Abstract
Germline mutations in the mismatch repair genes mutL homolog 1 (MLH1) and mutS homolog 2 (MSH2), MSH6, and postmeiotic segregation increased 2 (PMS2) lead to the development of hereditary nonpolyposis colorectal cancer (HNPCC). Diagnosis of HNPCC relies on the compilation of a thorough family history of cancer, documentation of pathological findings, tumor testing for microsatellite instability (MSI) and immunohistochemistry (IHC), and germline mutation analysis of the suspected genes. As a hallmark of HNPCC, microsatellite instability is widely accepted as a primary method for identifying individuals at risk for HNPCC. It serves as an excellent, easy-to-evaluate marker of mismatch repair deficiency. Recent improvements in MSI testing have significantly enhanced the accuracy and reduced its cost. Proficiency testing for MSI is available, and laboratory-to-laboratory reproducibility of such testing can be easily evaluated. In addition, the combination of microsatellite instability testing, MLH1 promoter methylation analysis, and BRAF (V600E) mutation analysis can distinguish a sporadic colorectal cancer from one associated with HNPCC, helping to avoid costly molecular genetic testing for germline mutations in mismatch repair genes. In this article, we discuss the development of MSI markers used for HNPCC screening and focus on the advantages and disadvantages of MSI testing in screening for HNPCC patients. We conclude that MSI is as sensitive and specific as IHC, given its excellent reproducibility and its potential capability to indicate mutations not be detected by IHC. MSI has been used and will continue to prevail as the primary screening tool for identifying HNPCC patients.
The diagnosis of hereditary nonpolyposis colorectal cancer (HNPCC) at the molecular level relies on the presence of a deleterious germline mutation in one of the mismatch repair (MMR) genes. Because cancer morbidity and mortality can be dramatically reduced by colonoscopic screening of individuals with the HNPCC syndrome and by prophylactic surgeries, molecular screening of colorectal cancer patients for HNPCC is now feasible.1,2,3,4 The challenge is to establish a strategy that is able to screen effectively for HNPCC. Microsatellite instability (MSI) in colorectal cancer was discovered in 1993 and was subsequently found to be present in colon cancer tissue from most HNPCC patients.5,6,7,8
Genotyping for microsatellite instability was initially used to screen for HNPCC,1,3 while immunohistochemistry (IHC) analysis of the MMR proteins has been more recently proposed as an alternative method for screening HNPCC.2 Two recent studies have indicated that microsatellite instability testing and immunohistochemistry are both highly effective strategies for selecting patients for molecular genetic testing (germline mutation analysis).2,9 However, it is unclear which approach should be used as the primary method for screening HNPCC. Here, we summarize both the early and more recent literature data on the use of MSI, discuss the molecular basis of microsatellite instability in MMR-deficient tumors, and outline the advantages and limitations of this methodology. Our analysis indicates that given several merits of MSI that IHC does not have (see Advantages of MSI, below), MSI is an excellent, easy to use marker for identifying HNPCC. Therefore, it is important that clinicians are aware of the pros and cons of these two tests as both are widely used in screening HNPCC cases.
Literature Review
Microsatellite Instability
Microsatellites are short, tandemly repeated DNA sequences of 1 to 6 bases scattered throughout the human genome;10,11 they can be affected by a form of genomic instability called microsatellite instability.5,6,8,12 MSI is a change in length of a microsatellite allele due to either insertion or deletion of repeating units during DNA replication and failure of the DNA mismatch repair system to correct these errors. MSI analysis has been used as a screening method to identify HNPCC patients and a subgroup of colorectal cancer patients for further genetic testing.
