SUMMARY
Cadherins are cell surface adhesion proteins important for tissue development and integrity. Type I and type II, or “classical”, cadherins form adhesive dimers via an interface formed through the exchange, or “swapping”, of the N-terminal β-strands from their membrane-distal EC1 domains. Here we ask which sequence and structural features in EC1 domains are responsible for β-strand swapping and whether members of other cadherin families also form similar strand-swapped binding interfaces. We first create a comprehensive database consisting of multiple alignments of each type of cadherin domain. We use the known three-dimensional structures of classical cadherins to identify conserved positions in multiple sequence alignments that appear to be crucial determinants of the cadherin domain structure. We further identify features that are unique to EC1 domains. On the basis of our analysis we conclude that all cadherin domains have very similar overall folds but, with the exception of classical and desmosomal cadherin EC1 domains, most of them do not appear to bind through a strand swapping mechanism. Thus, non-classical cadherins that function in adhesion are likely to use different protein-protein interaction interfaces. Our results have implications for the evolution of molecular mechanisms of cadherin-mediated adhesion in vertebrates.
Keywords: cadherins, adhesion, domain swapping, strand swapping, cell surface receptors
INTRODUCTION
The cadherins constitute a superfamily of Ca2+-binding cell surface transmembrane glycoproteins, many of which are known to mediate cell-cell adhesion in vertebrates and invertebrates1–5. Cadherin ectodomain regions include tandemly repeated ~100 amino acid domains called extracellular cadherin (EC) domains. With the exceptions of the seven-pass flamingo cadherins and truncated (T-) cadherin, which is GPI-anchored, cadherins are single-pass transmembrane proteins that contain a short cytoplasmic domain that indirectly links the EC domains to the cytoskeleton. Type I and type II, or classical, cadherins are cell-cell adhesion molecules that are present only in vertebrates, as are the closely related desmosomal cadherins, desmocollins and desmogleins. Other cadherin families include the clustered protocadherins, which appear only in vertebrates and regulate neuronal connectivity5; invertebrate cadherins such as the Drosophila melanogaster DE- and DN-cadherins, which are known to have an adhesive function6,7; the Dachsous and Fat families, which are present in vertebrates and invertebrates and appear to play a role in defining cell polarity8–11; and the seven-pass transmembrane flamingo cadherins, which are also present in both vertebrates and invertebrates and appear to regulate cell polarity12. Our focus in this work is on the nature of the adhesive interface formed between cadherin molecules. Specifically, we ask whether all cadherins bind to one another in a manner similar to that observed for type I and type II cadherins, or whether different binding interfaces are likely to be used by members of other cadherin subfamilies that are still incompletely characterized in structural terms.
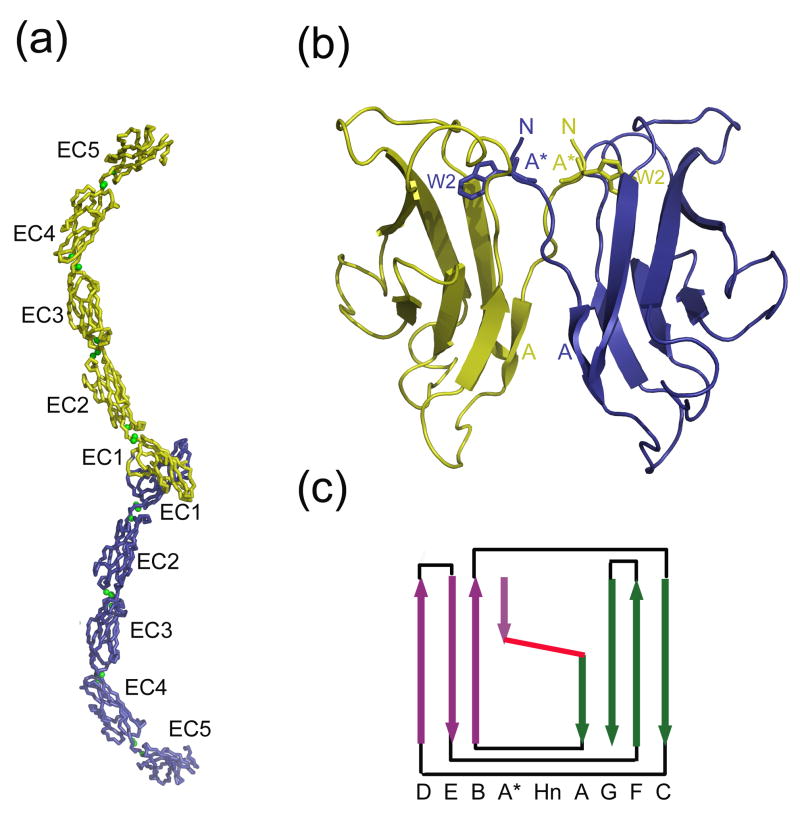
The three-dimensional structures of a number of type I and type II cadherin ectodomain adhesive regions have been determined by both X-ray crystallography and NMR, and the structure for the full ectodomain of Xenopus laevis C-cadherin has also been determined13–23. Table 1 contains a list of all available cadherin structures13–25. All type I and type II cadherins contain five EC domains that are connected via linker regions which bind Ca2+ ions (Figure 1a). EC domains adopt a Greek-key β-sandwich fold comprised of seven β-strands, similar to immunoglobulin variable domains (Figures 1b and c). One sheet of the β-sandwich contains strands D, E, and B, while the opposing sheet is formed by strands G, F, and C. As in immunoglobulin variable domains, the A strand is divided into two segments, termed the A* and A strands, which differ in their hydrogen-bonding patterns and sheet placement. The N-terminal segment (the A* strand) has three residues which form β-sheet hydrogen bonds with the B strand in sheet I, while the C-terminal segment (the A strand) hydrogen bonds to the G strand in sheet II. The two segments are separated by 2-3 residues that cross between the two β-sheets (Figure 1c). We refer to this 2-3 residue segment as the “hinge” due to its change of conformation in EC1 upon dimerization, which facilitates motion of the EC1 A* strand (see below). Although the hinge region is not mobile in non-EC1 domains, which do not dimerize, the hinge segment does cross between the two β-sheets in these domains as well. In most domains, the hinge residues do not form hydrogen bonds with either sheet; in type II EC1 domains, however, the hinge residues also hydrogen bond to the G strand of the same peptide chain.
Table 1.
Summary of cadherin domain structures included in this study
| Family | Protein | Domain | PDB ID | Range | Resolution (Ǻ) | Notes |
|---|---|---|---|---|---|---|
| Type I | E-cadherin | EC1 | 1FF516 | 1–99 | 2.93 | non-swapped; non-native extra residue§ (M) at N-terminus |
| EC2 | 1FF5 | 105–212 | 2.93 | |||
| Type I | E-cadherin | EC1 | 1Q1P21 | 2–99 | 3.20 | swapped; N-terminal residue disordered |
| EC2 | 1Q1P | 105–212 | 3.20 | |||
| Type I | E-cadherin | EC1 | 1EDH15 | 3–99 | 2.00 | non-swapped; N-terminal 2 residues disordered |
| EC2 | 1EDH | 105–212 | 2.00 | |||
| Type I | E-cadherin | EC1 | 2O7223 | 1–99 | 2.00 | swapped |
| EC2 | 2O72 | 105–212 | 2.00 | |||
| Type I | E-cadherin | EC1 | 1O6S19 | 1–99 | 1.80 | in complex with internalin; non-native extra 4 residues (GPLG) at N-terminus and D1S mutation |
| Type I | E-cadherin | EC1 | 1SUH13 | 1–99 | NMR | monomeric |
| Type I | C-cadherin | EC1 | 1L3W17 | 1–99 | 3.08 | swapped |
| EC2 | 1L3W | 105–212 | 3.08 | |||
| EC3 | 1L3W | 218–324 | 3.08 | |||
| EC4 | 1L3W | 329–431 | 3.08 | |||
| EC5 | 1L3W | 437–540 | 3.08 | |||
| Type I | N-cadherin | EC1 | 1NCI14 | 2–99 | 2.10 | swapped; non-native extra 2 residues (GS) at N-terminus; D27G mutation |
| Type I | N-cadherin | EC1 | 1NCG14 | 1–99 | 2.10 | swapped; D27G mutation |
| Type I | N-cadherin | EC1 | 1NCH14 | 1–99 | 2.10 | swapped; non-native extra 2 residues (GS) at N-terminus of one chain; D27G mutation |
| Type I | N-cadherin | EC1 | 1NCJ27 | 2–99 | 3.40 | monomeric; N-terminal residue disordered |
| EC2 | 1NCJ | 105–214 | 3.40 | |||
| Type I (non-swapping) | T-cadherin | EC1 | ---* | 1–98 | 1.10 | non-swapped; A1S mutation |
| Type I (non-swapping) | T-cadherin | EC1 | ---* | 1–98 | 2.90 | non-swapped; A1S mutation |
| EC2 | 104–216 | 2.90 | ||||
| Type II | mn-cadherin | EC1 | 1ZVN22 | 1–97 | 2.16 | swapped; non-native extra residue (G) inserted between the first two N-terminal residues |
| Type II | cadherin-11 | EC1 | 2A4C22 | 1–97 | 2.90 | swapped; non-native extra residue (S) at N-terminus |
| Type II | cadherin-11 | EC1 | 2A4E22 | 1–97 | 3.20 | swapped; non-native extra residue (S) at N-terminus |
| EC2 | 2A4E | 102–206 | 3.20 | |||
| Type II | cadherin-8 | EC1 | 1ZXK22 | 1–97 | 2.00 | swapped; G1S mutation |
| Type II | cadherin-8 | EC1 | 2A6222 | 1–97 | 4.50 | swapped; G1S mutation |
| EC2 | 2A62 | 103–206 | 4.50 | |||
| EC3 | 2A62 | 212–322 | 4.50 | |||
| Protocadherin | protocadherin-α4 | EC1 | 1WUZ25 | 1–98 | NMR | monomeric |
| Protocadherin | protocadherin-α14 | EC1 | 1WYJ** | 9–106 | NMR | monomeric; non-native extra 7 residues (GSSGSSG) at N-terminus |
| Protocadherin | protocadherin-7 | EC3 | 2YST** | 11–112 | NMR | monomeric; non-native extra 6 residues (SSGSSG) and native linker sequence (NDN) at N- terminus |
| Type I | N-cadherin | prodomain | 1OP420 | 33–124 | NMR | monomeric |
| Desmoglein | desmoglein-2 | EC1 | 2YQG** | 11–111 | NMR | monomeric; non-native extra 8 residues (GSSGSSGQ) and native protease site (KR) at N-terminus |
Non-native residues include residues seen in the crystal structure that are not found in the native protein sequence.
unpublished
to be published; coordinates available from the RCSB Protein Data Bank (http://www.rcsb.org/pdb/)
Figure 1.
