Abstract
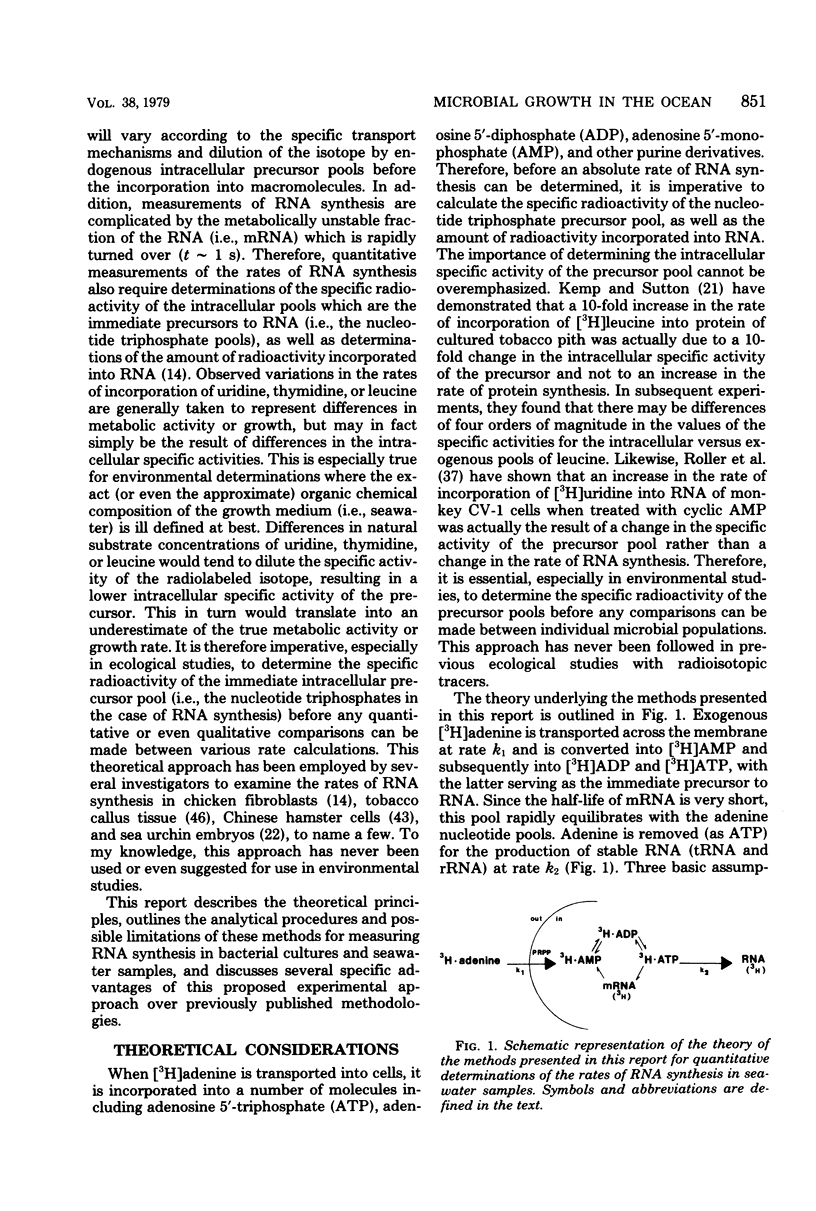
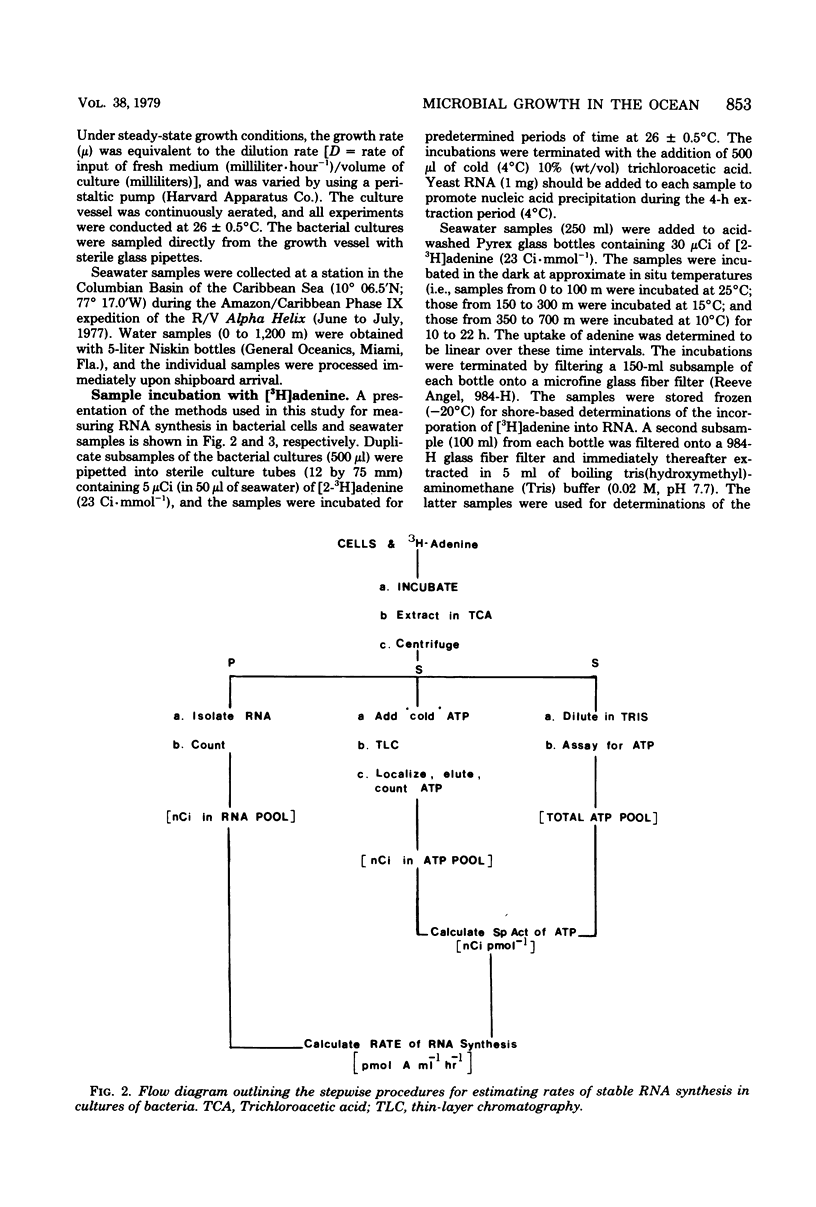
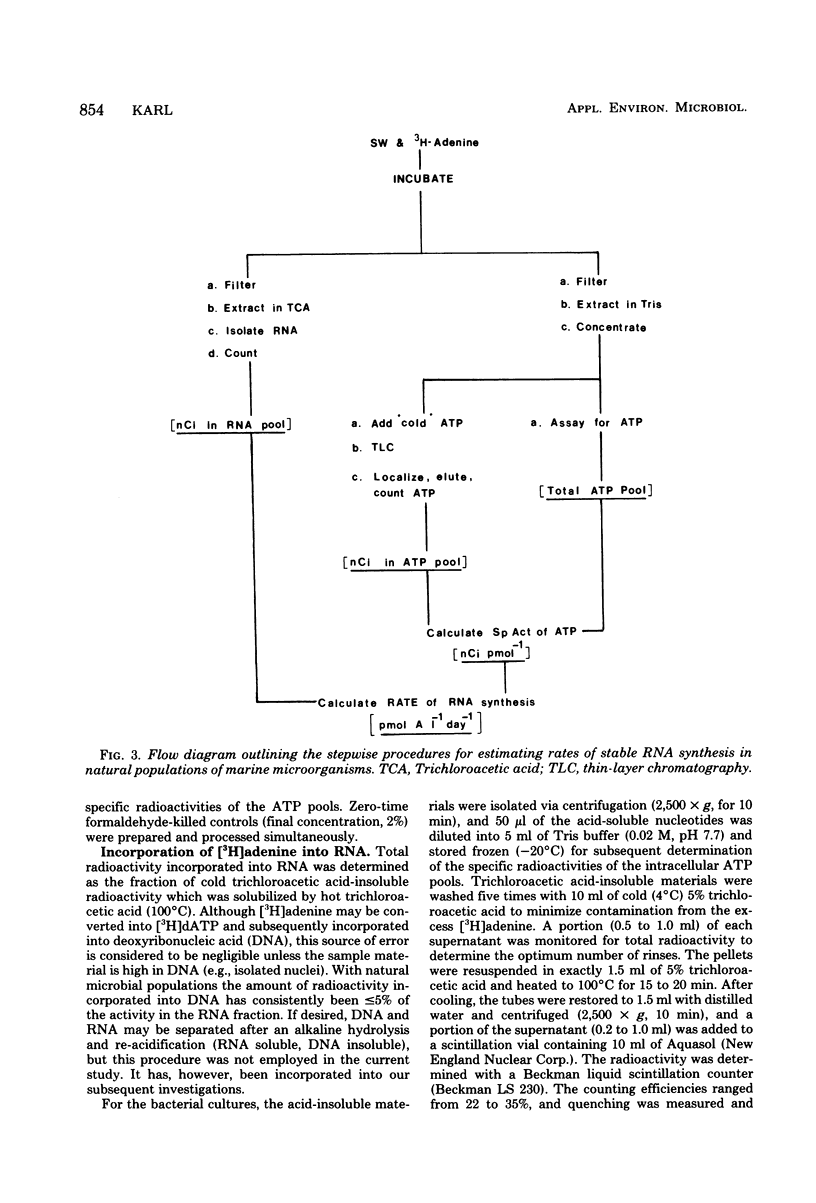
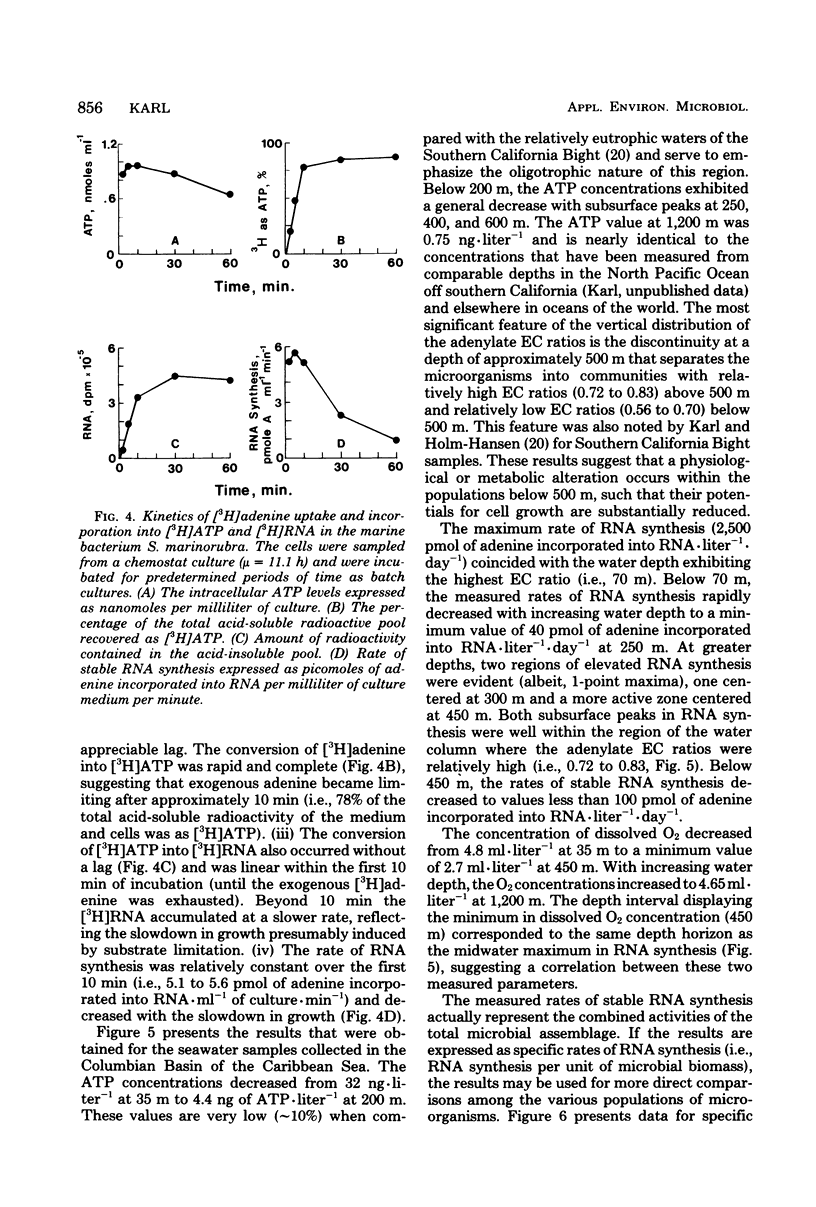
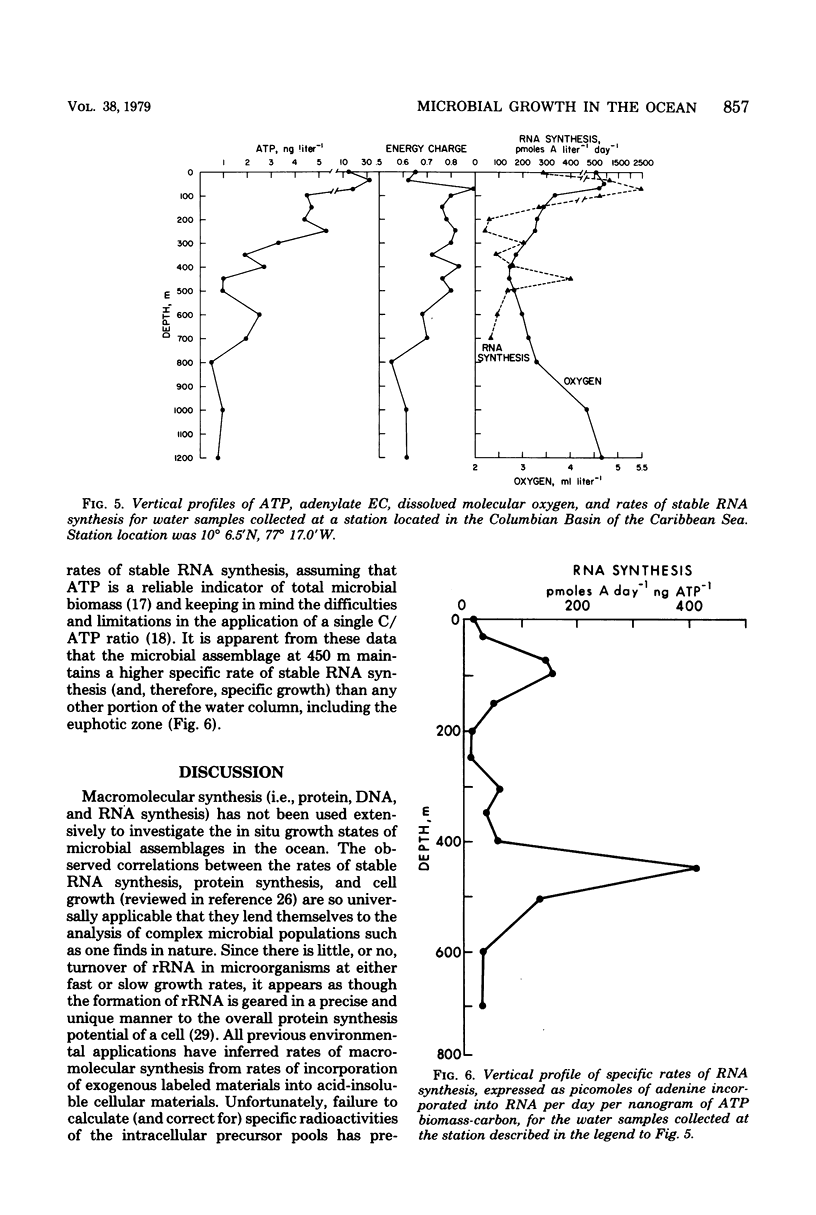
A relatively simple and extremely sensitive technique for measuring rates of stable ribonucleic acid (RNA) synthesis was devised and applied to bacterial cultures and seawater samples. The procedure is based upon the uptake and incorporation of exogenous radiolabeled adenine into cellular RNA. To calculate absolute rates of synthesis, measurements of the specific radioactivity of the intracellular adenosine 5′-triphosphate pools (precursor to RNA) and of the total amount of radioactivity incorporated into stable cellular RNA per unit time are required. Since the rate of RNA synthesis is positively correlated with growth rate, measurements of RNA synthesis should be extremely useful for estimating and comparing the productivities of microbial assemblages in nature. Adenosine 5′-triphosphate, adenylate energy charge, and rates of stable RNA synthesis have been measured at a station located in the Columbian Basin of the Caribbean Sea. A subsurface peak in RNA synthesis (and therefore growth) was located within the dissolved oxygen minimum zone (450 m), suggesting in situ microbiological utilization of dissolved molecular oxygen. Calculations of the specific rates of RNA synthesis (i.e., RNA synthesis per unit of biomass) revealed that the middepth maximum corresponded to the highest specific rate of growth (420 pmol of