Abstract
NADH:ubiquinone oxidoreductase (complex I) of the mitochondrial inner membrane is a multi-subunit protein complex containing eight iron–sulphur (Fe–S) clusters. Little is known about the assembly of complex I and its Fe–S clusters. Here, we report the identification of a mitochondrial protein with a nucleotide-binding domain, named Ind1, that is required specifically for the effective assembly of complex I. Deletion of the IND1 open reading frame in the yeast Yarrowia lipolytica carrying an internal alternative NADH dehydrogenase resulted in slower growth and strongly decreased complex I activity, whereas the activities of other mitochondrial Fe–S enzymes, including aconitase and succinate dehydrogenase, were not affected. Two-dimensional gel electrophoresis, in vitro activity tests and electron paramagnetic resonance signals of Fe–S clusters showed that only a minor fraction (∼20%) of complex I was assembled in the ind1 deletion mutant. Using in vivo and in vitro approaches, we found that Ind1 can bind a [4Fe–4S] cluster that was readily transferred to an acceptor Fe–S protein. Our data suggest that Ind1 facilitates the assembly of Fe–S cofactors and subunits of complex I.
Keywords: metal cofactor, mitochondria, myopathy, NTPase, oxidoreductase
Introduction
Iron–sulphur (Fe–S) clusters are ubiquitous and versatile cofactors of proteins mediating electron transfer, enzymatic catalysis and regulation of gene expression (Beinert, 2000). The most commonly found clusters are the rhombic [2Fe–2S] and the cubane [4Fe–4S] clusters. Despite the simple structure of Fe–S cofactors and spontaneous assembly of Fe–S proteins by chemical means, in living cells Fe–S protein maturation is an enzymatic process. Many of the genes involved have been identified over the past decade, and are markedly conserved from bacteria to higher eukaryotes (for reviews, see Balk and Lobréaux, 2005; Johnson et al, 2005; Lill and Mühlenhoff, 2008). At least three Fe–S cluster assembly systems can be distinguished. The NIF system is specialized for the maturation of nitrogenase in nitrogen-fixing bacteria. A second system called ISC (for iron–sulphur cluster assembly) is present in most bacteria, and is conserved in mitochondria. A third system termed SUF mediates Fe–S cluster assembly under iron-limiting and oxidative stress conditions. Each system, encoded by operons or gene clusters in bacteria but dispersed genes in eukaryotic genomes, consists of a cysteine desulphurase for the generation of sulphane sulphur (Nfs1 in Baker's yeast, Saccharomyces cerevisiae); a scaffold protein to which the sulphur is transferred and assembled with iron (Isu1 and Isu2 in S. cerevisiae mitochondria); and auxiliary proteins, the functions of which are less well understood, including frataxin that is thought to facilitate iron delivery.
The mitochondrial ISC machinery is also required for the assembly of Fe–S proteins in the cytosol and nucleus (Kispal et al, 1999; Lill and Mühlenhoff, 2008). An as yet unknown compound is exported through the ATP-binding cassette transporter of the mitochondria, Atm1, to assist iron–sulphur protein assembly in the cytosol and nucleus. The first known cytosolic Fe–S cluster assembly protein was the essential P-loop NTPase Cfd1, which was identified in a mutant screen in Baker's yeast (Roy et al, 2003). Two highly conserved cysteine residues in a CxxC motif (Cys201 and Cys204) were found to be important for its function. Cfd1 forms a stable complex with the related Nbp35 protein (Hausmann et al, 2005), and together they bind up to three [4Fe–4S] clusters. Rapid transfer of the labile Fe–S clusters to target Fe–S proteins in vitro and the requirement of Cfd1–Nbp35 for Fe–S cluster assembly in vivo demonstrated that the Cfd1–Nbp35 complex can function as a novel Fe–S scaffold in the yeast cytosol (Netz et al, 2007).
Cfd1 and Nbp35 are classified as P-loop GTPases based on a unique set of sequence and structural signatures (Leipe et al, 2002). The small subfamily of Mrp/NBP35-like proteins can be further divided into five groups based on sequence similarities and conserved domains. Nbp35 has a highly conserved N terminus that binds an Fe–S cluster (Hausmann et al, 2005), whereas Cfd1 lacks this domain (Figure 1A and Supplementary Figure S1). A third group of Mrp/NBP35-like proteins was identified in the model plant Arabidopsis in screens for photosynthesis mutants displaying high chlorophyll fluorescence. hcf101 mutant alleles failed to accumulate photosystem I, which has three [4Fe–4S] clusters in its reaction centre (Lezhneva et al, 2004; Stöckel and Oelmüller, 2004). The mutants also had decreased levels of the [4Fe–4S] protein ferredoxin–thioredoxin reductase, suggesting a specific role of HCF101 in the assembly of Fe–S proteins (Lezhneva et al, 2004). The HCF101 group has a conserved N-terminal domain of unknown function, designated DUF59. The fourth group of Mrp/NBP35-like proteins has an N-terminal sequence that is predicted to target the protein to mitochondria. Although this protein is not found in Baker's yeast, it is present in most eukaryotes, including plants, mammals and fungi. The fifth group of Mrp/NBP35-like sequences is found in bacteria. Whereas the Escherichia coli Mrp (MetG-related protein) lends its name to the subfamily, only the ApbC orthologue in Salmonella enterica has been studied experimentally. Initially identified as a step in the alternative pyrimidine biosynthetic pathway (Petersen and Downs, 1996), the ApbC protein was later implicated in Fe–S cluster assembly. Deletion of apbC affected aconitase and succinate dehydrogenase activities by 30–35%, and this decrease was additive with mutations in the alternative Fe–S scaffold protein iscA, suggesting that the gene products function in the same biochemical pathway (Skovran and Downs, 2003).
Figure 1.
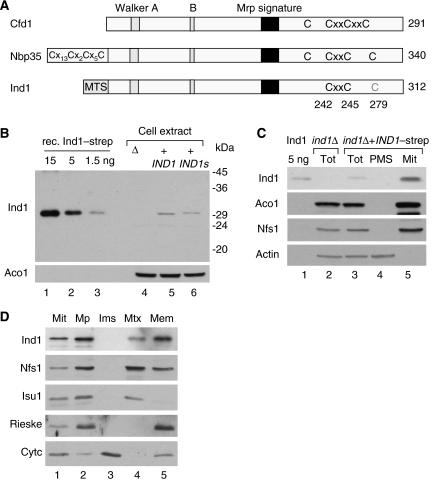
Ind1 is targeted to mitochondria. (A) Cartoon of Mrp/NBP35-like proteins found in Yarrowia lipolytica. Cfd1, YALI0E19074g; Nbp35, YALI0E02354g; Ind1, YALI0B18590g. The proteins are 40% similar in amino-acid sequence but differ in their N termini. MTS, mitochondrial targeting sequence. Conserved cysteine motifs are indicated in black. Cys279 in Ind1 is drawn in grey, as it is not conserved in the mitochondrial group. (B) Polyclonal antibodies raised against recombinant Ind1–strep recognized a 30–31 kDa protein in cell extracts (20 μg protein per lane) of Y. lipolytica expressing full-length IND1 or IND1–strep (IND1s) under the control of its own promoter from plasmid pUB4. (C) Immunoblot showing the mitochondrial localization of Ind1. Y. lipolytica cells expressing IND1–strep were treated with zymolyase, broken in a Dounce homogenizer (Tot=total lysate) and fractionated in mitochondria (Mit) and post-mitochondrial supernatant (PMS). Protein (20 μg) was separated by SDS–PAGE, blotted and labelled with antibodies against Ind1, the mitochondrial proteins aconitase (Aco1) or cysteine desulphurase (Nfs1) or cytosolic actin. (D) Immunodetection of Ind1 in the mitochondrial matrix, associated with membranes. Mitochondria (50 μg protein) of Y. lipolytica expressing IND1–strep were incubated in hypotonic buffer to swell the organelles and break the outer membrane, followed by 10 min incubation and centrifugation in the presence of 150 mM KCl to separate soluble and membrane-bound proteins of the intermembrane space (Ims) from mitoplasts (Mp). The pellet (Mp) was resuspended in hypotonic buffer and subjected to three rounds of freeze–thawing, followed by centrifugation to separate soluble matrix proteins (Mtx) and membranes (Mem). The volume of each mitochondrial fraction was adjusted to 50 μl in 1 × gel loading buffer, and 20 μl of each fraction was analysed by SDS–PAGE and immunoblotting to visualize Ind1, Nfs1, the Fe–S scaffold protein Isu1 (soluble matrix protein), Rieske Fe–S protein (integral membrane protein of complex III) and cytochrome c (Cytc; protein of the intermembrane space). Note that the separation of cytochrome c from the mitoplasts was incomplete in this experiment. Similar results were obtained for cells expressing Ind1 without Strep tag (not shown).
