Abstract
Changes in the promoter methylation of hTERT, the gene that encodes telomerase, a ribonucleoprotein responsible for replacing telomeric repeats, have been demonstrated in differentiating cells where hTERT is inhibited, suggesting epigenetic regulation of hTERT. All-trans retinoic acid (ATRA) induces differentiation in human leukemia cells and has had significant clinical success treating promyelocytic leukemia in what is termed ‘differentiation therapy’. It is thought that the inhibition of telomerase is a target of retinoids and is closely tied to the differentiated phenotype. This study demonstrates the epigenetic changes associated with ATRA-induced inhibition of telomerase activity, including the hypoacetylation and hypermethylation of the hTERT promoter. Further, we have found changes in the differential expression of the three DNA methyltransferases during ATRA-induced differentiation of HL60 human leukemia cells. These results suggest that alteration of DNA methylation may play a role in the activation of telomerase in cancer cells and that epigenetic mechanisms may represent a target for differentiation therapy mechanisms. We propose that epigenetic changes in the hTERT promoter represent a stable locking mechanism in the retionoid-induced suppression of telomerase acitivity.
Keywords: telomerase, epigenetics, DNA methyltransferases, methtylation, retinoids, histone acetylation
Introduction
Telomeres are 5‘-TTAGGG-3’ nucleotide repeats that cap the ends of linear chromosomes and serve to maintain chromosomal stability (1,2). Because between 30-150 nucleotides of telomeres are lost with each mitotic division (3), progressive telomere attrition can lead to end-to-end fusions, degradations that can signal a senescence response and/or lead to tumorigenic mutations (2). Telomerase is a ribonucleoprotein with reverse transcriptase capability that adds telomeric repeats to the 3’-end of the G-rich strand of telomeres. Thus, telomerase protects chromosomes from the telomeric attrition associated with the ‘end replication problem’.
It has been recently discovered that there are 3 components to the telomerase enzyme necessary for its activity. The RNA subunit, hTR, is ubiquitiously expressed while the catalytic portion, hTERT, is over-expressed in ∼90% of cancers. The final protein needed for telomerase activity is dyskerin, which is thought to stabilize hTR (4). It has been shown that hTERT is correlated with telomerase activity (5,6). This differential expression suggests regulation at the transcriptional level. In addition to the binding of various transcription factors, hTERT (reviewed in ref. 7) gene expression is regulated in part by chromatin remodeling via epigenetic modulation and methylation of CpG sequences (8). The hTERT promoter, which lacks both TATA and CAAT boxes (9), is CpG rich (10) and it is thought that regulation through CpG methylation may be a prominent mechanism of hTERT control.
It has long been accepted that telomerase is down-regulated in all but a few progenitor cell lines and furthermore, the reactivation of telomerase is responsible for the immortality associated with cancer cells (5). The addition of telomeric DNA by telomerase in cancer cells is thought to be a major cause in maintaining the indefinite proliferation of cancer cells.
There are three principle DNA methyltransferases (DNMTs) that catalyze the transfer of a methyl moiety from the cofactor S-adenosyl-L-methoinine (SAM) to cytosine, principally found in CpG palindrome sequences (11). DNMT1, the most abundant methyltransferase in mammalian cells (12), primarily has maintenance methylating activity that maintains methylation patterns by using hemimethylated DNA substrates (13), whereas DNMT3a and 3b are considered de novo methyltransferases and are extremely important in early development. Xie et al (14) demonstrated these three DNMTs of mammalian systems are encoded by separate genes controlled at the transcriptional level. The pivotal role of the DNMTs in cellular processes is demonstrated in studies that reveal the deletion of any one of the three DNMTs in mice is lethal (15,16). From previous studies, it has been found that DNMT expression is strikingly different in neoplastic cells compared to non-cancerous cell lines (17,18).
All-trans retinoic acid (ATRA) has been used with clinical success to induce differentiation in human leukemia cells. Retinoids, such as all-trans retinoic acid (ATRA), freely enter the cell membrane and are escorted to the nucleus by cytoplasmic binding proteins where they bind to a class of nuclear receptors related to vitamin D and thyroid hormone receptors. Although the mechanisms of the anti-cancer effects of retinoids are not fully understood, the reversal of the immortalized phenotype involves the inhibition of telomerase activity. Telomerase activity in cancer cells is associated with a high proliferative capacity and resistance to apoptosis. It is likely that the anti-cancer effects of retinoids are due to their ability to inhibit proliferation and induce apoptosis, which may be a result of the inhibition of telomerase activity. Telomerase activity is a hallmark of the immortal phenotype and its suppression in ‘differentiation therapy’ using retinoids is likely a key mechanism in maintaining the differentiated phenotype of human leukemia cells. Aspects of the normalization of the immortal phenotype include a reduction in cell proliferation, expression of differentiation markers, ability to undergo apoptosis, and the inhibition of telomerase.