Cadherin domain topology and architecture. (a) X-ray crystal structure of C-cadherin full length dimer. The two protomers are in yellow and blue, and the Ca2+ ions are in green. (b) Magnified view of the C-cadherin EC1-EC1 swapping interface. Trp2 of each protomer is shown in stick representation. For each protomer, the N-terminus (N), A* strand, W2, and A strand are labeled. (c) Schematic representation of C-cadherin EC2. Arrows indicate β strands. Sheet I of the β sandwich is shown in purple and sheet II in green. The hinge (denoted ‘Hn’) where the A*/A strand crosses β sheets is in red.
The A* and A strands play a critical role in the binding interface. In almost all structures that have been determined, cadherins form homodimers in such a way that the monomer-monomer interface is located entirely on the membrane-distal EC1 domain. The two interacting EC1 domains bind in a parallel fashion, with the interface formed through a reciprocal swap of the N-terminal A* strand that inserts the side chain of the conserved residue Trp2 into the core of the partner domain (Figure 1b, Supplementary Table 1). In type I cadherins17, almost the entire interface is formed from strand swapped residues, while type II cadherins22 form a larger interface that includes additional contacts. There are three examples of classical cadherin X-ray structures in which swapping is not observed (Table 1). Two of them, PDB codes 1FF516 and 1EDH15, involve constructs of E-cadherin with additional residues at the N-terminus of the EC1 domain (respectively, Met and Met-Arg). It is known that the presence of extra N-terminal residues can prevent cadherin adhesive function by interfering with strand swapping26, and indeed both of these proteins fail to form swapped dimers (in the case of 1EDH, electron density for critical interfacial residues is absent). Although N-cadherin EC1 constructs with additional N-terminal residues do form swapped dimers14, in those cases the non-native residues (Gly-Ser) may not have been as disruptive as the bulkier Met and Arg residues found in the E-cadherin non-swapping constructs. In the third non-swapping structure, the N-cadherin EC1-EC2 construct (1NCJ), the N-terminus is disordered, including the side chain of the critical interface residue W2.
There is a large body of evidence that indicates that the strand-swapped interface observed in structural studies corresponds to the adhesive interface that drives inter-cellular adhesion (see e.g. discussion in Patel et al., 2006). For example, a variety of mutations that disrupt the swapped interface have been shown to abrogate adhesion in cell aggregation studies26–29. Moreover, electron tomography studies of desmosomes30 suggest that the EC1-EC1 interface seen in the crystal structure of C-cadherin17 is indistinguishable at the resolution of these studies from the interface formed between trans cadherin dimers located on apposing cell surfaces. There is evidence that lateral cis-dimers also form between cadherins on the same cell surface16,31–33. Cross-linking experiments have demonstrated that the lateral dimer interface involves the same strand swapped interface seen in trans dimers28.
The central role of strand swapping in the formation of the adhesive interface between EC1 domains raises the question of whether there are unique features of these domains, relative to other EC domains. Here, we identify a number of EC1-specific structural features and, on this basis, suggest that other classical cadherin EC domains would not swap, even if their N-termini were free. Our analysis suggests that desmosomal cadherins also dimerize through strand swapping between their EC1 domains. However, other cadherins, including all cadherins from invertebrates, despite having a domain structure that is similar to that of classical cadherins, do not appear to exploit a swapping mechanism for dimerization and, consequently, those domains that dimerize will likely employ different interfaces than observed for classical cadherins.
Since strand swapping involves the N-terminal strand of the EC1 domain, a proper comparison to other domains requires that their N-termini be defined in a consistent way. This is not straightforward since a Ca2+-binding linker regions connects every two EC domains and a decision has to be made where one domain ends and the other begins. For this reason, many different definitions of a cadherin domain have been suggested and, in most cases, three-dimensional structure has not been used to define the N- and C- termini of each domain. Here, we provide a structure-based definition of cadherin domains which is somewhat different than those that have appeared in the literature and in widely used sequence databases. The N-terminus of each cadherin domain is precisely defined, and this in turn reveals important differences between EC1 domains and other EC domains. In addition, we offer a structural and functional interpretation of a number of cadherin sequence motifs and compile an extensive cadherin domain sequence database that is consistent with available structural information.
RESULTS
Defining a cadherin domain
Cadherins are typically identified through sequence motifs such as DxNDN that are associated with Ca2+ binding. However, these amino acids are located in a linker region between domains and cannot be clearly associated with a single domain. The SMART database34 omits the entire N-terminal region of all EC domains including EC1 (the A* and A strand and part of the B strand), while Pfam35 omits the A* and part of the A strand. Many studies properly define the N-terminal of the EC1 domain but extend the domain past the linker region to include residues that clearly belong to EC2. In the following, we have associated the Ca2+-binding inter-domain linker residues with the domain from which they originate but ensure that the linker sequence does not include residues that are part of the following domain. As will become evident, the proper definition of a domain boundary is a crucial step in identifying the determinants of swapping.
Cadherin domain sequence database
Our goal in this and the following section is to derive a multiple alignment and consensus sequence for most known cadherin domain classes. There are 9 known cadherin sub-families and we associate a separate class with each of the extracellular domains in a particular sub-family. For example, there are 5 domain classes associated with type I cadherins (EC1-EC5), 5 for type II, 5 desmocollin and 4 desmoglein domain classes, 34 fat and 9 flamingo domain classes, and 6 α, 6 β, and 6 γ protocadherin classes; 80 classes in all.
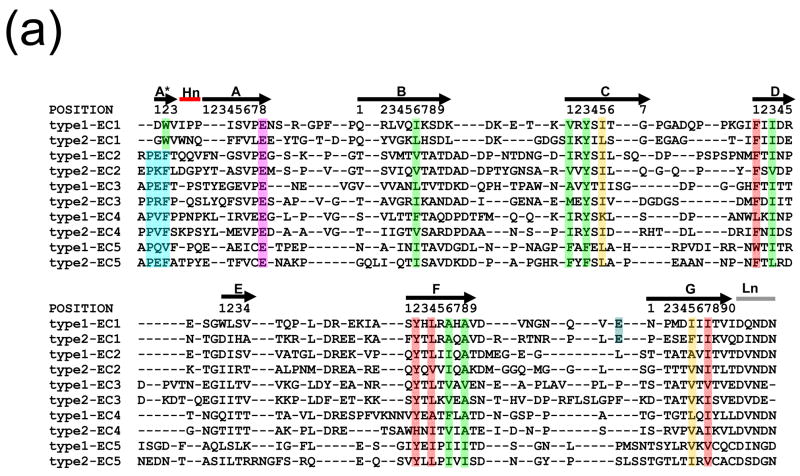
We begin by assembling a database of cadherin domains, and then we assign them to specific domain classes based on their relationship to well-annotated domains of that class. Finally, consensus sequences are constructed for each domain class. This step in our analysis is based entirely on sequence relationships. The database of cadherin domain sequences was derived by first using the Hidden Markov Model (HMM) for cadherin domains found in the SMART database34 to search the non-redundant sequence database UniRef10036. About 21,500 sequences belonging to about 3,305 proteins were retrieved in this way. Since the SMART domain omits the three N-terminal strands (the A*, A, and part of the B strands), each of these sequences was extended to include the omitted strands. Where present, the conserved Ca2+-binding motif DxNDN was used to define the linker region between domains. Visual inspection of all available structures of EC2-5 domains indicates that the N-terminus follows the Ca2+ linker sequence and usually has the sequence XPXF. The first two N-terminal residues (XP of the XPXF motif) are structurally part of the body of the domain but do not participate in β-sheet hydrogen bonding. The N-terminal A* strand generally consists of three residues starting with the XF residues of XPXF motif (see Figure 2a). In most previous studies the Pro residue of this motif was associated with domain 137–39, as was the highly conserved Phe. Yet the A* strand and the preceding two residues are at the N-terminus of each cadherin domain, and the Phe is aligned with the conserved Trp2 in EC1 that plays a crucial role in swapping (see below). In our database, the residue preceding the Pro is generally the N-terminal residue of the domain to which it is assigned.
Figure 2.
