNA editing, nuclear retention of the mRNA and its association with the protein p54nrb. Further, we show that inverted Alu elements can act in a similar fashion in their natural chromosomal context to silence the adjoining gene. For example, the Nicolin 1 gene expresses multiple mRNA isoforms differing in the 3′-UTR. One isoform that contains the inverted repeat is retained in the nucleus, whereas another lacking these sequences is exported to the cytoplasm. Taken together, these results support a novel role for Alu elements in human gene regulation.
Keywords: Alu elements, gene silencing, nuclear retention, RNA editing
Introduction
Alu elements are the most abundant SINEs present in the human genome, with up to 1.4 million copies and constituting over 10% of the genome (Lander et al, 2001). The Alu elements are not randomly distributed throughout the genome. Rather, they are frequently found in gene-rich regions, generally within noncoding segments of transcripts, such as in introns and untranslated regions (Versteeg et al, 2003). Most human pre-mRNAs contain a surprisingly high number of Alu elements (reviewed in DeCerbo and Carmichael, 2005). Surprisingly, the functional significance of these elements remains elusive. Recently, a growing body of evidence has suggested that Alu elements are involved in different biological processes. They are implicated in human genome evolution, by modifying genes through insertions, gene conversion and recombination (Hasler and Strub, 2006). The Alu elements can also disrupt promoter regions, change methylation status, insert new regulatory features and possibly influence the efficiency of initiation of translation (Deininger and Batzer, 1999; reviewed in Brosius, 1999). Alu elements can also interfere with alternative splicing, or be incorporated into exons and directly influence the open reading frame in a mature mRNA (Lev-Manor et al, 2003; reviewed in Eisenberg et al, 2005). More recently, bioinformatic analyses showed that Alu elements within 3′-UTRs can serve as potential targets of certain microRNAs (Smalheiser and Torvik, 2006).
Adenosine-to-inosine (A-to-I) RNA editing is recognized as a cellular mechanism for generating both RNA and protein isoform diversity (reviewed in Bass, 2002). Editing is catalysed in the nucleus by the ADAR enzymes and can be either highly site-selective or promiscuous, depending on the RNA targets. Optimal activity for promiscuous editing is seen with dsRNAs of at least 100 bp in length, resulting in editing of up to 50% of the A's on each strand (Bass and Weintraub, 1987, 1988; Nishikura, 1992; Bass, 2002). Curiously, the majority of A-to-I RNA editing events reported for humans are found within Alu elements (Athanasiadis et al, 2004; Blow et al, 2004; Kim et al, 2004; Levanon et al, 2004). Alu elements share a 300-nucleotide consensus sequence and have relatively high homology among subfamilies, as these elements arose relatively recently from the 7SL RNA gene through head-to-tail fusion and were amplified throughout the genome by transposition of RNA intermediates (Batzer and Deininger, 2002; reviewed in Hasler et al, 2007). Thus, owing to their abundance, many Alu elements are likely to form intramolecular long RNA duplexes with nearby inverted Alu sequences, and these structures could then serve as substrates for A-to-I RNA editing by ADAR. By comparing human mRNA and expressed sequence tag (EST) sequences to genomic sequences and searching for the clusters of A-to-G changes as an indicator, a large number of editing sites have been found in noncoding introns and untranslated regions of RNA sequences, with the majority of these editing sites residing within Alu elements (Kim et al, 2004; Levanon et al, 2004). More importantly, each edited Alu has a reverse-oriented partner nearby, which also appears to be edited. The extent of editing appears to depend on the distance between two inverted Alu repeats (Athanasiadis et al, 2004; Blow et al, 2004). In agreement with this prediction, the ability of two inverted repeated (IR) Alu elements to form an intramolecular dsRNA has been demonstrated by showing that both the sense and antisense strands of the Alu elements, but not flanking non-Alu sequences, have been extensively edited in the second intron of the CNNM3 gene as well as the sixteenth intron of the NFκB1 gene (Kawahara and Nishikura, 2006).
What are the consequences of these extensively edited IRAlu elements within a gene? It has been suggested that one major fate of hyperedited RNA in the nucleus is retention within that compartment by the p54nrb complex (Zhang and Carmichael, 2001). Editing within introns might not lead to significant effects on gene expression, as the introns are removed during mRNA maturation. However, this is not the case for IRAlu elements located in the 3′-UTR of a gene. Prasanth et al (2005) found that a novel 8-kb nuclear-retained CTN-RNA from the mouse cationic amino-acid transported 2 (mCAT2) gene contains an extended 3′-UTR sequence and this has inverted repeat SINE elements that can form duplex RNA structures that are highly A-to-I edited. mCAT2 encodes a protein involved in the uptake of extracellular arginine, the precursor to nitric oxide. Under normal situations, cells not only express a cytoplasmic form of mCAT2 mRNA that encodes a basal level of the arginine transporter, but also abundant levels of another form of CTN-RNA that contains the same open reading frame as mCAT2 but which is retained within the nucleus in association with the p54nrb complex. Under stress conditions, CTN-RNA appears to be cleaved within its 3′-UTR near an alternative polyadenylation signal to remove the SINE-associated retention elements. This RNA is rapidly exported to the cytoplasm, where it allows for increased production of the arginine transporter. This study demonstrated further that editing of the repetitive elements in CTN-RNA correlates strongly with nuclear retention. This raised the important question of whether retention is a common or general fate of RNAs that are highly edited in their 3′-UTR regions.
After a thorough analysis of the released human mRNA and EST sequences in the UCSC genome browser, we identified a set of 333 genes with IRAlu elements in their 3′-UTR regions (Supplementary Table 1). Importantly, mRNA and EST sequences corresponding to many of these IRAlu elements have been reported to be extensively edited. Here we asked whether a single pair of IRAlu elements in the 3′-UTR might have an important regulatory function on gene expression. To address this question, we first made a series of EGFP-fused IRAlus constructs and transfected them into HEK293 cells to investigate EGFP expression and the fates of the egfp-IRAlus RNA. We show that a single pair of IRAlu elements in the 3′-UTR of the egfp mRNA strongly represses EGFP expression. Further, this reduction is accompanied by significant nuclear retention of the mRNAs, likely by the p54nrb complex. Finally, we present evidence for nuclear retention of an endogenous mRNA with IRAlus in its 3′-UTR.
Results
Extensive editing of IRAlu elements in the 3′-UTRs of Nicolin 1 and Lin28
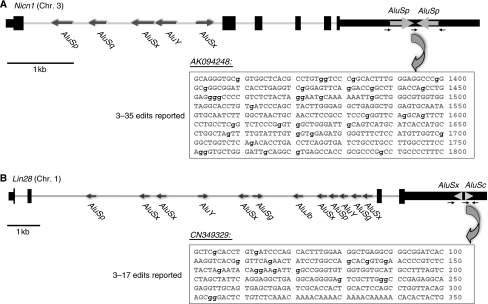
Two genes, Nicolin 1 (Nicn1) and Lin28, each of which has a pair of IRAlu elements in the 3′-UTR, were identified after analysis of a 3′-UTR database of the IRAlu elements (see Supplementary Table 1). Although originally cloned in 2002, the function of the NICN1 protein remains to be elucidated (Backofen et al, 2002). Analysis of the Nicn1 genomic sequence with RepeatMasker (http://www.repeatmasker.org) revealed that the 3′-UTR contains a pair of IR AluSp elements that are positioned within 70 bp of one another. Two of the four currently available cDNA sequences indicate that both the sense and antisense IRAluSp elements are highly edited. For instance, one of the sequenced cDNAs, AF538150, shows that 12 A's on the IRAluSps have been edited to I's, whereas another sequenced mRNA, AK094248, shows that there are 35 A's changed to I's, accounting for 20% of the total A's in the IRAluSp region (Figure 1A).
Figure 1.
