Abstract
To identify gene expression patterns in human dopamine (DA) neurons in the substantia nigra pars compacta (SNc) of male and female control and Parkinson disease (PD) patients, we harvested DA neurons from frozen SNc from 16 subjects (4 male PD, 4 female PD, 4 male and 4 female controls) using Laser Capture microcrodissection and microarrays. We assessed for enrichment of functional categories with a hypergeometric distribution. The data were validated with QPCR.
We observed that gender has a pervasive effect on gene expression in DA neurons. Genes upregulated in females relative to males are mainly involved in signal transduction and neuronal maturation, while in males some of the upregulated genes (alpha-synuclein and PINK1) were previously implicated in the pathogenesis of PD. In females with PD we found alterations in genes with protein kinase activity, genes involved in proteolysis and WNT signaling pathway, while in males with PD there were alterations in protein-binding proteins and copper-binding proteins.
Our data reveal broad gender-based differences in gene expression in human dopaminergic neurons of SNc that may underlie the predisposition of males to PD. Moreover, we show that gender influences the response to PD, suggesting that the nature of the disease and the response to treatment may be gender-dependent.
Introduction
Parkinson Disease (PD) is a progressive neurodegenerative disorder affecting up to 3% of individuals over the age of 65 years and it is associated with degeneration of dopamine (DA) neurons in the substantia nigra pars compacta (SNc) (Standaert and Young, 2005), resulting in loss of DA innervation to the caudate and putamen. The cause of most cases of PD is unknown. In familial forms of the disease, several genetic mutations have been identified, including alpha-synuclein (mutations and gene multiplications, ASYN), PINK1, Parkin, UCH-L1 and LRRK2 (Gwinn-Hardy, 2002). While these provided insight into some aspects of the disease, collectively they account for a very small fraction of the total number of cases of PD. The lack of strong monogenic risk factors in the majority of typical late-onset PD cases is supported by twin studies (Tanner, 2003) as well as a recent genome-wide SNP study (Maraganore et al., 2005). These findings have focused attention on more complex risk factors including environmental effects.
One of the strongest identified risk factors for development of PD is male gender. The prevalence of PD in males is higher in most populations studied, and data on disease incidence suggest that men have at least a two-fold greater risk of PD at all ages (Baldereschi et al., 2000; Van Den Eeden et al., 2003). Gender also influences the clinical features of the disease after onset, with females exhibiting more impairment of postural stability, depression, and reduced ability to conduct daily activities (Haaxma et al., 2006; Lyons et al., 1998; Rojo et al., 2003; Uitti et al., 2005). In contrast, men are more likely to exhibit REM behavior disorders and other sleep disturbances (Scaglione et al., 2005). Gender also interacts with other risk factors, attenuating the protective effect of caffeine in women (Ascherio et al., 2003) and altering the relationship between cigarette smoking, alleles of MAO B, and risk for PD (Kelada et al., 2002). These gender-specific characteristics of PD are also associated with gender-specific differences in DA metabolism in the basal ganglia, with higher densities of DA markers in healthy women, and greater preservation of dopamine uptake sites in women with PD (Kaasinen et al., 2001).
To identify biological factors involved in the pathogenesis of PD and how they are influenced by gender, we have used Affymetrix high-density gene arrays to study DA neurons isolated from postmortem human brain. We used LCM to isolate DA neurons from human post-mortem SN from male and female control and PD subjects. This allowed us to control for the profound loss of DA cells that occurs in PD, as it may present special challenges in studies of gene expression. In fact homogenates of the SNc are likely to contain relatively few neurons and a large number of glia. The microarray data was analyzed using several different bioinformatics strategies including a statistical approach to study the functional profile of the identified genes.
Subjects and Methods
Brain tissue
Frozen human brain tissue was obtained from the Harvard Brain Tissue Resource Center at McLean Hospital and from the Alzheimer Disease Research Center (ADRC) at Massachusetts General Hospital. The clinical characteristics are described in Table 1. Out of a total of 34 brains initially examined, we collected frozen substantia nigra from 16 subjects (4 male PD, 4 female PD, 4 male and 4 female controls) matched for age, post-mortem interval (PMI) and most importantly RNA quality (Table 1). The remaining 18 brains, 17.6% were unsuitable because they did not match the age range in consideration, 30% were unsuitable for the study because of poor RNA quality, and 5.8% could not be used because of RNA quality and a concomitant diagnosis of AD along with the diagnosis of PD.
Table 1. Vital Statistics of the cases used in this study.
Samples from a total of 16 human brains, 8 PDs (4 male and 4 female) and 8 controls (4 male and 4 female) were studied. These were selected from available material after screening total RNA extracted from a small region of the SN for integrity of 18S and 28S species using an Agilent bioanalyzer. We also matched the cases for age and post mortem interval (PMI) (PD Female age, 75 ± 4.4, PMI, 20.5 ± 2.5; PD Male age 77 ± 6.3, PMI 13.85 ± 1.7; Con Female age 68 ± 3.3, PMI 21.8 ± 0.8; Con Male age 70.2 ± 6.3, PMI 18.11 ± 3.6).
| Case | Age | Sex | PMI | Case | Age | Sex | PMI |
|---|---|---|---|---|---|---|---|
| Pdf1 | 81 | F | 17 | cf1 | 61 | f | <24 |
| Pdf2 | 84 | F | 24.08 | cf2 | n/a | f | >48 |
| Pdf3 | 69 | F | <24 | cf3 | 74 | f | 23.00 |
| Pdf4 | 66 | F | <24 | cf4 | 69 | f | 20.70 |
| pdm1 | 68 | m | 17.16 | Cm1 | 67 | m | 22.33 |
| pdm2 | 67 | m | 14.5 | Cm2 | 63 | m | 21.50 |
| pdm3 | 94 | m | 9.25 | Cm3 | 62 | m | 21.20 |
| pdm4 | 78 | m | 14.50 | Cm4 | 89 | m | 7.42 |
RNA quality
To assess RNA quality we dissected a small piece of SNc, extracted RNA with Tri reagent (Sigma, St. Louis, MO) according to specifications, and used an Agilent Bioanalyzer (Agilent, Palo Alto, CA) for total RNA (Buesa et al., 2004). Only the brains that showed good RNA integrity by the presence of sharp 18S and 28S peaks, and no signs of RNA degradation in the fluorograms as previously described (Buesa et al., 2004) were considered for subsequent LCM and microarray hybridization.
Laser Capture Microdissection (LCM)
All procedures were carried out under strict RNAse-free conditions. Eight-micron thick cryostat sections of human SNc were mounted on uncoated glass slides, and stored at −80°. The majority of human SNc DA neurons in the adult brain contain neuromelanin, thus these neurons can be visualized without staining, however we stained with methylene blue to help localizing the neuronal soma (Cantuti-Castelvetri et al., 2005b). Prior to capturing, the sections were thawed, fixed (70% ethanol, 45 secs), stained (0.1% methylene blue, 20 secs) and dehydrated (50, 70, 90, 100% ethanol, 5 secs each; xylenes, 5–10 min). LCM was performed with an Arcturus PixCell II instrument (Mountain View, CA).
Amplification and gene expression microarray analysis
RNA from 500 DA neurons from the SNc of each specimen were extracted using the Picopure™ RNA isolation kit and amplified using the RiboAmp™ RNA amplification kit (Arcturus). This kit is based on a T7-RNA polymerase method and can linearly amplify as little as 100 pg of total RNA. Samples that yielded products with an average size of 1Kbp and an absorbance λ260/280 between 1.85–2.1 were sent to the Harvard Partners Center for Genetics and Genomics (HPCGG) facility to generate biotinylated amplified RNA (aRNA) and be hybridized to GeneChip® Human X3P expression arrays (Affymetrix, Santa Clara, CA The X3P Array targets a total of 47,000 transcripts with 61,000 probe-sets.
