Abstract
We recently reported the introduction of oncogene-expressing avian retroviruses into somatic mammary cells in mice susceptible to infection by transgenic expression of tva, encoding the receptor for subgroup A avian leukosis-sarcoma virus (ALSV). Because ALSV-based vectors poorly infect nondividing cells, they are inadequate for studying carcinogenesis initiated from nonproliferative cells (e.g., stem cells). Lentivirus pseudotyped with the envelope protein of ALSV infects nondividing TVA-producing cells in culture but has not previously been tested for introducing genes in vivo. Here, we demonstrate that these vectors infected mammary cells in vivo when injected into the mammary ductal lumen of mice expressing tva under the control of the keratin 19 promoter. Furthermore, intraductal injection of this lentiviral vector carrying the polyoma middle T antigen gene induced atypical ductal hyperplasia and ductal carcinoma in situ-like premalignant lesions in 30 days and palpable invasive tumors at a median latency of 3.3 months. Induced tumors were a mixed epithelial/myoepithelial histologic diagnosis, occasionally displayed squamous metaplasia, and were estrogen receptor-negative. This work demonstrates the first use of a lentiviral vector to introduce oncogenes for modeling cancer in mice, and this vector system may be especially suitable for introducing genetic alterations into quiescent cells in vivo.
Introduction
The breast epithelium is composed of an inner layer of luminal epithelial cells and an outer layer of myoepithelial (or basal) cells [1]. The breast epithelium also harbors stem cells and their daughter multipotent progenitor cells, which are precursors to these more differentiated epithelial and myoepithelial cells; the exact number and precise location of mammary stem and multipotent progenitor cells in the epithelium are still controversial [2]. Accumulation of genetic and epigenetic mutations in the epithelium leads to breast cancer, and many mutations and estrogen signaling have been associated with breast cancer. However, it is still unclear how genetic and epigenetic lesions transform normal breast cells into cancerous cells in vivo, and whether certain cells in the breast are more vulnerable to carcinogenic insult than other cells.
Mouse models have been invaluable in our efforts to understand the cellular and molecular mechanisms of breast carcinogenesis [3]. Chemical carcinogenesis, virus- or transposon-mediated mutagenesis, and transgenic and knockout models have all helped advance our knowledge of breast cancer initiation and progression [3,4]. Recently, we reported a new method of creating mouse models of breast cancer using RCASBP/A (replication-competent ALV LTR with a splice acceptor, Bryan polymerase, subgroup A; referred to as RCAS henceforth) derived from the subgroup A avian leukosis-sarcoma virus (ALSV) to deliver oncogenes into a small number of mammary epithelial cells in transgenic mice expressing the viral receptor, tva, behind tissue-specific promoters [5]. Before our report, this viral method had been used to induce tumorigenesis after postnatal oncogene delivery to several other organs including the cancers of the brain [6,7], ovary [8], vascular endothelium [9], pancreas [10], liver [11], and other cell types [12,13] after the initial report using RCAS for introducing genes into myoblast cells in mice [14].
RCAS infect nondividing cells with a much reduced efficiency and cannot stably carry an insert larger than approximately 2.5 kb [15]. To overcome these limitations, Lewis et al. [16] reported a lentiviral vector that was pseudotyped with the envelope protein of ALSV subgroup A and demonstrated that this pseudotyped lentiviral vector could infect 293 cells and cell cycle-arrested mouse embryo fibroblasts that express tva. However, it is not known whether such a vector can be used for in vivo gene delivery and tumor initiation.
Here, we demonstrate that pseudotyped lentiviral vectors can infect mouse cells in vivo. Furthermore, delivery of lentiviral vectors encoding the mouse polyoma virus middle T antigen (PyMT) into the mammary glands of transgenic mice expressing tva under the control of the epithelial keratin 19 promoter resulted in the formation of invasive mammary tumors. Thus, ALSV-pseudotyped lentiviral vectors are suitable for tumor development studies in vivo.
Materials and Methods
Mice
Transgenic mice (on a mixture of FVB and 129 backgrounds) expressing tva under the control of the keratin 19 promoter have been reported [17]. These mice (K19-tva) also contained loxP sites inserted into the p53 locus for unrelated reasons [18]. Transgenic mice (on the FVB background) expressing tva from the MMTV LTR (MMTV-tva) have also been described [5]. All mice were kept on a diet of PicoLab standard rodent feed #5053 (Purina, St. Louis, MO). All procedures using mice were performed in compliance with an Institutional Animal Care and Use Committee-approved animal protocol.
Virus Production
To generate ALSV env-pseudotyped HIV virus for green fluorescent protein (GFP) expression (HIV(ALSV)-GFP), ∼50% confluent 293T cells grown in DMEM (Gibco, Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (Gibco) plus penicillin and streptomycin (Gibco) on 15-cm dishes precoated with 0.02% poly-lysine (BD Biosciences, Bedford, MA) were transfected by calcium phosphate precipitation with the following plasmids: 10 µg of HIV Δ8.9 plasmid (supplying the HIV gag and pol) [16], 15 µg of pCB6WTA(env) plasmid (providing the ALSV env) [16], and 10 µg of HIV backbone plasmid FG-12 (containing the GFP gene to be packaged in the virus; Figure W1) [19]. The transfected cells were washed and changed to fresh medium the following day. Viral medium was harvested 3 or 4 days after transfection. After centrifugation for 5 minutes at 500g to remove cell debris, the virus was concentrated by ultracentrifugation of the viral medium at 90,000g for 90 minutes. The resulting 100x viral concentrate was stored in aliquots at -80°C. The viral titer was determined by limiting dilution in NIH 3T3-tva cells; fluorescence microscopy was used to identify the highest dilution with infectious virus.