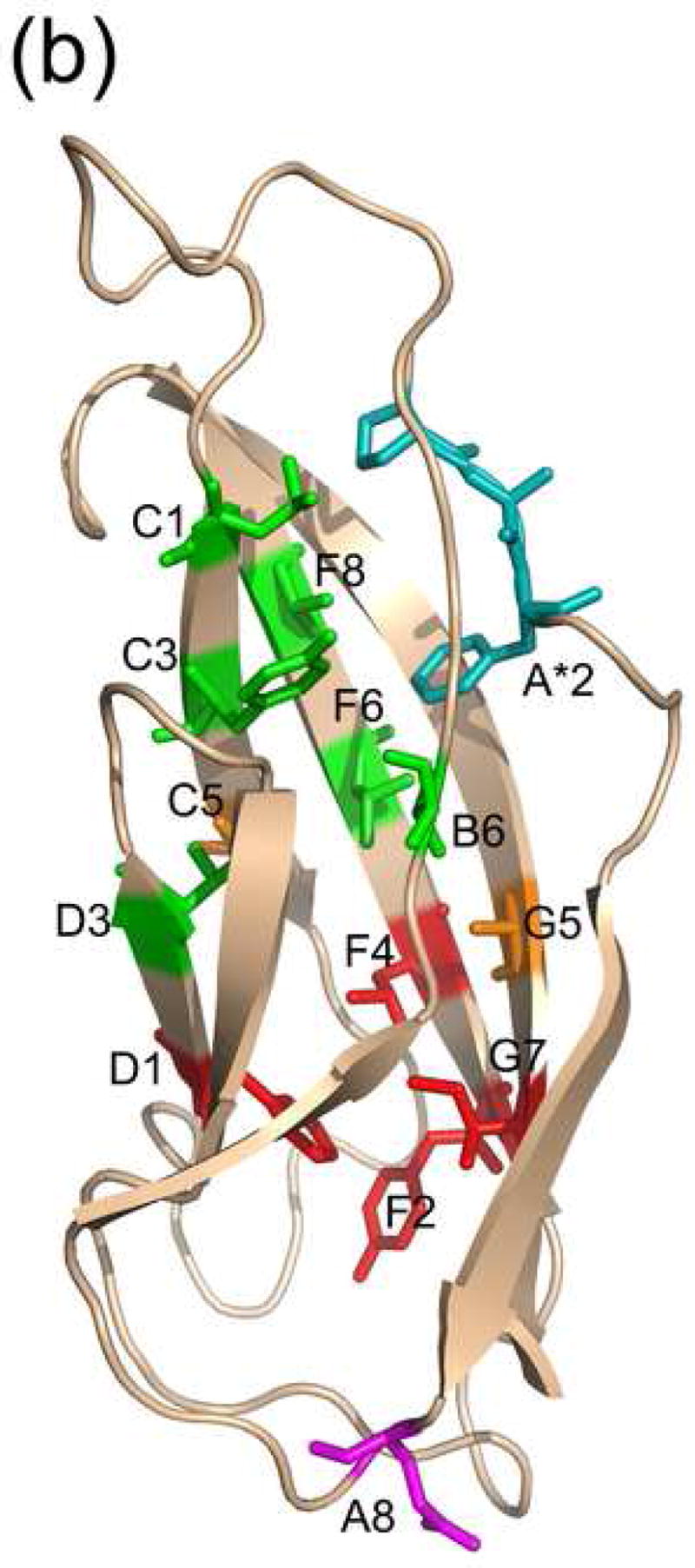
(a) Structure-based sequence alignment of cadherin consensus sequences. Positions that are conserved in terms of residue type are highlighted, with residues of cluster 1 in green and residues of cluster 2 in red. The two bridge positions are in orange. Residue positions are labeled according to their positions in this alignment, using the typeI-EC1 consensus sequence as a reference. The EC1-specific position E89 is shown in blue, and the PxF motif of ECs 2-5 is in aqua. The hinge is denoted ‘Hn’ and the linker ‘Ln’. (b) The locations in the structure of the conserved positions in C-cadherin EC2 (PDB code 1L3W) are shown in stick representation and colored as in (a).
All the domain sequences were pooled and filtered to remove redundancy using CD-HIT40 with a threshold of 98% sequence identity. Short hits (length <90) which are not full-length cadherin domains were also removed. Following the filtering step, 9788 domain sequences remain. In order to assign each sequence with a particular domain class (for example, one “class” would contain EC1 domains from type I cadherins, and another EC3 domains from desmosomal cadherins). To accomplish this, we first needed to obtain a representative seed sequence for each domain. We searched the SwissProt database for well-annotated sequences belonging to each of the 9 cadherin families. These were then partitioned into sequences for individual domains based on visual inspection of the linker regions. Where present, the DxNDN Ca2+-binding motif in the linker region defined the C-terminus of the previous domain (DxNE for the EC3-EC4 linker). For cases where the Ca2+-binding motif was absent, alignments to similar sequences where the motif was present were used to aid the domain definitions. The domains defined in this way were then clustered at the level of 60% sequence identity. For each domain class a multiple sequence alignment was created using T-COFFEE41. An HMM was constructed from the alignment and used to generate the representative sequence for each domain class using HMMER42 with default parameters. The 80 HMMs are available as supplementary data and are also on our website, http://luna.bioc.columbia.edu/honiglab/Cadherins/, as are the representative sequences.
Domain class-specific consensus sequences
Each of the 9788 sequences in our database was associated with one of the 80 domain classes if the sequence was greater than 50% identical to the corresponding representative seed sequence. If this criterion was met by more than one seed sequence, the query sequence was assigned to the domain with which it had the greater level of identity. The choice of 50% as a cutoff is somewhat arbitrary, but it is based on the fact that cadherin domains are typically 45–60% identical to homologous sequences of the same domain and 20–40% identical to sequences of other domains. Table 2 (supplemental) provides some of the relevant numbers for classical and desmosomal cadherins, but the results are quite similar for all cadherin domains (data not shown).
Of the 9788 domain sequences, 3868 could be assigned to one of the 80 domain classes based on the 50% sequence identity criterion. A multiple alignment of the sequences assigned to a particular domain class was carried out with T-COFFEE and a majority-rule consensus sequence was constructed by selecting the residue most frequently found at each position of the alignment. Note that in the previous section we constructed HMMs and representative seed sequences from a relatively small number of well-annotated SwissProt sequences and used them to assign sequences to individual domains. In this section we build “consensus sequences” from the entire set of sequences assigned to a particular domain class.
Structure-based alignment of consensus sequences of types I and II cadherin domains
The procedure described above allowed us to obtain consensus sequences for each of the 80 domain classes. We focus now on the ten classical cadherin domain classes and use structural information to align their consensus sequences to one another. This alignment will enable the identification of conserved positions that are common to all ten domain classes. A multiple structure alignment of 17 Type I and Type II cadherin domains was generated with Ska43, and this yielded a structure-based sequence alignment. The consensus sequence for the five type I cadherin domains (EC1-5) and 3 type II domains (EC1-3) for which structures are available were then aligned to the structure-based sequence alignment using an approach that has been described previously44. Since there are no available structures for the type II EC4 and EC5 domains, the HMAP program45 was used to align the consensus sequence of these domains to the structure-based alignment. Figure 2a shows the multiple alignment of the 10 consensus sequences. The residues that are highlighted are discussed in the following sections.
Sequence and structure determinants of the cadherin fold
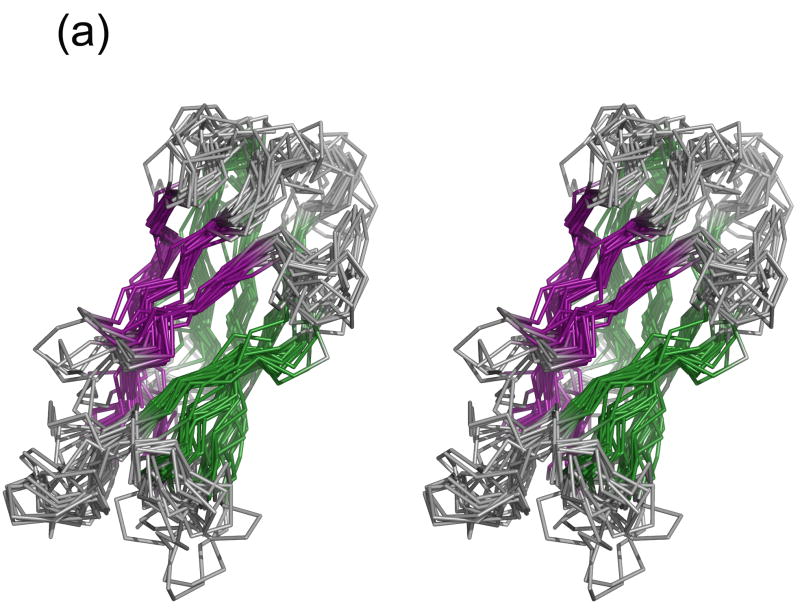
Figure 3a displays a multiple alignment of cadherin domains of known structure. Where multiple structures were available for the same domain (for example, E-cadherin EC2 and cadherin-8 EC1), the domain structure with the highest resolution and most native sequence (with the fewest number of non-native residues) was used. It is clear that the basic cadherin fold is well-conserved and that the core β-sheets superimpose quite well. The greatest variation is seen in the N-terminal region where EC1 domains, due to swapping, have different conformations, and in the loops. Most pairwise RMSDs are below 3 Å (Figure 3b), and most of the RMSDs calculated using only residues in β-sheets are below 2 Å (data not shown). Corresponding domains from the same family are structurally almost indistinguishable; for example, the three type I EC1s and type II EC1s have pairwise RMSDs of 1.2.–1.9 Å and 0.5–1.3 Å, respectively (for β-sheets, the ranges are 0.47–0.53 Å and 0.30–0.37 Å). Moreover, corresponding domains from different families are generally more similar than different domains within the same family; for example, type I EC1s are most closely related to type II EC1s, with pairwise RMSDs <2.2 Å; type I EC2s are most closely related to type II EC2s, with pairwise RMSDs <1.4 Å. Similarly, the recent NMR structure of desmoglein-2 EC1 (PDB 2YQG) is closest to the type I EC1s, with pairwise RMSDs of 2.2–2.3 Å. The two protocadherin EC1s are structurally similar to each other (2.3 Å RMSD) but are outliers relative to the classical cadherin domains, with RMSDs of 2.7–4.3 Å due primarily to differences in the loop regions. In contrast to the classical cadherin domains, which contain only β structure, protocadherin EC1s have an α-helix inserted between the B and C strands. When only the β-sheets are considered, the protocadherin EC1s are 1.04–3.08 Å distant from the classical domains, indicating that the β-sheet regions are structurally conserved.
Figure 3.