adenine incorporated into RNA·day−1) of all depths sampled, including the euphotic zone. The existence of an intermediate depth zone of active microbial growth may be an important site for nutrient regeneration and may serve as a source of reduced carbon for mesopelagic and deep sea environments.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkinson D. E., Fall L. Adenosine triphosphate conservation in biosynthetic regulation. Escherichia coli phosphoribosylpyrophosphate synthase. J Biol Chem. 1967 Jul 10;242(13):3241–3242. [PubMed] [Google Scholar]
- Atkinson D. E. The energy charge of the adenylate pool as a regulatory parameter. Interaction with feedback modifiers. Biochemistry. 1968 Nov;7(11):4030–4034. doi: 10.1021/bi00851a033. [DOI] [PubMed] [Google Scholar]
- Azam F., Beers J. R., Campbell L., Carlucci A. F., Holm-Hansen O., Reid F. M., Karl D. M. Occurrence and metabolic activity of organisms under the ross ice shelf, antarctica, at station j9. Science. 1979 Feb 2;203(4379):451–453. doi: 10.1126/science.203.4379.451. [DOI] [PubMed] [Google Scholar]
- Azam F., Hodson R. E. Dissolved ATP in the sea and its utilisation by marine bacteria. Nature. 1977 Jun 23;267(5613):696–698. doi: 10.1038/267696a0. [DOI] [PubMed] [Google Scholar]
- BUCHWALD M., BRITTEN R. J. Incorporation of ribonucleic acid bases into the metabolic pool and RNA of E. coli. Biophys J. 1963 Mar;3:155–166. doi: 10.1016/s0006-3495(63)86811-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandhorst B. P., Humphreys T. Synthesis and decay rates of major classes of deoxyribonucleic acid a5brandhorst BP, Humphreys T: Synthesis and decay rates of major classes of deoxyribonucleic acid-like ribonucleic acid in sea urchin embryos. Biochemistry. 