Organization and Alu characterization of Nicn1 and Lin28. The genomic sequences of Nicn1 (A) and Lin28 (B) are drawn to scale. Exons and UTRs are shown as black bars, with coding regions being thicker. The Alu elements present in these two genes are shown as gray arrows with the indicated orientations. There is a single pair of IRAlus in each of the 3′-UTRs of Nicn1 and Lin28. The small black arrows indicate the PCR primers, their directions and their relative positions for the cloning sequences on the pEGFP-C1 vector. AK094248 shows one of the highly edited mRNA sequences of Nicn1 in the UCSC genome browser, whereas CN349329 shows one of the highly edited mRNA sequences of Lin28. The edited residues are denoted in lowercase in bold.
Lin28 also exhibits extensive editing of the IRAlu elements in its 3′-UTR. The mRNA for this gene is very abundant in diverse types of undifferentiated cells (Balzer and Moss, 2007). LIN28 is known to be a regulator of the developmental timing in Caenorhabditis elegans (Horvitz et al, 1983; Ambros and Horvitz, 1984), suggesting that mammalian LIN28 has an important role in gene regulation during embryonic stem (ES) cell differentiation. Recent studies have shown that LIN28 colocalizes with mRNP complexes, P-bodies and stress granules in pluripotent cells, suggesting that it might influence the translation or stability of specific mRNAs during differentiation (Balzer and Moss, 2007) and that this protein appears to be one of four that together can reprogramme somatic cells to ES cells (Yu et al, 2007). In a recent work, this protein has been shown to affect microRNA processing in ES cells (Viswanathan et al, 2008). The long (3500 nt) 3′-UTR of Lin28 contains a single pair of AluSx and AluSc elements separated by 50 bp. All three mRNA sequences (BC028566, AF521099 and AK022519) sequenced across the 3′-UTR AluSx/AluSc region show a moderate level of A-to-I editing (3–5 A's are edited to I's). In addition, some of the available EST sequences show that AluSc, one of the paired IRAlus/AluSc, is highly edited. For example, clone CN349329 shows that 17 A's are edited to I's, which accounts for 23% of A's in this AluSc element (Figure 1B).
For both of the above-mentioned sequences or other sequences in our 3′-UTR Alu element database (see Supplementary Table 1), the extent of editing in the IRAlu elements is highly variable among different mRNA sequences or ESTs, with some of the cloned sequences showing no editing at all in the same Alu locus. Thus, although hyperediting occurs in IRAlu elements within 3′-UTRs, the extent of editing is variable and may be random or regulated in a yet unappreciated manner in different tissues and cell types.
IRAlus in the 3′-UTR of egfp mRNA suppresses EGFP expression
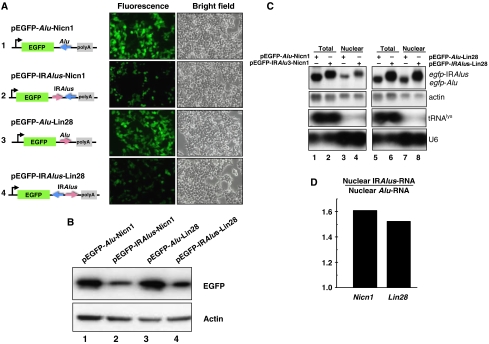
To investigate the effects of IRAlus within the 3′-UTRs of human genes, we utilized a simple EGFP expression system and the experimental flow outlined in Supplementary Figure 1. We amplified the single antisense AluSp, or the pair of IRAluSp elements from the 3′-UTR of Nicn1 (Figure 1A, black arrows indicate the position of primers used), and then inserted each into the egfp 3′-UTR region of the expression vector pEGFP-C1 to generate constructs 1 and 2 shown in Figure 2A. Next, we measured the EGFP expression level from each plasmid 44 h after transfection into HEK293 cells. The pair of IRAluSps derived from Nicn1 in the 3′-UTR significantly reduced EGFP fluorescence when compared with the single AluSp element. This repression effect was confirmed by western blotting analysis with anti-EGFP antibody (Figure 2B, lanes 1 and 2).
Figure 2.
IRAlus in the 3′-UTR of egfp mRNA suppress EGFP expression at a post-transcriptional level. (A) IRAlus in the 3′-UTR of egfp mRNA suppress EGFP expression. IRAlus and Alu were PCR-amplified from the 3′-UTR of either Nicn1 or Lin28 and then inserted separately into the 3′-UTR of egfp mRNA. HEK293 cells were transfected with the indicated plasmids and fluorescence pictures were taken 44 h after transfection. (B) The expression of EGFP from the same batch of transfected HEK293 cells as described in panel A was investigated by western blotting, by probing with anti-GFP antibody. Actin was used as the loading control. (C) IRAlus RNAs are retained in the nucleus. Total and nuclear RNAs were isolated from the same batch of transfected HEK293 cells used in panels A and B and then resolved on a denaturing agarose gel. Transcripts of egfp-tagged RNAs were probed with a Dig-labelled egfp fragment. Actin RNA was used as the loading control; tRNAlys and U6 snRNA were used as markers for nuclear/cytoplasmic RNA isolation. (D) Preferential retention of IRAlus RNAs within the nucleus compared with single Alu peers. Total and nuclear IRAlus RNAs, as well as Alu-containing RNAs, were quantified from panel C and normalized to the relative amount of actin mRNAs. The ratio was obtained by comparison of the normalized value of the nuclear-retained IRAlus RNA to those of the nuclear-retained Alu RNA.
What mechanism could account for this silencing phenomenon? There are about 30 human microRNAs that exhibit typical short-seed complementarity with a specific and highly conserved site within Alu elements (Smalheiser and Torvik, 2006). To test the possibility that microRNA regulation might contribute to the silencing of EGFP, we performed a microRNA northern blot with both antisense and sense AluSp probes to total RNAs collected from the pEGFP-AluSp- or pEGFP-IRAluSps-transfected HEK293 cells. However, we could detect no AluSp-related small RNAs in these experiments (data not shown), suggesting that a microRNA-related mechanism is unlikely to account for the regulation reported here.
We next asked whether A-to-I editing might lead to enhanced mRNA degradation and consequently lower gene expression. A recent study suggested an interaction between components of the ADAR and RNAi pathways by Tudor staphylococcal nuclease (Tudor-SN), which is a subunit of the RNA-induced silencing complex and specifically interacts with and promotes cleavage of model hyperedited dsRNA substrates containing multiple I·U and U·I pairs (Scadden, 2005). Thus, it is possible that highly A-to-I edited sites within the IRAluSps might be recognized by Tudor-SN, in turn reducing the level of EGFP message. To examine this possibility, we carried out northern blotting with a Dig-labelled egfp probe and found that the egfp-AluSp and egfp-IRAluSps RNAs were expressed at the same level (Figure 2C, lanes 1 and 2), and there was no apparent preferential degradation of RNA containing the IRAlus. This experiment also eliminated a difference in the transfection efficiencies of individual plasmids as the cause of differential EGFP expression.
As the Alu elements have diverged into more than 200 subfamilies (Price et al, 2004), we asked whether IRAlus pairs from different subfamilies also have a similar effect on gene expression. The 3′-UTR of Lin28 has a pair of IRAlu elements that belong to the AluSx and AluSc subfamilies, and these are shown to be extensively edited (Figure 1B). By using the same cloning strategy, we inserted into the egfp 3′-UTR of the vector pEGFP-C1 either the sense AluSc element or the pair of inverted AluSx/AluSc repeats from 3′-UTR of Lin28 (Figure 1B, black arrows indicate primers used) (Figure 2A, constructs 3 and 4). Transfection experiments with HEK293 cells once again showed strong repression of EGFP expression, by both fluorescence microscopy (Figure 2A) and immunoblotting (Figure 2B, lanes 3 and 4), when IRAluSx/AluSc were fused to the 3′-UTR of egfp. Northern blots further showed that the transcripts of egfp-AluSc and egfp-IRAluSx/AluScs are abundantly expressed (Figure 2C, lanes 5 and 6). These results indicate that a pair of inverted Alu repeats in the 3′-UTR of a gene can induce gene silencing, regardless of their subfamilies.