In this study, human promyelocytic leukemia cells (HL60) were differentiated into granulocytes using all-trans retinoic acid (ATRA). Proliferation, apoptosis, hTERT expression, acetylation of the hTERT promoter, the expression of the three DNMTs, and DNMT1 binding to the hTERT promoter were evaluated. This study sought to determine if epigenetic changes at the hTERT promoter and changes in the expression of the DNMTs are associated with retinoid-induced telomerase inhibition of HL60 cells.
Materials and methods
Assessment of proliferation and differentiation
HL60 cells (American Type Culture Collection, Manassas, VA) were grown in Iscoves media (Mediatech Inc., Herndon, VA) and supplemented with 20% fetal bovine serum and 1X APS (Ampicilin B, penicillin, and streptomycin; Gibco, Carlsbad, CA). The cells were treated with 2 μM of ATRA (Sigma Chemical Co., St. Louis, MO) based on dose-response analysis (data not shown). Cells were counted using a hemacytometer after trypan blue exclusion staining. Cellular differentiation was determined using fluorescence-activated cell sorting (FACS) analyses of CDllb expression, a differentiation marker for HL60 cells. Briefly, 1× 106 cells were washed with cold PBS, and incubated with 2 μg of fluorochrome-conjugated monoclonal antibody fluorescein isothiocyanate (FITC) against human CD11b antigen (Sigma). After the anti-body incubation, the cells were washed with PBS containing 1% bovine serum albumin and 0.1% NaN3 and fixed with 2% paraformaldehyde. The cells were analyzed on a Becton Dickinson FACScalibur flow cytometer (San Jose, CA) with CellQuest software.
Assessment of hTERT, telomerase activity and DNMT expression
RNA was extracted and purified using an RNeasy Mini Kit (Qiagen, Germantown, MD). Purified RNA (∼5 μg) was transcribed into cDNA using the SuperScript Pre-amplification System for First Strand cDNA Synthesis (Invitrogen, Carlsbad, CA). As the template, 2.5 μl of the cDNA was used for a polymerase chain reaction (PCR). The DNMT1, DNMT3a, DNMT3b, and GAPDH primers (18) amplified 335-, 551-, 190- and 550-bp products, respectively. The PCR products were resolved on a 2% agarose gel, stained with ethidium bromide (0.5 μg/ml), and visualized under UV light. To study telomerase activity in response to ATRA treatment, the Telomeric Repeat Amplification Protocol (TRAP) assay was performed as we have done previously (19). Samples underwent polymerase chain reaction (PCR) amplification for 36 cycles. The PCR product was visualized on a 10% polyacrylamide non-denaturing gel stained with SYBR Green (Molecular Probes, Eugene, OR) and analyzed with Kodak Digital Science software (Kodak, New Haven, CT).
Analysis of histone acetylation and DNMT1 binding to hTERT
In order to assess the histone acetylation status at the hTERT promoter, the EZ Chromatin Immunoprecipitation (EZ ChIP™) assay was used (Upstate Biotech, Charlottesville, VA) as we have done previously. Briefly, treated and non-treated controls were crosslinked using fresh 1% paraformaldehyde, harvested and lysed in 20% SDS lysis buffer. Crosslinked DNA was sheared using model 150 V/T Ultrasonic Homogenizer (Biologics, Inc., Manassas, VA) to ∼200-1000 bp in length. Sheared lysate was pre-cleared with a 50% Protein G agarose slurry and 10% of the supernatant was removed to serve as the input control. The pre-cleared lysate was incubated with antibodies specific for DNMT1 (Imgenex, San Diego, CA) overnight at 4°C with rotation. Antibody/antigen/DNA complex was collected using the Protein G agarose slurry and washed with salt buffers included in the kit. Protein/DNA complexes were eluted in elution buffer containing 1 M NaHCO3, 20% SDS lysis buffer, and distilled water. Protein/DNA crosslinks were reversed in 5 M NaCl at 65°C for 4 h. DNA was purified according to the protocol using spin columns provided in the kit (Upstate Biotech, Charlottesville, VA). Purified DNA was amplified by standard PCR with primers specific for a 553-bp fragment in the hTERT promoter: sense, 5‘-CTCCGTCCTCCCCTTCAC-3’ and anti-sense, 5‘-CAGCGCTGCCTGAAACTC-3’. PCR was performed under the following conditions: 94°C for 1 min followed by 30 cycles of 94°C for 15 sec, 52°C for 30 sec and 72°C for 2 min.