1971 Mar 2;10(5):877–881. doi: 10.1021/bi00781a023. [DOI] [PubMed] [Google Scholar]
- Brock T. D. Bacterial growth rate in the sea: direct analysis by thymidine autoradiography. Science. 1967 Jan 6;155(3758):81–83. doi: 10.1126/science.155.3758.81. [DOI] [PubMed] [Google Scholar]
- Cashel M., Lazzarini R. A., Kalbacher B. An improved method for thin-layer chromatography of nucleotide mixtures containing 32P-labelled orthophosphate. J Chromatogr. 1969 Mar 11;40(1):103–109. doi: 10.1016/s0021-9673(01)96624-5. [DOI] [PubMed] [Google Scholar]
- Dinauer M. C., LaMarca M. J. The size of the guanosine triphosphate pool active in the synthesis of stable RNA by stage 6 oocytes of Xenopus laevis. Exp Cell Res. 1976 Aug;101(1):122–126. doi: 10.1016/0014-4827(76)90420-1. [DOI] [PubMed] [Google Scholar]
- Edlin G., Broda P. Physiology and genetics of the "ribonucleic acid control" locus in escherichia coli. Bacteriol Rev. 1968 Sep;32(3):206–226. doi: 10.1128/br.32.3.206-226.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson C. P., Jr, Humphreys T. A simple and sensitive method for quantitative measurement of cellular RNA synthesis. Anal Biochem. 1971 Apr;40(2):254–266. doi: 10.1016/0003-2697(71)90384-8. [DOI] [PubMed] [Google Scholar]