To generate ALSV env-pseudotyped HIV virus expressing PyMT (HIV(ALSV)-PyMT), we first inserted the coding sequence for PyMT into the XhoI site of pOZ-FH-C [20] to obtain C-terminal FLAG- and hemagglutinin (HA)-tagged PyMT. EcoRV was added to both ends of this tagged PyMT insert using polymerase chain reaction primers (TCAGCTACGAAGATATCTACCGGTAAGGTGGACCATCCTCTAGA and GTTCTCGAGTCGATATCCACTAGTAAATCTCTCGATGCGGCCCTA). The resulting EcoRV fragment was inserted into the HIV vector (pCS-CG cppt) [16] that had been cut with NheI and XhoI (New England Biolabs, Ipswich, MA) and blunted with the Klenow fragment of DNA polymerase I. The resulting viral construct (Figure W1) was transfected along with the other helper plasmids into 293T cells to produce virus as described previously. The viral titer was determined by limiting dilution in NIH 3T3-TVA cells using immunofluorescence to identify the highest dilution point. Of note, addition of HA and FLAG tags did not alter the transforming potency of PyMT when assayed in cultured rodent fibroblasts (data not shown), but it is unknown whether this tagged PyMT might cause a different immune response than untagged PyMT in mice.
RCAS-GFP [5] was similarly titered as HIV(ALSV)-GFP. RCAS-PyMT was made by inserting the tagged PyMT into RCAS and was titered by limiting dilution in DF1 chicken fibroblasts followed by Western blot analysis.
Intraductal Injection
The up-the-teat injection method [21] was used with modifications. Mice were anesthetized, mammary ducts were exposed by removal of nipple ends, and a Hamilton syringe equipped with a 33-gauge needle (Hamilton, Reno, NV) was used to inject 10 µl of viral concentrate into the ductal lumen of glands #2, 3, and 4. After all injections into a given mouse were completed, an incision in the ventral skin was made to view the mammary ducts; trace bromophenol blue (Fisher, Fair Lawn, NJ) added to the concentrate allowed confirmation of injection success.
Flow Cytometry
Five days after intraductal injection of HIV(ALSV)-GFP or RCAS-GFP, infected mammary glands were excised, minced finely by scalpel, and digested in DMEM/F-12 medium containing 5% fetal bovine serum (Gibco), 171 U/ml collagenase (Worthington, Lakewood, NJ), 100 U/ml hyaluronidase (Sigma, St. Louis, MO), 5 µg/ml insulin (Biosource, Rockville, MD), 500 ng/ml hydrocortisone, and 10 ng/ml epidermal growth factor (Invitrogen, Carlsbad, CA) at 37°C for 1 hour. Cells were then digested in 0.25% trypsin EDTA (Gibco) for 1 minute, followed by digestion for 5 minutes in 2.4 U/ml dispase II (Roche, Penzbeg, Germany) + 0.1 mg/ml DNAse (Worthington). Red blood cells were lysed in NH4Cl, and cells were passed through a 40-µm filter (BD Falcon, Bedford, MA) before flow cytometry, which was performed on an EPICS XL-MCL flow cytometer (Beckman Coulter, Fullerton, CA). In some experiments, 0.5 µg/ml propidium iodide (Sigma) was added immediately before flow cytometry to identify dead cells. The resulting data were analyzed using the EXPO 32 ADC analysis software package (Beckman Coulter).
Immunohistochemistry
Mammary glands and tumors were fixed in 10% formalin overnight and embedded in paraffin. Paraffin blocks were sectioned at 3 µm thickness, deparaffinized by serial incubations in xylene (4x) and graded ethanol solutions (2x 100%; 2x 95%; 1x 75%; 1x 50%), and rinsed in ddH2O. Sections were either hematoxylin and eosin (H&E)-stained or used for immunohistochemical staining as follows: Antigen retrieval was performed by simmering in EDTA buffer (1 mM EDTA, 0.05% Tween 20, pH 8.0) for 15 minutes. Endogenous peroxidase activity was quenched in 3% H2O2 for 5 minutes. Tissue sections were blocked in either the M.O.M. mouse IgG blocking reagent or goat serum (Vector Labs, Burlingame, CA), and then incubated for 1 hour at room temperature with a primary antibody. Antibodies used include mouse anti-influenza HA (16B12 at 1:500 dilution; Covance, Princeton, NJ), rabbit anti-estrogen receptor α (ERα) (MC-20 at 1:500; Santa Cruz Biotechnology, Santa Cruz, CA), rat anti-keratin 8 (at 1:100; Developmental Studies Hybridoma Bank, Iowa City, IA), rabbit anti-keratin 5 (AF 138 at 1:1000; Covance), rabbit anti-keratin 14 (AF 64 at 1:100; Covance), rabbit anti-keratin 6 (PRB 169P at 1:200; Covance), mouse anti-α smooth muscle actin (1A4 at 1:100; Dako, Glostrup, Germany), mouse anti-p63 (63P02 at 1:100; NeoMarkers, Fremont, CA), and mouse anti-BrdU (B44 at 1:100; Becton Dickinson, Franklin Lakes, NJ). The reactions were amplified with either M.O.M. biotinylated antimouse IgG or Vectastain biotinylated antirabbit IgG, followed by detection using Vectastain Elite ABC Reagent with Nova Red as substrate (Vector Labs). Tissues were counterstained in hematoxylin. Sections were viewed using a microscope (objective, x20; DMLB; Leica Microsystems, Wetzlar, Germany), and images were captured with a digital camera (Optronics, Goleta, CA) using MagnaFire SP software (Optronics). Tumors were considered estrogen receptor-positive if 5% or more of total tumor cells produced ERα.