Analysis of apoptosis in response to ATRA treatment
The amount of apoptosis was measured using the Apoptag® Plus Peroxidase In Situ Apoptosis Detection kit (Chemicon International, Temecula, CA). Briefly, the harvested HL60 cells were fixed in 1% paraformaldehyde, quenched in endogenous peroxidase, treated with anti-digoxigenin peroxidase conjugate, and stained with methyl green dye. Cells under-going apoptosis stained brown while non-apoptotic cells stained green. Apoptosis was also determined using FACS. According to the protocol of Vibrant Apoptosis Detection Kit (Invitrogen), 1×106 cells were washed with cold PBS, and incubated with Alexa 488 and with propidium iodine (PI). The cells were analyzed on a Becton Dickinson FACScalibur flow cytometer (San Jose, CA) with CellQuest software.
Bisulfite modification of DNA
Methylation of the hTERT promoter was analyzed using the MethylEasy™ DNA bisulphite modification kit (Human Genetic Signatures, Macquarie Park, Australia) according to the manufacturer’s protocol. In brief, DNA from ATRA-treated and non-treated controls was extracted using the DNeasy Tissue Kit (Qiagen, Valencia, CA). Up to 4 μg of DNA were treated with 3 M NaOH and incubated at 37°C for 15 min. Bisulphite modification was carried out at 55°C overnight to ensure complete conversion of non-methylated cytosines. Samples were subjected to nested PCR for two rounds using the following primers specific for a 158-bp region of the hTERT promoter: First round sense; 5-GTTTTTAGGGTTTTTATATTATGG-3’, first round anti-sense, 5-AACTAAAAAATAAAAAAACAAAAC-3’, for second round PCR amplification sense; 5-GGGTTATTTTATAGTTTAGGT-3’, and second round antisense, 5-AATCCCCAATCCCTCC-3’. PCR was performed under the following conditions: 95°C for 3 min followed by 30 cycles of 95°C for 1 min, 50°C for 2 min, and 72°C for 2 min then 1 additional incubation at 72°C for 10 min. Amplified DNA was purified using the QIAquick PCR purification kit (Qiagen, Valencia, CA). Amplified segments were sequenced using the 3730 DNA Sequencer (Applied Biosystems, Foster City, CA).
Results
Inhibition of proliferation and induction of differentiation
HL60 cells were cultured with 2 μM ATRA as determined by dose-response analysis (data not shown) for 12 days. The ATRA-treated cells underwent morphological changes characteristic of granulocytes, becoming larger and more internally complex with segmented nuclei (Fig. 1A). Clumping is apparent by day 6 and dominant by day 12 of ATRA treatment (Fig. 1A). Cells were counted after exclusion staining with trypan blue dye on days 3, 6, 9, and 12 of treatment using a hemacytometer. As shown in Fig. 1B, untreated controls exponentially doubled every two days while the 2 μM ATRA treated cells showed marked reduction in proliferation by day 6, continuing to day 12. By day 12 the untreated cells increased by 1075%, whereas the ATRA treated cells reduced in number. In addition to morphological changes associated with differentiation, the expression of CD11b - a granulocyte differentiation marker - increased from 2.22 to 90.6% by day 12 of treatment (Fig. 2).
Figure 1.
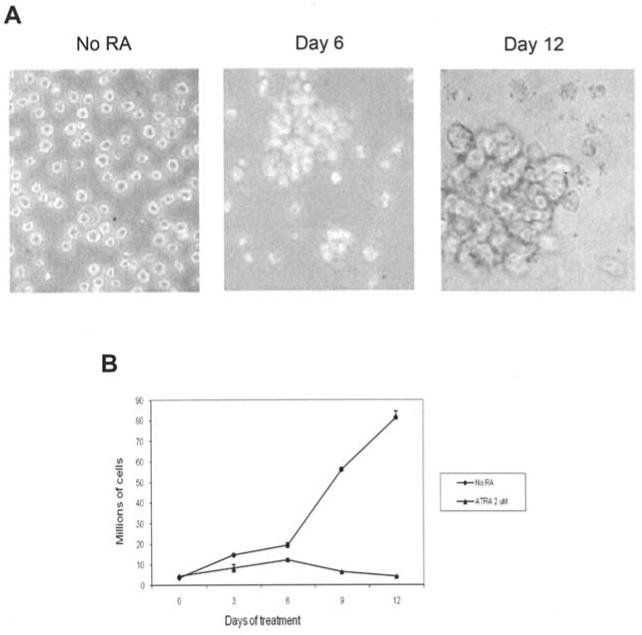
Growth and morphological effects of ATRA treatment of HL60 cells. (A) Morphological changes in HL60 cells treated with 2 μM all-trans retinoic acid. Photographs were taken at magnification x200. Cells treated with 2 μM ATRA become larger and more internally complex as they differentiate into granulocytes. Clumping begins on day 6 and is dominant by day 12 of ATRA treatment. (B) Effects of 2 μM all-trans retinoic acid on HL60 proliferation. Cells were counted on a hemocytometer after trypan blue exclusion staining. Bars represent standard error of the mean from three separate experiments.