Real-Time quantitative PCR
To validate the results of the microarray in silico analysis we harvested additional samples of DA SNpc neurons from the same cases and analyzed them by real-time PCR (QPCR) without prior RNA amplification. Short synthetic (18–20 mer) PCR primers were designed to amplify small (100–300 bp) amplicons for candidate mRNAs using Primer3 software (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). First-strand cDNA synthesis was carried out on RNA extracted from the dissected cells with an Invitrogen (Carlsbad, CA) Superscript™ first-strand synthesis kit. QPCR was carried out in a 96 well plate using an iCycler® (BioRad, Hercules, CA), and SYBR® Green PCR Master Mix (Applied Biosystems, Foster City, CA) with the following protocol: 6 min hot start, at 95°C, denaturation 95°C. 30sec, annealing 60°C, 30sec, elongation 72°C, for 45 cycles, followed by eighty 0.5°C increases in temperature (starting at 55°C) to collect melting curve data. For each primer set we used a curve with known concentrations of cDNA that was used to calculate the primers’ efficiency, and to quantitate products. All the QPCR results were calculated with the ΔΔCt method (Fink et al., 1998; Livak and Schmittgen, 2001) and normalized to number of LCM cells in the PCR reaction. Then following transcripts were considered for validation: Actin gamma (NM_001614, forward: TCTGTGGCTTGGTGAGTCTG; reverse: GTCCTACGGCTTGGACTTTC, 164bp, annealing temp. 60°C), beta catenin (NM_001904, forward: AAACAGGAGGGATGGAAGG, reverse: ATACCACCCACTTGGCAGAC, 152 bp, annealing temp. 60°C; KLK6 (NM_006303, forward: CGTGATGAGTGAGGACTTGG, reverse: AGCCACTGCCTTATGGAGAC, 254 bp, annealing temp. 60°C). The sequences for primers for alpha-synuclein and heatshock protein 90 were previously published (Cantuti-Castelvetri et al., 2005a).
Statistical Methods
Microarray analysis
Chips were developed, scanned, and normalized using global scaling. All quality control parameters calculated by Affymetrix GCOS Software were monitored. The images of the chips were analyzed to find spotted or damaged array regions and the graphical analysis of all chips through this step provides evidence that data is of good quality (Quackenbush, 2002).
The overall present call rate was 23.84%±5.18% (Controls 24%±6.5%; PD 23.6%±3.76%; males 27.87%±1.9%; females 19.8%±4). The variability of the overall background, noise, and raw present signal was less than 45% pre-normalization, and the spike in controls less than 20%.
All data were normalized using the GeneChip Robust MultiChips Analysis (GCRMA) algorithm (Cope et al., 2004), performed with ArrayAssistLite (Stratagene). Any probes associated with transcripts no longer listed in the Entrez Gene, the Entrez Nucleotide, or the Unigene databases (http://www.ncbi.nlm.nih.gov/entrez) were excluded from the analysis, as were sequences associated with warnings in the same databases.
Selection of differentially expressed probes
The probe-sets with intensity lower than background level (log2 5.63 ± 0.46 SD) in all samples were filtered out (intensity log2 ≤ 4), leaving 17,838 probe-sets after filtering. The remaining probe-sets (intensity log2 ≥ 4) were used for further statistical analysis. The average of the groups PD, Control, Female, and Male, as well as the 4 subgroups female PD, female Control, male PD, and male Control were used to calculate the expression for each gene. Tests were computed with two-tailed unpaired t-test, multiclass analysis, blocks and unpaired 2 classes analyses using the Significance Analysis of Microarrays (SAM) algorithms computed with the SAM 2.0 plug-in for Excel (Efron and Tibshirani, 2002; Larsson et al., 2005; Troyanskaya et al., 2001; Tusher et al., 2001).
We defined genes as being regulated by the disease when all of these criteria were met: a) the ratio PD/Control was either up- or down-regulated at least two fold, b) the difference between the two groups was statistically significant (p ≤0.05), c) and the difference was independent of the gender of the subjects (p>0.05 for the ratio Female/Male, and ratios female PD/female Control and male PD/male Control both at least up- or down-regulated 2 fold).
We defined genes as gender-specific when all of the following criteria were met: a) the ratio female/male was either up- or down-regulated at least two fold, b) the difference between the two groups was statistically significant (p ≤0.05), c) and the difference was independent of the disease status of the subjects (p>0.05 for the ratio PD/Control, and ratios PD female/PD male and Control female/Control male both at least up- or down-regulated 2 fold).
We defined genes as female PD-specific, genes which: a) are not represented in the Disease or the Gender specific lists, b) the ratio female PD/female Control was either up- or down-regulated at least two fold, and c) the difference between the two groups was statistically significant (p ≤0.05). Similarly, we defined genes as specific for male PD, genes which: a) are not represented in the Disease or the Gender specific lists, b) the ratio male PD/male Control was either up- or down-regulated at least two fold, and c) the difference between the two groups was statistically significant (p ≤ 0.05).
To control for the variance within each probe-set considered in the analysis we also calculated false discovery rates (q) with SAM for the 30,000 first probe-sets ranked by average signal of the 4 groups (Efron and Tibshirani, 2002; Larsson et al., 2005; Troyanskaya et al., 2001; Tusher et al., 2001). This parameter is influence by the variability of the data set as it compares the relative difference of the samples to the distribution of the dataset after random permutations of the sample set (Efron and Tibshirani, 2002; Larsson et al., 2005; Troyanskaya et al., 2001; Tusher et al., 2001).
Clustering and Self Organizing Maps
K-means clustering and Self-Organizing Maps (SOM) were used to cluster the statistically significant genes by similarity of profiles on the normalized averages of the subgroups of samples and hierarchical clustering on median normalized samples using cosine correlation with complete linkage was performed (Fig 1); on all samples using SpotFire DecisionSite for Functional Genomics 8.0 (Eisen et al., 1998).
Figure 1. Hierarchical clustering.