Protein Analysis
Frozen tumor samples were ground to powder in liquid nitrogen, and proteins were solubilized using M-PER protein extraction reagent (Pierce, Rockford, IL) with the addition of a complete protease inhibitor cocktail (Roche). Protein concentration was determined using the bicinchoninic acid method (Pierce), and 30 µg of total protein per sample was run on a sodium dodecyl sulfate gel. Samples were transferred to nitrocellulose using a Trans-Blot SD apparatus (Bio-Rad, Hercules, CA), blocked overnight, and probed with a mouse anti-HA antibody (16B12 at 1:500 dilution; Covance) for 1 hour, followed by incubation with an horseradish peroxidase-linked goat antimouse secondary antibody (# 1858413 at 1:5000 dilution; Pierce) for 1 hour. Horseradish peroxidase was detected using Supersignal Pico chemoluminescent substrate (Pierce). Blots were stripped using Restore buffer (Pierce) for 15 minutes at 37°C, blocked and probed as previously mentioned with a mouse anti-β-actin antibody (1:5000 dilution; Sigma). Quantitation was performed using the FlourChem 8000 software package (Alpha Innotech, San Leandro, CA).
DNA Purification and Southern Hybridization
Tumor DNA (20 µg), purified by proteinase K digestion and phenol-chloroform extraction, was digested by BamHI, separated on a 0.7% agarose gel, and transferred to Hybond-N membrane (GE Healthcare Bio-Sciences Corp, Piscataway, NJ). After UV cross-linking and prehybridization, the membrane was hybridized overnight at 68°C to a [α-32P]dCTP (PerkinElmer, Waltham, MA)-labeled probe transcribed from a 2.4-kb template DNA fragment, excised from the HIV HA-PyMT plasmid using BamHI and EcoRV. The RadPrime DNA Labeling System (Invitrogen) was used following the manufacturer's instructions. After washing under stringent conditions, the radioactive bands on the blot were detected using BioMax MR Film (Kodak, Rochester, NY).
Coimmunofluorescence
Sectioning, deparaffinization, and antigen retrieval were performed as for immunohistochemical staining previously discussed. After 1 hour of blocking in goat serum, sections were incubated for 1 hour at room temperature in a primary antibody solution that contained mouse anti-HA and rabbit anti-keratin 14 (as for immunohistochemistry), and then for 30 minutes at room temperature in a secondary antibody solution that contained Alexa 568-conjugated goat antirabbit (A11011 at 1:200; Molecular Probes, Invitrogen) and fluorescein-conjugated horse antimouse (Fl-2000 at 1:100; Vector Labs). Stained slides were mounted in Vectashield mounting medium with 4′-6-diamidino-2-phenylindole (Vector Labs) and examined under a microscope (Axioskop 2 plus; Zeiss, Thornwood, NJ). Photomicrographs were taken under the x40 objective using an AxioCam digital camera and AxioVision 4.0 software (Zeiss).
Results
Infection of Mouse Mammary Cells Using a Pseudotyped Lentivirus
Keratin 19 (K19) is expressed selectively in epithelial cells in several organs including the mammary gland and pancreas [17]. K19 is a general epithelial cell marker in the mammary gland, although it might also be expressed in mammary bipotential progenitor cells that give rise to both more mature epithelial cells and myoepithelial cells [22,23]. It is not expressed in myoepithelial cells and seems to be absent in mammary stem cells [23]. Transgenic mice expressing tva from the K19 promoter has been reported [17]. TVA produced in transgenic K19-tva mice confers susceptibility to RCAS infection in pancreatic ductal cells [17]. Using immunohistochemical staining for TVA, we found that TVA was produced in nearly all epithelial cells in the ducts and in the body cells of terminal end buds in the mammary glands of 7-week-old K19-tva transgenic mice (Figure 1, B and E). This is in contrast to the patchy pattern of expression in MMTV-tva transgenic mice (Figure 1, C and F) [5].
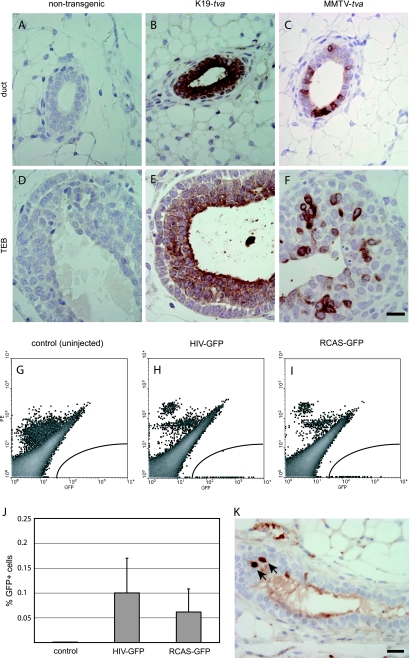
Figure 1.
Lentiviral infection in K19-tva transgenic mice. (A–F) Immunohistochemical staining for TVA in mammary gland sections from pubertal K19-tva transgenic mice. Staining results of both ducts and terminal end buds (TEBs) are shown. Genotypes are indicated at the top. Scale bar, 20 µm. (G–I) The left #2 to 4 mammary glands of six adult K19-tva transgenic mice were injected with HIV(ALSV)-GFP (104 IU/gland), and an equal amount of RCAS-GFP was injected into each of the contralateral glands. Infection efficiency was determined by FACS for GFP 5 days after injection. Shown are representative FACS plots of mammary cells from uninjected (G), HIV(ALSV)-GFP-injected (H), and RCAS-GFP-injected (I) K19-tva transgenic mice. No phycoerythrin (PE) was added to the cells; the PE channel (Y-axis) was used to segregate autofluorescing cells (high in both PE and GFP channels) from GFP+ cells (high GFP and no PE). (J) Quantitation of GFP+ cells 5 days after injection (n = 6 mice, mean ± SD). (K) Immunohistochemical staining for GFP in mammary gland sections from adult K19-tva transgenic mice harvested 4 days after injection of HIV(ALSV)-GFP (105 IU) into the ductal lumen. Arrows indicate positively stained cells. Scale bar, 20 µm.