IRAlus derived from an intron also repress EGFP expression when placed in the 3′-UTR of egfp
To exclude the possibility that some unique sequences present in the IRAlus pairs or the sequences between the IRAlus pairs from the 3′-UTRs of Nicn1 and Lin28 might be responsible for silencing of EGFP, we tested the effect on gene expression of IRAlus from an intronic region. The second intron of the Apobec3G gene has one pair of IRAluSp elements separated by 121 bp (Supplementary Figure 2). Several mRNA and EST sequences have shown that there are 9–14 A-to-I edits in the sense AluSq element. By inserting either only the sense AluSq element or the pair of IRAluSqs from the second intron of Apobec3G (Supplementary Figure 2A, black arrows indicate primers used) into the 3′-UTR (Supplementary Figure 2B, constructs 2 and 3), we carried out experiments similar to those described above. When positioned in the 3′-UTR of egfp, the IRAluSqs from Apobec3G, but not the single AluSq, led to striking repression of EGFP expression as measured by fluorescence microscopy (Supplementary Figure 2B) and western blotting (Supplementary Figure 2C, lanes 2 and 3). Yet, the transcripts of egfp-AluSq and egfp-IRAluSqs were expressed at similar levels (Supplementary Figure 2D, lanes 2 and 3).
To address the question of whether a single copy of an Alu element in the 3′-UTR affects gene expression, we compared egfp and egfp-AluSq transcripts and their ability to translate into the protein EGFP. A single Alu element in the 3′-UTR of egfp exerted no obvious repression, as EGFP protein levels and transcript levels from the egfp and egpf-AluSq plasmids are almost equal (Supplementary Figure 2C and D, lanes 1 and 2). To rule out any cell type-specific effects, we also examined the ability of IRAlu elements in the 3′-UTR to repress gene expression in HeLa cells and COS7 cells, using the same experimental strategies. A similar silencing effect on EGFP expression was observed in both cell lines, and we have never observed any difference in gene expression between the parent plasmid pEGFP and derivatives that contained only single Alu elements, whether from Lin28, Nicn1 or Apobec3G (data not shown).
Silencing mechanism: egfp-IRAlus mRNAs are retained within the nucleus
After fractionating cytoplasmic and nuclear RNAs from transfected HEK293 cells, we observed that IRAlus-containing RNAs appear to be preferentially retained in the nucleus in comparison with those having a single Alu element. As shown in Figure 2C (lanes 3 and 4), the IRAluSps from Nicn1 caused a 1.6-fold greater nuclear retention of the EGFP mRNA when compared with the corresponding egfp-AluSp after normalization to the amount of nuclear-retained actin mRNA as control (Figure 2D, Nicn1). These data were consistent with the repression of EGFP expression (0.5-fold) that we observed in experiments reported above. A similar level of nuclear retention was observed for the Lin28-derived IRAluSx/AluSc (Figure 2C, lanes 7 and 8) and the IRAluSqs elements from the Apobec3G intron 2 when these elements were placed in the 3′-UTR of egfp (Supplementary Figure 2C, lanes 5 and 6). There was a 1.5-fold increased nuclear retention of egfp-IRAluSx/AluSc compared with egfp-AluSc (Figure 2D, Lin28) and a 1.6-fold increased nuclear retention of egfp-IRAluSqs compared with egfp-AluSq (data not shown). Again these two sets of data were consistent with the data on EGFP gene silencing (0.4-fold). On the other hand, there is no significant difference in nuclear/cytoplasmic distribution of egfp-Alu mRNA compared with control (Supplementary Figure 3C, lanes 4 and 5), consistent with our finding that a single Alu element in the 3′-UTR does not affect EGFP gene expression.
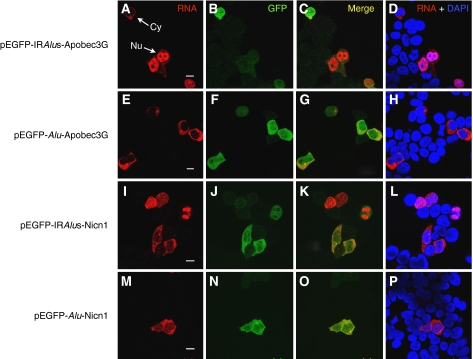
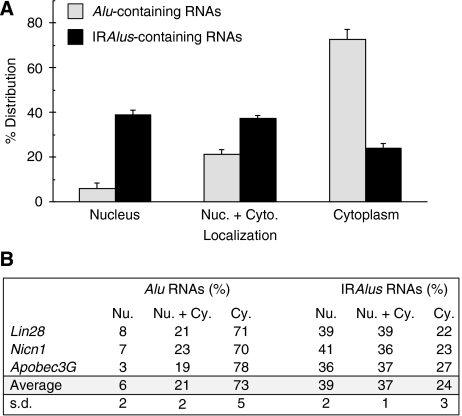
We next performed RNA in situ hybridization experiments with a Dig-labelled antisense fragment of egfp to directly visualize the subcellular distribution of transgene RNAs (Figure 3). The egfp-Alu RNAs and egfp-IRAlus RNAs produced in transfected HEK293 cells have distinct localization patterns, consistent with the data presented in Figure 2. Constructs with a single Alu within the 3′-UTR express RNAs that are almost completely localized to the cytoplasm, where they are efficiently translated (Figure 3E–H and M–P). On the other hand, mRNAs with inverted Alu repeats in their 3′-UTRs show a more variable pattern, with some cells showing mRNAs in the cytoplasm (where they are efficiently translated), but others in which the mRNA is retained in the nucleus (Figure 3A–D and I–L). Importantly, cytoplasmic localization correlates strongly with EGFP expression. Although RNAs from each transgene exhibited heterogeneous localization patterns within cells (completely localized to the cytoplasm in some cells, completely localized to the nucleus in other cells and localized to both the cytoplasm and nucleus in some cells; see Supplementary Figure 3), the overall patterns of localization differed dramatically, as revealed by the statistical analysis shown in Figure 4. The majority (73%) of the transfected cells that host the Alu-containing RNAs exhibited exclusive localization in the cytoplasm, but only about 24% of the cells transfected with the egfp-IRAlus vector showed such a distribution. Strikingly, 39% of the cells transfected with egfp-IRAlus constructs showed a distinct and punctate nuclear localization (see Supplementary Figure 3), whereas only 6% of the cells transfected with pEGFP-Alu showed this pattern. Finally, in some cells, RNA was present at almost equal levels in both compartments (21% for egfp-Alu and 37% for egfp-IRAlus). In these cells, the nuclear pattern was not punctate, but more diffuse throughout this compartment (see Supplementary Figure 3 for an example of this pattern). Taken together, these data strongly support our conclusion that the mechanism by which IRAlus-containing RNAs repress gene expression is by nuclear retention.
Figure 3.
Nuclear retention of IRAlus-containing RNAs correlates with silencing of EGFP expression. RNA in situ hybridization (A, E, I, M) was performed with Dig-labelled antisense egfp probe (red) for each different transfection with plasmids encoding either IRAlus or Alu-RNA, and representative images are shown. No signals were detected with the Dig-labelled sense strand egfp fragment (data not shown). EGFP was visualized using anti-GFP antibodies (B, F, J, N). The white arrows in panel A identify cells in which mRNA is in either the cytoplasm or nucleus. Note that when RNA expression in the cytoplasm is higher, in these cells the expression of GFP is also higher. When the RNA is retained in the nucleus, GFP expression is reduced. Panels C, G, K and O merge the RNA and GFP signals, and panels D, H, L and P merge the RNA signal with nuclear DAPI staining. Scale bars 10 μm.
Figure 4.