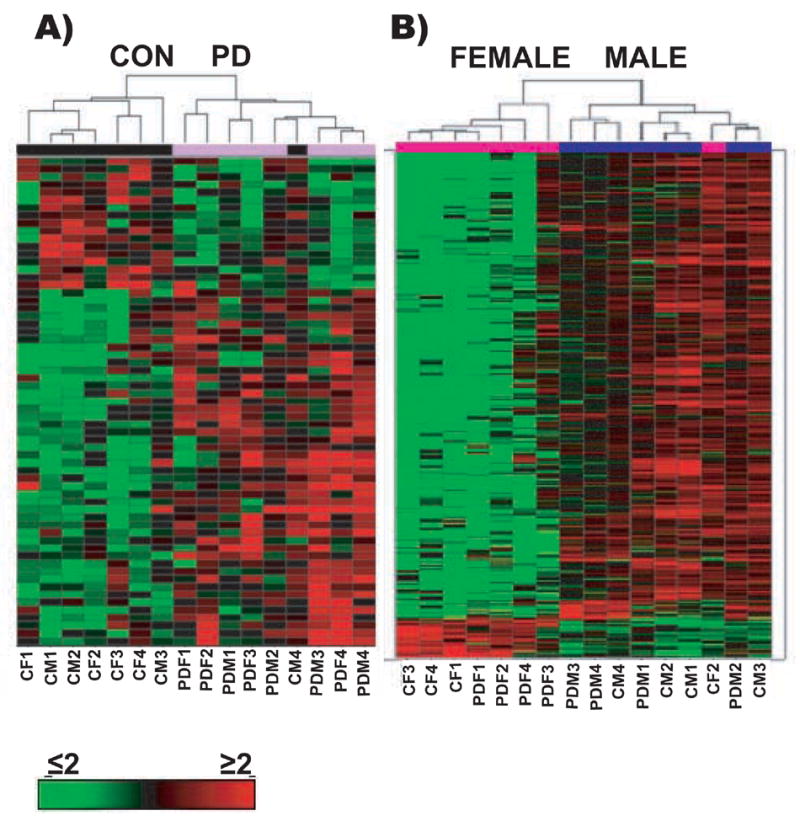
Hierarchical clustering on median normalized samples using cosine correlation with complete linkage was performed on all samples to determine the gene clustering and to better visualize differences in expression profiles between the PD patients and healthy control subjects regardless of their gender (panel A, Disease); and between the female and the male subjects regardless of their disease status (panel B, Gender). This approach discriminates well between control and disease cases, with co-clustering of 7 of 8 control cases, and 7 of 8 females.
Functional Profiling
To determine the biological processes in which the selected genes are involved we used the Gene Ontology (GO) annotations (Ashburner et al., 2000). GO annotations for each probe-set on the Affymetrix X3P array, were extracted from GeneBank (ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/gene2go.gz). The representation of each functional category is dependent on the biology of the system, and the number of regulated genes in the system and the number of genes belonging to each functional category represented on the array. Thus, for each term belonging to the biological process of the GO we consider the null hypothesis that regulation is equivalent to random sampling from the overall population of genes. To do so, we use the hypergeometric distribution: . This formula computes the probability of observing exactly k regulated genes belonging to a given GO term containing a total of u genes, given that we have r regulated genes in the experiment and n genes on the array.
By computing the probability of observing at least k regulated genes, given by the formula we obtain the level of significance pk at which we can reject the null hypothesis of random regulation for a GO term containing u genes.
To compute the functional analyses for each experimental group we used only those significantly different probe-sets that had a q≤11%. We considered the genes, but not the probe-sets, to calculate which categories were overrepresented in our datasets, so that genes represented by more than one probe-set would actually be counted only once. We considered as ‘significantly overrepresented’ categories that were significant at p≤0.01 and that were populated by at least 3% of the regulated genes in that experimental group. Moreover in the data presentation we do not trim any category that is ‘significantly overrepresented’ even when they are very broad (to navigate the categories and the hierarchical relationship between categories see QuickGO: http://www.ebi.ac.uk/ego/).
Results
Microarray Results
Gender has a broad impact on gene expression patterns in SNc DA neurons
We evaluated the impact of gender on the gene expression patterns of laser-picked DA neurons of the SNc by comparing all the male cases against all of the female cases, regardless of the disease status (see methods). The genes differentially expressed between male and female subjects but not different between controls and PD cases were identified as gender-specific (supplemental table 1). We identified 124 probe-sets representing approximately 120 genes upregulated in the females relative to males, and over 2060 probe-sets (representing approximately 2000 genes) upregulated in the males relative to females. Not surprisingly the genes with the largest magnitude difference in expression were X- and Y-chromosome linked, i.e. XIST (423 fold, p ≤10E-11) in the females, and RPS4Y in males (31.6 fold, p ≤2.0E-13), both of which have previously been reported as differentially expressed by gender in the mammalian brain (Eriksson et al., 1999; Vawter et al., 2004), and another Y linked gene DDX3Y which was overexpressed in males (111 fold, p ≤8.8E-15).
The majority of the gender-specific genes identified, however, were not linked to sex chromosomes, and have not previously been implicated in gender differences in human brain.
To analyze the biological roles of the differentially expressed genes we used the annotations contained in the databases of the Gene Ontology Consortium, and calculated the probability that particular functional categories were over-represented in our data set with a hypergeometric distribution (see methods), using the 1870 probe-sets with a multiclass q value<0.11. The functional categories considered significantly overrepresented in the genes upregulated in males or in the female subjects considered in this study had to be significant at alpha ≤0.01 and represented by at least 3% of the significantly regulated genes.
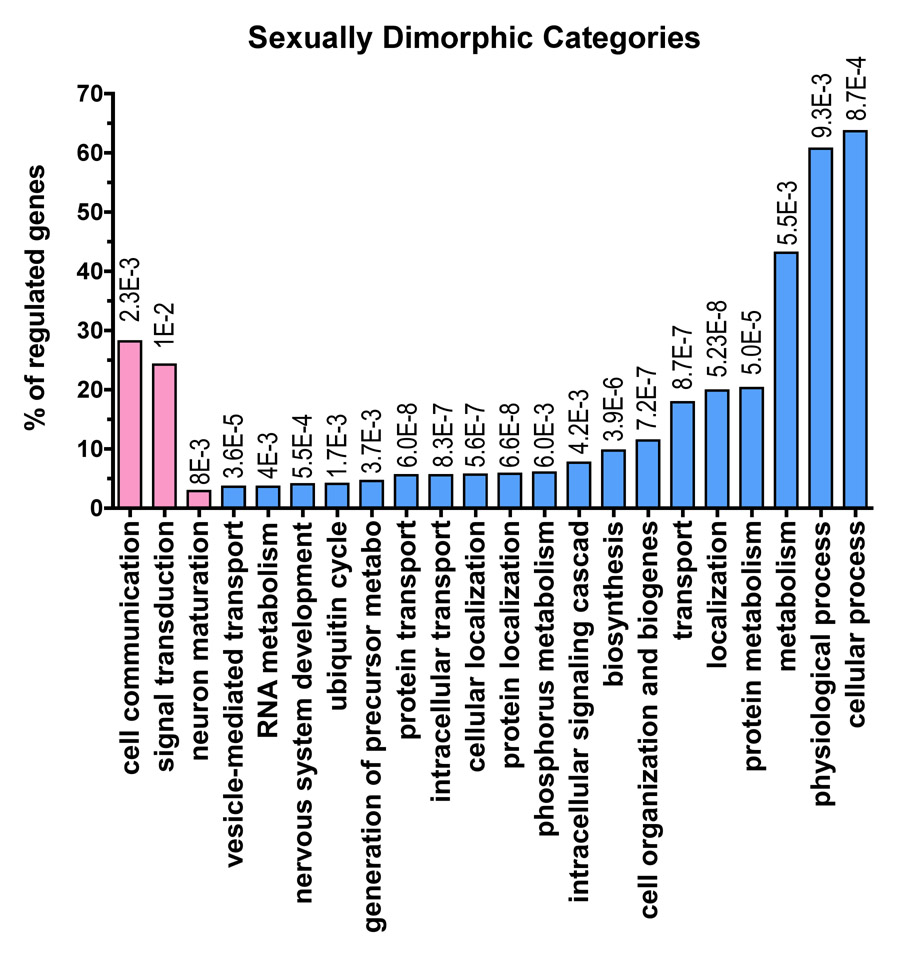
With this approach, we found genes influenced by gender to represent a wide variety of metabolic processes within the cell (Fig. 2). Interestingly, genes upregulated in the female subjects are mainly involved in signal transduction, and neuronal maturation, while in the ones upregulated in males we found many other different types of upregulated genes, of extreme interest is that in the genes upregulated in the males we see genes related to previously implicated in the mechanisms of PD pathogenesis, such as oxidative phosphorylation (genes contained in the phosphorous metabolism category). We also observed upregulation in males of several transcripts directly related to known ‘genetic’ mechanisms of PD, including ASYN (Gwinn-Hardy, 2002), and PINK1 (Gwinn-Hardy, 2002).