Figure 2.
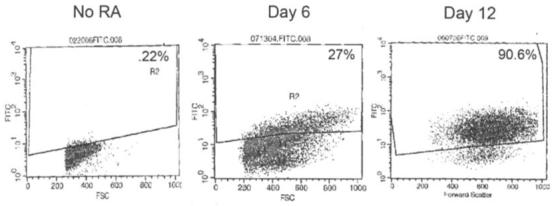
Cellular differentiation in response to ATRA treatment of HL60 cells. Flow-activated cell sorting (FACS) analysis of HL60 cells after incubation with fluorochrome conjugates of monoclonal antibody against human CD11b antigen. CD11b served as a marker of granulocyte differentiation. The values indicate the percentage of CD11b-positive differentiated cells.
Down-regulation of hTERT mRNA expression and inhibition of telomerase activity
The effect of 2 μM ATRA on the expression of hTERT was examined. As shown in Fig. 3A, day 3 is the last day of treatment with detectable hTERT mRNA levels indicating that ATRA is a potent inhibitor of hTERT gene expression in HL60 cells. The effect of 2 μM ATRA treatment on telomerase activity was examined with the TRAP method. Fig. 3B shows the laddering of oligonucleotide templates in 6-bp increments due to the addition of hexomeric repeats by the telomerase enzyme. The telomerase positive signal (the laddering) was suppressed by day 6 of treatment for HL60 cells treated with 2 μM ATRA.
Figure 3.
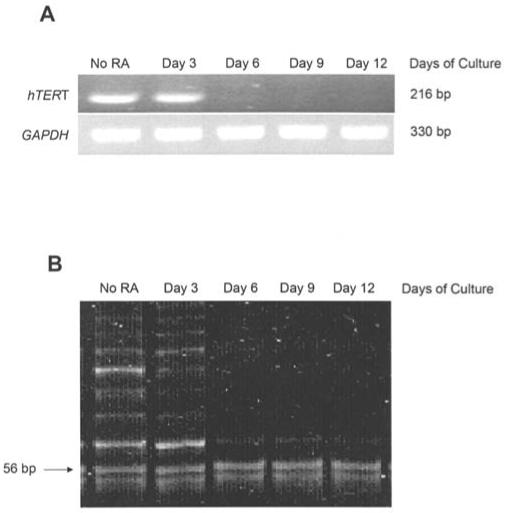
Changes in hTERT gene expression and telomerase activity following ATRA treatment. (A) hTERT RT-PCR analysis of HL60 cells treated with 2 μM all-trans retinoic acid (ATRA). hTERT cDNA was visualized as a band at 216 bp. These findings are representative of triplicate gels from three individual experiments. (B) Telomerase activity determined using the telomere repeat amplification protocol (TRAP). The laddering is a telomerase-positive result. The 56-bp band served as an internal control for the assay. This gel is representative of triplicate gels from three individual experiments.
Induction of apoptosis and changes in viability
To analyze the reduction in cellular viability, cells stained with trypan blue were compared to the total number of cells counted. Viability in response to ATRA treatment showed a time-dependent reduction with the least amount of viable cells at day 12 (Fig. 4B). Two apoptosis assays were used to investigate the induction of apoptosis by 2 μM ATRA. Fig. 4C shows photographs of HL60 cells, incubated with anti-digoxigenin peroxidase conjugate, and stained with methyl green dye. The cells undergoing apoptosis stained brown, while the non-apoptotic cells stained green (Fig. 4C). Cells with no ATRA treatment had a low number of apoptosis (∼1%; Fig. 4A). In the first 6 days of treatment there was a gradual increase in the number of apoptotic cells. By day 9 of treatment, however, there was a considerable amount of apoptosis. The greatest number of apoptosis was seen on day 12 of treatment (28.6%; Fig. 4A).