Although S. cerevisiae has proved to be an excellent model organism for unravelling the biochemical pathways of Fe–S protein maturation in the mitochondria and cytosol, it is lacking respiratory complex I (proton pumping NADH:ubiquinone oxidoreductase), a multi-subunit Fe–S-containing protein complex. Complex I contains 8 Fe–S clusters in mitochondria and 8–9 Fe–S clusters in bacteria. From X-ray crystallographic studies of the peripheral arm of complex I of the bacterium Thermus thermophilus, it was deduced that seven clusters form an ‘electrical wire' transferring electrons from the substrate NADH, through FMN, to ubiquinone (Hinchliffe and Sazanov, 2005). Little is known about the assembly of complex I or the insertion of its Fe–S clusters. Only five assembly factors of complex I have been identified to date, of which three, CIA30, CIA84 and B12.7L, are conserved from fungi to man (Küffner et al, 1998; Ogilvie et al, 2005; Vogel et al, 2005, 2007; Dunning et al, 2007; Saada et al, 2008). These proteins have been classified as molecular chaperones, as they assist in the protein folding and/or assembly of intermediates but are not part of the fully assembled complex. Furthermore, it was shown that a functional sulphurtransferase associated with complex I in the yeast Yarrowia lipolytica was not required for Fe–S cluster assembly on complex I (Abdrakhmanova et al, 2006). It is therefore likely that the cysteine desulphurase Nfs1 and other ISC assembly components are involved in Fe–S insertion into complex I. In support of this, two recent reports described patients with strongly decreased levels of the mitochondrial scaffold protein ISCU who displayed a general deficiency in mitochondrial Fe–S proteins. The biochemical symptoms included a decrease in the activity and subunit abundance of complex I, although this effect was minor compared with the strongly affected succinate dehydrogenase and aconitase enzymes (Mochel et al, 2008; Olsson et al, 2008 and references therein). Also, frataxin may be involved in providing iron to complex I, as deletion of the bacterial frataxin homologue cyaY resulted in 30% less of the protein complex (Pohl et al, 2007).
The presence of a putative, mitochondrial Mrp/NBP35-like protein in eukaryotes with the exception of Baker's yeast led us to investigate the function of this protein in the yeast Y. lipolytica, with particular attention to complex I. Here, we describe a deletion mutant of the gene called IND1 that lacked approximately 80% of complex I activity due to a similar decrease in the abundance of complex I. Survival of the mutant was dependent on expression of an alternative NADH dehydrogenase transgene. Moreover, aconitase and succinate dehydrogenase activities were not affected, indicating that Ind1 is specifically involved in the assembly of complex I. Ind1 has a CxxC motif that is essential for its function. Similar to the cytosolic scaffold proteins Cfd1–Nbp35, Ind1 bound an Fe–S cluster that could be transferred to an Fe–S apo-protein, suggesting that Ind1 performs a specific role in assembling Fe–S clusters in complex I.
Results
Y. lipolytica expresses an Mrp/NBP35-like protein targeted to mitochondria
The genome of the obligate respiratory yeast Y. lipolytica encodes three P-loop NTPases classified as Mrp/NBP35-like proteins (Leipe et al, 2002). Two of these, YALI0E02354 and YALI0E19074, bear high sequence similarity and domain organization to the S. cerevisiae proteins Nbp35 and Cfd1, respectively (Supplementary Figure S2). The third P-loop NTPase, YALI0B18590, carries the Mrp family signature and the conserved cysteine motif (CxxC) found in Nbp35 and Cfd1 (Figure 1A and Supplementary Figure S2). In addition, YALI0B18590, which we have named IND1 for iron–sulphur protein required for NADH-dehydrogenase, encodes a protein with an N terminus comprising a putative mitochondrial targeting signal. To investigate expression of IND1, antibodies were raised against the recombinant Ind1 protein and applied to immunoblots of total extracts of Y. lipolytica cells expressing full-length IND1, with or without a C-terminal Strep tag, under the control of its own promoter. A protein of 30–31 kDa was detected in cells expressing IND1(–strep) (Figure 1B, lanes 5 and 6), which is smaller than expected from the full-length protein (34.365 kDa including Strep tag). Affinity purification of Ind1–strep from Y. lipolytica cell extract and N-terminal sequencing gave the sequence SxE(N), corresponding with cleavage after Arg34, thus giving a calculated molecular mass of 30.550 kDa including Strep tag. These results are consistent with the cleavage site predicted by TargetP (Emanuelsson et al, 2000), and with the observed molecular mass.
To determine the subcellular localization of Ind1, cells were fractionated in mitochondria and post-mitochondrial supernatant. Equal amounts of protein were separated by SDS–PAGE and subjected to immunoblotting. Ind1 was detected in the mitochondrial fraction but not in the post-mitochondrial supernatant of cells expressing Ind1–strep (Figure 1C, lanes 3–5). The purity of the mitochondrial fraction was confirmed by probing the same nitrocellulose membrane for the mitochondrial proteins aconitase (Aco1) and cysteine desulphurase (Nfs1), and for cytosolic actin. To establish in which mitochondrial subcompartment Ind1 is localized, mitochondria were further fractionated into mitoplasts, intermembrane space, matrix and membranes. Comparison with marker proteins showed that Ind1 is located in the matrix and is mostly membrane bound (Figure 1D).
Taken together, these data show that the predicted N-terminal targeting sequence of Ind1 is cleaved and that the mature protein is localized to the mitochondrial matrix with a large fraction bound to the inner membrane.