- Hochstadt-Ozer J., Stadtman E. R. The regulation of purine utilization in bacteria. II. Adenine phosphoribosyltransferase in isolated membrane preparations and its role in transport of adenine across the membrane. J Biol Chem. 1971 Sep 10;246(17):5304–5311. [PubMed] [Google Scholar]
- Hochstadt J. The role of the membrane in the utilization of nucleic acid precursors. CRC Crit Rev Biochem. 1974 Mar;2(2):259–310. doi: 10.3109/10409237409105449. [DOI] [PubMed] [Google Scholar]
- Kemp J. D., Sutton D. W. Protein Metabolism in Cultured Plant Tissues: III. Changes in the Rate of Protein Synthesis, Accumulation, and Degradation in Cultured Pith Tissue. Plant Physiol. 1972 Apr;49(4):596–601. doi: 10.1104/pp.49.4.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijima S., Wilt F. H. Rate of nuclear ribonucleic acid turnover in sea urchin embryos. J Mol Biol. 1969 Mar 14;40(2):235–246. doi: 10.1016/0022-2836(69)90472-0. [DOI] [PubMed] [Google Scholar]
- Larock P. A., Lauer R. D., Schwarz J. R., Watanabe K. K., Wiesenburg D. A. Microbial biomass and activity distribution in an anoxic, hypersaline basin. Appl Environ Microbiol. 1979 Mar;37(3):466–470. doi: 10.1128/aem.37.3.466-470.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leick V. Ratios between contents of DNA, RNA and protein in different micro-organisms as a function of maximal growth rate. Nature. 1968 Mar 23;217(5134):1153–1155. doi: 10.1038/2171153a0. [DOI] [PubMed] [Google Scholar]