Figure 2. Functional profiling of the gender-specific genes.
124 probe-sets (~ 120 genes) upregulated in the females relative to males, and over 2060 probe-sets (~2000 genes) upregulated in the males relative to females were assigned to functional categories using the Gene Ontology Database. For each functional grouping, the probability that the number of genes observed was greater than chance was calculated according to the hypergeometric distribution (see methods). Only the probe-sets with a multiclass q value<0.11 were used for functional analysis (1870 probe-sets). The table shows the categories identified as significant (p <0.01, # genes in category ≥3% of total regulated genes) for the biological process: Pk= p value; k/u= number of regulated genes observed (k) over the total number of genes in the GO class (u). The regulated genes can be represented under more than one functional term as they can be implicated in one or more biological processes. A list of the genes regulated by gender can be found as supplementary Table 1. The functional categories overrepresented in the gene sets upregulated in the females are shown with the pink bars and the functional categories overrepresented in the gene sets upregulated in males are shown with the light blue bars. At the top of each bar is shown the p value for each functional category. In this figure we show all of the functional categories that are significantly different according to our method. We show both more specific and informative categories such as ‘ubiquitin cycle’ and more broad categories such as ‘cellular process’. Cellular process in only one of eighteen different sub-categories under the parent term ‘biological process’. Some of the categories at the same hierarchical level as ‘cellular process’ are ‘biological adhesion’, ‘reproductive process’ or ‘response to stimulus’ (to search categories and their meaning and hierarchical relationship see QuickGO: http://www.ebi.ac.uk/ego/).
Parkinson Disease affects the expression of a small number of genes in the dopaminergic neurons of the substantia nigra
We identified genes that were differentially expressed in controls and PD cases irrespective of gender (supplementary table 2). We found only 149 probe-sets that were altered by the disease but not gender. Of these, 35 probe-sets (35 genes) were downregulated in PD with respect to control, and 114 probe-sets (110 genes) were upregulated.
Functional analysis similar to that performed for the gender group was performed on the 112 probe-sets with a multiclass q value<0.11. This approach revealed that genes affected by disease but not gender were involved several cellular processes such as transmission of nerve impulse, synaptic transmission, cell-cell signaling, establishment of localization, localization, transport (Fig 3). Among the genes that exhibit gender-independent altered expression in PD are: neuronal beta-catenin, which has been implicated in Alzheimer disease and synaptic plasticity (Fuentealba et al., 2004; Goda, 2002); PAEL-R, a protein accumulated in Lewy bodies (Murakami et al., 2004) and a potential parkin substrate (Nakahara et al., 2003; Yang et al., 2003); kallikrein 6, involved in ASYN degradation (Iwata et al., 2003); and transferrin, which may be related to iron deposition observed in PD brain (Morris et al., 1994).
Figure 3. Functional profiling of the Disease-specific genes: Genes common to the two genders.
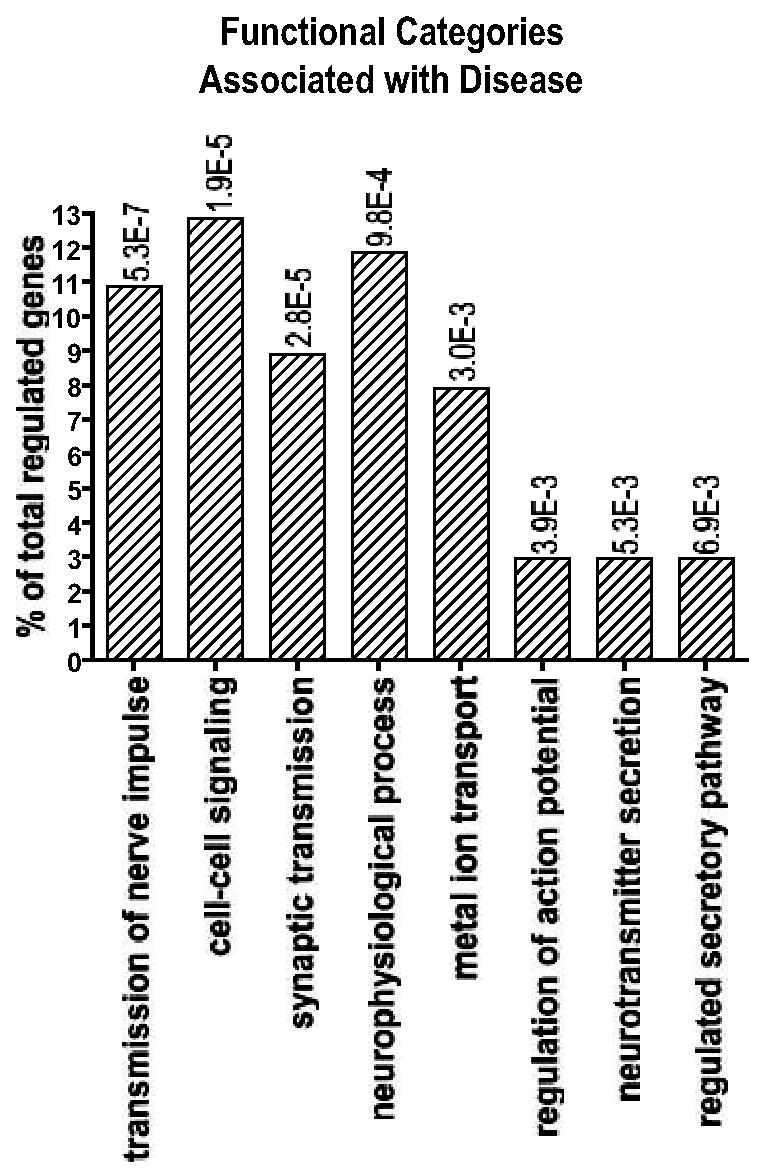
Of 149 probe-sets (35 probe-sets/genes were downregulated in PD, and 114 probe-sets/110 genes were upregulated in PD with respect of controls) regulated by PD, but not by gender, 112 probe-sets with a multiclass q value<0.11 were analyzed using the Gene Ontology Database with a process similar to that used for the gender-specific genes. The table shows the categories identified as significant (p <0.01, # genes in category ≥3% of total regulated genes) for the biological process: Pk= p value; k/u= number of regulated genes observed (k) over the total number of genes in the GO class (u). A list of the disease-specific genes common to the two genders can be found as supplementary Table 2.
Gender influences the response to Parkinson Disease
Finally we identified genes altered only in females with PD (supplementary table 3) by comparing females with PD against control females.
This analysis revealed 288 probe-sets differentially regulated in females with PD. Of these 102 probe-sets were downregulated (100 with a multiclass q-value<0.11), and 186 probe-sets upregulated (169 with a multiclass q-value<0.11). Functional analysis similar to that performed for the effect of gender alone revealed that DA neurons from females with PD compared to female controls showed alterations in genes with kinase and protein kinase activity, genes involved in proteolysis and genes related to the WNT signaling pathway (Fig. 4).
Figure 4. Functional profiling of the Disease-specific genes: Gender specific effects of PD.