Subcellular distribution of IRAlus RNA. HEK293 cells were transfected with the plasmids described in Figures 2 and 3 and RNA in situ hybridization was performed with a Dig-labelled antisense egfp probe as in Figure 3. (A) A total of 200 transfected cells were recorded randomly by confocal microscopy following each different transfection, and the percentage of each distinct localization pattern of IRAlus-RNA or Alu-RNA was recorded. The average percentage of each pattern was calculated by the mean of IRAlus or Alu-RNA that comes from different genomic locus (Nicn1, Lin28 and Apobec3G). Signals that were exclusively nuclear are labelled ‘Nucleus'. Cells with RNA distributed both in the nucleus and cytoplasm are labelled ‘Nuc.+Cyto'. Cells with exclusively cytoplasmic signals are labelled ‘Cytoplasm'. The criteria used for assignment are illustrated in Supplementary Figure 3. The results are graphed as the sum of results from the various plasmids. (B) Overall results and statistical analysis of distribution patterns are tabulated. s.d., standard deviation.
It was next considered important to rule out possible effects of IRAlus on translation. The data in Figure 3 clearly show that when IRAlus-containing egfp transcripts are in the cytoplasm, they appear to be efficiently translated (see, for example, white arrows in panel A). To confirm this, we compared sucrose gradient polysome profiles of cytoplasmic egfp-IRAlus transcripts and egfp transcripts. The cytoplasmic levels of egfp-IRAlus transcripts were lower than those of egfp-Alu transcripts, consistent with our fractionation and in situ hybridization results, whereas the polysome profiles were indistinguishable (Supplementary Figure 4).
RNA editing and association with p54nrb correlate with nuclear retention of IRAlus-containing RNAs
To determine whether IRAlus-containing RNAs are edited and whether such editing correlates with nuclear retention, we first used RT–PCR to amplify the region spanning the downstream Alu element from RNA isolated from cells transfected with construct pEGFP-IRAlus-Nicn1. As expected, DNA sequencing confirmed frequent and promiscuous editing. Out of the 35 clones examined, 29 showed A-to-G transitions indicative of ADAR editing, with levels of editing ranging from 2 to 40% of the adenosine residues converted to inosines (Supplementary Figure 5). As we examined only one of the two Alus, it is quite likely that this is a gross underestimate of the true extent of editing in these transcripts. Next, we carried out similar sequencing studies, but on RNAs isolated from either cytoplasmic or nuclear fractions (Supplementary Figure 6). The results confirmed that IRAlus-containing RNAs isolated from the nuclei are more highly edited than those isolated from the cytoplasm.
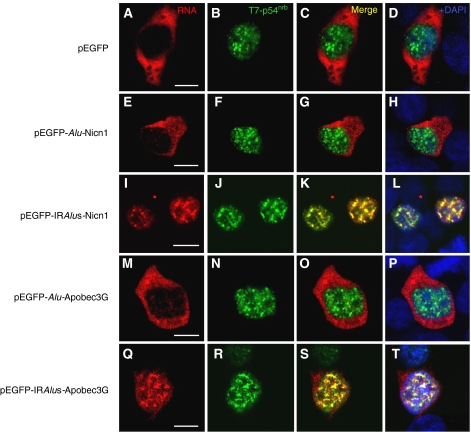
As described above, approximately 40% of the transfected cells show a distinct subnuclear localization pattern of the IRAlus-containing RNAs. We therefore asked whether this localization pattern matched that of any nuclear proteins. In a number of repeated experiments, we were unable to demonstrate significant colocalization of the nuclear-retained RNAs with a variety of other nuclear factors (PSP1α, SC35 (Supplementary Figure 7); Drosha, data not shown). However, as the IRAlus that we have studied have been shown to be highly A-to-I edited in cells, we hypothesized that such edited transcripts may associate with p54nrb. p54nrb is found both in paraspeckles and elsewhere throughout the nucleoplasm, and has many roles in the regulation of RNA metabolism (Karhumaa et al, 2000; Straub et al, 2000; Zhang and Carmichael, 2001; Fox et al, 2002; Peng et al, 2002; Ishitani et al, 2003; Kameoka et al, 2004; Bladen et al, 2005; Kaneko et al, 2007). Importantly, it is the first RNA-binding protein described that exhibits a strong affinity for inosine-containing RNAs (Zhang and Carmichael, 2001). It is thus possible that edited IRAlu elements in the 3′-UTR of egfp associate with p54nrb, which in turn leads to nuclear retention. To test this, we investigated the colocalization of the transfected IRAlus-containing RNAs and epitope T7-tagged p54nrb in HEK293 cells. For the majority of the p-C1 vector-transfected cells, egfp mRNA alone is distributed only in the cytoplasm (Figure 5A) and shows no colocalization with T7-p54nrb (Figure 5B–D). The localization pattern of the single AluSp (from Nicn1) present at the 3′-UTR of the egfp was very similar to this control pattern. A total of 71% of the transfected HEK293 cells showed a clear cytoplasmic localization of egfp-AluSp, and again did not colocalize with T7-p54nrb (Figure 5E). Although a small fraction of transfected cells (8%) showed nuclear localization, in these cells the nuclear pattern was indistinct, lacking obvious punctate staining (data not shown). In sharp contrast, the transfected HEK293 cells that expressed nuclear-retained egfp-IRAluSps RNAs showed almost complete colocalization of the hybridization signal with T7-p54nrb (Figure 5I–L). Additional colocalization was performed using serial sections and confocal microscopy, and again demonstrated that the egfp-IRAluSps RNA and T7-p54nrb colocalized with each other throughout the fixed cells (data not shown). The same colocalization pattern of T7-p54nrb was also observed with other IRAlus-containing RNA constructs. For instance, 41% of the cells transfected with the plasmid pEGFP-AluSx/AluSc-Lin28 showed the punctate nuclear localization pattern of egfp-AluSx/AluSc RNAs, and these also colocalized with T7-p54nrb (Supplementary Figure 8). Finally, we studied the RNA colocalization patterns for T7-p54nrb and the Apobec3G intron-derived IRAluSqs element fused to the 3′-UTR of egfp. Again, 36% of the transfected cells showed a distinct nuclear localization pattern, and in these cells the RNA colocalized with T7-p54nrb (Figure 5Q–T), whereas the majority (78%) of the egfp-AluSq RNAs showed only a unique cytoplasmic localization without nuclear localization signals (Figure 5M–P). These colocalization studies further reinforce that even IRAlus from intronic loci can lead to silencing by sequestering RNAs within the nucleus likely in association with p54nrb.
Figure 5.
IRAlus-RNA is retained in the nucleus and colocalizes with T7-p54nrb. RNA in situ hybridization was performed with Dig-labelled antisense egfp probe (red) for each different transfection with plasmids encoding either IRAlus or Alu-RNA, and representative images are shown (A, E, I, M, Q). No signals were detected with the Dig-labelled sense strand egfp fragment (data not shown). Co-transfected T7-p54nrb was visualized with anti-T7 antibody (green; B, F, J, N, R). Panels C, G, K, O and S merge the RNA and T7-p54nrb signals, and panels D, H, L, P and T merge the RNA and protein signals with nuclear DAPI staining. Scale bars, 10 μm.
p54nrb is associated with endogenous IRAlus RNAs, including Nicn1 RNA
As the IRAlus RNAs colocalize with T7-p54nrb in the nucleus, we next asked whether IRAlus RNA is associated with endogenous p54nrb. As shown in Figure 6A, even though the endogenous p54nrb staining pattern appears less punctate than that of the transfected T7-p54nrb, it is still nucleoplasmic and excluded from the nucleolus (Figure 6A). Remarkably, the transfected nuclear-retained egfp-IRAluSqs-Apobec3G RNA is also excluded from nucleoli by RNA in situ analysis, and shows a nuclear pattern similar to that of endogenous p54nrb. Importantly, a direct association of p54nrb and RNA was demonstrated by immunoprecipitation (IP) using p54nrb antibody. For this experiment, we made a construct with two copies of the MS2 coat protein binding site positioned downstream of egfp-IRAluSqs-Apobec3G in the pEGFP-C1 construct (Supplementary Figure 9A). After transfection into HEK293 cells, p54nrb-associated RNP complexes were immunoprecipitated with the p54nrb antibodies (Supplementary Figure 9B). Total RNA was isolated from the immunoprecipitates and a specific reverse primer located within the MS2 sequence was used to discriminate transcripts from the transfected construct from those from the endogenous loci of IRAluSqs (Supplementary Figure 9A). Full-length IRAluSqs-MS2 RNAs were detected in the immunoprecipitated complexes but not in the mock IP, when reverse transcription was performed under stringent conditions (Supplementary Figure 9C, lane 4).