To test whether ALSV env-pseudotyped lentivirus could infect TVA+ mammary cells, we first ascertained that it could infect TVA+ primary mouse mammary cells ex vivo. A total of 5 x 106 infectious units (IU) of HIV(ALSV)-GFP were added to primary cells 1 day after preparation from the mammary glands of K19-tva mice. Three days later, GFP was observed in approximately 10% of the cells (data not shown), demonstrating that TVA+ primary mammary cells in culture are susceptible to infection by pseudotyped lentivirus. As expected, this virus did not infect mammary cells prepared from nontransgenic mice or mammary tumor cells prepared from transgenic mice that expressed neu (ErbB2) from the MMTV promoter [24] (data not shown).
To determine whether a pseudotyped lentiviral vector could infect mammary epithelial cells in vivo, we intraductally injected 104 IU of HIV(ALSV)-GFP viral concentrate into the left #2, 3, and 4 mammary glands in six female K19-tva transgenic mice at 11 weeks of age. Five days after injection, we collected the infected glands, made single-cell suspensions, and used flow cytometry to quantify the number of GFP+ cells. (Sample FACS plots are shown in Figure 1, G–I.). We found that 0.1 ± 0.07% of the cells (approximately 400 cells per gland) were positive for GFP (Figure 1J). (Because infected cells may proliferate or die during these 5 days, the initial number of infected cells may be more or less.) This result demonstrates that the ALSV-pseudotyped lentivirus can infect mammary cells in tva transgenic mice.
To compare the infection efficiency of the HIV(ALSV) and RCAS vectors, we also injected the contralateral three glands in all six mice with an equivalent titer of RCAS-GFP viral concentrate and quantified the fraction of GFP+ cells (Figure 1I). The percentage of GFP+ cells in RCAS-GFP-infected glands was 0.06 ± 0.05%, not significantly different (P = .26) from that achieved by the pseudotyped lentiviral vector (Figure 1J). Two additional, similar experiments gave an equivalent result, suggesting that at the level of viral dosage tested, HIV(ALSV) and RCAS vectors are similar in their ability to infect mammary epithelial cells in adult K19-tva transgenic mice.
To verify the tissue specificity of infection in K19-tva transgenic mice using the intraductal injection method, we intraductally injected 105 IU of HIV(ALSV)-GFP viral concentrate into all #2, 3, and 4 mammary glands in four female K19-tva transgenic mice at 14 weeks of age. Four days after injection, we collected the mammary glands, hearts, intestines, lungs, pancreas, skins, and salivary glands of injected mice. We confirmed by immunohistochemistry that HIV (ALSV)-GFP could infect mammary cells in vivo; only cells bordering the ductal lumen were observed to produce GFP (Figure 1K), which correlates with the pattern of TVA+ cells (Figure 1A). We did not detect GFP+ cells in all the other organs examined (data not shown), including skin and salivary gland that have been reported to express tva in this transgenic mouse line [17]. Therefore, infection by the HIV(ALSV) vector introduced by intraductal injection is restricted to TVA-producing cells of the mammary glands.
Early Mammary Lesions Formed by HIV(ALSV)-PyMT
Having shown that HIV(ALSV) could infect murine mammary cells in vivo, we next investigated the suitability of pseudotyped lentiviral vectors for inducing mammary tumors. PyMT is a potent transforming oncogene whose gene product activates multiple signaling pathways including those mediated by Src, Shc, and phosphatidylinositol 3-kinase, and it has been used to induce mammary tumors [5,25,26]. Therefore, we tested whether infection by HIV(ALSV)-PyMT (Figure W1) could transform mammary cells in vivo. We intraductally injected 103 IU of HIV(ALSV)-PyMT into both #4 mammary glands of three female K19-tva transgenic mice (9–11 weeks old), harvested infected glands at 30 days after injection, and examined their paraffin sections for histopathologic changes by H&E. We observed atypical hyperplasia and ductal carcinoma in situ-like lesions in five of six infected mammary glands (Figure 2A); solid cell proliferation with central necrosis was also observed in one infected gland. We confirmed that PyMT was produced in these lesions by immunohistochemical staining for the HA-tag (Figure 2B). As expected, HA staining was absent in the normal-appearing ducts adjacent to these lesions (Figure 2B, blue arrows), and neither early lesions nor HA staining was observed in noninjected glands (Figure 2, C and D). Therefore, we conclude that PyMT delivered by this pseudotyped lentiviral vector can induce premalignant changes in tva transgenic mice.
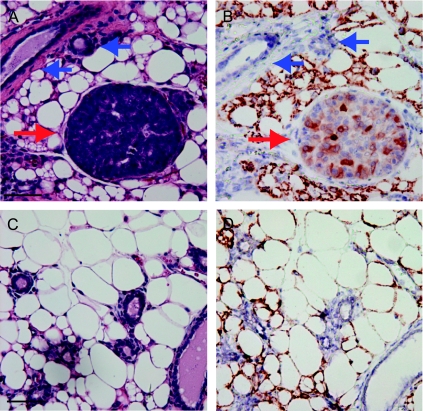
Figure 2.
Early lesions in HIV(ALSV)-PyMT-injected mammary glands. Three adult K19-tva transgenic mice were intraductally injected with 103 IU HIV(ALSV)-PyMT. Mice were killed 30 days after injection, and mammary glands were sectioned and stained by H&E for histologic examination. One or more early lesions were observed in all mice (red arrow) (A). Adjacent sections were examined for HA-tagged PyMT by immunohistochemistry (B); many cells are positively stained (red arrow). Note that HA was not detected in adjacent normal-appearing ducts (blue arrows). Sections from noninjected glands had neither early lesions (C) nor cells producing PyMT (D). Scale bar, 100 µm.