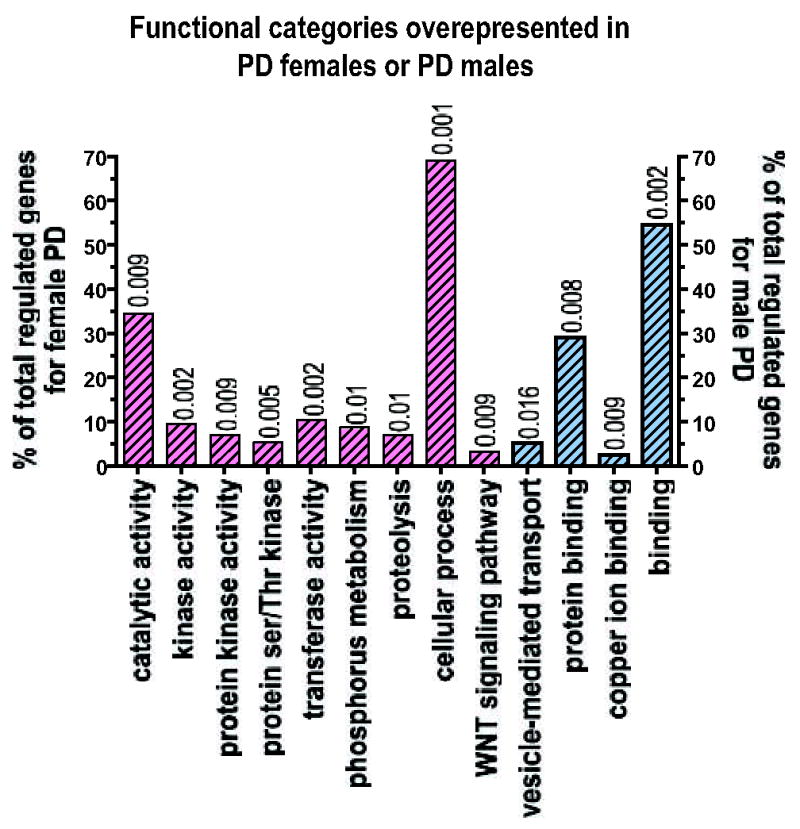
To analyze the functional categories associated with PD females or PD males compared to their respective controls using the Gene Ontology Database with a process similar to that used for the gender-independent genes, we used only probe-sets with a multiclass q value<0.11), i.e. 269 probe-sets out of 289 for the females with PD and 198 probe-sets out of a total of 292. The figure shows the categories for the biological processes and molecular function identified as significant (p <0.01, # genes in category ≥3% of total regulated genes). The pink bars with diagonal bands represent the categories associated with female PD and the light blue bars with diagonal bands represent the functional categories associated with male PD. The complete list of genes regulated in female PD with respect to female controls can be found as supplementary Table 3 and the complete list of genes regulated in male PD with respect to male controls can be found as supplementary Table 4. In this figure we show all of the functional categories that are significantly different according to our method. We show both more specific and informative categories such as ‘ubiquitin cycle’ and more broad categories such as ‘cellular process’. Cellular process in only one of eighteen different sub-categories under the parent term ‘biological process’. Some of the categories at the same hierarchical level as ‘cellular process’ are ‘biological adhesion’, ‘reproductive process’ or ‘response to stimulus’ (to search categories and their meaning and hierarchical relationship see QuickGO: http://www.ebi.ac.uk/ego/).
Genes altered specifically in males with PD (supplementary table 4) were determined by comparing males with PD against control males. In males with PD 292 probe-sets were differentially regulated, 232 probe-sets downregulated (161 with a multiclass q-value<0.11), 60 probe-sets upregulated (38 with a multiclass q-value<0.11). In the males affected by PD, a small percentage of the genes belonged to proteins involved in vesicle-mediated transport, although this category is represented by 5.3% of the regulated genes, only showed a trend to significance (p=0.016), most of the remaining regulated genes were protein-binding proteins and copper-binding proteins (Fig 4).
QPCR Array Validation
To validate the results from the microarray analysis, we harvested additional DA cells and examined mRNA abundance of a specific subset of genes using real-time PCR (QPCR) without prior RNA amplification. We used cells from the same male and female control subjects from the array study to test actin-gamma, ASYN, and HSP90. The levels of each of these three transcripts determined with QPCR were in good accordance with the array results (Table 2).
Table 2. QPCR validation of the arrays.
Neurons harvested by LCM from the same male and female control subjects used in the arrays, were used to test actin-gamma, ASYN, and HSP90. The levels of these genes determined with QPCR were in good accordance with the array results. Cells captured from the control cases as well as male and female PD subjects were used to test three other targets: beta-catenin, and KLK6. QPCR confirmed the array data for KLK6 and beta-catenin. Beta catenin’s decrease in the PD cases as determined by PCR was not as dramatic as in the arrays.
| DISEASE | |||
|---|---|---|---|
| Ratio PD/CON | KLK6 | Beta-Catenin | |
| ARRAY | 2.70 | 2.02E-01 | |
| PCR (cumulative) | 3.48 | 7.16E-01 | |
| PDM/CM | 3.63 | 5.82E-01 | |
| PDF/CF | 1.81 | 8.68E-01 | |
| GENDER | |||
| Ratio male/female | Actin-Gamma | HSP90 | ASYN |
| PCR | 3.19 | 3.55 | 3.86 |
| ARRAY | 2.96 | 2.64 | 5.56 |
Cells dissected from control cases and PD subjects were used to test two additional targets: beta-catenin, and kallikrein 6 (KLK6). QPCR confirmed the array data for KLK6 and beta-catenin, although beta catenin’s decrease in the PD cases as determined by PCR was not as dramatic as in the arrays. We noted that KLK6 and beta-catenin also show more pronounced changes in males with PD than females with PD, suggesting that the influence of gender on gene expression may be even more pervasive than the array analysis indicates.
Discussion
The present study shows that gene expression patterns in dopaminergic nigral neuron are different in men and women, and that the effect of PD on gene expression in dopaminergic neurons is strongly influenced by gender. Although no prior array study has addressed gender differences in neurodegenerative diseases, there is substantial evidence for gender dimorphisms in gene expression in other human brain regions (Vawter et al., 2004). Not surprisingly, some of the most strongly dimorphic genes in dopaminergic neurons are those present on X and Y chromosomes, such as XIST and RPSR4Y. The role of these genes in gender-specific development has been studied extensively, but they have gained attention only recently (Dewing et al., 2006). In addition, there are a large number of genes that exhibit gender-specific expression, which is not related to their chromosomal localization.
Our array data is in line with several studies of the anatomy and structure of central dopaminergic systems suggesting that the effects of gender are pervasive. Gender-based differences are noted in monkeys (Leranth et al., 2000) and rats (Beyer et al., 1991) with female animals showing higher numbers of dopaminergic cells in the substantial nigra. These differences seem to be determined by factors other than gonadal hormones, as they are noticeable before the beginning of the critical period of sexual differentiation (Beyer et al., 1991). Dopaminergic neurons from male and female rat embryos dopaminergic cultured in identical hormonal environments show gender-related dimorphism (Raab et al., 1995; Reisert and Pilgrim, 1991). Gonadal hormones, however seem to have a role in maintaining these gender based differences. In female African Green monkeys, which normally have larger numbers of TH-positive neurons in the SN than male monkeys, ovarectomy reduces the numbers of TH-positive cells to a level comparable to males. Estrogens administration shortly after the ovarectomy can restore the numbers of TH neurons, but prolonged estrogen deprivation seems to produce irreversible loss of TH neurons (Leranth et al., 2000). Sexual dimorphism in the nigrostriatal system has also been demonstrated in normal humans using PET with the dopamine transporter ligand FP-CIT (Lavalaye et al., 2000; Staley et al., 2001), the cocaine analog TRODAT-1 (Mozley et al., 2001) and the dopamine precursor [18F]fluorodopa (Laakso et al., 2002). These studies have shown than the density of dopamine uptake sites in the caudate and putamen of women is greater than that of men, although both genders show age-related decline.