IND1 is implicated with complex I
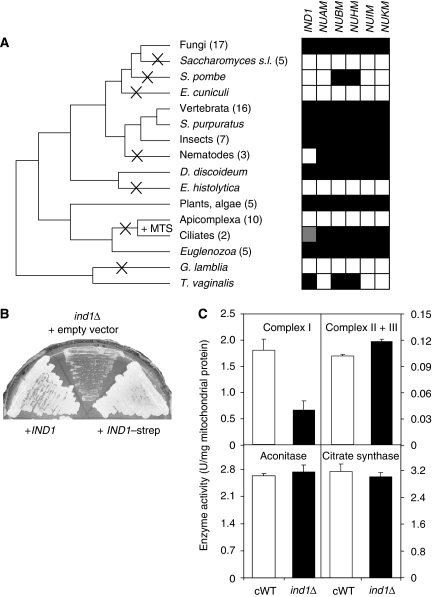
What is the function of Ind1? Because of sequence similarity to Nbp35, Cfd1 and the bacterial Mrp/ApbC, Ind1 may have a role in Fe–S protein assembly. Moreover, the absence of Ind1 in S. cerevisiae suggested a specific mitochondrial target Fe–S protein that is not present in S. cerevisiae: complex I (NADH:ubiquinone oxidoreductase). We therefore examined the presence of IND1 and complex I genes among 77 published eukaryotic genomes, using PSI-Blast searches to find homologues and phylogenetic analyses to determine orthology relations (Gabaldón et al, 2005). There is a strong genomic link between IND1 and complex I genes, as they occur together in 53 genomes, and are both absent from 18 genomes. Furthermore, assuming a standard eukaryotic phylogeny, complex I genes and IND1 have clearly co-evolved, as they have been lost concertedly five times in evolution, whereas in Schizosaccharomyces pombe IND1 has been lost with a subset of complex I genes (Figure 2A). This indicates a functional link between IND1 and complex I. Nevertheless, there are a few exceptions to the pattern: two ciliates appear to have replaced IND1 with an IND1 paralogue that has gained a mitochondrial targeting signal (Figure 2A and Supplementary Figures S1 and S4). Furthermore, the nematodes have a complete set of complex I genes without IND1, and Trichomonas vaginalis has an IND1 with the same subset of complex I genes as S. pombe. The reverse, a genome that has IND1 without having any complex I genes was not found, consistent with the hypothesis that Ind1 is required for the formation of complex I.
Figure 2.
IND1 is phylogenetically and functionally linked to complex I. (A) Coevolution of IND1 and genes for Fe–S protein subunits of complex I. A cross indicates the loss of IND1 in that lineage, and +MTS indicates the gain of a mitochondrial targeting signal in an IND1 paralogue. White and black squares represent the absence or presence, respectively, of the gene indicated above each column. Grey indicates the presence of an IND1 paralogue with a mitochondrial targeting signal. Abbreviated species names: Saccharomyces s.l. Saccharomyces sensu lato; S. (Schizosaccharomyces) pombe; E. (Encephalitozoon) cuniculi; S. (Strongylocentrotus) purpuratus; D. (Dictyostelium) discoideum; E. (Entamoeba) histolytica; G. (Giardia) lamblia; T. (Trichomonas) vaginalis. (B) Growth of Y. lipolytica strain ind1Δ in which the IND1 gene was deleted (centre, transformed with an empty plasmid) and the ind1Δ strain expressing the wild-type IND1 gene (left), or IND1 fused to the Strep tag coding sequence (right), expressed under the control of the IND1 promoter from the pUB4 plasmid. (C) Activities of Fe–S enzymes in purified mitochondria from ind1Δ cells (black bars) or the IND1-complemented wild type (cWT, white bars). Complementation with IND1 or IND1–strep gave similar results (not shown). Complex I was measured as NADH:HAR oxidoreductase activity in alamethicin-permeabilized mitochondria. Complex II plus III activity was measured following the electron transfer from succinate to cytochrome c in intact mitochondria. Aconitase activity was assayed following cis-aconitate consumption. Citrate synthase activity served as a non-Fe–S enzyme control. Error bars represent the standard deviation, n=3.
We then investigated whether Ind1 is functionally required for complex I activity. A deletion mutant was obtained in Y. lipolytica by replacing the entire coding sequence of IND1 with a URA3 marker using homologous recombination (Supplementary Figure S3). The haploid strain used contains the Y. lipolytica NDH2 transgene fused to a mitochondrial targeting sequence (NDH2i), resulting in the expression of a version of the alternative NADH dehydrogenase on the matrix side of the inner mitochondrial membrane, bypassing the absolute requirement for respiratory complex I (Kerscher et al, 2001). The absence of IND1 and its expression in the deletion mutant was confirmed by PCR (Supplementary Figure S3) and immunoblotting (Figure 1B). The resulting ind1Δ (NDH2i) strain (in the following abbreviated as ind1Δ) was viable, but grew slowly. Growth of the ind1Δ strain was restored following complementation with a plasmid expressing the full-length IND1 gene, with or without Strep tag, under the control of its own promoter (Figure 2B). Backcrossing the ind1Δ strain with a strain lacking NDH2i resulted in spores that had either NDH2i, or IND1, or both genes (Supplementary Figure S5). These data indicated that IND1 is essential for viability and is required for NADH oxidation in the mitochondrial matrix.
Next, we assayed the activities of several key Fe–S enzymes in mitochondria purified from ind1Δ cells. No significant differences were found between ind1Δ and the complemented wild-type strain (cWT) for the enzyme activities of aconitase, succinate dehydrogenase (complex II) and cytochrome bc1 complex (complex III), or the non-Fe–S enzyme citrate synthase (Figure 2C). In contrast, electron transfer from NADH to the artificial electron acceptor hexaammineruthenium(III) chloride (HAR), known to accept electrons from the 51-kDa NUBM subunit of complex I, was significantly decreased in the ind1Δ mutant. These results indicate that Ind1 is required for normal function specifically of complex I, as suggested by the co-evolution of Ind1 and complex I genes in eukaryotes.
Cys242 and Cys245 are essential for the function of Ind1
To analyse NADH oxidation by complex I in more detail, we isolated unsealed mitochondrial membranes and measured the electron transfer rate from NADH to HAR, as well as electron transfer from deamino-NADH (dNADH) to n-decylubiquinone (DBQ) that is sensitive to the complex-I-specific inhibitor 2-decyl-4-quinazolinyl amine (DQA). Whereas the NADH:HAR activity involves only the primary electron acceptor FMN bound to the 51-kDa subunit, the DQA-sensitive dNADH:DBQ oxidoreductase activity involves all cofactors of the peripheral arm, therefore reflecting the physiological NADH:ubiquinone oxidoreductase activity of complex I. Comparison of the two activities has been useful in the past to identify impaired electron transport due to mutations in the ubiquinone-binding pocket (Tocilescu et al, 2007). We found that both the NADH:HAR and the DQA-sensitive dNADH:DBQ activity of complex I were decreased by approximately 70% in mitochondrial membranes isolated from cells lacking Ind1 (Table I).
Table 1.