Tumor Formation by HIV(ALSV)-PyMT Virus
Having demonstrated the formation of early lesions after infection by HIV(ALSV)-PyMT, we used this vector to induce tumors in K19-tva transgenic mice. We injected 21 adult mice with HIV(ALSV)-PyMT (103 IU/gland, six glands/mouse). Injection success rate as determined by visualization of the ductal tree immediately after injection varied from 0/6 to 6/6 glands per mouse; only the 13 mice with two or more successful injections were monitored for tumor formation. Ten of these mice developed palpable tumors within 1 year in at least one gland, and another one died after 9 months for nontumor reasons. Median tumor latency was 3.3 months (Figure 3A). All nine successfully injected glands in the remaining two mice were collected after 1 year and examined for histopathologic changes, but no early lesions were identified and no HA tag could be found in at least five representative sections prepared from each of these nine glands, suggesting that the infected cells had either died or failed to express PyMT from the provirus. In the tumors that developed in injected glands, PyMT could be detected in the tumor cells by immunohistochemical staining (Figure 4A), and tumors were never found in noninjected gland #1 or #5, nor in noninjected control K19-tva transgenic mice. Therefore, we conclude that pseudotyped lentiviral vectors can be used to introduce oncogenes for modeling breast cancer in mice.
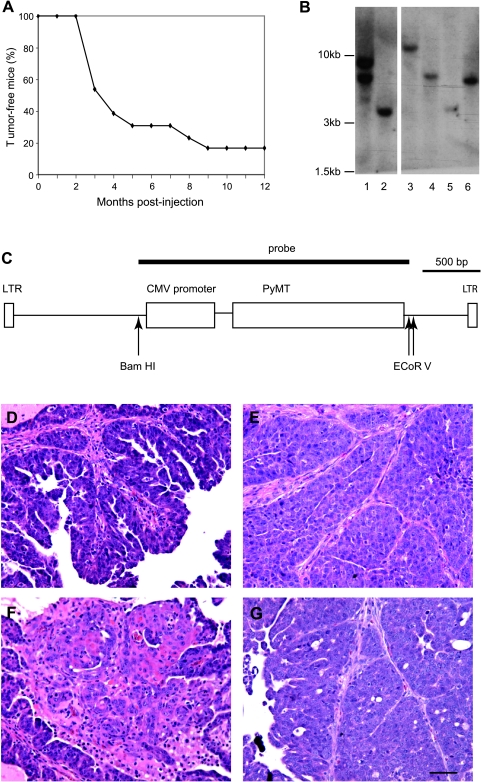
Figure 3.
Tumor induction by HIV(ALSV)-PyMT. K19-tva transgenic mice were intraductally injected with 103 IU of HIV(ALSV)-PyMT into the #2 to 4 mammary glands. (A) Kaplan-Meier curve showing tumor-free survival of infected mice (n = 13). One mouse died of natural causes and was censored at 9 months after injection. (B–C) Clonality analysis of HIV(ALSV)-PyMT virus-induced mammary tumors in K19-tva transgenic mice. Genomic DNA from 6 HIV(ALSV)-PyMT-induced tumors was subjected to Southern blot analysis after digestion with BamHI (B); molecular weights are indicated (left). A diagram of the HIV(ALSV)-PyMT provirus is shown (C); the radioactive probe was transcribed from the indicated 2.4-kb BamHI-EcoRV fragment. (D–G) Representative photomicrographs showing both papillary adenomyoepithelioma/carcinoma (D and E) and squamous cell-like (F) regions of tumors induced by HIV(ALSV)-PyMT. For comparison, an H&E-stained section from a tumor induced by RCAS-PyMT in a K19-tva transgenic mouse is shown (G); RCAS-PyMT-induced tumors were reminiscent of papillary adenomyoepithelioma/carcinomas. Scale bar, 50 µm.
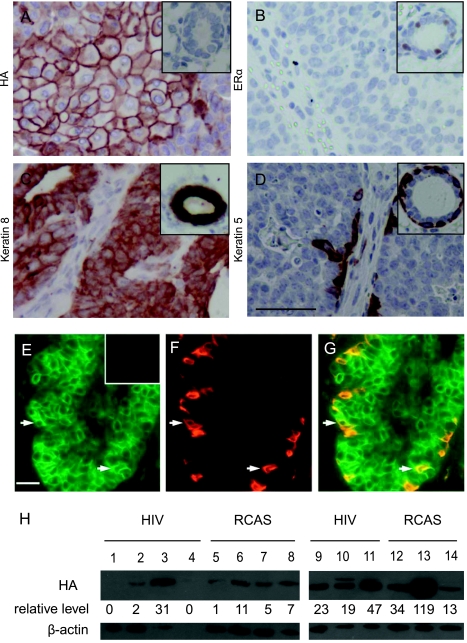
Figure 4.
Characterization of HIV(ALSV)-PyMT-induced mammary tumors. Paraffin sections from HIV(ALSV)-PyMT-induced tumors were analyzed by immunohistochemistry for HA-tagged PyMT (A), ERα (B), keratin 8 (C), and keratin 5 (D). In each case, the insert shows a representative staining of a normal mammary gland section. Scale bar, 25 µm; in all sections, n = 15 tumors. (E–G) HIV(ALSV)-PyMT-induced tumors contain myoepithelial cells of infected cell origin. Representative photomicrographs of coimmunofluorescent staining of paraffin sections from HIV(ALSV)-PyMT-induced mammary tumors for cells producing HA-tagged PyMT (E) and the myoepithelial marker keratin 14 (F). Arrows indicate myoepithelial cells positive for both keratin 14 and PyMT (G). Insert: negative HA staining in a tumor from an MMTV-Wnt-1 transgenic mouse. Scale bar, 25 µm; n = 5. (H) Western blot analysis of PyMT in tumors. Sample lysates from tumors induced by the indicated PyMT-expressing vectors were probed for the HA tag in PyMT (top panel), with β-actin serving as a loading control (bottom panel). Quantitation of relative HA signal intensity (corrected for total protein) is indicated below each lane (arbitrary units).