- McCarthy B. J., Britten R. J. The Synthesis of Ribosomes in E. coli: I. The Incorporation of C-Uracil into the Metabolic Pool and RNA. Biophys J. 1962 Jan;2(1):35–47. doi: 10.1016/s0006-3495(62)86839-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R. R., Keller S. J. DNA polymerase activity: Factors affecting counting efficiency of radioactive DNA on filter paper discs. Anal Biochem. 1972 Mar;46(1):332–337. doi: 10.1016/0003-2697(72)90424-1. [DOI] [PubMed] [Google Scholar]
- NEIDHARDT F. C., MAGASANIK B. Studies on the role of ribonucleic acid in the growth of bacteria. Biochim Biophys Acta. 1960 Jul 29;42:99–116. doi: 10.1016/0006-3002(60)90757-5. [DOI] [PubMed] [Google Scholar]
- Neidhardt F. C. The regulation RNA synthesis in bacteria. Prog Nucleic Acid Res Mol Biol. 1964;3:145–181. doi: 10.1016/s0079-6603(08)60741-2. [DOI] [PubMed] [Google Scholar]
- Nierlich D. P. Radioisotope uptake as a measure of synthesis of messenger RNA. Science. 1967 Dec 1;158(3805):1186–1188. doi: 10.1126/science.158.3805.1186. [DOI] [PubMed] [Google Scholar]
- Nierlich D. P. Regulation of ribonucleic acid synthesis in growing bacterial cells. I. Control over the total rate of RNA synthesis. J Mol Biol. 1972 Dec 30;72(3):751–764. doi: 10.1016/0022-2836(72)90189-1. [DOI] [PubMed] [Google Scholar]
- Nierlich D. P., Vielmetter W. Kinetic studies on the relationship of ribonucleotide precursor pools and ribonucleic acid synthesis. J Mol Biol. 1968 Feb 28;32(1):135–147. doi: 10.1016/0022-2836(68)90151-4. [DOI] [PubMed] [Google Scholar]