Complex I activities in mitochondrial membranes from Y. lipolytica IND1 mutants
| Strain | NADH:HAR activity | DQA-sensitive dNADH:DBQ activity | ||
|---|---|---|---|---|
| U mg−1 | % of wild type | U mg−1 | % of wild type | |
| ind1Δ+pUB4 | 0.43±0.01 | 28 | 0.23±0.01 | 28 |
| ind1Δ+pUB4-IND1 | 1.56±0.05 | 100 | 0.83±0.02 | 100 |
| ind1Δ+pUB4-IND1–strep | 1.47±0.08 | 94 | 0.82±0.01 | 99 |
| C242A | 0.39±0.02 | 25 | 0.19±0.01 | 23 |
| C242E | 0.32±0.02 | 21 | 0.17±0.01 | 20 |
| C242S | 0.47±0.03 | 30 | 0.21±0.01 | 25 |
| C245A | 0.45±0.02 | 29 | 0.23±0.01 | 28 |
| C245E | 0.31±0.02 | 20 | 0.15±0.01 | 18 |
| C245S | 0.32±0.02 | 21 | 0.17±0.01 | 20 |
| C279A | 1.25±0.06 | 80 | 0.67±0.02 | 81 |
| C279E | 0.25±0.01 | 16 | 0.12±0.01 | 14 |
| C279S | 1.50±0.07 | 96 | 0.83±0.02 | 100 |
| Activities are given as mean±standard deviation (n=5). | ||||
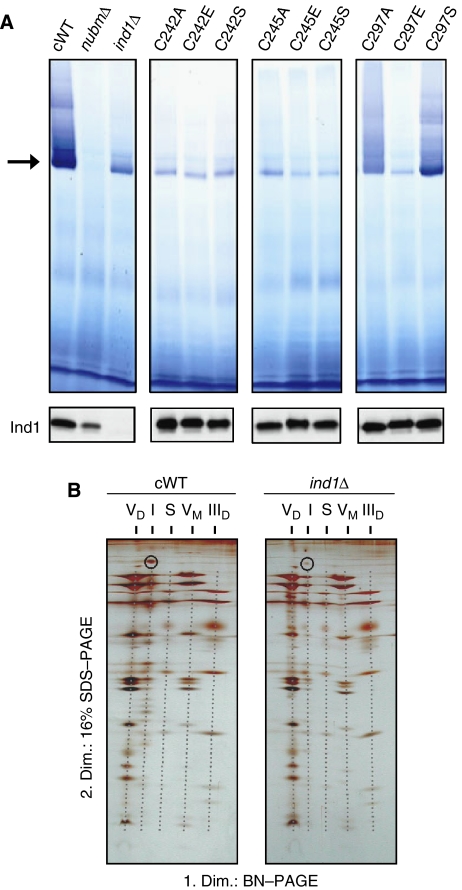
We then investigated the importance of the CxxC motif in Ind1 (Figure 1A) using site-directed mutagenesis of Cys242 and Cys245 to alanine, glutamate or serine. All single amino-acid changes resulted in a decrease in NADH:HAR and DQA-dependent dNADH:DBQ activity (Table I). The magnitude of the decrease was similar to that observed in the deletion mutant, demonstrating that the CxxC motif is essential for the function of Ind1. In contrast, mutations of Cys279, which is not evolutionary conserved (Supplementary Figure S4), to Ala or Ser had no effect on complex I activity. Changing Cys279 to Glu did abolish most of complex I activity, possibly because this change affected protein folding. Immunoblot analysis showed that all mutant Ind1 proteins were stably expressed (Figure 3A, lower panels).
Figure 3.
Deletion of IND1 leads to a major decrease in fully assembled complex I. (A) In-gel staining of complex I in mitochondrial membranes (upper panels) and immunostaining of Ind1 protein (lower panels). Mitochondrial membrane proteins were separated by BN–PAGE and incubated with NADH and NBT to visualize NADH dehydrogenase activity. Monomeric complex I (with a molecular mass of 947 kDa) is marked by an arrow. Activity-stained bands of higher molecular mass represent supercomplexes containing complex I. cWT (ind1Δ+pUB4-IND1); nubmΔ, a strain lacking the gene encoding the 51-kDa NUBM subunit of complex I; ind1Δ carrying the pUB4 plasmid; C242A–C279S, point mutations that exchange cysteines at positions 242, 245 or 279 in pUB4-IND1. (B) Two-dimensional (BN/SDS) PAGE analysis of mitochondrial membranes from complemented wild-type (cWT) and ind1Δ cells, stained with silver. The following multiprotein assemblies are indicated by dashed grey lines: VD and VM, dimeric and monomeric forms of complex V; I, complex I; S, an incompletely characterized supercomplex that contains complex III, IIID, dimeric form of complex III. The 75-kDa NUAM subunit of complex I is circled.
We concluded from these enzyme assays that the function of Ind1 is required for full activity of complex I and that this function strictly depends on the cysteine residues of the conserved CxxC motif.
ind1 mutants have minor amounts of fully assembled complex I
The parallel decrease in NADH:HAR and dNADH:DBQ activities in the ind1Δ strain is indicative of a decrease in the amount, rather than a dysfunction, of complex I. To investigate this further, we analysed the amount and integrity of the 40-subunit protein complex using blue-native polyacrylamide gel electrophoresis (BN–PAGE) in combination with SDS–PAGE. First, mitochondrial membranes were subjected to BN–PAGE to separate the intact respiratory complexes, and stained using NADH and nitroblue tetrazolium (NBT) as a complex I-specific stain. The intensity of the major activity associated with monomeric, fully assembled complex I (Figure 3A, arrow) was strongly decreased in membranes from the ind1Δ strain and the C242/C245 cysteine mutants compared with the complemented wild type (cWT). However, a fraction of intact, active complex I was still present in the ind1 mutants, in contrast to the mutant lacking the 51-kDa subunit (nubmΔ), which did not have any complex I staining at all. In the IND1 mutants, no novel activities of smaller molecular mass were detected, indicating that subcomplexes containing a functional NBT reduction site (within the 51-kDa subunit) were absent.
Second, the assembly and subunit composition of complex I and other respiratory complexes were investigated by separating the individual subunits in the second dimension using SDS–PAGE and silver staining. This revealed that the abundance of all complex I subunits was strongly diminished in the ind1Δ mutant (Figure 3B). None of the other respiratory complexes were affected, in accordance with normal activities of complexes II and III (Figure 2C). Again, no subcomplexes of complex I were found. Two-dimensional gel electrophoresis of mitochondrial membranes of the C242 and C245 mutants showed similar results as for the deletion mutant (not shown). Semi-quantitative analysis of the complex I stain and the silver-stained polypeptides showed that the amount of fully assembled complex I was decreased by approximately 80% in agreement with the activity measurements (Table I).
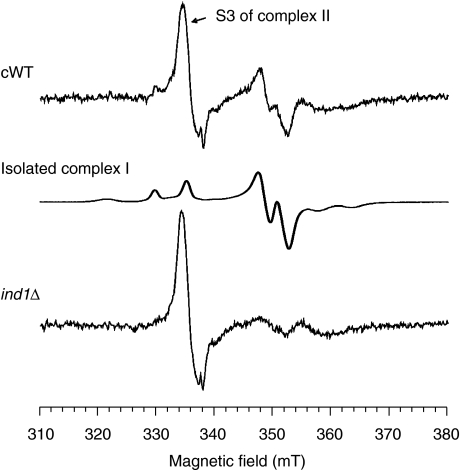
The low levels of fully assembled complex I in the ind1Δ mutant strain could be confirmed using electron paramagnetic resonance (EPR) spectroscopy (Figure 4). The typical EPR signals of the Fe–S clusters of complex I, mainly appearing in the region of 350 mT (g∼1.93) and the characteristic gz component of the cluster N2 spectrum at 330 mT (g∼2.05), were largely absent in the ind1Δ mutant strain. On the other hand, the prominent EPR signal of the S3 [3Fe–4S]1+ cluster of complex II was unaffected. By comparing cluster N2 signal intensities of the two samples, complex I content in the mutant was estimated to be ∼20% of the WT sample. A similar result was obtained by comparison of cluster N1 signal intensities at 40 K, conditions that allow the detection of [2Fe–2S] clusters only (data not shown). Addition of the stronger reductant sodium dithionite clearly diminished the S3 signal intensity in both samples due to reduction of S3 to the [3Fe–4S]0 state, which is EPR silent. In contrast, complex-I EPR signals did not alter in the ind1Δ mutant sample, indicating that there was no shift in redox midpoint potentials that prevented complete reduction of the Fe–S clusters by NADH (data not shown).
Figure 4.
Deletion of IND1 results in significantly diminished EPR signals of complex I. EPR spectra of mitochondrial membranes from complemented wild type (cWT, top) and ind1Δ cells (bottom). A spectrum of isolated complex I is shown for comparison (middle). Samples were treated with 2 mM NADH, frozen and EPR spectra were recorded at 12 K. Instrument parameters: microwave frequency 9.47 GHz, modulation amplitude 0.64 mT, modulation frequency 100 kHz, microwave power 5 mW.