Figure 4.
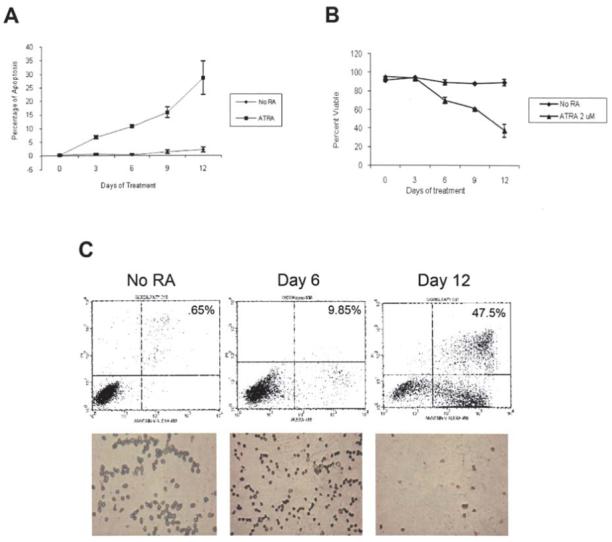
Cell viability and apoptosis in response to ATRA treatment. (A) Summary graph of all-trans retinoic acid (ATRA)-induced apoptosis. Percentage of apoptosis determined by Apoptag® Apoptosis assay for HL60 cells treated with 2 μM ATRA. (B) Effects of 2 μM all-trans retinoic acid on HL60 cell viability. Cells were counted on a hemocytometer after trypan blue exclusion staining. Bars represent standard error of the mean from three separate experiments. (C) Fluorescence-activated cell sorting (FACS) analysis of apoptosis after staining with Annexin V-conjugated Alexafluor-488 and propidium iodide. The lower right quadrants contain apoptotic cells. The values indicate representative percentages of apoptosis taken from three separate experiments. Analysis of apoptosis was determined by Apoptag® Apoptosis assay for HL60 cells treated with 2 μM all-trans retinoic acid (ATRA). Brown cells are apoptotic and green cells negative for apoptosis.
A second apoptosis assay was performed using FACS analysis of treated HL60 cells after incubation with Annexin V-conjugated Alexafluor-488 and propidium iodide. The bottom right quadrants indicate cells undergoing apoptosis. As seen in Fig. 4C, there was a time-dependent induction of apoptosis in response ATRA treatment with the largest level of apoptosis seen on day 12 (47%).
Changes in mRNA expression of the three DNA methyltransferases
Reverse transcription polymerase chain reaction (RT-PCR) depicted marked changes in the gene expression of the three DNMTs in ATRA-treated HL60 cells. Fig. 5A depicts changes in DNMT1, DNMT3a, DNMT3b, and GAPDH mRNA transcripts in HL60 cells exposed to 2 μM ATRA. DNMT1 expression progressively declined from day 3 to day 12 of treatment. DNMT3b expression was abruptly suppressed on day 6 of treatment. On the other hand, DNMT3a expression abruptly increased from day 6 to day 12 of treatment. By day 12 expression of DNMT1 and 3b decreased by 87 and 81% respectively, whereas the expression of DNMT3a increased to 2687% of control levels (Fig. 5B). The expression of GAPDH, the ubiquitous control, remained constant through the treatment. All RT-PCR assays were preformed in triplicate.
Figure 5.
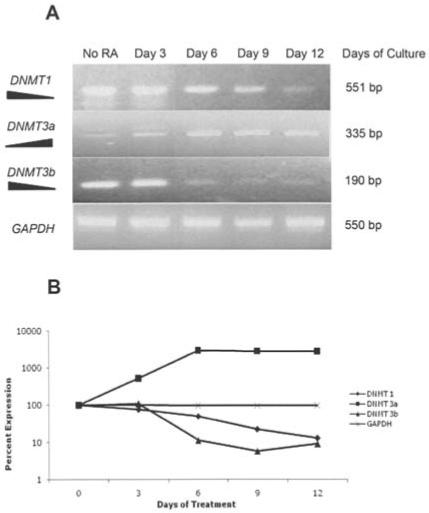
DNA methyltransferase expression following ATRA treatment. (A) RT-PCR analysis of gene expression of the three DNA methyltransferases (DNMTs) after treatment with 2 μM all-trans retinoic acid. Both DNMT1 and DNMT3b decrease during treatment with the highest amount of down-regulation at day 12. DNMT3a expression is increased with treatment of HL60 cells with ATRA. (B) Line graph summarizing the change in expression resulting from ATRA treatment. GAPDH was used as a PCR and loading control.