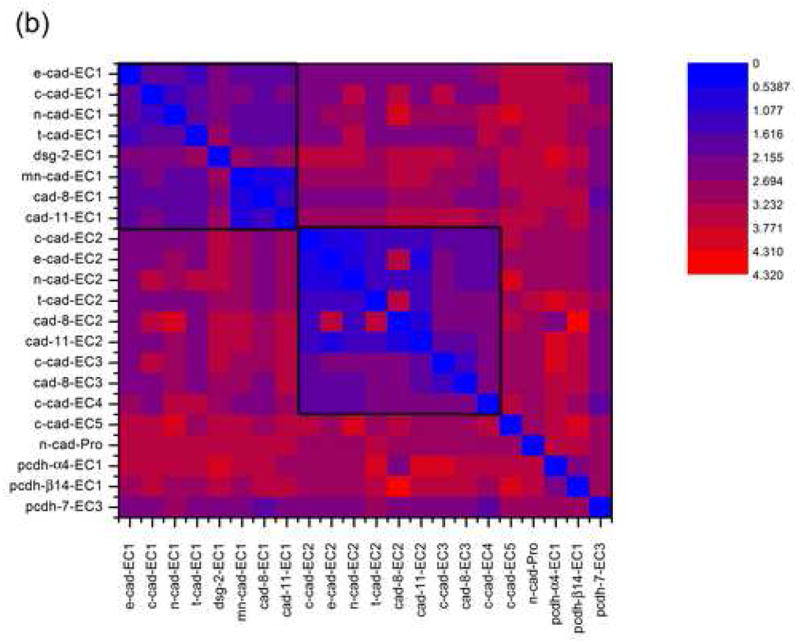
Structural alignment of 22 cadherin domains. (a) Ribbon representation of the aligned domains, in stereo. The β strands of sheet I are shown in purple and of sheet II in green. The structural alignment was constructed using Ska43. (b) Contour plot of all-against-all RMSD values for cadherin domains. The boxed regions indicate EC1s (upper left) and ECs 2, 3, and 4 (center).
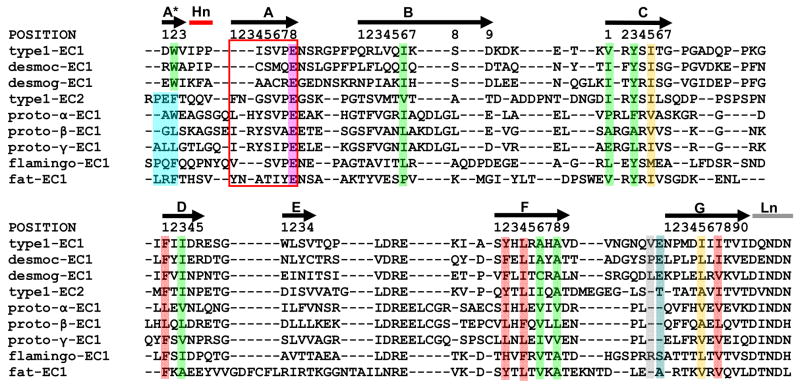
In order to identify determinants of the cadherin fold, we analyzed the consensus sequences of the ten classical cadherin domain classes and their multiple sequence alignments (Figure 2a) by looking for conserved sequence signals at each position in the alignment. These signals were based on a grouping of amino acids based on physical properties that was introduced by Ptitsyn and Ting46. The residue types are: nonpolar (LIVMFYWA); acidic and amide (DENQ); basic (KRH); hydroxyl (ST); proline (P); cysteine (C); and glycine (G). Conserved positions were identified in two stages: First, each column of the global consensus sequence alignment (Figure 2a) was examined to see if the same residue type was present in 9 of the 10 consensus sequences at that position, with no gaps allowed. If this criterion was satisfied, the corresponding position of each type I and type II domain class-specific multiple sequence alignment in our cadherin database was checked. If the same residue type was present in the majority of sequences (>50%) for each domain class separately, the column was considered to be a position whose properties are common to all classical cadherin EC domains, and was labeled as a “conserved” position.
This procedure identifies fourteen conserved positions, thirteen of which are hydrophobic (Figure 2). Eleven of the positions are especially conserved, with the same residue type present in >70% of the sequences of each separate alignment. Based on their positions in the β-strands, these positions are labeled A*2, B6, C1, C3, D1, D3, F2, F4, F6, F8, G5, and G7. The remaining two conserved positions, A8 and C5, are found in 50–70% of the sequences of each separate alignment. All of the conserved hydrophobic residues are buried in the protein interior with average solvent accessible surfaces (SAS) <20 Å2. The single polar conserved position is a Glu in the AB loop that forms part of the interdomain Ca2+ binding site at the four junctions between the five cadherin domains. However, this Glu is also present in EC5 even though it does not bind Ca2+. Some of these thirteen positions have been seen in previous work37,47; however, our use of structure-based definition of a domain adds A*2, F6, and F8 as conserved domain positions.
The conserved nonpolar residues form a network of contacts that define the hydrophobic core of a cadherin domain. The contacts delineate two independent clusters of core residues within each domain, with core residues G5 and C5 bridging the two clusters (Figure 2b). In the first cluster, which consists of seven residues (A*2, B6, D3, C1, C3, F6, and F8), twelve pairwise contacts are conserved in at least 14 of the 17 cadherin domain structures. In the second cluster, four residues (D1, F2, F4, and G7) make five conserved contacts. Most of the contacts are present at the interface between the two β-sheets and are closely related to the contacts between the core residues of immunoglobulin domains48.
Determinants of swapping in the EC1 domain
Although all classical cadherin repeats share a common core, only the N-terminal domain (EC1) forms the strand-swapped dimer interface. There is presently little direct evidence as to whether other EC domains would form swapped dimers if their N-termini were free, but a study of the E-cadherin EC2 domain does show that it is monomeric in solution49, and our own results show that E-cadherin EC2-EC3 domain constructs are monomeric (unpublished). Our goal in this section is to determine whether there are differences between the EC1 domain and all other EC domain classes that suggest why only EC1 domains form a strand-swapped dimer. The conservation patterns shown in Figure 2a suggest that there are a number of features that are unique to EC1. These include:
Trp2 - There is a Trp residue at position 2 in all EC1 domains, whereas other EC domains have another hydrophobic residue at that position, usually a Phe. It is known that the mutation of Trp2 to an Ala eliminates cell-cell adhesion, whereas mutation to a Phe significantly reduces the extent of adhesion, suggesting that the dimerization free energy is severely reduced by the mutation27. The Trp binding pocket in the strand-swapped dimer is part of one of the two hydrophobic clusters described above. It is formed by residues B6 (Ile24 in EC1), C3 (Tyr36), F6 (Ser78), and F8 (Ala80). In monomer forms of EC1, and in domains EC2-5, Trp2 (or Phe4) inserts into the corresponding pocket in its own domain. Type II cadherins have a second conserved tryptophan, Trp4, that also participates in swapping interactions.
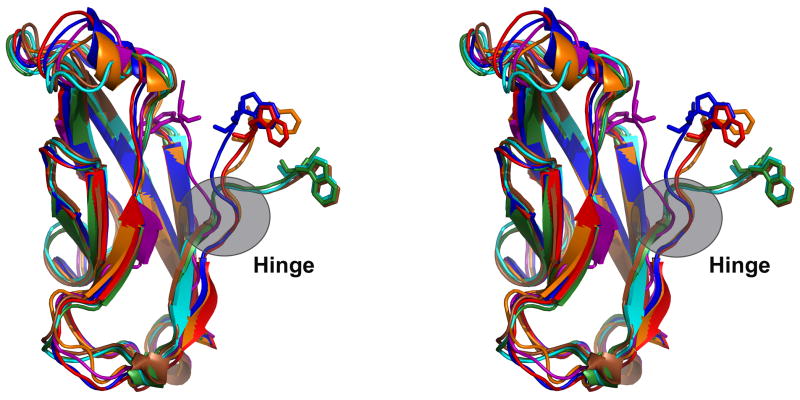
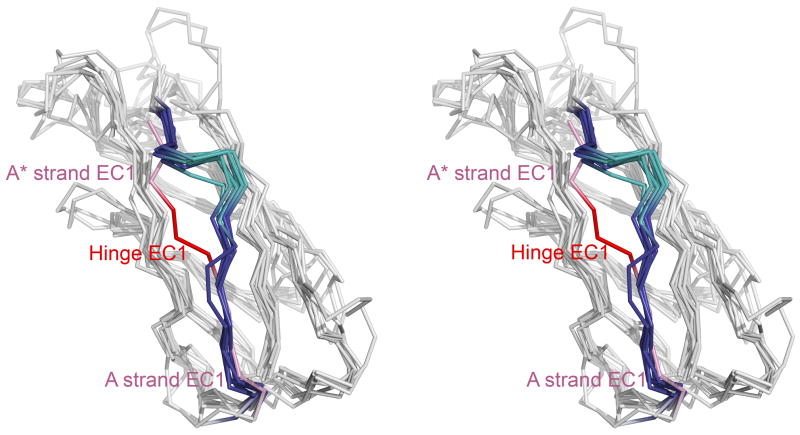
Length of the A strand and strain - The A strand of EC1s is shorter than in other domains. Compared to EC1, ECs 2-4 have two extra residues in the A strand that form additional hydrogen bonds with the G strand (Figure 2a). EC5 domains appear to have only one extra residue. The three residues preceding the A strand (positions 3–5) in EC1 form the hinge that links the swapping N-terminus to the rest of the domain (Figure 4). As a consequence of the longer A strand, the hinge region in ECs 2-5 is positioned higher in the structure than in EC1 and forms a bulge (Figure 5). It is possible that the shorter strand in EC1 destabilizes the monomer by inducing strain in closed conformations, where Trp2 tucks into a pocket in its own monomer. Indeed, it has been observed in other systems that shortening an N-terminal strand leads to the formation of swapped dimers50–52.