Taken together, these results show that in the absence of Ind1, the amount of fully assembled complex I was decreased by ∼80%, and that no subcomplexes were formed. Thus, the presence of Ind1 significantly facilitates complex I assembly.
Ind1 binds a transferable Fe–S cluster
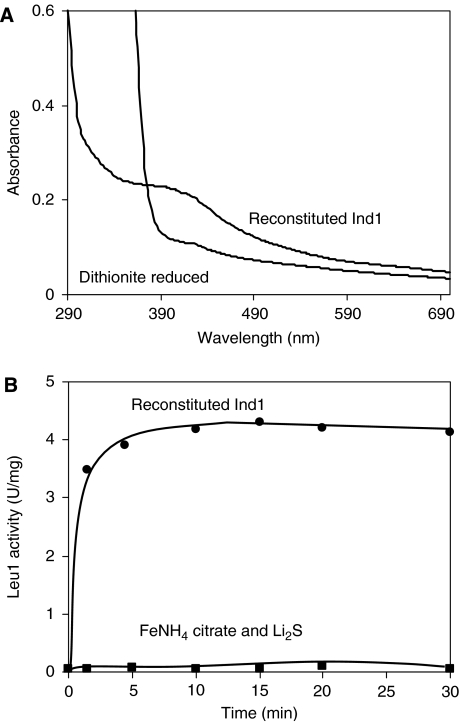
Earlier we showed that the CxxC motif is essential for the function of Ind1 (Table I and Figure 3A). As this motif is frequently found in Fe–S-binding sequences, we investigated whether Ind1 can bind an Fe–S cluster. First, Fe–S binding to the recombinant Ind1–strep was analysed. The protein was isolated from E. coli extracts as a colourless, non-[Fe–S]-containing protein following Strep-Tactin affinity purification. Metal ions or a cluster could have been lost due to inherent lability, especially as the purification was carried out in the presence of normal oxygen levels. Therefore, chemical reconstitution of Ind1 was attempted under strictly anaerobic conditions. Reconstituted and desalted Ind1 was brown in colour and had a non-structured UV–Vis spectrum with a shoulder around 400 nm (Figure 5A). Dithionite reduction led to 40–50% bleaching of the visible chromophore. These features are common for [4Fe–4S] cluster-containing proteins. The [2Fe–2S] content was low, as typical absorbance peaks of 410, 460 and 560 nm were absent (Orme-Johnson and Orme-Johnson, 1982). Upon air exposure, absorbance bands at 415, 532 and 606 nm became detectable before onset of protein precipitation and loss of visible chromophores (Supplementary Figure S6A), indicating the formation of an unknown cluster species during breakdown of the [4Fe–4S] cluster. The UV–Vis data were corroborated by a weak EPR signal for dithionite-reduced Ind1 in the g=2 region, with similar g values as reconstituted Cfd1 from S. cerevisiae (Supplementary Figure S6B). Our observations suggest that under low oxygen conditions (i.e. inside the mitochondria) a [4Fe–4S] cluster can be loaded onto Ind1.
Figure 5.
Ind1 binds a labile Fe–S cluster in vitro. (A) Binding of an [4Fe–4S] cluster to Ind1 in vitro as revealed by UV–Vis spectroscopy. Recombinant Ind1–strep (0.85 mg/ml, 26 μM) was reconstituted and desalted as described in Materials and methods. UV–Vis spectra were recorded in 100 mM Tris–HCl pH 9.0, 100 mM NaCl in the presence and absence of 2 mM sodium dithionite (as indicated). (B) Ind1 can efficiently transfer an Fe–S cluster to a model substrate, apo-isopropylmalate isomerase (Leu1). Reconstituted Ind1 (4 μM) was mixed with reduced apo-Leu1 (4 μM) under anaerobic conditions and Leu1 enzyme activity was measured at regular intervals (circles). As a control, activation of 4 μM apo-Leu1 with ferric ammonium citrate and Li2S at concentrations identical to those present in Ind1 was carried out (squares).
In a recent study (Netz et al, 2007), it has been shown that the cytosolic Cfd1–Nbp35 complex mediates transfer of transiently bound Fe–S clusters to the apoform of isopropylmalate isomerase (Leu1), a target Fe–S protein. As the apoform of complex I is not available as an in vitro target for Fe–S cluster transfer experiments, the Leu1 activation assay was used to assess the ability of Ind1 to transfer its Fe–S cluster to potential target proteins. Reconstituted and desalted Ind1 harbouring a [4Fe–4S] cluster was added to apo-Leu1 and cluster transfer was followed by the increase of enzymatic activity upon conversion of apo-Leu1 to the [4Fe–4S]-containing form. Cluster transfer from Ind1 occurred rapidly and efficiently in vitro (Figure 5B), and was comparable to the results with the Cfd1–Nbp35 complex (Netz et al, 2007). At the earliest time point that we were able to measure (1.5 min), more than 80% of the maximal Leu1 activity was obtained. In contrast, chemical reconstitution with the same concentration of ferric iron and sulphide hardly led to activation of apo-Leu1 (Figure 5B). These in vitro data suggest a scenario in which Ind1 assists with Fe–S cluster transfer to complex I.
In vivo 55Fe binding to Ind1 requires Nfs1 and Isu1 in S. cerevisiae
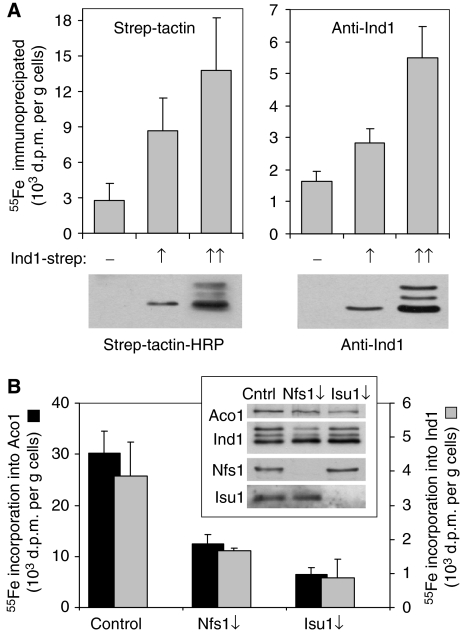
To test whether the in vitro studies are relevant for the function of Ind1 in the living cell, we investigated whether Ind1 binds Fe–S in vivo. For this purpose, we exploited a previously developed radiolabelling assay (Kispal et al, 1999). As the expression level of Ind1 in Y. lipolytica was too low for the detection limits of this assay, full-length Ind1–strep protein was overproduced under the control of an intermediate or strong promoter in S. cerevisiae. The protein was correctly targeted to mitochondria (not shown) and processed to its mature size, although processing intermediates were observed at high expression levels (Figure 6A, lower panels). Cells were labelled with 55Fe, followed by immunoprecipitation of Ind1–strep protein from the cell extract using either Strep-Tactin or Ind1 antibodies coupled to protein A-sepharose. Immunoprecipitation was confirmed by immunoblotting (not shown). The amounts of 55Fe co-precipitated from cell extracts increased significantly with the amounts of Ind1–strep in the cell extract (Figure 6A), indicating that Ind1 bound iron in vivo.
Figure 6.