- Pace N. R. Structure and synthesis of the ribosomal ribonucleic acid of prokaryotes. Bacteriol Rev. 1973 Dec;37(4):562–603. doi: 10.1128/br.37.4.562-603.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roller B. A., Hirai K., Defendi V. Effect of cyclic AMP on nucleoside metabolism. III. Effect on uridine incorporation in monkey (CV-1) cells. Biochim Biophys Acta. 1974 Nov 6;366(4):402–410. doi: 10.1016/0005-2787(74)90038-0. [DOI] [PubMed] [Google Scholar]
- Rutherford C. L., Wright B. E. Nucleotide metabolism during differentiation in Dictyostelium discoideum. J Bacteriol. 1971 Oct;108(1):269–275. doi: 10.1128/jb.108.1.269-275.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHAECHTER M., MAALOE O., KJELDGAARD N. O. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J Gen Microbiol. 1958 Dec;19(3):592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- Salser W., Janin J., Levinthal C. Measurement of the unstable RNA in exponentially growing cultures of Bacillus subtilis and Escherichia coli. J Mol Biol. 1968 Jan 28;31(2):237–266. doi: 10.1016/0022-2836(68)90442-7. [DOI] [PubMed] [Google Scholar]
- Stambrook P. J., Sisken J. E. The relationship between rates of ( 3 H)uridine and ( 3 H)adenine incorporation into RNA and the measured rates of RNA synthesis during the cell cycle. Biochim Biophys Acta. 1972 Sep 29;281(1):45–54. doi: 10.1016/0005-2787(72)90186-4. [DOI] [PubMed] [Google Scholar]
- Summerton J. E. Measurement of the pool size and synthesis rate of the metabolically unstable fraction of RNA in Escherichia coli by a method independent of hybridization efficiency and unaffected by precursor compartmentation. J Mol Biol. 1976 Jan 15;100(2):127–140. doi: 10.1016/s0022-2836(76)80144-1. [DOI] [PubMed] [Google Scholar]