The absence of a proline near the N-terminus - Domains EC2-EC5 have a conserved Pro2 that is absent in EC1 domains (Figure 2a). This Pro is part of a PXF motif that is found in most classical cadherin domains, though some contain variants; for example, many type I cadherins have a PXP in EC5. The presence of the extra proline in EC2-5 may play a role in inhibiting swapping. In these domains, residues of the B and G strands form a pocket into which the Pro inserts and which may serve to anchor the A and A* strands (Figure 6a) so as to inhibit swapping. In EC1, the proline “anchor” is missing and the proline pocket is filled instead by a C-terminal residue, Val88. Val88 is part of an extended FG loop in the EC1 G strand that has no structural equivalent in ECs 2-5 (Figure 6b and c). It appears then that EC1 has added residues in the FG loop that fill a hydrophobic pocket which is filled by Pro2 in other EC domains.
Glu89 - EC1 domains have a conserved Glu at position 89 that is not present in other domains (Figure 2a). Glu89, like the neighboring Val88, is located in the FG loop of EC1. The Glu89 side chain forms a salt bridge with the charged amino terminus as part of the swapping interface. Mutations of EC1 that destroy the salt bridge either by replacing Glu89 with Ala or by extending the N-terminus interfere with adhesion26. Given the critical contribution of the salt bridge to adhesion, the presence of a negatively charged residue that is capable of forming the salt bridge seems likely to be required for swapping.
Figure 4.
Stereo view of alternate conformations of the A* strand. The E-cadherin EC1 monomer (PDB 1O6S) with W2 inserted into its pocket (purple) was aligned using Ska43 to a set of EC1 dimer protomers with W2 extended toward its partner molecule: E-cadherin (2O72, blue), N-cadherin (1NCG, orange), C-cadherin (1L3W, red), cadherin-8 (1ZXK, green), cadherin-11 (2A4C, cyan), and mn-cadherin (1ZVN, brown). The hinge connecting the swapping A* strand to the rest of the domain is indicated.
Figure 5.
Multiple structure alignment of cadherin domains. Domains in the alignment include E-cadherin EC1 (PDB 1O6S), N-cadherin EC2 (1NCJ), E-cadherin EC2 (2O72), C-cadherin EC2 (1L3W), cadherin-11 EC2 (2A4E), cadherin-8 EC2 (2A62), C-cadherin EC3 (1L3W), cadherin-8 EC3 (2A62), and C-cadherin EC4 (1L3W). The A* strand, hinge, and A strand are highlighted (E-cadherin EC1 A* and A strands in pink and hinge in red; ECs 2-4 A* and A strands in blue and hinge in teal). C-cadherin EC5 (1L3W) was excluded for clarity of visualization, since its A strand has a slightly different orientation. The view is in stereo.
Figure 6.
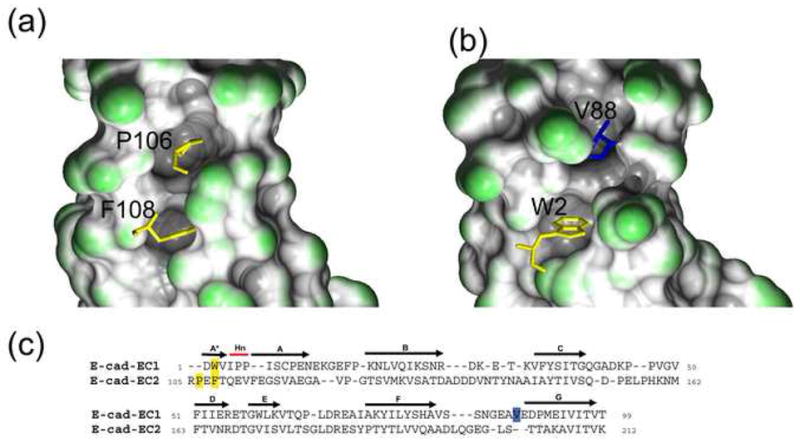
Pocket filling residues in EC2 and EC1. (a) In EC2, P106 of the A* strand PxF motif fills a pocket in the molecule. This interaction is shown with the PxF motif in stick representation and the molecular surface of the pocket drawn. The A* and A strand residues were not included in building the molecular surface. (b) In EC1, the equivalent pocket is filled by a residue of the G strand, V88. The A*, A strand, and A87-V88 residues were not included in building the molecular surface so as to highlight the filled pocket. (c) Structure-based sequence alignment of E-cadherin EC1 and E-cadherin EC2 (PDB 1FF5). N-terminal residues – P106 and F108 in EC2 and W2 in EC1 – are indicated in yellow. V88, in the EC1 G strand, is in blue.
Non-classical cadherins
The sequence relationships in supplementary Table 2, an alignment of consensus sequences (Figure 2a), and the conservation pattern of the residues in the hydrophobic core (data not shown) strongly suggest that all the cadherin domains included in this work have structures that closely resemble the domain structure of classical cadherins. Here we consider whether the individual EC1 domains are likely to dimerize through a strand-swapped interface. Figure 7 contains an alignment of the consensus sequences of EC1 domains for which no structures have been determined with the consensus type I EC1 and EC2 sequences. This alignment will be used as a basis for determining whether non-classical cadherins form strand-swapped interfaces. While the alignment demonstrates that all of the non-classical cadherin domains share the overall fold as well as the conserved core positions with the classical domains, significant differences arise in the regions that contribute to swapping (Figure 7).
Figure 7.
Sequence alignment of type I EC1 and EC2 consensus sequences with consensus sequences from non-classical cadherin EC1s. The alignment was generated by aligning each non-classical cadherin consensus sequence to the type I EC1 consensus sequence using HMAP45. The pairwise alignments were merged and the resulting multiple sequence alignment is displayed. The alignment columns corresponding to the core positions of the classical cadherin domains are colored as in Figure 2. The EC1-specific positions V88 (gray) and E89 (blue) are highlighted, and the region aligned to the PxF motif of EC2 is in aqua. The A strand is boxed in red.
Desmosomal cadherins
As can be seen in Figure 7, all of the swapping determinants can be found in the desmocollin and desmoglein EC1 domains. Specifically, a) there is a conserved Trp at position 2, b) the three conserved pocket positions whose sidechains directly contact Trp2 – Ile24 (B6), Tyr36 (C3), and Ala80 (F8) – are conserved, c) there is no Pro near the N-terminus and the extended FG loop containing Val88 is present, d) the A strand is the same length as in classical cadherin EC1s, and e) Glu89 is conserved. The recent NMR structure of desmoglein-2 EC1 (PDB 2YQG) confirms the conservation of these structural features in desmogleins. It therefore seems quite likely that desmosomal cadherins dimerize through a strand-swapping mechanism.
Fat and flamingo cadherins
In contrast, fat and flamingo EC1 domains appear to resemble type I EC2 domains more than they resemble EC1 domains and thus are not predicted to swap. They have more residues in the A strand, a Phe instead of a Trp that aligns with Trp2, and an extra nonpolar residue that aligns with the N-terminal Pro in EC2 domains. In addition, the FG loop is not extended. It is interesting that fats do have a Glu residue that is nearly aligned with Glu89 in EC1, but that is the only swapping determinant that may be present.
Clustered protocadherins
Although two NMR structures of protocadherin EC1 domains are available, the N-terminal conformations are still uncertain. The structure of protocadherin-β14 (1WYJ) has seven residues at the N-terminus that are due to a cloning artifact and do not appear in the native sequence. As a result the N-terminal conformation is distorted. The correct N-terminus has been shown to be crucial for the formation of a strand-swapped dimerization interface in classical cadherins21,26, and this may be true of protocadherins as well. The N-terminal sequence of the protocadherin-α4 structure (1WUZ) is GNSQ, as predicted by signal peptide prediction algorithms25. In contrast, results from a high-throughput mass spectrometry screen suggest that the N-terminus of protocadherin-α2 may contain four additional residues N-terminal to the GNSQ sequence (AWEAGNSQ)53. In our analysis we have used the structural alignment of 1WYJ and 1WUZ to the classical cadherin structures to guide the alignment of the corresponding EC1 consensus sequences, but we have manually adjusted the N-terminal regions to be in accord with the sequence determined from mass spectrometry analysis. In the N-terminal cadherin domains of all 3 protocadherin families, most of the core positions are conserved. Structural alignment of the classical cadherin domains with the NMR structures of protocadherins-α4 and -β14 confirm that the positions of the core positions are equivalent in these molecules. Position C1 is an exception; it is not hydrophobic in 2 of the 3 protocadherin families.
Our analysis suggests that protocadherins do not form strand-swapped dimers. The A strand is longer than in EC1 domains of classical cadherins and there is no residue that aligns with Glu89. On the other hand, about two thirds of the α-protocadherins contain potential N-terminal Trps, as do some members of the γ-protocadherin family. However, the Trp is not universally conserved in these families as it is in the classical cadherins, and it is rarely present in the β-protocadherins. Moreover, although the three swapping pocket residues are hydrophobic in the clustered protocadherins, the sidechains in the protocadherins tend to be bigger than in the classical cadherins: For example, α protocadherins tend to have valine at position F8 instead of alanine. As a result, the pockets formed by these positions in the clustered protocadherins are smaller and less likely to accommodate the large Trp sidechain. Moreover, all three families of protocadherins have an α-helix between the B and C strands which is absent in classical cadherins. A highly conserved leucine in this helix (L26 in 1WUZ and L34 in 1WYJ) partially fills the hydrophobic pocket in both protocadherin NMR structures, further reducing the cavity volume.