Ind1 binds an Fe–S cluster in vivo. (A) Wild-type S. cerevisiae cells carrying an empty plasmid (−) or plasmids for intermediate (↑) or high (↑↑) expression of Y. lipolytica IND1–strep were radiolabelled with 55Fe. Cells were harvested after 2 h and, following preparation of a cell extract, Ind1–strep was immunoprecipitated with either Strep-Tactin or Ind1 antibodies coupled to sepharose A. The 55Fe associated with the protein was quantified by scintillation counting. The expression levels of Ind1–strep were confirmed by immunoblotting (lower panels). (B) Iron-55 labelling and immunoprecipitation of Ind1 or endogenous aconitase (Aco1) upon depletion of the cysteine desulphurase Nfs1 (Nfs1↓) or the scaffold protein Isu1 (Isu1↓). Wild-type S. cerevisiae (control), Gal-NFS1 or Gal-ISU1/isu2Δ strains were transformed with a plasmid to overexpress Y. lipolytica IND1–strep, and grown on glucose to downregulate the expression of NFS1 or ISU1 as indicated. Expression of Aco1, Ind1, Nfs1 and Isu1 was visualized by immunoblotting (inset). The scintillation values were corrected for the average background signal obtained from experiments with cells containing an empty plasmid (for Ind1; 1.6 × 103 d.p.m./g cells, see A), or with aco1Δ cells (for aconitase; 1.3 × 103 d.p.m./g cells). Error bars represent the standard deviation, n=3–5.
To investigate whether 55Fe binding to Ind1 was dependent on the cysteine desulphurase Nfs1 and the scaffold proteins Isu1/Isu2 and thus represents binding of an Fe–S cluster, we expressed Ind1–strep at high levels in S. cerevisiae cells in which the expression of NFS1 or ISU1 can be regulated (Kispal et al, 1999; Gerber et al, 2003). Ind1 antibodies were used for immunoprecipitation, because, for unknown reasons, the C-terminal Strep tag was cleaved off upon downregulation of NFS1 or ISU1, but the Ind1 protein itself was stable (Figure 6B and data not shown). Upon depletion of either Nfs1 or Isu1, the amount of 55Fe associated with Ind1 was significantly decreased. A similar decline in 55Fe incorporation was observed under these conditions for mitochondrial aconitase (Aco1), a control Fe–S protein known to be dependent on Nfs1 and Isu1 (Figure 6B). These data show that 55Fe binding to Ind1 is dependent on two central components of the ISC assembly system in the S. cerevisiae mitochondrial environment, thus indicating the presence of an Fe–S cluster on Ind1 in vivo.
Discussion
We have investigated the function of a mitochondrial P-loop NTPase, called Ind1, in the obligate respiratory yeast Y. lipolytica. Our data showed that Ind1 is specifically required for effective assembly of complex I, NADH:ubiquinone oxidoreductase. We first identified Ind1 as a protein sequence with high sequence similarity to the S. cerevisiae P-loop NTPases Cfd1 and Nbp35, which form a heterotetramer and mediate the transfer of Fe–S clusters to cytosolic Fe–S proteins (Roy et al, 2003; Hausmann et al, 2005; Netz et al, 2007). Most eukaryotes possess a Cfd1-like protein with an N-terminal sequence predicted to target the protein to mitochondria (Figure 2A and Supplementary Figure S1). Indeed, we confirmed that the Y. lipolytica Ind1 protein was localized exclusively in mitochondria (Figure 1C). Co-evolution of Ind1 and complex I (Figure 2A) suggested a role for Ind1 in the assembly and/or function of complex I.
The impact of IND1 deletion on complex I was investigated at different levels, including in vitro enzyme assays, complex integrity in BN–PAGE gels, subunit abundance and composition in two-dimensional gel electrophoresis, as well as EPR spectroscopy of the Fe–S cofactors (Figures 2C, 3, 4 and Table I). All methods concurrently revealed a decrease in complex I content of approximately 80% in the ind1Δ mutant compared with the cWT strain. The deleterious effect was specific for complex I, as none of the other respiratory complexes were affected, either at the level of protein assembly (complexes III and V; Figure 3B), Fe–S cluster assembly (complex II; Figure 4) or activity (complexes II and III; Figure 2C). This specificity is different from that observed for the bacterial homologue ApbC in Salmonella enterica, in that lesions in the latter decreased complex II (succinate dehydrogenase) activity by 34% (Skovran and Downs, 2003). The NADH:ubiquinone oxidoreductase activity was not analysed in the apbC mutant and it will be interesting to do so to establish whether the ApbC and Ind1 have overlapping functions.
Our data indicate that Ind1 mediates an ATP or GTP-dependent, Fe–S-requiring step in complex I assembly, but the details remain to be investigated. Ind1 has not been identified previously as a protein associated with complex I, despite thorough proteomics studies (reviewed in Hirst et al, 2003; Brandt, 2006). Therefore, Ind1 would qualify as a true assembly factor, which is consistent with its low abundance (Figure 1B). Future protein-binding studies, as well as immunodetection of Ind1 in assembly intermediates may establish the precise role of Ind1 in complex I assembly. It is interesting to note that small amounts of complex I (∼20% of cWT) were still assembled in the absence of Ind1. Clearly, Ind1 facilitates a particular step in the assembly process that can be either bypassed or is also carried out by other proteins at low efficiency. The remaining complex I activity was not sufficient for survival during sporulation in the absence of ND2i (Supplementary Figure S5), suggesting that a certain threshold of complex I activity is required during this developmental stage.
Our in vitro and in vivo data established the ability of Ind1 to bind a [4Fe–4S] cluster (Figures 5 and 6). Cys242 and Cys245 were demonstrated to be essential for the function of Ind1 using site-directed mutagenesis (Figure 3A and Table I), and may form the ligands of the Fe–S cluster. It remains to be investigated whether other, non-cysteine residues function as additional ligands, or whether the cubane cluster is coordinated by ligands from two Ind1 polypeptides forming a homodimer, as is the case for NifH (Georgiadis et al, 1992). Further in vitro data showed that the Fe–S cluster is oxygen labile and can readily be transferred to an Fe–S apoprotein (Figure 5B). Although these properties suggest a scaffold function of Ind1, we found that it could not replace the scaffold protein Isu1 and vice versa. First of all, Fe–S cluster loading on Ind1 occurred downstream of Isu1 in S. cerevisiae mitochondria (Figure 6B). It needs to be investigated, however, whether the same is true in Y. lipolytica. Second, overexpression of IND1 could not rescue growth of Isu1-depleted Gal-ISU1/isu2Δ S. cerevisiae cells on glucose (data not shown). Third, overexpression of Y. lipolytica ISU1 using the inducible POX2 promoter did not increase the levels of assembled complex I in the ind1Δ mutant (data not shown). These results suggested that Ind1 performs a molecular function that is clearly different from the generic Fe–S scaffold protein Isu1.
The identification of a novel assembly factor of complex I is of major clinical importance, as mutations in numerous genes encoding complex I subunits are causative of several human diseases. Only a handful of complex I assembly factors have been identified thus far and many complex I deficiencies remain to be elucidated at the molecular level (Janssen et al, 2006). We have preliminary evidence that the human orthologue of Ind1 (GeneID: 80224) could partially rescue the growth and complex I activity of the Y. lipolytica ind1Δ strain (data not shown). Moreover, RNAi-mediated downregulation of human IND1 expression in HeLa cells resulted in a ∼70% decrease of complex I activity (A Sheftel et al, manuscript in preparation). Thus, further molecular characterization of Ind1 in the model organism Y. lipolytica or in human cell lines will be of vital importance for our understanding of the assembly process of complex I, and the numerous mitochondrial diseases associated with complex I deficiency.