The relatively long latency in these mice suggested that these tumors were clonal in origin. To determine their clonality, DNA prepared from six tumors were digested with BamHI to generate a unique fragment for each proviral integration in the cellular genome. This fragment was subsequently detected by a radioactively labeled PyMT probe (Figure 3C). Five tumors yielded single bands (Figure 3B), demonstrating their clonal origin. The sixth tumor harbored two insertion sites (Figure 3B, lane 1). We cannot conclude whether this tumor originated from a single cell infected by two viruses or from two singly infected cells. Because the two bands are similar in intensity, we suspect that this tumor arose from a doubly infected cell. Double infection occurs in TVA+ mammalian cells, including mammary epithelial cells, using RCAS vectors [27] (W. Bu, unpublished observations).
Characterization of Mammary Tumors Induced by HIV(ALSV)-PyMT
Histologically, HIV(ALSV)-PyMT-induced mammary tumors were predominantly papillary adenomyoepithelioma/carcinomas, with occasional foci of squamous cell metaplasia in 7 of 16 cases (Figure 3, D–F). The tumors were highly proliferative; 9.9 ± 2.5% of tumor cells incorporated 5-bromo-2′-deoxyuridine (BrdU) 2 hours after BrdU injection (n = 5), much higher than the 0.8 ± 0.5% seen in epithelial cells from normal mammary glands of adult K19-tva mice (n = 3, data not shown).
Using immunohistochemical staining for the epithelial cell marker keratin 8 and the myoepithelial cell markers keratin 5, p63, and α-smooth muscle actin, we found that HIV(ALSV)-PyMT-induced tumors were composed of both luminal cells and myoepithelial cells (Figures 4, C–D, and W2). Using coimmunofluorescent staining, we found positive HA staining in many of the keratin 14+ myoepithelial tumor cells (Figure 4, E–G), indicating that they were progeny of the HIV(ALSV)-PyMT-infected cells. As expected, HA was also detected in the keratin 8+ luminal cells, which comprised the bulk of the tumor mass (data not shown). The cellular heterogeneity of these tumors strongly suggests an origin in bipotential progenitor cells, although we cannot rule out the remote possibility that the initial infection may have occurred in a terminally differentiated epithelial cell that in the course of oncogenesis dedifferentiated, producing progeny in both lineages.
The finding that HIV(ALSV)-PyMT-induced tumors contain mixed differentiated cell populations is in contrast to the poorly differentiated tumors arising in MMTV-PyMT transgenic mice [26], possibly reflecting the consequences of oncogene expression in the developing mammary gland in these transgenic mice. In this respect, our lentivirus-induced tumors are reminiscent of tumors initiated by RCAS-PyMT in MMTV-tva transgenic mice, which also contain mixed cell populations [5]. However, RCAS-PyMT-induced tumors in MMTV-tva transgenic mice lacked squamous metaplasia. In addition, whereas most of these RCAS-PyMT-induced tumors produced ERα (Z. Du and Y. Li, unpublished observations), tumors induced by HIV(ALSV)-PyMT lacked ERα (Figure 4B). To test whether these differences may have been caused by the difference in the tva transgene or the genetic background of the two mouse lines, by intraductal injection, we infected 12 K19-tva transgenic mice with RCAS-PyMT (105 IU per gland, two or three glands per mouse). Palpable tumors developed within 4 to 6 weeks in all infected mice, comparable to the latency observed in MMTV-tva transgenic mice [5]. Like tumors arising in RCAS-PyMT-infected MMTV-tva transgenic mice, RCAS-PyMT-induced tumors arising in K19-tva transgenic mice were adenomyoepithelioma/carcinomas containing both epithelial and myoepithelial lineages (Figure 3G), produced ERα in 6 of 13 tumors, and had no squamous metaplasia (data not shown). To examine whether differences in PyMT expression levels could explain the differences between HIV- and RCAS-PyMT-induced tumors, we used Western blot analysis to assay samples from seven tumors induced by each vector for relative levels of PyMT. We noted great variability of PyMT levels between individual tumors (Figure 4H). However, there was no consistent trend toward higher PyMT levels with one vector that could easily explain our observed differences. Therefore, even in the same tva transgenic line, differences in ER status and squamous metaplasia existed between tumors induced by HIV(ALSV)-PyMT and those induced by RCAS-PyMT.
Discussion
We here demonstrate that pseudotyped lentiviral vectors can infect mouse mammary cells in vivo and can serve as a vehicle for tumor induction. This method shares the advantages of the RCAS-TVA technology [28], namely, oncogene expression at a specific time, tumor initiation in a background of normal cells, targeting of specific populations of cells for oncogenesis through the choice of a promoter regulating tva expression, and relative ease of testing different putative oncogenes for a role in tumor formation. However, the HIV (ALSV) vector is more suitable for infecting nondividing cells and can carry a much larger DNA insert, allowing the introduction of two or more genes in one vector. This larger cloning capability is especially useful for testing oncogenic collaboration.
Because cell proliferation is low in adult mammary glands, we anticipated that HIV(ALSV) vectors would infect more cells in tva transgenic mice than RCAS viruses of comparable titers. However, we only saw a trend toward higher infection using HIV(ALSV)-GFP that did not reach significance (Figure 1). It is likely that, at the viral titer used, we did not saturate the number of TVA+ proliferating cells in the gland with the RCAS virus. Indeed, the 0.8% proliferation rate in the mammary epithelium in adult K19-tva transgenic mice is still 13-fold higher than the 0.06% infection efficiency observed with the RCAS vector (Figure 1J). The use of higher titer viral stocks may demonstrate the higher number of mammary cells in vivo susceptible to infection by HIV(ALSV) relative to RCAS viruses, as has been shown in cultured epithelial cells and fibroblasts [16].