Figure 6.
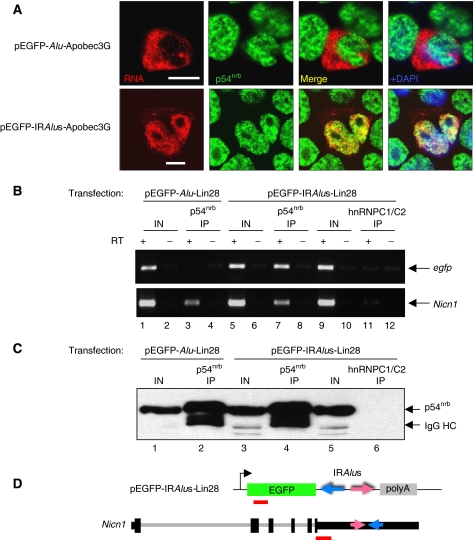
IRAlus RNA and endogenous Nicn1 associate with p54nrb. (A) Colocalization of IRAlus RNA and endogenous p54nrb. HEK293 cells were transfected with plasmids encoding Alu RNA (upper panel) and IRAlus RNA (lower panel), and RNA in situ hybridization was carried out with Dig-labelled antisense egfp probe (red). Endogenous p54nrb was visualized with anti-p54nrb antibody (green). Nuclei were stained with DAPI. Scale bars, 10 μm. (B) IP from pEGFP-Alu-Lin28- or pEGFP-IRAlus-Lin28-transfected HEK293 cells using anti-p54nrb antibody or anti-hnRNPC1/C2 antibody (mock IP). RT–PCR of egfp from the IP using hexamer primers showed amplification only in the pEGFP-IRAlus-Lin28-transfected cells by anti-p54nrb IP, but not by IP with anti-hnRNPC1/C2 antibody or by IP in the pEGFP-Alu-Lin28-transfected cells by anti-p54nrb IP (upper panel). Endogenous Nicn1 RNA was associated with p54nrb in both transfections, but did not associate with hnRNP C1/C2 (lower panel). (C) Western blotting using anti-p54nrb antibody was used with extracts from the sample in panel B to confirm the specificity of IP. (D) Schematic representation of the fragments (red bars) in the pEGFP-C1 vector and the endogenous Nicn1 that were analysed by PCR amplification in panel B.
To investigate whether the IRAlus in other transfected plasmids could also bind to p54nrb, we did similar IPs of p54nrb containing RNP complexes from HEK293 cells transiently expressing egfp-AluSc-Lin28 or egfp-IRAluSx/AluSc-Lin28 (Figure 6B and C). RT–PCR was carried out using egfp-specific primers (Figure 6D). In these studies, anti-p54nrb did not immunoprecipitate egfp-AluSc-Lin28 mRNA (Figure 6B, lane 3). A control IP experiment using anti-hnRNPC1/C2 antibodies did not immunoprecipitate egfp-IRAluSx/AluSc-Lin28 mRNA, excluding nonspecific binding of egfp-IRAluSx/AluSc-Lin28 mRNAs (Figure 6B, lane 11). As shown in Figure 6B (lane 7), p54nrb interacts strongly with the egfp-IRAluSx/AluSc-Lin28 mRNA, demonstrating the association of p54nrb with IRAlus within the nucleus.
To study whether endogenous Nicn1 mRNA is also associated with p54nrb, RT–PCR was carried out with primers that are located upstream of the IRAluSp elements in the 3′-UTR of the Nicn1 gene (Figure 6D). Endogenous Nicn1 mRNA was consistently specifically bound to p54nrb in both pEGFP-AluSc-Lin28- and pEGFP-IRAluSx/AluSc-Lin28-transfected cells (Figure 6B, lanes 3 and 7). Association of Nicn1 mRNA and p54nrb was also detected in HeLa cells (data not shown). Control IP with the hnRNP C1/C2 antibodies failed to immunoprecipitate Nicn1 mRNA (Figure 6B, lane 11).
Direct evidence that editing correlates with nuclear retention was obtained in additional experiments. As discussed above (Supplementary Figure 6), RNAs containing IRAlus isolated from nuclei are edited to a significantly greater extent than those isolated from the cytoplasm. This confirms that nuclear retention correlates with editing. To obtain direct evidence that edited Alus are generally recognized by p54nrb, we immunoprecipitated RNP complexes containing p54nrb or control protein hnRNP C1/C2 (Supplementary Figure 10) and characterized the associated Alu-containing RNAs (Supplementary Figure 11). When the RNAs isolated from immunoprecipitates were examined for specific transcripts known to contain or lack IRAlus, those with IRAlus were found to be associated with p54nrb whereas those lacking them were not (Supplementary Figure 11A). When more general levels of Alu editing were examined in these RNA pools by using degenerate primers, we found that p54nrb is often associated with hyperedited Alus, whereas hnRNP C is not. Thus, 11% of the examined clones immunoprecipitated with anti-p54nrb antibody contained highly edited sequences (>10% A-to-I conversions), whereas only a few of those examined in the nuclear RNA fraction or immunoprecipitated with anti-hnRNP C1/C2 antibody contained highly edited sequences (Supplementary Figure 11B). Taken together, these findings indicate that endogenous RNAs containing IRAlus, as well as Nicn1 mRNA in particular, might utilize the p54nrb mechanism to regulate their expression through editing and nuclear retention.
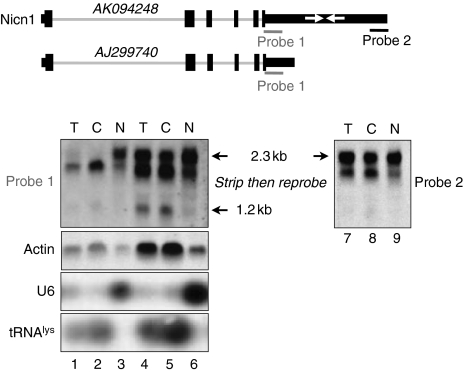
Finally, we asked whether NICN1 expression is also regulated in a manner similar to that of the murine mCAT2 gene, with a nuclear-retained transcript and a shortened cytoplasmic message. The results presented in Figure 7 support this prediction. We performed northern blot analyses with two Nicn1 probes within the 3′-UTR, one lying upstream (probe 1) and the other downstream (probe 2) of the IRAluSps (Figure 7, top). Probe 1 revealed three Nicn1 transcripts, one corresponding to the reported full-length mRNA (clone AK094248), a slightly shorter transcript (consistent with several cDNAs containing deletions in the region of the IRAlus) and a third even smaller transcript of approximately 1200 nt, consistent with the polyadenylated cDNA AJ299740. Importantly, the full-length mRNA partitions mostly to the nucleus (Figure 7, bottom panel), whereas the shorter transcript is almost exclusively cytoplasmic. The short transcript is not seen with probe 2, confirming that it lacks IRAlus. This picture is in conformity with our hypothesis that the edited Alu sequences help to sequester the Nicn1 mRNA in the nucleus.
Figure 7.
Characterization of Nicn1. Northern blot analysis of Nicn1 RNA in nuclear and cytoplasmic fractions from HEK293 cells revealed that a 1.2 kb band was enriched in the cytoplasmic fraction. Two probes from the 3′-UTR of Nicn1 were used for the northern blot. Probe 1 (red bar) is located upstream of the IRAlus whereas probe 2 (green bar) is located downstream of IRAlus. tRNAlys and U6 were used as makers for the nuclear and cytoplasmic RNA fractionation. In the bottom panel, total (T), cytoplasmic (C) and nuclear (N) RNAs were analysed by northern blotting. Lanes 1–3 are the same samples as lanes 4–6, except that a less amount of the samples was loaded onto the gel. Following hybridization with probe 1, the membrane representing lanes 4–6 was stripped and re-probed with probe 2 (lanes 7–9).