The magnitude of the influence of gender on gene expression is considerable, however the authors are confident that this difference reflects the biology of the system studied and not the reflection of a bias in the sample. Recent studies (Preece and Cairns, 2003; Preece et al., 2003) have suggested an effect of gender on quality of the RNA and total RNA amount extracted from post mortem. In one study, specimens originated from female patients yielded worse quality RNA compared to males (Preece and Cairns, 2003), in another study specimens originated from female patients yielded less mRNA per 10μg of total RNA extracted (Preece et al., 2003). In our current study we only used cases with optimal and comparable RNA quality for specimens originated from both male and females. Moreover we hybridized on the arrays equal amounts of mRNA per sample, making sure that the changes we see cannot be attributed to the differences in absolute mRNA content observed in the two genders. At this point in time it is difficult to speculate why there is such a disparity in the number of genes upregulated in the male subjects compared to the number of genes upregulated in the female subjects, but we do hope that the present study can lay the foundation for a deeper understanding of gender differences in the brain, and of the greater sensitivity of males to PD.
In addition to revealing the important effect of gender on the biology of human dopaminergic neurons, our study provides some insight into the mechanisms of PD. It is important to recognize that our study is necessarily limited to analysis of those dopaminergic neurons that survive at the end stage of PD, and therefore may not be representative of the abnormalities that arise in the neurons, which are lost to the disease process. In the pool of surviving neurons, we observed that the disease itself modifies only a modest number of genes. Several of these are plausibly linked to the mechanisms of neurodegeneration, including beta-catenin, kallikrein 6, and transferrin. Further study of these candidates in model systems will be required before a causal relationship of any of these to PD can be established.
It should not come as surprise that PD men and women show a different response at the mRNA level, considering that differences in the frequency and phenotype of the disease is well established, and gender differences have also been demonstrated in animal models of PD. In MPTP models of PD, female mice have been reported to be more resistant to the dopamine-depleting effects of the toxin (Disshon and Dluzen, 2000; Dluzen, 1996; Dluzen et al., 1996; Dluzen et al., 2003; Freyaldenhoven et al., 1996; Miller et al., 1998; Nishino et al., 1998; Tamas et al., 2005). Similarly, male rats show greater susceptibility to toxicity of 6-OHDA when measured either by depletion of dopaminergic neurons or by behavioral recovery (Cass et al., 2005; Datla et al., 2003; Gillies et al., 2004; Murray et al., 2003; Tamas et al., 2005). Interestingly, these gender differences are preserved also in the interactions between environmental factors, gender, and dopaminergic toxins in animals. For example, prenatal exposure to pesticides enhances the vulnerability of males but not females to dopaminergic neurotoxicants later in life (Barlow et al., 2004).
Our array study could be showing the biological basis for such differences.
Several factors have previously been implicated in the male prevalence of PD. In model systems, a protective effect of estrogens can be demonstrated in females but not males (Leranth et al., 2000; Murray et al., 2003). In humans, postmenopausal estrogen replacement does not alter the risk in women (Ascherio et al., 2003), although premature loss of estrogens may be associated with an increased risk of PD (Martignoni et al., 2002; Ragonese et al., 2004). Non-hormonal gender factors are likely involved as well, a view supported by strong linkage to the X chromosome in several genome-wide studies of PD susceptibility (Pankratz et al., 2003; Scott et al., 2001). The differences in gene expression, which we have observed likely, reflect both hormonal and non-hormonal effects.
A strength of this study is that we have used LCM to directly examine gene expression in pure dopaminergic neurons. Prior studies of gene expression in human PD have been limited to studies of homogenates of nigral tissue (Grunblatt et al., 2004; Hauser et al., 2003; Mandel et al., 2005; Miller et al., 2005). Since the loss of DA neurons in PD is profound, and there is replacement by glia, such studies may not be as informative with respect to the metabolism of the dopamine neurons themselves. In addition, none of these prior studies have directly examined the effect of gender, and in most of the studies males were over-represented in the PD sample, precluding a gender analysis. It is also possible that comparing tissue homogenates of control cases (rich in neurons) and PD patients (rich in active glia) masks a potential gender effect that could be intrinsic to the neurons and not to the glia, suggesting that the function of the SN may be sexually dimorphic but not the ‘immunological’ response to the disease.
Another strength of the study is our conservative statistical approach. We have based our main conclusions on the analysis of broad patterns of expression rather than on individual genes. The modest sample size, which is feasible with our experimental approach, provides only moderate confidence in the significance of observed changes in individual genes, but stronger statistical power to detect differences in the differences in groups of genes as defined by their ontology (Rockett and Hellmann, 2004). This allowed us to control better for the issue of the scarcity of human nigral tissue with the necessary characteristics for successful LCM isolation of RNA from dopamine neurons (absence of freezing artifact, and preservation of RNA) and the need to match the groups for gender, age, and postmortem interval. Furthermore, the profound loss of DA neurons in PD requires us to capture neurons from a large volume of tissue, which limits the study to PD cases where the nearly the entire substantia nigra is available for study.
Male gender is a strong risk factor for PD, and our data suggest that this may result from underlying differences in the metabolic properties of dopaminergic neurons in males and females. Our data also indicate that that the response of DA neurons to PD is affected by gender, with activation of very different pathways. Further analysis of the biological substrate for these gender based differences may lead to a deeper understanding of the disease and to neuroprotective strategies useful in both men and women, and is critical to the design of clinical studies intended to test the effectiveness of putative neuroprotective treatments.