Introduction of PyMT to mammary cells using the HIV(ALSV)-PyMT vector resulted in papillary adenomyoepithelioma/carcinomas quite similar to those arising in RCAS-PyMT-infected mammary glands (Figure 3) but very different from those arising in MMTV-PyMT transgenic mice [26]. Although the tumors induced by the RCAS and HIV(ALSV) vectors share many similarities, one difference between the tumors is the presence of occasional foci resembling squamous cell metaplasia in HIV(ALSV)-PyMT-initiated, but not in RCAS-PyMT-initiated tumors (Figure 3). In addition, the tumors induced by these vectors display different ER status. We do not know the reasons for these differences. One possible explanation for the presence of regions of squamous differentiation is that the HIV virus infected a broader range of cells, including those with greater developmental plasticity.
The tumor latency was much longer in mice infected by HIV (ALSV)-PyMT (Figure 3A) than in mice infected by RCAS-PyMT [5]. The longer latency in the former mice is likely due to the much smaller viral dosage (103 vs 105 IU) used to infect the mice, leading to the infection of presumably a few cells (vs several thousand cells in RCAS-PyMT-infected mice). Whereas PyMT seems to be sufficient to transform a subset of somatic mammary cells in mice based on the extremely short latency (median = 2 weeks) in MMTV-tva mice infected by RCAS-PyMT (107 IU per gland, leading to the infection of approximately 3000 cells [5]), other mammary cells may not be converted to malignancy after PyMT is expressed because a much longer latency (median = 2 months) was observed when 1000-fold less RCAS-PyMT virus was used to infect the same line of mice (Z. Du and Y. Li, unpublished observations). Therefore, the HIV (ALSV)-PyMT-injected mice that either developed tumors very late or never developed tumors might have received infection in only a small number of the mammary cells that were more resistant to tumor induction by PyMT. In addition, the longer latency in HIV(ALSV)-PyMT-infected mice may also be caused by the down-regulation of the cytomegalovirus promoter driving PyMT expression in the lentiviral vector. The cytomegalovirus promoter, although apparently more active in cultured cells than the RCAS LTR, has been shown to be silenced in hematopoietic and other tissues in mice [29,30].
In conclusion, this work demonstrates that lentiviral vectors can be used as a vehicle to introduce oncogenes in specific TVA+ mouse cells for cancer induction and expands the versatility of the TVA technology for functional in vivo testing of candidate genes, especially larger genes that cannot be expressed from RCAS. This vector system may also be better suited for the introduction of oncogenes into quiescent stem cells or highly differentiated postmitotic cells.
Supplementary Material
Acknowledgments
The authors thank David Baltimore and Andy Leavitt for the FG-12 vector plasmids, Andy Leavitt for the antibody to TVA, Zhijun Du for technical assistance with intraductal injections, Cassandra Horne for assistance with flow cytometry, Jian Huang and the Breast Center Path Core (supported by P01 CA30195) for tissue processing, Gladys Morrison for assistance with immunohistochemistry, Richard Sutton and Roland Wolkowicz for advice on the use of lentiviral vectors, and Gary Chamness for helpful comments on the manuscript. M.J. is a post doctoral fellow in the laboratory of Harold Varmus.
Abbreviations
- ALSV
avian leukosis-sarcoma virus
- BrdU
5-bromo-2′-deoxyuridine
- ERα
estrogen receptor α
- GFP
green fluorescent protein
- HA
hemagglutinin
- IU
infectious unit
- K19
keratin 19
- PyMT
polyoma middle T antigen
- RCAS
replication-competent avian leukosis virus with splice acceptor
Footnotes
This work was supported by National Institutes of Health grant R01 CA113869 (to Y. L.); S. K. S. was supported by the NIH T32 Translational Breast Cancer Research Training grant CA090221; B. S. was supported by R01CA34722 and P01CA50661; and B. C. L. was supported by a Career Development Award in the Biomedical Sciences from the Burroughs Wellcome Fund.
This article refers to supplementary materials, which are designated by Figures W1 and W2 and are available online at www.neoplasia.com.