Discussion
A significant proportion of human pre-mRNAs are A-to-I edited owing to the formation of duplex structures from inverted repeats of the conserved Alu sequences (Athanasiadis et al, 2004; Blow et al, 2004; Kim et al, 2004; Levanon et al, 2004). In most cases, editing is confined to IRAlus that are located within introns and UTR regions and not within coding regions. Our bioinformatic analysis of the available human cDNA and EST databases (Supplementary Table 1) has identified 333 genes that contain IRAlu elements in their 3′-UTRs, and many of these Alu sequences have already been shown to be highly susceptible to editing. To address the functional significance of Alu repeats in 3′-UTRs, here we show that the presence of a single pair of inverted Alu elements in the 3′-UTR of the egfp gene consistently reduces EGFP expression, and that much of this repression can be accounted for by A-to-I editing and the retention of the IRAlus-containing mRNAs within the nucleus by the p54nrb complex. This silencing effect is observed with several Alu subfamily members tested and is independent of the flanking regions that naturally surround the Alu elements. Importantly, we show that the endogenous Nicn1 mRNA, which carries IRAluSps elements in its 3′-UTR, is also associated with the p54nrb complex (Figure 6B). Northern blots show that there are multiple isoforms of Nicn1 RNA in the cell, and the larger one, containing IRAluSps elements, is enriched in the nucleus (Figure 7). Thus, IRAlus present in 3′-UTRs may act in general to retain mRNAs in the nucleus.
Several lines of evidence support the notion that nuclear retention is mediated by p54nrb. Fluorescence studies show that p54nrb colocalizes with our IRAlus-containing reporter RNAs (Figures 3 and 5). Using antibodies directed against a number of other nuclear factors, we have been unable to find evidence for the colocalization of the nuclear-retained reporter RNAs with any protein other than p54nrb. IP experiments further demonstrate that p54nrb associates with IRAlus-containing RNAs, but not with transcripts containing a single Alu element (Figure 6B). Also, hyperedited RNAs are preferentially retained in the nucleus (Supplementary Figure 6) and p54nrb IP enriches for RNAs with hyperedited Alu elements (Supplementary Figure 11).
What is the significance of Alu-mediated regulation? As seen from the human cDNA and EST databases (Athanasiadis et al, 2004; Blow et al, 2004; Kim et al, 2004; Levanon et al, 2004) as well as our own data, A-to-I editing within IRAlus elements is both promiscuous and variable, ranging from few to many A-to-I changes in individual messages. mRNA isoforms that lack or contain only low levels of inosines appear to be exported to the cytoplasm, whereas more highly edited RNA isoforms are selectively retained in the nucleus. Thus, editing might serve to modulate gene expression of IRAlus-containing mRNAs by titrating the amount of mRNA that is allowed to reach the cytoplasm. We do not yet know whether different cells or tissues differ in their relative editing efficiencies of IRAlus, but it is well known that ADAR1 and ADAR2 are expressed in a tissue-specific and developmentally controlled manner in mammals. For instance, the expression of both ADAR1 and ADAR2 is higher in the brain, and editing levels in this organ appear to be higher as well (Bass, 2002; Blow et al, 2004). We predict that genes with IRAlus in their 3′-UTRs may exhibit enhanced nuclear retention in the brain. Also, ADAR activity is enhanced strongly by inflammation, interferon treatment and immune stimulation (Liu et al, 1997; George and Samuel, 1999; Rabinovici et al, 2001; Yang et al, 2003; George et al, 2005). This provides another mechanism of regulation through enhanced nuclear retention of hyperedited mRNAs involving Alu repeats.
An alternative possibility is that cells or tissues might respond to different external or growth-related signals or stimuli by influencing the level of editing. Editing within IRAlus in 3′-UTRs of genes could provide an additional layer of gene regulation by sequestering otherwise mature mRNAs within the nucleus, and these might be available for export by a rapid response pathway. The mechanism would mirror that of the mouse CTN-RNA, which is retained in the nucleus until cell stress occurs and is then cleaved to remove its 3′-UTR nuclear retention signal (inverted repeats of a murine SINE), which contains numerous A-to-I editing sites. The truncated message is then transported efficiently to the cytoplasm for translation (Prasanth et al, 2005). That such desequestration might be a general mechanism is hinted by the different nuclear/cytoplasmic localizations of Nicn1 mRNA isoforms. Although the function of NICN1 remains to be elucidated, it will be of interest to determine whether the structure or cytoplasmic accumulation of Nicn1 mRNA responds to alcohol intake of other cellular stress, as this gene has been reported to be upregulated in alcoholics (Mulligan et al, 2006).
Alternative 3′-end formation may also participate in the regulation of genes with IRAlus in their 3′-UTRs. It has become increasingly evident in the past several years that many mammalian genes contain multiple polyadenylation signals (Lee et al, 2007). A common interpretation for the significance of alternative 3′-end processing is that it changes cis-acting elements in the 3′-UTR that can regulate the stability or translation of the mRNA. Our data suggest yet another function: if these signals flank sequences that promote A-to-I editing, then the choice of polyadenylation signal would strongly influence the ability of the mRNA to be exported to the cytoplasm. Another possible mechanism of gene regulation of this type is the inclusion by splicing of alternative 3′-UTRs. Two interesting examples of this are caspase 8 and caspase 10, which lie adjacent to one another on chromosome 2 (see Supplementary Table 1). These genes utilize two different 3′UTRs; the upstream 3′-UTR contains IRAlus, whereas the downstream 3′-UTR does not. One might hypothesize that the inclusion of these alternative 3′-UTRs would affect the level of expression of the encoded proteins, perhaps regulated in response to cellular stress.
Yet another potential application of the nuclear retention of the IRAlus-containing mRNA-mediated gene silencing could be in gene regulation during human ES cell differentiation. LIN28 is involved in the regulation of developmental timing in C. elegans, and although the exact mechanism is unknown in humans, microarray analysis showed that it is primarily expressed in pluripotent cells (Richards et al, 2004). Also, it has recently been shown that this protein is among a small group of factors that can reprogramme somatic cells to ES cell characteristics (Yu et al, 2007). It is possible that the nuclear retention mechanism described here may regulate the expression of LIN28 during embryonic development. However, we do not yet know whether IRAlus can be edited in these cells and whether the p54nrb pathway is functional or whether any alternate pathway is used.
We still do not understand whether there is any functional relationship among genes that have inverted Alus in their 3′-UTRs, although there may be some interesting connections apparent from gene ontology analysis. For example, some zinc-finger transcription factors and apoptosis-related genes appear to be over-represented in our database (Supplementary Table 1). However, the mechanism we describe here suggests a provocative new function of the Alu elements that perhaps confers an evolutionary advantage. Finally, we have noticed that genes listed in GenBank with IRAlus in their 3′-UTRs generally include multiple cDNAs or ESTs that appear to be missing all or part of the inverted repeat structure. Rather than resulting from splicing, we suspect that these sequences reflect cloning artefacts, as the deletion junctions are always flanked by short repeated sequences and not consensus splicing signals (JN DeCerbo and GG Carmichael, unpublished).
Materials and methods
Plasmid construction and cell culture
The Alu and IRAlus elements from Nicn1, Lin28 and Apobec3G were PCR-amplified using specific 5′ and 3′ primers (Supplementary Table 2) and ligated into the expression vector pEGFP-C1 (BD Biosciences Clontech) at the BglII and HindIII sites. To construct pEGFP-IRAlus-ms2, two copies of the MS2 coat protein binding site were introduced into pEGFP-IRAlus-Apobec3G with HindIII and SalI. Primers are given in Supplementary Table 2. HEK293 cells were cultured in DMEM supplemented with 10% fetal bovine serum and transfection was performed by the standard calcium phosphate procedure.