Materials and methods
Yeast strains and growth
The haploid Y. lipolytica GB10 strain (ura3-302, leu2-270, lys11-23, NUGM-Htg2, NDH2i, MatB) was used to generate an IND1 deletion mutant, ind1Δ. The IND1 gene (systematic name YALI0B18590g, GeneID: 2907193) is intron-less and is predicted to encode an open reading frame of 312 amino-acid residues. The open reading frame plus 27 bp upstream and 60 bp downstream were replaced by the URA3 marker gene using the homologous recombination strategy described in Abdrakhmanova et al (2006). More than 440 transformants were screened and the correct gene replacement was confirmed by PCR. The IND1 gene was reintroduced in the ind1Δ mutant to obtain the cWT. For this purpose, the entire IND1 gene, including 1329 nucleotides upstream of the start codon containing the endogenous promoter, was cloned into the pUB4 plasmid (Kerscher et al, 2001), generating pUB4-IND1. A C-terminal Strep tag was added to the IND1 open reading frame, creating pUB4-IND1–strep. pUB4-IND1 was used for site-directed mutagenesis using PCR. Y. lipolytica cells were grown in rich medium containing 1% (w/v) glucose (YPD). Hygromycin B (50 μg/ml) was added for the initial selection of cells transformed with the pUB4 plasmid.
S. cerevisiae strain W303-1A (MATa, ura3-1, ade2-1, trp1-1, his3-11,15, leu2-3112) served as WT. The Gal-NFS1 and Gal-ISU1/isu2Δ strains and conditions for depletion of the respective proteins were described previously (Kispal et al, 1999; Gerber et al, 2003). The following yeast plasmids were used: pRS416 containing the MET25 promoter for intermediate expression of IND1–strep, and pRS426 with the TDH3 promoter for high expression (Mumberg et al, 1995).
Gel electrophoresis
Separation of the components of the respiratory chain in mitochondrial membranes of Y. lipolytica was carried out using BN–PAGE (Schägger, 2003). Total protein (500 μg) was solubilized with 3.0 g/g digitonin and 500 mM amino caproic acid. The resulting solubilized mitochondrial membranes were separated on non-denaturing 4–13% gradient gels. Individual lanes were cut out, incubated for 30 min in 1% SDS and applied on denaturing 16% polyacrylamide Tricine-SDS gels for resolution in the second dimension. Gels were stained with silver. For in-gel detection of the NADH dehydrogenase activity of complex I, unfixed BN–PAGE gels were incubated for about 5 min in a solution containing 3 mM NBT and 120 μM NADH. To stop the reaction, gels were incubated in 50% methanol and 10% acetic acid.
EPR spectroscopy
X-band EPR spectra were recorded with a Bruker ESP 300E spectrometer equipped with a HP 53159A frequency counter (Hewlett Packard), an ER 035 M NMR gaussmeter (Bruker, BioSpin) and a liquid helium continuous flow cryostat (Oxford Instruments). Mitochondrial membranes, at a final concentration of 25 mg protein/ml, were treated with 2 mM NADH. Samples were frozen in cold isopentane/methylcyclohexane (5:1, ∼120 K) and stored in liquid nitrogen. Typically, spectra were recorded at temperatures of 40 K to analyse binuclear clusters only or at 12 K to analyse both binuclear and tetranuclear clusters. A highly concentrated sample of isolated complex I (∼86 mg/ml) reduced by NADH was used as a reference.
Chemical reconstitution and transfer of Fe–S clusters
The IND1–strep open reading frame, excluding the first 36 codons and starting with a MetAUG, was cloned into the pET15b vector (Novagen) for overexpression of the protein following the manufacturer's instructions. The protein was purified using Strep-Tactin (IBA, Göttingen, Germany) and eluted in 100 mM Tris–HCl pH 9.0, 100 mM NaCl (buffer A), 2.5 mM desthiobiotin. Freshly prepared, recombinant apo-Ind1–strep was chemically reconstituted in an anaerobic chamber (Coy Laboratory Products, Ann Arbor, MI). In a typical experiment, 20 μM of Ind1 in buffer A was treated with 5 mM DTT (final concentration) for 1 h at 15°C. The reconstitution was started by addition of 10 molar equivalents of ferric ammonium citrate and Li2S and the mixture was incubated for 3 h at 15°C. To remove non-bound iron and sulphide, reconstituted Ind1 was passed through a Sephadex G25 gel filtration column using buffer A containing 1 mM DTT. The assembly of an Fe–S cluster on apo-Ind1 was ascertained by UV–Vis spectroscopy (Jasco V-550).
Chemically reconstituted Ind1 harbouring an [Fe–S] cluster was tested for the ability to transfer its cluster to the apoform of yeast isopropylmalate isomerase (apo-Leu1) in vitro, as described by Netz et al (2007).
Miscellaneous methods
The following published methods were used: isolation of intact mitochondria (Galkin et al, 2006) and unsealed mitochondrial membranes (Kerscher et al, 1999); enzyme assays of aconitase, succinate dehydrogenase and citrate synthase (Kispal et al, 1999); complex I activity assays (Tocilescu et al, 2007). Polyclonal antibodies were raised against Ind1–strep purified from E. coli (Harlow and Lane, 1998). Antisera against Aco1, Nfs1 and Isu1 have been described previously (Kispal et al, 1999; Gerber et al, 2003; Gelling et al, 2008). The monoclonal antibody against Arabidopsis actin was from Affinity BioReagents, Golden, CO, USA. Labelling with Strep-Tactin horse radish peroxidase (IBA) was carried out according to the manufacturer's instructions. Radiolabelling, cell lysis and immunoprecipitation of the Fe–S reporter proteins of interest were as described previously (Kispal et al, 1999). Error bars represent the standard deviation of the mean value.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Acknowledgments
We thank Gianpiero Vigani, Gudrun Beyer and Martin Stümpfig for technical assistance and Rudolf K Thauer (Max-Planck-Institute, Marburg) for access to the EPR spectrometer. This research was funded by the Royal Society (JB), the Danish Research Agency and the Carlsberg Foundation (KB), Deutsche Forschungsgemeinschaft: SFB 628 and EXC 115 (UB), SFB 593, Gottfried-Wilhelm Leibniz program, and GRK 1216 (RL), Fonds der chemischen Industrie (RL) and the Netherlands Genomics Initiative (Horizon programme) (MAH).