References
- 1.Wiseman BS, Werb Z. Stromal effects on mammary gland development and breast cancer. Science. 2002;296:1046–1049. doi: 10.1126/science.1067431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stingl J, Raouf A, Eirew P, Eaves CJ. Deciphering the mammary epithelial cell hierarchy. Cell Cycle. 2006;5:1519–1522. doi: 10.4161/cc.5.14.2983. [DOI] [PubMed] [Google Scholar]
- 3.Vargo-Gogola T. Modelling breast cancer: one size does not fit all. Nat Rev Cancer. osen JM, (2007;7:659–672. doi: 10.1038/nrc2193. [DOI] [PubMed] [Google Scholar]
- 4.Collier LS, Largaespada DA. Transposable elements and the dynamic somatic genome. Genome Biol. 2007;8(Suppl 1):S5. doi: 10.1186/gb-2007-8-s1-s5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du Z, Podsypanina K, Huang H, McGrath A, Toneff MJ, Bogoslovskaia E, Zhang X, Moraes RC, Fluck MM, Allred DC, et al. Introduction of oncogenes into mammary glands in vivo with an avian retroviral vector initiates and promotes carcinogenesis in mouse models. Proc Natl Acad Sci USA. 2006;103:17396–17401. doi: 10.1073/pnas.0608607103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holland EC, Varmus HE. Basic fibroblast growth factor induces cell migration and proliferation after glia-specific gene transfer in mice. Proc Natl Acad Sci USA. 1998;95:1218–1223. doi: 10.1073/pnas.95.3.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holland EC, Hively WP, DePinho RA, Varmus HE. A constitutively active epidermal growth factor receptor cooperates with disruption of G1 cell-cycle arrest pathways to induce glioma-like lesions in mice. Genes Dev. 1998;12:3675–3685. doi: 10.1101/gad.12.23.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orsulic S, Li Y, Soslow RA, Vitale-Cross LA, Gutkind JS, Varmus HE. Induction of ovarian cancer by defined multiple genetic changes in a mouse model system. Cancer Cell. 2002;1:53–62. doi: 10.1016/s1535-6108(01)00002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montaner S, Sodhi A, Molinolo A, Bugge TH, Sawai ET, He Y, Li Y, Ray PE, Gutkind JS. Endothelial infection with KSHV genes in vivo reveals that vGPCR initiates Kaposi's sarcomagenesis and can promote the tumorigenic potential of viral latent genes. Cancer Cell. 2003;3:23–36. doi: 10.1016/s1535-6108(02)00237-4. [DOI] [PubMed] [Google Scholar]
- 10.Lewis BC, Klimstra DS, Varmus HE. The c-myc and PyMT oncogenes induce different tumor types in a somatic mouse model for pancreatic cancer. Genes Dev. 2003;17:3127–3138. doi: 10.1101/gad.1140403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis BC, Klimstra DS, Socci ND, Xu S, Koutcher JA, Varmus HE. The absence of p53 promotes metastasis in a novel somatic mouse model for hepatocellular carcinoma. Mol Cell Biol. 2005;25:1228–1237. doi: 10.1128/MCB.25.4.1228-1237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pao W, Klimstra DS, Fisher GH, Varmus HE. Use of avian retroviral vectors to introduce transcriptional regulators into mammalian cells for analyses of tumor maintenance. Proc Natl Acad Sci USA. 2003;100:8764–8769. doi: 10.1073/pnas.1133333100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu SL, Huang YJ, Liang FP, Huang YF, Chuang CF, Wang SW, Yao JW. Malignant transformation of an epithelial cell by v-Src via tv-a-mediated retroviral infection: a new cell model for studying carcinogenesis. Biochem Biophys Res Commun. 2005;338:830–838. doi: 10.1016/j.bbrc.2005.10.045. [DOI] [PubMed] [Google Scholar]
- 14.Federspiel MJ, Bates P, Young JA, Varmus HE, Hughes SH. A system for tissue-specific gene targeting: transgenic mice susceptible to subgroup A avian leukosis virus-based retroviral vectors. Proc Natl Acad Sci USA. 1994;91:11241–11245. doi: 10.1073/pnas.91.23.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes SH. The RCAS vector system. Folia Biol (Praha) 2004;50:107–119. doi: 10.14712/fb2004050030107. [DOI] [PubMed] [Google Scholar]
- 16.Lewis BC, Chinnasamy N, Morgan RA, Varmus HE. Development of an avian leukosis-sarcoma virus subgroup A pseudotyped lentiviral vector. J Virol. 2001;75:9339–9344. doi: 10.1128/JVI.75.19.9339-9344.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morton JP, Mongeau ME, Klimstra DS, Morris JP, Lee YC, Kawaguchi Y, Wright CV, Hebrok M, Lewis BC. Sonic hedgehog acts at multiple stages during pancreatic tumorigenesis. Proc Natl Acad Sci USA. 2007;104:5103–5108. doi: 10.1073/pnas.0701158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marino S, Hoogervoorst D, Brandner S, Berns A. Rb and p107 are required for normal cerebellar development and granule cell survival but not for Purkinje cell persistence. Development. 2003;130:3359–3368. doi: 10.1242/dev.00553. [DOI] [PubMed] [Google Scholar]
- 19.Qin XF, An DS, Chen IS, Baltimore D. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc Natl Acad Sci USA. 2003;100:183–188. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakatani Y, Ogryzko V. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 2003;370:430–444. doi: 10.1016/S0076-6879(03)70037-8. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen D-A, Beeman NG, Lewis M, Schaack J, Neville MC. Intraductal injection into the mouse mammary gland. In: Ip MM, Asch BB, editors. Methods in Mammary Gland Biology and Breast Cancer Research. New York, NY: Kluwer Academic; 2000. pp. 259–270. [Google Scholar]
- 22.Gudjonsson T, Villadsen R, Nielsen HL, Ronnov-Jessen L, Bissell MJ, Petersen OW. Isolation, immortalization, and characterization of a human breast epithelial cell line with stem cell properties. Genes Dev. 2002;16:693–706. doi: 10.1101/gad.952602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 24.Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci USA. 1992;89:10578–10582. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Politi K, Kljuic A, Szabolcs M, Fisher P, Ludwig T, Efstratiadis A. “Designer” tumors in mice. Oncogene. 2004;23:1558–1565. doi: 10.1038/sj.onc.1207275. [DOI] [PubMed] [Google Scholar]
- 26.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holland EC, Celestino J, Dai C, Schaefer L, Sawaya RE, Fuller GN. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat Genet. 2000;25:55–57. doi: 10.1038/75596. [DOI] [PubMed] [Google Scholar]
- 28.Orsulic S. An RCAS-TVA-based approach to designer mouse models. Mamm Genome. 2002;13:543–547. doi: 10.1007/s00335-002-4003-4. [DOI] [PubMed] [Google Scholar]
- 29.Zhang F, Thornhill SI, Howe SJ, Ulaganathan M, Schambach A, Sinclair J, Kinnon C, Gaspar HB, Antoniou M, Thrasher AJ. Lentiviral vectors containing an enhancer-less ubiquitously acting chromatin opening element (UCOE) provide highly reproducible and stable transgene expression in hematopoietic cells. Blood. 2007;110:1448–1457. doi: 10.1182/blood-2006-12-060814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellis J. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum Gene Ther. 2005;16:1241–1246. doi: 10.1089/hum.2005.16.1241. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.