Supplementary Material
Acknowledgments
Supported by the MGH/MIT Morris Udall Center of Excellence in PD Research (NIH NS38372), and the APDA Advanced Center for Parkinson Research at MGH. The brain tissue used in this study was provided by the Massachusetts Alzheimer’s Disease Research Center Neuropathology Core (NIH AG005134) and by Harvard Brain Tissue Resource Center (supported in part by PHS R24-MH 068855). Running title: Gender and Parkinson Disease
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- Ascherio A, et al. Caffeine, postmenopausal estrogen, and risk of Parkinson’s disease. Neurology. 2003;60:790–5. doi: 10.1212/01.wnl.0000046523.05125.87. [DOI] [PubMed] [Google Scholar]
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldereschi M, et al. Parkinson’s disease and parkinsonism in a longitudinal study: two-fold higher incidence in men. ILSA Working Group. Italian Longitudinal Study on Aging. Neurology. 2000;55:1358–63. doi: 10.1212/wnl.55.9.1358. [DOI] [PubMed] [Google Scholar]
- Barlow BK, et al. A fetal risk factor for Parkinson’s disease. Dev Neurosci. 2004;26:11–23. doi: 10.1159/000080707. [DOI] [PubMed] [Google Scholar]
- Beyer C, et al. Dopamine content and metabolism in mesencephalic and diencephalic cell cultures: sex differences and effects of sex steroids. J Neurosci. 1991;11:1325–33. doi: 10.1523/JNEUROSCI.11-05-01325.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buesa C, et al. DNA chip technology in brain banks: confronting a degrading world. J Neuropathol Exp Neurol. 2004;63:1003–14. doi: 10.1093/jnen/63.10.1003. [DOI] [PubMed] [Google Scholar]
- Cantuti-Castelvetri I, et al. Alpha-Synuclein and Chaperones in Dementia With Lewy Bodies. J Neuropathol Exp Neurol. 2005a;64:1058–1066. doi: 10.1097/01.jnen.0000190063.90440.69. [DOI] [PubMed] [Google Scholar]
- Cantuti-Castelvetri I, et al. Somatic mitochondrial DNA mutations in single neurons and glia. Neurobiol Aging. 2005b;26:1343–55. doi: 10.1016/j.neurobiolaging.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Cass WA, et al. Reductions in spontaneous locomotor activity in aged male, but not female, rats in a model of early Parkinson’s disease. Brain Res. 2005;1034:153–61. doi: 10.1016/j.brainres.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Cope LM, et al. A benchmark for Affymetrix GeneChip expression measures. Bioinformatics. 2004;20:323–31. doi: 10.1093/bioinformatics/btg410. [DOI] [PubMed] [Google Scholar]
- Datla KP, et al. Differences in dopaminergic neuroprotective effects of estrogen during estrous cycle. Neuroreport. 2003;14:47–50. doi: 10.1097/00001756-200301200-00009. [DOI] [PubMed] [Google Scholar]
- Dewing P, et al. Direct regulation of adult brain function by the male-specific factor SRY. Curr Biol. 2006;16:415–20. doi: 10.1016/j.cub.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Disshon KA, Dluzen DE. Estrogen reduces acute striatal dopamine responses in vivo to the neurotoxin MPP+ in female, but not male rats. Brain Res. 2000;868:95–104. doi: 10.1016/s0006-8993(00)02329-5. [DOI] [PubMed] [Google Scholar]
- Dluzen DE. Effects of testosterone upon MPTP-induced neurotoxicity of the nigrostriatal dopaminergic system of C57/B1 mice. Brain Res. 1996;715:113–8. doi: 10.1016/0006-8993(95)01566-3. [DOI] [PubMed] [Google Scholar]
- Dluzen DE, et al. Estrogen as a neuroprotectant against MPTP-induced neurotoxicity in C57/B1 mice. Neurotoxicol Teratol. 1996;18:603–6. doi: 10.1016/0892-0362(96)00086-4. [DOI] [PubMed] [Google Scholar]
- Dluzen DE, et al. Gender differences in methamphetamine-induced mRNA associated with neurodegeneration in the mouse nigrostriatal dopaminergic system. Neuroendocrinology. 2003;77:232–8. doi: 10.1159/000070278. [DOI] [PubMed] [Google Scholar]
- Efron B, Tibshirani R. Empirical Bayes methods and false discovery rates for microarrays. Genet Epidemiol. 2002;23:70–86. doi: 10.1002/gepi.1124. [DOI] [PubMed] [Google Scholar]
- Eisen MB, et al. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson A, et al. Isolation of sex-specific cDNAs from fetal mouse brain using mRNA differential display and representational difference analysis. Brain Res Mol Brain Res. 1999;74:91–7. doi: 10.1016/s0169-328x(99)00265-x. [DOI] [PubMed] [Google Scholar]
- Fink L, et al. Real-time quantitative RT-PCR after laser-assisted cell picking. Nat Med. 1998;4:1329–33. doi: 10.1038/3327. [DOI] [PubMed] [Google Scholar]
- Freyaldenhoven TE, et al. The dopamine-depleting effects of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine in CD-1 mice are gender-dependent. Brain Res. 1996;735:232–8. doi: 10.1016/0006-8993(96)00598-7. [DOI] [PubMed] [Google Scholar]
- Fuentealba RA, et al. Signal transduction during amyloid-beta-peptide neurotoxicity: role in Alzheimer disease. Brain Res Brain Res Rev. 2004;47:275–89. doi: 10.1016/j.brainresrev.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Gillies GE, et al. Sex dimorphisms in the neuroprotective effects of estrogen in an animal model of Parkinson’s disease. Pharmacol Biochem Behav. 2004;78:513–22. doi: 10.1016/j.pbb.2004.04.022. [DOI] [PubMed] [Google Scholar]
- Goda Y. Cadherins communicate structural plasticity of presynaptic and postsynaptic terminals. Neuron. 2002;35:1–3. doi: 10.1016/s0896-6273(02)00765-1. [DOI] [PubMed] [Google Scholar]
- Grunblatt E, et al. Gene expression profiling of parkinsonian substantia nigra pars compacta; alterations in ubiquitin-proteasome, heat shock protein, iron and oxidative stress regulated proteins, cell adhesion/cellular matrix and vesicle trafficking genes. J Neural Transm. 2004;111:1543–73. doi: 10.1007/s00702-004-0212-1. [DOI] [PubMed] [Google Scholar]
- Gwinn-Hardy K. Genetics of parkinsonism. Mov Disord. 2002;17:645–56. doi: 10.1002/mds.10173. [DOI] [PubMed] [Google Scholar]
- Haaxma CA, et al. Gender Differences in Parkinson’s Disease. J Neurol Neurosurg Psychiatry. 2006 doi: 10.1136/jnnp.2006.103788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser MA, et al. Genomic convergence: identifying candidate genes for Parkinson’s disease by combining serial analysis of gene expression and genetic linkage. Hum Mol Genet. 2003;12:671–7. [PubMed] [Google Scholar]
- Iwata A, et al. Alpha-synuclein degradation by serine protease neurosin: implication for pathogenesis of synucleinopathies. Hum Mol Genet. 2003;12:2625–35. doi: 10.1093/hmg/ddg283. [DOI] [PubMed] [Google Scholar]
- Kaasinen V, et al. Increased frontal [(18)F]fluorodopa uptake in early Parkinson’s disease: sex differences in the prefrontal cortex. Brain. 2001;124:1125–30. doi: 10.1093/brain/124.6.1125. [DOI] [PubMed] [Google Scholar]
- Kelada SN, et al. Gender difference in the interaction of smoking and monoamine oxidase B intron 13 genotype in Parkinson’s disease. Neurotoxicology. 2002;23:515–9. doi: 10.1016/s0161-813x(02)00061-x. [DOI] [PubMed] [Google Scholar]
- Laakso A, et al. Sex differences in striatal presynaptic dopamine synthesis capacity in healthy subjects. Biol Psychiatry. 2002;52:759–63. doi: 10.1016/s0006-3223(02)01369-0. [DOI] [PubMed] [Google Scholar]