DE- and DN-cadherin
For the D. melanogaster proteins DE- and DN-cadherin, the number of cadherin domains in the extracellular region as well as the precise N-termini have been the subject of some uncertainty. Oda et al.6 suggested that the DE-cadherin sequence contains six cadherin repeats, with the first repeat cleaved off by a protease6. However, Hill et al.54 contend that DE-cadherin has eight cadherin repeats, where the proposed cleavage site is in the middle of the second repeat, and that there is no cleavage upon maturation. Similarly, the number of cadherin repeats predicted in DN-cadherin ranges from 16–1954,55. In this case, a hypothetical internal cleavage site is found in the last cadherin repeat55. Despite this uncertainty, for both DE- and DN-cadherin, both the first cadherin repeat (which may be cleaved off) and the N-terminal repeat of the proposed mature protein align much better to classical cadherin EC2 domains than they do to EC1. None of the swapping determinants are present, suggesting that these adhesion molecules form a different dimerization interface than do classical cadherins.
DISCUSSION
The major goal of this paper is to determine whether the non-classical cadherins have dimerization interfaces similar to those formed by classical cadherins. Specifically, we address the question of whether non-classical cadherins exploit the β-strand swapping mechanism used by classical cadherins to form an interface. Since the presence of strand swapping is known to be extremely sensitive to the length and characteristics of the sequence in the N-terminal region of the EC1 domain, we used structural information to define the N-termini of other domains. We find that all current definitions of cadherin domains leave out the N-terminal residues, and some databases omit entire N-terminal strands. This problem highlights the need to use structural information, where available, to make domain assignments. We have compiled a new and comprehensive database of cadherin domains and have generated HMMs and consensus sequences for each. Our classified cadherin sequences are available as supplementary data and at http://luna.bioc.columbia.edu/honiglab/Cadherins/.
Based on the known structures of type I and type II cadherin domains, we have identified a number of sequence features that are unique to EC1 domains. These include the length of the A strand, the presence of a Trp residue in position 2 in EC1 domains, and a Glu at position 89 that forms a salt-bridge with the N terminus. In addition, domains EC2-5 have an extra residue near their N-terminus, usually a Pro, that inserts into a hydrophobic pocket in its own domain and in this way appears to inhibit swapping. We assume that EC2-5 are prototypes of domains that do not strand swap. The central finding of this work is that, with the exception of desmosomal cadherins, all other cadherin EC1 domains more closely resemble classical cadherin domains EC2-5 than they do EC1. This is true at the sequence level (Table 2 supplemental), but it is most evident by comparing sequences in the N-terminal region, where it is clear that the unique features of classical EC1s are absent in all non-classical EC1 domains.
Our analysis thus suggests that non-classical cadherins do not use a strand swap binding mechanism. Those that function in cell adhesion are therefore likely to associate through different interfaces than the one formed between EC1 domains of classical cadherins. This is somewhat surprising, since the domains themselves are very similar in structure and might be expected to interact in similar ways. EC1 superimposes structurally with other domains quite well (Figure 3a) and shares the same set of conserved positions that we have shown make up the hydrophobic core of all cadherin domains. These core residues include the Ala in the HAV motif that has often been thought to be a signature motif from cadherins. However, the His and Val are not generally conserved, and the motif itself has no obvious structural or functional significance. Indeed, the side chains of the His and Val do not even face the binding pocket. In contrast, the Ala corresponds to core position F8 and participates in interactions at the interface, where it forms part of the hydrophobic pocket and contacts the swapping Trp in EC1.
The hydrophobic core of EC1 must be maintained for adhesion to occur. Mutations at positions in the first hydrophobic cluster, at positions C3 (Tyr36), F6 (Ser78), and F8 (Ala80) in EC1 abolish adhesion. Mutations in the second cluster either abolish adhesion, as with position D1 (Phe51), or diminish it - position F2 (Tyr74)56. In contrast, mutations at position C3 or D1 in EC2 reduce adhesion only slightly, which is consistent with the fact that EC2 does not participate directly in the adhesive interface. The second cluster appears to be evolutionarily conserved, presumably due to its structural role in defining the hydrophobic core of the domain. The first cluster plays an additional, functional role: in EC1 domains, the cluster serves as the binding interface for the strand that swaps from a partner chain. The generation of a functional motif in EC1 from an internal structural motif is a striking example of how strand swapping can be used in the evolution of new oligomeric interfaces57.
A similar phenomenon has been observed in the evolution of dimerization in the Cro family of bacteriophage transcription factors. λ Cro forms a domain-swapped homodimer via a ‘ball-and-socket’ interface in which a Phe from one subunit inserts into a hydrophobic pocket on the partner subunit58. Dimeric λ Cro can bind specifically to its cognate operator sites and repress transcription. In contrast to λ Cro, which dimerizes as a free protein, the homologous P22 Cro domain forms a stable monomer in solution and dimerizes only when bound to DNA. It has been shown that the Cro dimer interface evolved from an intramolecular interaction that was part of the hydrophobic core in the ancestral Cro domain58. Here, we propose that a similar process occurred in the evolution of the swapped cadherin interface.
Strand swapping or, more generally, domain swapping, might be expected to be energetically unfavorable since, in the simplest definition of the process, interactions in the dimer replace corresponding interactions in the monomer. Moreover, the loss of translational and rotational entropy will always oppose dimer formation. It has been observed that interfaces that are formed via domain swapping tend to have lower binding affinities than comparably sized interfaces between subunits that do not swap59, but nevertheless, in many cases swapping is energetically favorable. In such instances, there must be favorable interactions present in the dimer that are absent in the monomer or, conversely, that strain in the monomer is removed upon dimer formation. The latter mechanism has been exploited in a number of protein design experiments either by shortening hinge regions in a way that inhibits the formation of monomeric conformations, or by adding prolines to a hinge where its limited conformational freedom can induce strain in a monomer conformation50,52,60–62. Type I cadherins appear to exploit both mechanisms since the hinge region in their EC1 domains is shorter than in other EC domains, and there are two prolines in the hinge whose possible importance has recently been discussed23. Type II cadherins do not contain prolines in the hinge region, however, in common with type I cadherins, the hinge region is significantly shorter than in other EC domains.
The nearest structural neighbors to cadherins are the set of I-type immunoglobulins, which have the same overall topology63. Interestingly, truncated constructs of I-type immunoglobulin domains in the Trk family of nerve growth factor receptors have also been shown to swap A strands64,65. The Trk dimers differ from cadherins in exchanging the full length of their A strands, with the hinge located at the AB loop, and the hinges in homologous Trk dimers can adopt different orientations65. In the case of the Trk family, the swapping domain is the fifth, membrane-proximal Ig domain of the extracellular region; it does not swap in the native protein since it is not at the N-terminus, but the truncated domain is capable of swapping. In contrast, the ability of cadherin EC1 to swap cannot be due only to its position at the N-terminus: truncated EC2 constructs have been shown to be monomeric49, and the EC2-like prodomain, which is cleaved in the mature protein, is incapable of mediating adhesion20,49. Rather, there must be specific structural features of EC1 that distinguish it from other cadherin domains and allow it to swap.
The exclusive presence of a Trp2 in all classical cadherin EC1 domains is difficult to understand. It replaces a nonpolar residue, usually a Phe, that occupies the same position in EC2-5 and, as mentioned above, a Trp to Phe mutation in EC1 weakens cell-cell adhesion27. Trp is less hydrophobic than Phe66, but if its interactions in the monomer form are identical to those in the dimer, this should not affect the binding affinity. It is possible that the greater size of a Trp, and the presence of polar atoms in its ring system, make it more difficult to accommodate in the monomer conformation and that, together with the shortened hinge loop, destabilizes the monomer with respect to the dimer. Despite uncertainties as to the physical basis for the role of Trp2, this residue provides a clear sequence marker for EC1 domains.
Our study does not provide evidence as to the possible role of Ca2+ in strand swapping. It is known that Ca2+ is required for the formation of adhesive interactions between cadherins on apposing cells; when Ca2+ is depleted, only lateral interactions between molecules on the same cell surface can form16,28,68. This observation has been attributed to the role of Ca2+ in rigidifying cadherin ectodomains in a conformation that facilitates trans interactions. In contrast, when the relative orientation of cadherin domains is not restricted, the protein can fold back on itself and can easily form lateral interactions with molecules on the same cell surface16,18,69. It is less clear whether, in addition, Ca2+ binding directly facilitates strand swapping. Haussinger et al.21 have found that the dimerization of E-cadherin EC1-EC2 domain constructs is enhanced by Ca2+ binding, suggesting a possible role in the swapping process; however, the molecular mechanism through which this might occur is still obscure.
As discussed above, EC1 domains of desmosomal cadherins are very similar to EC1 domains of classical cadherins, while all other non-classical cadherin domains are more similar to EC2-5. This suggests that the other non-classical cadherins dimerize through another interface and that the strand-swapped interface formed by EC1 domains represents an evolutionary development that is observed only in vertebrates. There is little direct information as to the nature of a second interface, but structural studies of cadherins with additional residues at their N-termini, which do not form strand-swapped dimers, (PDB codes 1FF516 and 1EDH15) may provide a clue. These proteins form an interface in the Ca2+ binding region that lies between their EC1 and EC2 domains and, based on the “X”-like shape of the four domains in the dimer, we have designated this as the “X-interface”. The physiological relevance, if any, of this interface is unknown, but it is of interest that T-cadherin dimerizes through the formation of an X-interface but not with the standard strand swapping mechanism (unpublished). A third type of interface has been observed in crystal structures of C-cadherin17 and E-cadherin15,16,23 in which residues from the C, D, and F strands of EC1 interact with the lower parts of the B, D, and E strands of EC2. Whether this interface has physiological relevance or arises solely from crystal contacts is currently unclear.