Epigenetic modifications and DNMT1 binding at the hTERT promoter
To assess the binding of DNMT1 to the hTERT promoter in response to ATRA treatment, we used chromatin immunoprecipitation (ChIP) analysis (Fig. 6A). As shown in Fig. 6A, ATRA treatment increased DNMT1 binding at the hTERT promoter in HL60 cells through day 9. It was shown that the highest amount of DNMT1 binding occurred at day 9. However, by day 12 binding of DNMT1 was reduced to levels similar to that seen at day 6. Next, we investigated changes in the amount of histone acetylation at the hTERTpromoter using ChIP after treatment with ATRA with antibodies specific for histone H3-K9. As seen in Fig. 6B, at day 3 of treatment histone acetylation at the hTERT promoter was reduced to ∼58% of levels observed with no ATRA added. By day 6 of treatment, the intensity of the band for the hTERT gene associated with acetylated histones was decreased to ∼21% of the no ATRA control, and at days 9 and 12 histone acetylation at the hTERT promoter was completely ablated. Input DNA expression remained similar throughout treatment (Fig. 6A and B).
Figure 6.
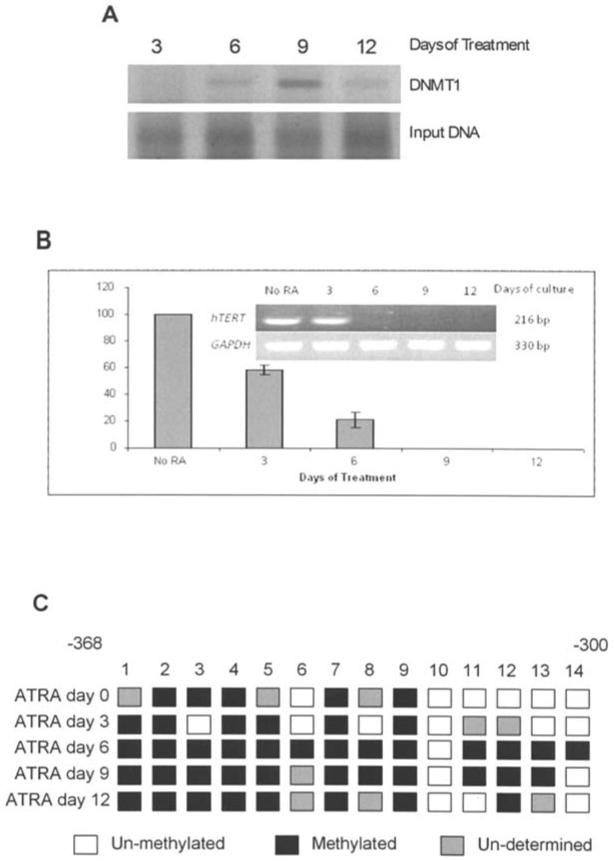
Methylation and DNMT1 binding at the hTERT promoter. (A) Chromatin immunoprecipitation of DNMT1 bound to the hTERT promoter. ChIP analysis revealed changes in the binding of DNMT1 binding with day 6 showing the highest level of binding. (B) ChIP analysis of histone acetylation at the hTERT promoter illustrating a marked decrease in acetylated H3-lys 9 by day 12. Pre-cleared lysate (1%) was taken before immunoprecipitation and used as the input control. (C) Sodium bisulfite conversion was used to analyze the methylation status of 14 CpG sites contained in the hTERT promoter. By day 6 the promoter becomes hypermethylated resulting in loss of expression of hTERT.
Another major epigenetic control mechanism is DNA methylation at gene promoter regions. To analyze the effects of ATRA treatment on methylation at the hTERT promoter we used the sodium bisulfite conversion of DNA. In Fig. 6C DNA methylation increased through day 6 of treatment. However, by day 12 DNA methylation at the region assayed was reduced to levels similar to that seen at day 3 (Fig. 6C).
Discussion
In this study we have examined the epigenetic regulation of hTERT, the catalytic subunit of telomerase, in human leukemia cells treated with all-trans retinoic acid (ATRA). Instead of a gradual attenuation of hTERT transcription, we demonstrate an abrupt inhibition of telomerase preceding a decrease in proliferation or the induction of cellular differentiation. The inhibition of telomerase was associated with down-regulation of hTERT transcription. This switch effect suggests stringent regulatory mechanisms that precede terminal differentiation. Along with the inhibition of telomerase activity, the decrease in proliferation and the re-established ability of HL60 cells to undergo apoptosis as a result of the ATRA treatment represent a return to the differentiated phenotype. HL60 differentiation was further verified by the increase in CD11b, a differentiation marker of HL60 cells, seen during the treatment. ATRA treatment both reduced cellular proliferation and induced apoptosis in the HL60 cells, but only after the inhibition of telomerase activity. These observations suggest that the decrease in proliferation and increase in apoptosis seen in retinoid treatments could be due to the inhibition of telomerase as an early event of the initiation of the differentiated phenotype.