References
- Abdrakhmanova A, Dobrynin K, Zwicker K, Kerscher S, Brandt U (2006) Functional sulfurtransferase is associated with mitochondrial complex I from Yarrowia lipolytica, but is not required for assembly of its iron–sulfur clusters. FEBS Lett 579: 6781–6785 [DOI] [PubMed] [Google Scholar]
- Balk J, Lobréaux S (2005) Biogenesis of iron–sulfur proteins in plants. Trends Plant Sci 10: 324–331 [DOI] [PubMed] [Google Scholar]
- Beinert H (2000) Iron–sulfur proteins: ancient structures, still full of surprises. J Biol Inorg Chem 5: 2–15 [DOI] [PubMed] [Google Scholar]
- Brandt U (2006) Energy converting NADH:quinone oxidoreductase (complex I). Annu Rev Biochem 75: 69–92 [DOI] [PubMed] [Google Scholar]
- Dunning CJR, McKenzie M, Sugiana C, Lazarou M, Silke J, Connelly A, Fletscher JM, Kirby DM, Thorburn DR, Ryan MT (2007) Human CIA30 is involved in the early assembly of mitochondrial complex I and mutations in its gene cause disease. EMBO J 26: 3227–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Gabaldón T, Rainey D, Huynen MA (2005) Tracing the evolution of a large protein complex in the eukaryotes, NADH:ubiquinone oxidoreductase (complex I). J Mol Biol 348: 857–870 [DOI] [PubMed] [Google Scholar]
- Galkin A, Dröse S, Brandt U (2006) The proton pumping stoichiometry of purified mitochondrial complex I reconstituted into proteoliposomes. Biochim Biophys Acta 1757: 1575–1581 [DOI] [PubMed] [Google Scholar]
- Gelling C, Dawes IW, Richhardt N, Lill R, Mühlenhoff U (2008) Mitochondrial Iba57p is required for Fe/S cluster formation on aconitase and activation of radical SAM enzymes. Mol Cell Biol 28: 1851–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiadis MM, Komiya H, Chakrabarti P, Woo D, Kornuc JJ, Rees DC (1992) Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii. Science 257: 1653–1659 [DOI] [PubMed] [Google Scholar]
- Gerber J, Mühlenhoff U, Lill R (2003) An interaction between frataxin and Isu1/Nfs1 that is crucial for Fe/S cluster synthesis on Isu1. EMBO Rep 4: 906–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D (1998) Using Antibodies: a Laboratory Manual. New York: Cold Spring Harbor Laboratory Press [Google Scholar]
- Hausmann A, Aguilar Netz DJ, Balk J, Pierik AJ, Mühlenhoff U, Lill R (2005) The eukaryotic P loop NTPase Nbp35: an essential component of the cytosolic and nuclear iron–sulfur protein assembly machinery. Proc Natl Acad Sci USA 102: 3266–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchliffe P, Sazanov LA (2005) Organization of iron–sulfur clusters in respiratory complex I. Science 309: 771–774 [DOI] [PubMed] [Google Scholar]
- Hirst J, Carroll J, Fearnley IM, Shannon RJ, Walker JE (2003) The nuclear encoded subunits of complex I from bovine heart mitochondria. Biochim Biophys Acta 1604: 135–150 [DOI] [PubMed] [Google Scholar]
- Janssen RJ, Nijtmans LG, van den Heuvel LP, Smeitink JA (2006) Mitochondrial complex I: structure, function and pathology. J Inherit Metab Dis 29: 499–515 [DOI] [PubMed] [Google Scholar]
- Johnson DC, Dean DR, Smith AD, Johnson MK (2005) Structure, function, and formation of biological iron–sulfur clusters. Annu Rev Biochem 74: 247–281 [DOI] [PubMed] [Google Scholar]
- Kerscher S, Eschemann A, Okun PM, Brandt U (2001) External alternative NADH:ubiquinone oxidoreductase redirected to the internal face of the mitochondrial inner membrane rescues complex I deficiency in Yarrowia lipolytica. J Cell Sci 114: 3915–3921 [DOI] [PubMed] [Google Scholar]
- Kerscher S, Okun JG, Brandt U (1999) A single external enzyme confers alternative NADH:ubiquinone oxidoreductase activity in Yarrowia lipolytica. J Cell Sci 112: 2347–2354 [DOI] [PubMed] [Google Scholar]
- Kispal G, Csere P, Prohl C, Lill R (1999) The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J 18: 3981–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küffner R, Rohr A, Schmiede A, Krüll C, Schulte U (1998) Involvement of two novel chaperones in the assembly of mitochondrial NADH:ubiquinone oxidoreductase (complex I). J Mol Biol 283: 409–417 [DOI] [PubMed] [Google Scholar]
- Leipe DD, Koonin EV, Aravind L (2002) Evolution and classification of P-loop kinases and related proteins. J Mol Biol 333: 781–815 [DOI] [PubMed] [Google Scholar]
- Lezhneva L, Amann K, Meurer J (2004) The universally conserved HCF101 protein is involved in assembly of [4Fe–4S]-cluster-containing complexes in Arabidopsis thaliana chloroplasts. Plant J 37: 174–185 [DOI] [PubMed] [Google Scholar]
- Lill R, Mühlenhoff U (2008) Maturation of iron–sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu Rev Biochem, (in press) [DOI] [PubMed] [Google Scholar]
- Mochel F, Knight MA, Tong WH, Hernandez D, Ayyad K, Taivassalo T, Andersen PM, Singleton A, Rouault TA, Fischbeck KH, Haller RG (2008) Splice mutation in the iron–sulfur cluster scaffold protein ISCU causes myopathy with exercise intolerance. Am J Hum Genet 82: 652–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122 [DOI] [PubMed] [Google Scholar]
- Netz DJ, Pierik AJ, Stümpfig M, Mühlenhoff U, Lill R (2007) The Cfd1–Nbp35 complex acts as a scaffold for iron–sulfur protein assembly in the yeast cytosol. Nat Chem Biol 3: 278–286 [DOI] [PubMed] [Google Scholar]
- Ogilvie I, Kennaway NG, Shoubridge EA (2005) A molecular chaperone for mitochondrial complex I assembly is mutated in a progressive encephalopathy. J Clin Invest 115: 2784–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson A, Lind L, Thornell LE, Holmberg M (2008) Myopathy with lactic acidosis is linked to chromosome 12q23.3–24.11 and caused by an intron mutation in the ISCU gene resulting in a splicing defect. Hum Mol Genet, (in press) [DOI] [PubMed] [Google Scholar]
- Orme-Johnson WH, Orme-Johnson AR (1982) Iron–sulfur proteins: the problem of determining cluster type. In Iron–Sulfur Proteins, Spiro TG (ed). New York: Wiley [Google Scholar]
- Petersen L, Downs DM (1996) Mutations in apbC (mrp) prevent function of the alternative pyrimidine biosynthetic pathway in Salmonella typhimurium. J Bacteriol 178: 5676–5682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl T, Walter J, Stolpe S, Soufo JH, Grauman PL, Friedrich T (2007) Effects of the deletion of the Escherichia coli frataxin homologue CyaY on the respiratory NADH:ubiquinone oxidoreductase. BMC Biochem 8: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Solodovnikova N, Nicholson T, Antholine W, Walden WE (2003) A novel eukaryotic factor for cytosolic Fe–S cluster assembly. EMBO J 22: 4826–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saada A, Edvardson S, Rapoport M, Shaag A, Amry K, Miller C, Lorberboum-Galski H, Elpeleg O (2008) C6ORF66 is an assembly factor of mitochondrial complex I. Am J Hum Genet 82: 32–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H (2003) Blue native electrophoresis. In Membrane Protein Purification and Crystallization: a Practical Guide, Hunte C, von Jagow G, Schägger H (eds), pp 105–130. San Diego: Academic Press [Google Scholar]
- Skovran E, Downs DM (2003) Lack of the ApbC or ApbE protein results in a defect in Fe–S cluster metabolism in Salmonella enterica serovar Typhimurium. J Bacteriol 185: 98–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöckel J, Oelmüller R (2004) A novel protein for photosystem I biogenesis. J Biol Chem 279: 10243–10251 [DOI] [PubMed] [Google Scholar]
- Tocilescu MA, Fendel U, Zwicker K, Kerscher S, Brandt U (2007) Exploring the ubiquinone binding cavity of respiratory complex I. J Biol Chem 282: 29514–29520 [DOI] [PubMed] [Google Scholar]
- Vogel RO, Janssen RJ, Ugalde C, Grovenstein M, Huijbens RJ, Visch H-J, van den Heuvel LP, Willems PH, Zeviani M, Smeitink JA, Nijtmans LG (2005) Human mitochondrial complex I assembly is mediated by NDUFAF1. FEBS Lett 272: 5317–5326 [DOI] [PubMed] [Google Scholar]
- Vogel RO, Janssen RJ, van den Brand MAM, Dieteren CEJ, Verkaart S, Koopman WJH, Willems PHGM, Pluk W, van den Heuvel LP, Smeitink JA, Nijtmans LG (2007) Cytosolic signaling protein Ecsit also localizes to mitochondria where it interacts with chaperone NDUFAF1 and functions in complex I assembly. Genes Dev 21: 615–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6