In earlier work, we have shown that strand swapping can provide the basis for explaining how a family of very similar molecules such as the type I and type II cadherins can form highly specific molecular interfaces that have weak binding affinities59. We demonstrated that low absolute dimerization affinities, which result from strand swapping, make it possible for even small differences between homophilic and heterophilic affinities, in combination with multiple interactions on the cell surface, to result in highly specific homophilic cell-cell adhesion59. The use of closely related cadherin paralogs to mediate cell-cell adhesion appears to be a phenomenon that is restricted largely to vertebrates. Invertebrates have far fewer cadherins than do vertebrates and the paralogs are less similar to each other. For example, humans have at least five type I cadherins and thirteen type II cadherins, while Drosophila have only three cadherins that contain the classical cytoplasmic domain, one of which is multiply spliced54,67. Moreover, while the N-terminal domains of human N-cadherin and E-cadherin are 60% identical in sequence, the equivalent domains of D. melanogaster N-cadherin and E-cadherin are only 28% identical. We speculate that the evolution in vertebrates of large families of closely related cadherins occurred in concert with the formation of a strand-swapped interface.
METHODS
Multiple sequence alignments were carried out with the T-coffee program41. Hidden Markov Models were constructed using HMMER42. The HMAP profile-profile alignment program was used to align cadherin domains for which no structure is available to domains of known structure. HMAP weights scores at each position in an alignment based on secondary structure information derived either from known structures or from predicted secondary structure. The Ska program43 was used to obtain structure-based alignments. All structural representations were prepared with PyMOL (http://pymol.sourceforge.net), except for Figure 6, which was prepared using GRASP243.
Supplementary Material
Acknowledgments
We thank Jeremie Vendome and Lucy Forrest for valuable discussions and critical reading of the manuscript. For helpful suggestions on sequence and structure analysis, we thank Donald Petrey and Christopher Tang. This work was supported by NIH grants GM30518 and U54-CA121852.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Takeichi M. Morphogenetic roles of classic cadherins. Curr Opin Cell Biol. 1995;7:619–27. doi: 10.1016/0955-0674(95)80102-2. [DOI] [PubMed] [Google Scholar]
- 2.Vleminckx K, Kemler R. Cadherins and tissue formation: integrating adhesion and signaling. Bioessays. 1999;21:211–20. doi: 10.1002/(SICI)1521-1878(199903)21:3<211::AID-BIES5>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 3.Pla P, Moore R, Morali OG, Grille S, Martinozzi S, Delmas V, Larue L. Cadherins in neural crest cell development and transformation. J Cell Physiol. 2001;189:121–32. doi: 10.1002/jcp.10008. [DOI] [PubMed] [Google Scholar]
- 4.Gooding JM, Yap KL, Ikura M. The cadherin-catenin complex as a focal point of cell adhesion and signalling: new insights from three-dimensional structures. Bioessays. 2004;26:497–511. doi: 10.1002/bies.20033. [DOI] [PubMed] [Google Scholar]
- 5.Takeichi M. The cadherin superfamily in neuronal connections and interactions. Nat Rev Neurosci. 2007;8:11–20. doi: 10.1038/nrn2043. [DOI] [PubMed] [Google Scholar]
- 6.Oda H, Uemura T, Harada Y, Iwai Y, Takeichi M. A Drosophila homolog of cadherin associated with armadillo and essential for embryonic cell-cell adhesion. Dev Biol. 1994;165:716–26. doi: 10.1006/dbio.1994.1287. [DOI] [PubMed] [Google Scholar]
- 7.Yonekura S, Ting C, Neves G, Hung K, Hsu S, Chiba A, Chess A, Lee C. The variable transmembrane domain of Drosophila N-cadherin regulates adhesive activity. Mol Cell Biol. 2006;26:6598–608. doi: 10.1128/MCB.00241-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moeller MJ, Soofi A, Braun GS, Li X, Watzl C, Kriz W, Holzman LB. Protocadherin FAT1 binds Ena/VASP proteins and is necessary for actin dynamics and cell polarization. EMBO J. 2004;23:3769–79. doi: 10.1038/sj.emboj.7600380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matakatsu H, Blair SS. Interactions between Fat and Dachsous and the regulation of planar cell polarity in the Drosophila wing. Development. 2004;131:3785–94. doi: 10.1242/dev.01254. [DOI] [PubMed] [Google Scholar]
- 10.Rock R, Schrauth S, Gessler M. Expression of mouse dchs1, fjx1, and fat-j suggests conservation of the planar cell polarity pathway identified in Drosophila. Dev Dyn. 2005;234:747–55. doi: 10.1002/dvdy.20515. [DOI] [PubMed] [Google Scholar]
- 11.Matakatsu H, Blair S. Separating the adhesive and signaling functions of the Fat and Dachsous protocadherins. Development. 2006;133:2315–24. doi: 10.1242/dev.02401. Epub 2006 May 10. [DOI] [PubMed] [Google Scholar]
- 12.Casal J, Lawrence P, Struhl G. Two separate molecular systems, Dachsous/Fat and Starry night/Frizzled, act independently to confer planar cell polarity. Development. 2006;133:4561–72. doi: 10.1242/dev.02641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Overduin M, Harvey T, Bagby S, Tong K, Yau P, Takeichi M, Ikura M. Solution structure of the epithelial cadherin domain responsible for selective cell adhesion. Science. 1995;267:386–9. doi: 10.1126/science.7824937. [DOI] [PubMed] [Google Scholar]
- 14.Shapiro L, Fannon A, Kwong P, Thompson A, Lehmann M, Grubel G, Legrand J, Als-Nielsen J, Colman D, Hendrickson W. Structural basis of cell-cell adhesion by cadherins. Nature. 1995;374:327–37. doi: 10.1038/374327a0. [DOI] [PubMed] [Google Scholar]
- 15.Nagar B, Overduin M, Ikura M, Rini JM. Structural basis of calcium-induced E-cadherin rigidification and dimerization. Nature. 1996;380:360–4. doi: 10.1038/380360a0. [DOI] [PubMed] [Google Scholar]
- 16.Pertz O, Bozic D, Koch AW, Fauser C, Brancaccio A, Engel J. A new crystal structure, Ca2+ dependence and mutational analysis reveal molecular details of E-cadherin homoassociation. EMBO J. 1999;18:1738–47. doi: 10.1093/emboj/18.7.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boggon T, Murray J, Chappuis-Flament S, Wong E, Gumbiner B, Shapiro L. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 2002;296:1308–13. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- 18.Haussinger D, Ahrens T, Sass H, Pertz O, Engel J, Grzesiek S. Calcium-dependent homoassociation of E-cadherin by NMR spectroscopy: changes in mobility, conformation and mapping of contact regions. J Mol Biol. 2002;324:823–39. doi: 10.1016/s0022-2836(02)01137-3. [DOI] [PubMed] [Google Scholar]
- 19.Schubert W, Urbanke C, Ziehm T, Beier V, Machner M, Domann E, Wehland J, Chakraborty T, Heinz D. Structure of internalin, a major invasion protein of Listeria monocytogenes, in complex with its human receptor E-cadherin. Cell. 2002;111:825–36. doi: 10.1016/s0092-8674(02)01136-4. [DOI] [PubMed] [Google Scholar]
- 20.Koch AW, Farooq A, Shan W, Zeng L, Colman DR, Zhou MM. Structure of the neural (N-) cadherin prodomain reveals a cadherin extracellular domain-like fold without adhesive characteristics. Structure. 2004;12:793–805. doi: 10.1016/j.str.2004.02.034. [DOI] [PubMed] [Google Scholar]
- 21.Haussinger D, Ahrens T, Aberle T, Engel J, Stetefeld J, Grzesiek S. Proteolytic E-cadherin activation followed by solution NMR and X-ray crystallography. EMBO J. 2004;23:1699–708. doi: 10.1038/sj.emboj.7600192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel S, Ciatto C, Chen C, Bahna F, Rajebhosale M, Arkus N, Schieren I, Jessell T, Honig B, Price S, Shapiro L. Type II cadherin ectodomain structures: implications for classical cadherin specificity. Cell. 2006;124:1255–68. doi: 10.1016/j.cell.2005.12.046. [DOI] [PubMed] [Google Scholar]
- 23.Parisini E, Higgins JM, Liu JH, Brenner MB, Wang JH. The Crystal Structure of Human E-cadherin Domains 1 and 2, and Comparison with other Cadherins in the Context of Adhesion Mechanism. J Mol Biol. 2007;373:401–11. doi: 10.1016/j.jmb.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Umitsu M, Morishita H, Murata Y, Udaka K, Akutsu H, Yagi T, Ikegami T. 1H, 13C and 15N resonance assignments of the first cadherin domain of Cadherin-related neuronal receptor (CNR)/protocadherin alpha. J Biomol NMR. 2005;31:365–6. doi: 10.1007/s10858-005-2450-4. [DOI] [PubMed] [Google Scholar]
- 25.Morishita H, Umitsu M, Murata Y, Shibata N, Udaka K, Higuchi Y, Akutsu H, Yamaguchi T, Yagi T, Ikegami T. Structure of the cadherin-related neuronal receptor/protocadherin-alpha first extracellular cadherin domain reveals diversity across cadherin families. J Biol Chem. 2006;281:33650–63. doi: 10.1074/jbc.M603298200. [DOI] [PubMed] [Google Scholar]
- 26.Harrison OJ, Corps EM, Kilshaw PJ. Cadherin adhesion depends on a salt bridge at the N-terminus. J Cell Sci. 2005;118:4123–30. doi: 10.1242/jcs.02539. [DOI] [PubMed] [Google Scholar]
- 27.Tamura K, Shan W, Hendrickson W, Colman D, Shapiro L. Structure-function analysis of cell adhesion by neural (N-) cadherin. Neuron. 1998;20:1153–63. doi: 10.1016/s0896-6273(00)80496-1. [DOI] [PubMed] [Google Scholar]