The absence of a CAAT or TATA box in the hTERT promoter along with the presence of numerous CpG islands suggests a possible role for DNA methylation in the regulation of hTERT transcription. Aberrant promoter methylation is a common molecular alteration in human neoplasias (20). Previous studies have correlated changes in hTERT promoter methylation with telomerase expression (21).
Aberrant expression of the (DNMTs) is associated with their immortal phenotype (22,23). To determine if the increase in hTERT promoter methylation is caused by changes in the transcription of the three DNA methyltransferases (DNMTs), the expression of the DNMTs was examined over 12 days of 2 μM ATRA treatment (Fig. 5). The decrease in DNMT1 expression is likely tied to the reduction of proliferative potential in differentiating cells. Its down-regulation in ATRA-induced differentiation is consistent with previous studies that link its aberrant elevation to the immortalized phenotype (22). Interestingly, the increase in DNMT3a is likely the mechanism for the observed increase in promoter methylation of hTERT during ATRA-induced differentiation (Fig. 6C). Thus, the retinoids signal an early transient inhibition of hTERT expression and an increase in the de novo methylator DNMT3a, which in turn may methylate the hTERT promoter serving to stabilize the hTERT repression. Furthermore, DNMT3a has been shown to bind to the histone deacetylases, which in turn may lead to deacetylation of the histones associated with the hTERT promoter. The deacetylation of histones further reduces hTERT repression. The observation that the increase in DNMT3a expression and the deacetylation of the hTERT promoter occur after hTERT inhibition suggest epigenetic changes - and their resulting effect on chromatin remodeling - are secondary, locking mechanisms that stabilize hTERT inhibition and thus maintain suppression of telomerase activity in differentiated cells.
We also observed changes in the binding of DNMT1 to the hTERT promoter. The increase in binding at day 9 indicates that the pattern of methylation established by the up-regulation in DNMT3a (Fig. 6C) is maintained by DNMT1. However, illustrated in Fig. 6A, there is a decrease in DNMT1 binding at day 12. This may be due to the regulation of DNMT1 gene expression itself, since it has been shown that DNMT1 is under promoter methylation control and may also be silenced by the ATRA-induced hypoacetylation (24,25).
The results from this study suggest changes in the gene expression of the DNMTs may play an important role in the regulatory events of terminal differentiation induced by ATRA in HL60 cells. We also suggest that the epigenetic changes associated with hTERT down-regulation my not be the mechanism of action for the initial reduction in transcription, and may be more associated with a mechanism for maintaining hTERT in a silenced state during differentiation.
Acknowledgements
This work was supported in part by grants from the National Cancer Institute and the American Institute for Cancer Research.
References
- 1.Greider CW, Blackburn EH. Telomeres, telomerase and cancer. Sci Am. 1996;274:92–97. doi: 10.1038/scientificamerican0296-92. [DOI] [PubMed] [Google Scholar]
- 2.Greider CW. Telomeres. Curr Opin Cell Biol. 1991;3:444–451. doi: 10.1016/0955-0674(91)90072-7. [DOI] [PubMed] [Google Scholar]
- 3.Vaziri H, Schachter F, Uchida I, Wei L, Zhu X, Effros R, Cohen D, Harley CB. Loss of telomeric DNA during aging of normal and trisomy 21 human lymphocytes. Am J Hum Genet. 1993;52:661–667. [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen SB, Graham ME, Lovrecz GO, Bache N, Robinson PJ, Reddel RR. Protein composition of catalytically active human telomerase from immortal cells. Science. 2007;315:1850–1853. doi: 10.1126/science.1138596. [DOI] [PubMed] [Google Scholar]