Protein translation efficiency and RNA nuclear retention analysis
Nuclear RNA isolation was performed as described (Hwang et al, 2007) with some modifications. Briefly, cells growing in 10 cm dishes were rinsed twice with ice-cold PBS 44 h after transfection, harvested in 5 ml ice-cold PBS by scraping and centrifuged at 1000 r.p.m. for 5 min. Cell pellets were resuspended by gentle pipetting in 200 μl lysis buffer A (10 mM Tris (pH 8.0), 140 mM NaCl, 1.5 mM MgCl2, 0.5% Igepal, 2 mM vanadyl ribonucleoside complex (VRC; Invitrogen)) and incubated on ice for 5 min. During the incubation, one-tenth of the lysate was collected for immunoblots to quantify the total translated EGFP with either Alu or IRAlus in the 3′-UTR of egfp, one-fifth of the lysate was added to 1 ml Trizol for total RNA purification and used to quantify the total egfp-Alu or egfp-IRAlus mRNA by northern blotting, whereas the rest of the lysate was centrifuged at 1000 g for 3 min at 4°C to pellet the nuclei. To obtain pure nuclear RNA, the nuclear pellets were subjected to two additional washes with 160 μl lysis buffer A and were then resuspended in 100 μl lysis buffer A followed by extraction with Trizol. For northern blotting, Dig-labelled egfp and Dig-labelled U6 were made by the DIG-High Prime DNA Labeling and Detection Starter Kit (Roche); Dig-labelled antisense tRNAlys was made using T7 RNA polymerase with the DIG Northern Starter Kit (Roche), and the actin probe used in the experiment was provided with the kit. Equal amounts of total or nuclear RNA were loaded onto each well and northern blotting was carried out according to the manufacturer's manual. The exogenous EGFP translation efficiency of egfp-IRAlus or egfp-Alu was normalized to each endogenous actin translation efficiency of actin mRNA, and calculated using the following formula: translation efficiency of egfp-IRAlus or egfp-Alu=(EGFP (total protein/total RNA))/(actin (total protein/total RNA)). The ratio of nuclear-retained egfp-IRAlus or egfp-Alu to total RNA was also normalized to each nuclear and total actin mRNA and calculated using the following formula: nuclear-retained egfp-IRAlus or egfp-Alu=(egfp (nuclear RNA/total RNA))/[actin (nuclear RNA/total RNA)). For the northern blot analysis of endogenous Nicn1, total, nuclear and cytoplasmic RNAs from HEK293 cells were loaded on a denaturing gel. After electrophoresis and transfer, the membrane was first probed with a Dig-labelled probe 1 (Figure 7). The membrane was then stripped and re-probed with a Dig-labelled antisense RNA probe 2 (Figure 7). A further stripping was performed, followed by probing for actin. The hybridization and stripping were performed according to the manufacturer's protocol (DIG Northern Starter Kit, Roche).
Fluorescent in situ hybridization and confocal microscopy
To detect egfp-IRAlus RNA in situ, a Dig-labelled antisense egfp probe was made using T7 RNA polymerase (DIG Northern Starter Kit, Roche). For the colocalization studies, 24 h after transfection, HEK293 cells were replaced on glass bottom culture dishes (P35G-1.0-14-C, MatTek Corporation), and 20 h later, cells were rinsed briefly in PBS and fixed in 3.6% formaldehyde and 10% acetic acid in PBS for 20 min at room temperature and then permeabilized with 0.5% Triton X-100 and 2 mM VRC for 5 min at room temperature. After 3 × 10 min PBS washes, cells were precipitated with 70% ethanol at 4°C overnight. Hybridization was performed with the Dig-labelled antisense egfp probe in a moist chamber at 50°C for 16 h. The following detections were carried out with primary sheep anti-Dig antibody (1:500, Roche) and secondary Alexa555-conjugated donkey anti-sheep IgG (1:500, Invitrogen). EGFP was detected with Alexa488-conjugated anti-GFP antibody (1:400, Invitrogen). Endogenous p54nrb was detected with mouse anti-p54nrb (1:50, BD Biosciences) and co-transfected T7-p54nrb was detected with mouse anti-T7 (1:500, Novagen), followed by the secondary antibody Alexa488 anti-mouse IgG (1:500, Invitrogen). Images were taken with a Zeiss LSM 510 microscope. For statistical analyses of the Alu RNA or IRAlus RNA pattern, more than 200 transfected cells were counted randomly under the microscope after each indicated transfection.
RNA–protein complex IP and RT–PCR
Cells growing in 15 cm dishes were rinsed twice with ice-cold PBS 44 h after transfection, harvested in 10 ml ice-cold PBS by scraping and then centrifuged at 1000 r.p.m. for 5 min. Then, the cell pellets were resuspended in 1 ml lysis buffer B (50 mM Tris, pH 7.4, 150 mM NaCl, 0.05% Igepal, 1 mM PMSF, 1 mM aprotinin, 1 mM leupeptin and 2 mM VRC) and subjected to two rounds of gentle sonication. After the lysates were centrifuged at 12 000 r.p.m. for 15 min, the supernatants were precleared with protein A/G beads (Santa Cruz) in lysis buffer B with a supplement of 10 μg yeast tRNA (Sigma). Then, the precleared lysates were used for IP with either p54nrb or hnRNP C1/C2 (Santa Cruz) antibodies. IP was carried out for 3 h at 4°C. The beads were washed five times with the same lysis buffer B, followed by extraction with buffer C (100 mM Tris, pH 6.8, 4% SDS, 12% β-mercaptoethanol and 20% glycerol) at room temperature for 10 min. One-third of the IP material was used for immunoblotting and the other two-thirds was used for RNA extraction with Trizol. For RT–PCR, each RNA sample was treated with DNase I (Ambion, DNA-free™ kit) and then reverse transcription was performed with random hexamers (SuperScript II, Invitrogen). The resulting material was used for PCR amplification using egfp- or Nicn1-specific primer pairs. To amplify the full-length IRAluSps-ms2, stringent conditions were used to denature the DNase I-treated total, p54nrb-immunoprecipitated and mock-immunoprecipitated RNAs along with the ms2-specific primers at 80°C for 5 min, and then the first strand cDNA was synthesized at 55°C for 50 min according to the manufacturer's instructions (SuperScript™ III, Invitrogen). The resulting material was used for PCR using IRAluSps-ms2-specific primers.
RNA editing analysis
HEK293 cells were transfected with pEGFP-IRAlus-Nicn1. Total, nuclear and cytoplasmic RNAs were isolated 44 h after transfection. After treatment with DNase I (Ambion, DNA-free™ kit), the egfp-AluSps first strand was reverse transcribed with Thermo-Script (Invitrogen) with a gene-specific primer located downstream of the IRAluSps on the pEGFP-C1 vector at 55°C for 50 min. The resultant cDNA was then amplified by PCR with Taq DNA polymerase (Invitrogen) and PCR products were subcloned using the TOPO TA cloning kit (Invitrogen). The editing frequency was determined by sequencing more than 100 individual clones containing the appropriately sized inserts (Agencourt).
Supplementary Material
Supplementary Figures 1–11
Supplementary Figure Legends
Supplementary Table 1
Supplementary Table 2
Acknowledgments
We thank H Lu, K Prasanth and T Vedakumar for advice with microscopy and in situ hybridization, A Gabriel for useful discussions before initiating the current project, K Morris, A Das, J Zhou and D Moschenross for helpful comments on the manuscript and Li Yang for useful advice throughout the project. This work was supported by grants GM066816 and CA04382 from the NIH and from the State of CT Stem Cell Initiative.