- 28.Troyanovsky R, Sokolov E, Troyanovsky S. Adhesive and lateral E-cadherin dimers are mediated by the same interface. Mol Cell Biol. 2003;23:7965–72. doi: 10.1128/MCB.23.22.7965-7972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harrison OJ, Corps EM, Berge T, Kilshaw PJ. The mechanism of cell adhesion by classical cadherins: the role of domain 1. J Cell Sci. 2005;118:711–21. doi: 10.1242/jcs.01665. [DOI] [PubMed] [Google Scholar]
- 30.He W, Cowin P, Stokes D. Untangling desmosomal knots with electron tomography. Science. 2003;302:109–13. doi: 10.1126/science.1086957. [DOI] [PubMed] [Google Scholar]
- 31.Tomschy A, Fauser C, Landwehr R, Engel J. Homophilic adhesion of E-cadherin occurs by a co-operative two-step interaction of N-terminal domains. Embo J. 1996;15:3507–14. [PMC free article] [PubMed] [Google Scholar]
- 32.Norvell S, Green K. Contributions of extracellular and intracellular domains of full length and chimeric cadherin molecules to junction assembly in epithelial cells. J Cell Sci. 1998;111:1305–18. doi: 10.1242/jcs.111.9.1305. [DOI] [PubMed] [Google Scholar]
- 33.Takeda H, Shimoyama Y, Nagafuchi A, Hirohashi S. E-cadherin functions as a cis-dimer at the cell-cell adhesive interface in vivo. Nat Struct Biol. 1999;6:310–2. doi: 10.1038/7542. [DOI] [PubMed] [Google Scholar]
- 34.Letunic I, Copley R, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–60. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finn R, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy S, Sonnhammer E, Bateman A. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–51. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Mazumder R, O’Donovan C, Redaschi N, Suzek B. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–91. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hatta K, Nose A, Nagafuchi A, Takeichi M. Cloning and expression of cDNA encoding a neural calcium-dependent cell adhesion molecule: its identity in the cadherin gene family. J Cell Biol. 1988;106:873–81. doi: 10.1083/jcb.106.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimoyama Y, Tsujimoto G, Kitajima M, Natori M. Identification of three human type-II classic cadherins and frequent heterophilic interactions between different subclasses of type-II classic cadherins. Biochem J. 2000;349:159–67. doi: 10.1042/0264-6021:3490159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noonan J, Grimwood J, Schmutz J, Dickson M, Myers R. Gene conversion and the evolution of protocadherin gene cluster diversity. Genome Res. 2004;14:354–66. doi: 10.1101/gr.2133704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–9. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 41.Notredame C, Higgins D, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–17. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 42.Eddy SR. Hidden Markov models. Curr Opin Struct Biol. 1996;6:361–5. doi: 10.1016/s0959-440x(96)80056-x. [DOI] [PubMed] [Google Scholar]
- 43.Petrey D, Honig B. GRASP2: visualization, surface properties, and electrostatics of macromolecular structures and sequences. Methods Enzymol. 2003;374:492–509. doi: 10.1016/S0076-6879(03)74021-X. [DOI] [PubMed] [Google Scholar]
- 44.Al-Lazikani B, Sheinerman F, Honig B. Combining multiple structure and sequence alignments to improve sequence detection and alignment: application to the SH2 domains of Janus kinases. Proc Natl Acad Sci U S A. 2001;98:14796–801. doi: 10.1073/pnas.011577898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang C, Xie L, Koh I, Posy S, Alexov E, Honig B. On the role of structural information in remote homology detection and sequence alignment: new methods using hybrid sequence profiles. J Mol Biol. 2003;334:1043–62. doi: 10.1016/j.jmb.2003.10.025. [DOI] [PubMed] [Google Scholar]
- 46.Ptitsyn O, Ting K. Non-functional conserved residues in globins and their possible role as a folding nucleus. J Mol Biol. 1999;291:671–82. doi: 10.1006/jmbi.1999.2920. [DOI] [PubMed] [Google Scholar]
- 47.Kister A, Roytberg M, Chothia C, Vasiliev J, Gelfand I. The sequence determinants of cadherin molecules. Protein Sci. 2001;10:1801–10. doi: 10.1110/ps.37001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chothia C, Gelfand I, Kister A. Structural determinants in the sequences of immunoglobulin variable domain. J Mol Biol. 1998;278:457–79. doi: 10.1006/jmbi.1998.1653. [DOI] [PubMed] [Google Scholar]
- 49.Prasad A, Housley N, Pedigo S. Thermodynamic stability of domain 2 of epithelial cadherin. Biochemistry. 2004;43:8055–66. doi: 10.1021/bi049693c. [DOI] [PubMed] [Google Scholar]
- 50.Green S, Gittis A, Meeker A, Lattman E. One-step evolution of a dimer from a monomeric protein. Nat Struct Biol. 1995;2:746–51. doi: 10.1038/nsb0995-746. [DOI] [PubMed] [Google Scholar]
- 51.Murray A, Lewis S, Barclay A, Brady R. One sequence, two folds: a metastable structure of CD2. Proc Natl Acad Sci U S A. 1995;92:7337–41. doi: 10.1073/pnas.92.16.7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kelley B, Chang L, Bewley C. Engineering an obligate domain-swapped dimer of cyanovirin-N with enhanced anti-HIV activity. J Am Chem Soc. 2002;124:3210–1. doi: 10.1021/ja025537m. [DOI] [PubMed] [Google Scholar]
- 53.Gevaert K, Goethals M, Martens L, Van DJ, Staes A, Thomas G, Vandekerckhove J. Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat Biotechnol. 2003;21:566–9. doi: 10.1038/nbt810. [DOI] [PubMed] [Google Scholar]
- 54.Hill E, Broadbent I, Chothia C, Pettitt J. Cadherin superfamily proteins in Caenorhabditis elegans and Drosophila melanogaster. J Mol Biol. 2001;305:1011–24. doi: 10.1006/jmbi.2000.4361. [DOI] [PubMed] [Google Scholar]
- 55.Iwai Y, Usui T, Hirano S, Steward R, Takeichi M, Uemura T. Axon patterning requires DN-cadherin, a novel neuronal adhesion receptor, in the Drosophila embryonic CNS. Neuron. 1997;19:77–89. doi: 10.1016/s0896-6273(00)80349-9. [DOI] [PubMed] [Google Scholar]
- 56.Kitagawa M, Natori M, Murase S, Hirano S, Taketani S, Suzuki S. Mutation analysis of cadherin-4 reveals amino acid residues of EC1 important for the structure and function. Biochem Biophys Res Commun. 2000;271:358–63. doi: 10.1006/bbrc.2000.2636. [DOI] [PubMed] [Google Scholar]
- 57.Bennett MJ, Schlunegger MP, Eisenberg D. 3D domain swapping: a mechanism for oligomer assembly. Protein Sci. 1995;4:2455–68. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Newlove T, Atkinson K, Van DL, Cordes M. A trade between similar but nonequivalent intrasubunit and intersubunit contacts in Cro dimer evolution. Biochemistry. 2006;45:6379–91. doi: 10.1021/bi052541c. [DOI] [PubMed] [Google Scholar]
- 59.Chen C, Posy S, Ben-Shaul A, Shapiro L, Honig B. Specificity of cell-cell adhesion by classical cadherins: Critical role for low-affinity dimerization through beta-strand swapping. Proc Natl Acad Sci U S A. 2005;102:8531–6. doi: 10.1073/pnas.0503319102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murray AJ, Head JG, Barker JJ, Brady RL. Engineering an intertwined form of CD2 for stability and assembly. Nat Struct Biol. 1998;5:778–82. doi: 10.1038/1816. [DOI] [PubMed] [Google Scholar]
- 61.Simeoni F, Masotti L, Neyroz P. Structural role of the proline residues of the beta-hinge region of p13suc1 as revealed by site-directed mutagenesis and fluorescence studies. Biochemistry. 2001;40:8030–42. doi: 10.1021/bi002812d. [DOI] [PubMed] [Google Scholar]
- 62.Barrientos LG, Louis JM, Botos I, Mori T, Han Z, O’Keefe BR, Boyd MR, Wlodawer A, Gronenborn AM. The domain-swapped dimer of cyanovirin-N is in a metastable folded state: reconciliation of X-ray and NMR structures. Structure. 2002;10:673–86. doi: 10.1016/s0969-2126(02)00758-x. [DOI] [PubMed] [Google Scholar]
- 63.Harpaz Y, Chothia C. Many of the immunoglobulin superfamily domains in cell adhesion molecules and surface receptors belong to a new structural set which is close to that containing variable domains. J Mol Biol. 1994;238:528–39. doi: 10.1006/jmbi.1994.1312. [DOI] [PubMed] [Google Scholar]
- 64.Wiesmann C, Ultsch M, Bass S, de VA. Crystal structure of nerve growth factor in complex with the ligand-binding domain of the TrkA receptor. Nature. 1999;401:184–8. doi: 10.1038/43705. [DOI] [PubMed] [Google Scholar]
- 65.Ultsch M, Wiesmann C, Simmons L, Henrich J, Yang M, Reilly D, Bass S, de VA. Crystal structures of the neurotrophin-binding domain of TrkA, TrkB and TrkC. J Mol Biol. 1999;290:149–59. doi: 10.1006/jmbi.1999.2816. [DOI] [PubMed] [Google Scholar]
- 66.Wolfenden R, Radzicka A. How hydrophilic is tryptophan? TIBS. 1986;11:69–70. [Google Scholar]
- 67.Ting C, Yonekura S, Chung P, Hsu S, Robertson H, Chiba A, Lee C. Drosophila N-cadherin functions in the first stage of the two-stage layer-selection process of R7 photoreceptor afferents. Development. 2005;132:953–63. doi: 10.1242/dev.01661. [DOI] [PubMed] [Google Scholar]
- 68.Klingelhofer J, Laur OY, Troyanovsky RB, Troyanovsky SM. Dynamic interplay between adhesive and lateral E-cadherin dimers. Mol Cell Biol. 2002;22:7449–58. doi: 10.1128/MCB.22.21.7449-7458.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pokutta S, Herrenknecht K, Kemler R, Engel J. Conformational changes of the recombinant extracellular domain of E-cadherin upon calcium binding. Eur J Biochem. 1994;223:1019–26. doi: 10.1111/j.1432-1033.1994.tb19080.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.