- 5.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 6.Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 7.Poole JC, Andrews LG, Tollefsbol TO. Activity, function, and gene regulation of the catalytic subunit of telomerase (hTERT) Gene. 2001;269:1–12. doi: 10.1016/s0378-1119(01)00440-1. [DOI] [PubMed] [Google Scholar]
- 8.Tzukerman M, Selig S, Skorecki K. Telomeres and telomerase in human health and disease. J Pediatr Endocrinol Metab. 2002;15:229–240. doi: 10.1515/jpem.2002.15.3.229. [DOI] [PubMed] [Google Scholar]
- 9.Cong YS, Wen J, Bacchetti S. The human telomerase catalytic subunit hTERT: organization of the gene and characterization of the promoter. Hum Mol Genet. 1999;8:137–142. doi: 10.1093/hmg/8.1.137. [DOI] [PubMed] [Google Scholar]
- 10.Takakura M, Kyo S, Kanaya T, Hirano H, Takeda J, Yutsudo M, Inoue M. Cloning of human telomerase catalytic subunit (hTERT) gene promoter and identification of proximal core promoter sequences essential for transcriptional activation in immortalized and cancer cells. Cancer Res. 1999;59:551–557. [PubMed] [Google Scholar]
- 11.Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci USA. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoder JA, Soman NS, Verdine GL, Bestor TH. DNA (cytosine-5)-methyltransferases in mouse cells and tissues. Studies with a mechanism-based probe. J Mol Biol. 1997;270:385–395. doi: 10.1006/jmbi.1997.1125. [DOI] [PubMed] [Google Scholar]
- 13.Tollefsbol TO, Hutchison CA. Control of methylation spreading in synthetic DNA sequences by the murine DNA methyltransferase. J Mol Biol. 1997;269:494–504. doi: 10.1006/jmbi.1997.1064. [DOI] [PubMed] [Google Scholar]
- 14.Xie S, Wang Z, Okano M, Nogami M, Li Y, He WW, Okumura K, Li E. Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene. 1999;236:87–95. doi: 10.1016/s0378-1119(99)00252-8. [DOI] [PubMed] [Google Scholar]
- 15.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 16.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 17.Melki JR, Warnecke P, Vincent PC, Clark SJ. Increased DNA methyltransferase expression in leukemia. Leukemia. 1998;12:311–316. doi: 10.1038/sj.leu.2400932. [DOI] [PubMed] [Google Scholar]
- 18.Mizuno S, Chijiwa T, Okamura T, Akashi K, Fukumaki Y, Niho Y, Sasaki H. Expression of DNA methyltransferases DNMT1, 3A, and 3B in normal hematopoiesis and in acute and chronic myelogenous leukemia. Blood. 2001;97:1172–1179. doi: 10.1182/blood.v97.5.1172. [DOI] [PubMed] [Google Scholar]
- 19.Saldanha SN, Andrews LG, Tollefsbol TO. Analysis of telomerase activity and detection of its catalytic subunit, hTERT. Anal Biochem. 2003;315:1–21. doi: 10.1016/s0003-2697(02)00663-2. [DOI] [PubMed] [Google Scholar]
- 20.Jones PA. DNA methylation errors and cancer. Cancer Res. 1996;56:2463–2467. [PubMed] [Google Scholar]
- 21.Guilleret I, Yan P, Grange F, Braunschweig R, Bosman FT, Benhattar J. Hypermethylation of the human telomerase catalytic subunit (hTERT) gene correlates with telomerase activity. Int J Cancer. 2002;101:335–341. doi: 10.1002/ijc.10593. [DOI] [PubMed] [Google Scholar]
- 22.Casillas MA, Jr, Lopatina N, Andrews LG, Tollefsbol TO. Transcriptional control of the DNA methyltransferases is altered in aging and neoplastically-transformed human fibroblasts. Mol Cell Biochem. 2003;252:33–43. doi: 10.1023/a:1025548623524. [DOI] [PubMed] [Google Scholar]
- 23.Lopatina N, Haskell JF, Andrews LG, Poole JC, Saldanha SN, Tollefsbol TO. Differential maintenance and de novo methylating activity by three DNA methyltransferases in aging and immortalized fibroblasts. J Cell Biochem. 2002;84:324–334. doi: 10.1002/jcb.10015. [DOI] [PubMed] [Google Scholar]
- 24.Slack A, Cervoni N, Pinard M, Szyf M. Feedback regulation of DNA methyltransferase gene expression by methylation. Eur J Biochem. 1999;264:191–199. doi: 10.1046/j.1432-1327.1999.00603.x. [DOI] [PubMed] [Google Scholar]
- 25.Kishikawa S, Ugai H, Murata T, Yokoyama KK. Roles of histone acetylation in the Dnmt1 gene expression. Nucleic Acids Res. 2002;2(Suppl):209–210. doi: 10.1093/nass/2.1.209. [DOI] [PubMed] [Google Scholar]