The DNA Mismatch Repair System and HNPCC
DNA MMR is an effective post-replication mechanism. Most errors that occur during DNA replication are immediately corrected by the 3′ to 5′ exonuclease activity of DNA polymerase. It is estimated that 99.9% of the mutations that escape the proofreading activity of DNA polymerase (DNA polymerase slippage) are repaired by the DNA MMR system, particularly single-bp mismatches and “loop outs” of unpaired bases.13 The replication machinery slips more frequently on repetitive sequences than on non-repetitive sequences, so microsatellite instability occurs in the repetitive sequences in MMR-deficient cells. The causes of MMR defects are: i) germline mutations in any one of the five DNA MMR genes—mutS homolog 2 (MSH2), mutL homolog 1 (MLH1), MSH6, and, infrequently, postmeiotic segregation increased 2 or 1 (PMS2 or PMS1), causing HNPCC14; and ii) somatic inactivation of MLH1 caused by promoter hypermethylation in approximately 15% of sporadic colorectal cancer.5,8,15
In MMR-deficient cells, genes that contain a microsatellite in their coding regions are more prone to frameshift mutations. Mutations in key genes that regulate cell growth and apoptosis ultimately lead to dysregulated cell proliferation and/or cell death, which further speeds the evolution of colorectal cancer.16 One example is the well studied frameshift mutations in the TGF-βRII gene, which commonly occurs in colorectal cancer but not in endometrial cancer. In most colorectal cancers, the polyadenine tract mutations affect both alleles of TGF-βRII, suggesting that TGF-βRII functions as a tumor suppressor during colorectal cancer development and is a critical target of inactivation in mismatch repair-deficient tumors.17,18,19 Similar frameshift mutations in coding microsatellites also occur in other genes involved in growth control and apoptosis (TCF4, IGFIIR, BAX, and RIZ), as well as in genes involved in DNA mismatch repair itself (MSH6, MSH3, and MSH2).14
MSI as a Marker for HNPCC Screening
The original (1997) Bethesda guidelines20,21 proposed a panel of five microsatellite markers for the uniform analysis of MSI in HNPCC. This panel, which is referred to as the Bethesda panel, included two mononucleotide (BAT-25 and BAT-26) and three dinucleotide (D5S346, D2S123, and D17S250) repeats. Samples with instability in two or more of these markers are defined as MSI-High (MSI-H), whereas those with one unstable marker are designated as MSI-Low (MSI-L). Samples with no detectable alterations are MSI-stable (MSS). Because mononucleotide markers appear to be more sensitive than dinucleotide markers for the detection of MSI-H, limitations in the original panel resulting from inclusion of dinucleotide repeats were addressed at a 2002 National Cancer Institute workshop, and revised recommendations for MSI detection were proposed. The revision mainly recommends testing a secondary panel of mononucleotide markers, such as BAT-40, to exclude MSI-L in cases in which only the dinucleotide repeats are mutated.22 According to the revised Bethesda guidelines, strategies based on MSI testing were effective in identifying MLH1/MSH2 mutation carriers (sensitivity 81.8% and specificity 98.0%).9
Advantages of MSI
Microsatellite Instability Serves as an Excellent, Easy-to-Evaluate Marker of MMR Deficiency, and Recent Improvements in MSI Testing Significantly Enhance Accuracy and Reduce Cost
A hallmark of tumors in HNPCC is microsatellite instability. Typically half or more of all microsatellites have mutations (contraction or elongation) in the tumor cells; therefore, microsatellite instability serves as an excellent, easy-to-evaluate marker of mismatch repair deficiency. Since both HNPCC and MSI are caused by MMR defects, MSI can be used as a surrogate marker of HNPCC and has been widely accepted as a primary method for identifying individuals at risk for HNPCC.
As mentioned under Literature Review, a recent follow-up NCI workshop recognized the limitations of the original Bethesda panel20,21 due to the inclusion of dinucleotide repeats, which are less sensitive and less specific than mononucleotide repeats for identification of cancers with MMR deficiency.22 To improve the accuracy of MSI testing using the Bethesda panel of MSI markers, a panel of five mononucleotide markers was developed and incorporated into a multiplex fluorescence assay: the Promega (Madison, WI) MSI Analysis System.23 These mononucleotide repeat markers are quasi-monomorphic; that is, almost all individuals are homozygous for the same common allele for a given marker. The use of monomorphic markers simplifies data interpretation. The added pentanucleotide repeat markers ensure that the tumor and matching normal specimen are from the same individual.23,24 The Promega system can help resolve cases of MSI-L into either MSI-H or MSS.25 The representative electropherograms of the Promega MSI Analysis System are shown in Figure 1. The microsatellite markers included in the Bethesda panel20,21,22 and the Promega MSI Analysis System,25,26 as well as other commonly used MSI markers, are summarized in Table 1.