References
- Ambros V, Horvitz HR (1984) Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226: 409–416 [DOI] [PubMed] [Google Scholar]
- Athanasiadis A, Rich A, Maas S (2004) Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol 2: e391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backofen B, Jacob R, Serth K, Gossler A, Naim HY, Leeb T (2002) Cloning and characterization of the mammalian-specific nicolin 1 gene (NICN1) encoding a nuclear 24 kDa protein. Eur J Biochem/FEBS 269: 5240–5245 [DOI] [PubMed] [Google Scholar]
- Balzer E, Moss EG (2007) Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA Biol 4: 16–25 [DOI] [PubMed] [Google Scholar]
- Bass BL (2002) RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem 71: 817–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass BL, Weintraub H (1987) A developmentally regulated activity that unwinds RNA duplexes. Cell 48: 607–613 [DOI] [PubMed] [Google Scholar]
- Bass BL, Weintraub H (1988) An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 55: 1089–1098 [DOI] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL (2002) Alu repeats and human genomic diversity. Nat Rev 3: 370–379 [DOI] [PubMed] [Google Scholar]
- Bladen CL, Udayakumar D, Takeda Y, Dynan WS (2005) Identification of the PSF–p54(nrb) complex as a candidate DNA double-strand break rejoining factor. J Biol Chem 280: 5205–5210 [DOI] [PubMed] [Google Scholar]
- Blow M, Futreal PA, Wooster R, Stratton MR (2004) A survey of RNA editing in human brain. Genome Res 14: 2379–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J (1999) RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene 238: 115–134 [DOI] [PubMed] [Google Scholar]
- DeCerbo J, Carmichael GG (2005) SINES point to abundant human editing. Genome Biol 6: 216–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger PL, Batzer MA (1999) Alu repeats and human disease. Mol Genet Metab 67: 183–193 [DOI] [PubMed] [Google Scholar]
- Eisenberg E, Nemzer S, Kinar Y, Sorek R, Rechavi G, Levanon EY (2005) Is abundant A-to-I RNA editing primate-specific? Trends Genet 21: 77–81 [DOI] [PubMed] [Google Scholar]
- Fox AH, Lam YW, Leung AK, Lyon CE, Andersen J, Mann M, Lamond AI (2002) Paraspeckles. A novel nuclear domain. Curr Biol 12: 13–25 [DOI] [PubMed] [Google Scholar]
- George CX, Samuel CE (1999) Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc Natl Acad Sci USA 96: 4621–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George CX, Wagner MV, Samuel CE (2005) Expression of interferon-inducible RNA adenosine deaminase ADAR1 during pathogen infection and mouse embryo development involves tissue-selective promoter utilization and alternative splicing. J Biol Chem 280: 15020–15028 [DOI] [PubMed] [Google Scholar]
- Hasler J, Samuelsson T, Strub K (2007) Useful ‘junk': Alu RNAs in the human transcriptome. Cell Mol Life Sci 64: 1793–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasler J, Strub K (2006) Alu elements as regulators of gene expression. Nucleic Acids Res 34: 5491–5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvitz HR, Sternberg PW, Greenwald IS, Fixsen W, Ellis HM (1983) Mutations that affect neural cell lineages and cell fates during the development of the nematode Caenorhabditis elegans. Cold Spring Harbor Symp Quant Biol 48 (Part 2): 453–463 [DOI] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, Mendell JT (2007) A hexanucleotide element directs microRNA nuclear import. Science 315: 97–100 [DOI] [PubMed] [Google Scholar]
- Ishitani K, Yoshida T, Kitagawa H, Ohta H, Nozawa S, Kato S (2003) p54(nrb) acts as a transcriptional coactivator for activation function 1 of the human androgen receptor. Biochem Biophys Res Commun 306: 660–665 [DOI] [PubMed] [Google Scholar]
- Kameoka S, Duque P, Konarska MM (2004) p54(nrb) associates with the 5′ splice site within large transcription/splicing complexes. EMBO J 23: 1782–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Rozenblatt-Rosen O, Meyerson M, Manley JL (2007) The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination. Genes Dev 21: 1779–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karhumaa P, Parkkila S, Waheed A, Parkkila AK, Kaunisto K, Tucker PW, Huang CJ, Sly WS, Rajaniemi H (2000) Nuclear NonO/p54nrb protein is a nonclassical carbonic anhydrase. J Biol Chem 275: 16044–16049 [DOI] [PubMed] [Google Scholar]
- Kawahara Y, Nishikura K (2006) Extensive adenosine-to-inosine editing detected in Alu repeats of antisense RNAs reveals scarcity of sense–antisense duplex formation. FEBS Lett 580: 2301–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A (2004) Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res 14: 1719–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Lee JY, Yeh I, Park JY, Tian B (2007) PolyA_DB 2: mRNA polyadenylation sites in vertebrate genes. Nucleic Acids Res 35: D165–D168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev-Manor G, Sorek R, Shomron N, Ast G (2003) The birth of an alternatively spliced exon: 3′-splice site selection in Alu exons. Science 300: 1288–1291 [DOI] [PubMed] [Google Scholar]
- Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, Olshansky M, Rechavi G, Jantsch MF (2004) Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol 22: 1001–1005 [DOI] [PubMed] [Google Scholar]
- Liu Y, George CX, Patterson JB, Samuel CE (1997) Functionally distinct double-stranded RNA-binding domains associated with alternative splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase. J Biol Chem 272: 4419–4428 [DOI] [PubMed] [Google Scholar]
- Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, Crabbe JC, Blednov YA, Grahame NJ, Phillips TJ, Finn DA, Hoffman PL, Iyer VR, Koob GF, Bergeson SE (2006) Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc Natl Acad Sci USA 103: 6368–6373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K (1992) Modulation of double-stranded RNAs in vivo by RNA duplex unwindase. Ann NY Acad Sci 660: 240–250 [DOI] [PubMed] [Google Scholar]
- Peng R, Dye BT, Perez I, Barnard DC, Thompson AB, Patton JG (2002) PSF and p54nrb bind a conserved stem in U5 snRNA. RNA 8: 1334–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL (2005) Regulating gene expression through RNA nuclear retention. Cell 123: 249–263 [DOI] [PubMed] [Google Scholar]
- Price AL, Eskin E, Pevzner PA (2004) Whole-genome analysis of Alu repeat elements reveals complex evolutionary history. Genome Res 14: 2245–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovici R, Kabir K, Chen M, Su Y, Zhang D, Luo X, Yang JH (2001) ADAR1 is involved in the development of microvascular lung injury. Circ Res 88: 1066–1071 [DOI] [PubMed] [Google Scholar]
- Richards M, Tan SP, Tan JH, Chan WK, Bongso A (2004) The transcriptome profile of human embryonic stem cells as defined by SAGE. Stem Cells (Dayton, Ohio) 22: 51–64 [DOI] [PubMed] [Google Scholar]
- Scadden AD (2005) The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat Struct Mol Biol 12: 489–496 [DOI] [PubMed] [Google Scholar]
- Smalheiser NR, Torvik VI (2006) Alu elements within human mRNAs are probable microRNA targets. Trends Genet 22: 532–536 [DOI] [PubMed] [Google Scholar]
- Straub T, Knudsen BR, Boege F (2000) PSF/p54(nrb) stimulates ‘jumping' of DNA topoisomerase I between separate DNA helices. Biochemistry 39: 7552–7558 [DOI] [PubMed] [Google Scholar]
- Versteeg R, van Schaik BD, van Batenburg MF, Roos M, Monajemi R, Caron H, Bussemaker HJ, van Kampen AH (2003) The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes. Genome Res 13: 1998–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI (2008) Selective blockade of microRNA processing by Lin-28. Science 320: 97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JH, Luo X, Nie Y, Su Y, Zhao Q, Kabir K, Zhang D, Rabinovici R (2003) Widespread inosine-containing mRNA in lymphocytes regulated by ADAR1 in response to inflammation. Immunology 109: 15–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R et al. (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318: 1917–1920 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Carmichael GG (2001) The fate of dsRNA in the nucleus. A p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell 106: 465–475 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1–11
Supplementary Figure Legends
Supplementary Table 1
Supplementary Table 2