- Larsson O, et al. Considerations when using the significance analysis of microarrays (SAM) algorithm. BMC Bioinformatics. 2005;6:129. doi: 10.1186/1471-2105-6-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavalaye J, et al. Effect of age and gender on dopamine transporter imaging with [123I]FP-CIT SPET in healthy volunteers. Eur J Nucl Med. 2000;27:867–9. doi: 10.1007/s002590000279. [DOI] [PubMed] [Google Scholar]
- Leranth C, et al. Estrogen is essential for maintaining nigrostriatal dopamine neurons in primates: implications for Parkinson’s disease and memory. J Neurosci. 2000;20:8604–9. doi: 10.1523/JNEUROSCI.20-23-08604.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lyons KE, et al. Gender differences in Parkinson’s disease. Clin Neuropharmacol. 1998;21:118–21. [PubMed] [Google Scholar]
- Mandel S, et al. Gene Expression Profiling of Sporadic Parkinson’s Disease Substantia Nigra Pars Compacta Reveals Impairment of Ubiquitin-Proteasome Subunits, SKP1A, Aldehyde Dehydrogenase, and Chaperone HSC-70. Ann N Y Acad Sci. 2005;1053:356–75. doi: 10.1196/annals.1344.031. [DOI] [PubMed] [Google Scholar]
- Maraganore DM, et al. High-resolution whole-genome association study of Parkinson disease. Am J Hum Genet. 2005;77:685–93. doi: 10.1086/496902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martignoni E, et al. Parkinson’s disease and reproductive life events. Neurol Sci. 2002;23(Suppl 2):S85–6. doi: 10.1007/s100720200082. [DOI] [PubMed] [Google Scholar]
- Miller DB, et al. The impact of gender and estrogen on striatal dopaminergic neurotoxicity. Ann N Y Acad Sci. 1998;844:153–65. [PubMed] [Google Scholar]
- Miller RM, et al. Robust dysregulation of gene expression in substantia nigra and striatum in Parkinson’s disease. Neurobiol Dis. 2005 doi: 10.1016/j.nbd.2005.07.010. [DOI] [PubMed] [Google Scholar]
- Morris CM, et al. Transferrin receptors in the parkinsonian midbrain. Neuropathol Appl Neurobiol. 1994;20:468–72. doi: 10.1111/j.1365-2990.1994.tb00997.x. [DOI] [PubMed] [Google Scholar]
- Mozley LH, et al. Striatal dopamine transporters and cognitive functioning in healthy men and women. Am J Psychiatry. 2001;158:1492–9. doi: 10.1176/appi.ajp.158.9.1492. [DOI] [PubMed] [Google Scholar]
- Murakami T, et al. Pael-R is accumulated in Lewy bodies of Parkinson’s disease. Ann Neurol. 2004;55:439–42. doi: 10.1002/ana.20064. [DOI] [PubMed] [Google Scholar]
- Murray HE, et al. Dose- and sex-dependent effects of the neurotoxin 6-hydroxydopamine on the nigrostriatal dopaminergic pathway of adult rats: differential actions of estrogen in males and females. Neuroscience. 2003;116:213–22. doi: 10.1016/s0306-4522(02)00578-x. [DOI] [PubMed] [Google Scholar]
- Nakahara T, et al. Effect of the neurotoxic dose of methamphetamine on gene expression of parkin and Pael-receptors in rat striatum. Parkinsonism Relat Disord. 2003;9:213–9. doi: 10.1016/s1353-8020(02)00052-4. [DOI] [PubMed] [Google Scholar]
- Nishino H, et al. Estrogen protects against while testosterone exacerbates vulnerability of the lateral striatal artery to chemical hypoxia by 3-nitropropionic acid. Neurosci Res. 1998;30:303–12. doi: 10.1016/s0168-0102(98)00010-8. [DOI] [PubMed] [Google Scholar]
- Pankratz N, et al. Genome-wide linkage analysis and evidence of gene-by-gene interactions in a sample of 362 multiplex Parkinson disease families. Hum Mol Genet. 2003;12:2599–608. doi: 10.1093/hmg/ddg270. [DOI] [PubMed] [Google Scholar]
- Preece P, Cairns NJ. Quantifying mRNA in postmortem human brain: influence of gender, age at death, postmortem interval, brain pH, agonal state and inter-lobe mRNA variance. Brain Res Mol Brain Res. 2003;118:60–71. doi: 10.1016/s0169-328x(03)00337-1. [DOI] [PubMed] [Google Scholar]
- Preece P, et al. An optimistic view for quantifying mRNA in post-mortem human brain. Brain Res Mol Brain Res. 2003;116:7–16. doi: 10.1016/s0169-328x(03)00208-0. [DOI] [PubMed] [Google Scholar]
- Quackenbush J. Microarray data normalization and transformation. Nat Genet. 2002;32(Suppl):496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- Raab H, et al. Effects of sex and estrogen on tyrosine hydroxylase mRNA in cultured embryonic rat mesencephalon. Brain Res Mol Brain Res. 1995;33:157–64. doi: 10.1016/0169-328x(95)00125-c. [DOI] [PubMed] [Google Scholar]
- Ragonese P, et al. Risk of Parkinson disease in women: effect of reproductive characteristics. Neurology. 2004;62:2010–4. doi: 10.1212/wnl.62.11.2010. [DOI] [PubMed] [Google Scholar]
- Reisert I, Pilgrim C. Sexual differentiation of monoaminergic neurons--genetic or epigenetic? Trends Neurosci. 1991;14:468–73. doi: 10.1016/0166-2236(91)90047-x. [DOI] [PubMed] [Google Scholar]
- Rockett JC, Hellmann GM. Confirming microarray data--is it really necessary? Genomics. 2004;83:541–9. doi: 10.1016/j.ygeno.2003.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo A, et al. Depression in Parkinson’s disease: clinical correlates and outcome. Parkinsonism Relat Disord. 2003;10:23–8. doi: 10.1016/s1353-8020(03)00067-1. [DOI] [PubMed] [Google Scholar]
- Scaglione C, et al. REM sleep behaviour disorder in Parkinson’s disease: a questionnaire-based study. Neurol Sci. 2005;25:316–21. doi: 10.1007/s10072-004-0364-7. [DOI] [PubMed] [Google Scholar]
- Scott WK, et al. Complete genomic screen in Parkinson disease: evidence for multiple genes. Jama. 2001;286:2239–44. doi: 10.1001/jama.286.18.2239. [DOI] [PubMed] [Google Scholar]
- Staley JK, et al. Sex differences in [123I]beta-CIT SPECT measures of dopamine and serotonin transporter availability in healthy smokers and nonsmokers. Synapse. 2001;41:275–84. doi: 10.1002/syn.1084. [DOI] [PubMed] [Google Scholar]
- Standaert GS, Young AB. Treatment of the central nervous system degenerative disorders. In: Brunton LL, editor. Goodman & Gilaman’s The Pharmacological Basis of Therapeutics. McGraw-Hill; New York, NY: 2005. pp. 527–545. [Google Scholar]
- Tamas A, et al. Age and gender differences in behavioral and morphological outcome after 6-hydroxydopamine-induced lesion of the substantia nigra in rats. Behav Brain Res. 2005;158:221–9. doi: 10.1016/j.bbr.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Tanner CM. Is the cause of Parkinson’s disease environmental or hereditary? Evidence from twin studies. Adv Neurol. 2003;91:133–42. [PubMed] [Google Scholar]
- Troyanskaya O, et al. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–5. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- Tusher VG, et al. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uitti RJ, et al. Defining the Parkinson’s disease phenotype: initial symptoms and baseline characteristics in a clinical cohort. Parkinsonism Relat Disord. 2005;11:139–45. doi: 10.1016/j.parkreldis.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Van Den Eeden SK, et al. Incidence of Parkinson’s disease: variation by age, gender, and race/ethnicity. Am J Epidemiol. 2003;157:1015–22. doi: 10.1093/aje/kwg068. [DOI] [PubMed] [Google Scholar]
- Vawter MP, et al. Gender-specific gene expression in post-mortem human brain: localization to sex chromosomes. Neuropsychopharmacology. 2004;29:373–84. doi: 10.1038/sj.npp.1300337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, et al. Parkin suppresses dopaminergic neuron-selective neurotoxicity induced by Pael-R in Drosophila. Neuron. 2003;37:911–24. doi: 10.1016/s0896-6273(03)00143-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.