Figure 1.
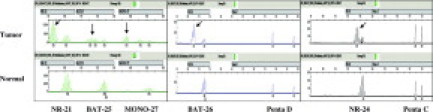
Representative electropherograms of the Promega MSI Analysis System (Version 1.1) generated using GeneMapper 3.7 Analysis Software. The shifted alleles are indicated by an arrow. Green: electropherogram showing the peaks of 2′,7′-dimethoxy-4,5-dichloro-6-carboxyfluorescein (JOE)-labeled loci, NR-21, BAT-25, and MONO-27. Blue: electropherogram showing peaks of the fluorescein-labeled loci, BAT-26 and Penta D. Black: electropherogram showing the peaks of tetramethyl rhodamine (TMR)-labeled loci, NR-24 and Penta C.
Table 1.
The Loci Information for Commonly Used MSI Markers
| Microsatellite marker | Repeat type | Chromosomal location (gene near marker/GenBank number) | Repeat motif*24,52 | Bethesda panel20,21,22 | Promega kit24,25 |
|---|---|---|---|---|---|
| BAT-25 | Mononucleotide | 4q12 (c-kit, intron 16) | TTTT.T.TTTT.(T)7.A(T)25 | X | X |
| BAT-26 | Mononucleotide | 2p16.3-p21 (hMSH2 gene, intron 5) | (T)5 . . . . . . (A)26 | X | X |
| NR-21 | Mononucleotide | 14q11.2 (SLC7A8, 5′UTR) | (T)21 | X | |
| NR-24 | Mononucleotide | 2q11.2 (ZNF2, 3′UTR) | (T)24 | X | |
| MONO-27 | Mononucleotide | 2p21 (MAP4K3, intron 13) | (A)27 | X | |
| BAT-40 | Mononucleotide | 1p13.1 (HSD3B2) | TTTT.TT . . (T)7 . . . . . . | ||
| . . . . TTTT.(T)40 | |||||
| D2S123 | Dinucleotide | 2p16 (MSH2) | (CA)13TA(CA)15(T/GA)7 | X | |
| D5S346 | Dinucleotide | 5q21/22 (APC) | (CA)26 | X | |
| D17S250 | Dinucleotide | 17q11.2-q12 (BRCA1) | (TA)7 . . . . . . . . . . . . . . . | X | |
| . . . . . . . . . . (CA)24 | |||||
| Penta C | Pentanucleotide | 21q22.3 (AL138752) | (AAAAG)3–15 | X | |
| Penta D | Pentanucleotide | 9p12-13.3 (AC003656) | (AAAAG)2–17 | X | |
| MYCL1 | Complex | 1p34.3 (MYCL1) | GAAAA(GAAA)2TAAA(A/G)10 | ||
| GAAAGA(GAAA)14 GAAA | |||||
| (GAAAA)8GAAAAA(GAAAA)2 |
Non-repetitive nucleotides are indicated as dots.
Since the Promega MSI Analysis System utilizes a multiplex fluorescence assay, PCR for all five mononucleotide markers and two pentanucleotide nucleotide markers can be performed in a single reaction. The size of the amplified products can be easily visualized using capillary electrophoresis. The cost of MSI testing is significantly reduced. In addition, in situations where the availability of representative tumor sections is limited, MSI testing can be done on one tumor section, whereas at least four tumor sections are required for IHC.
Proficiency Testing for MSI Is Available and the Reproducibility of MSI Testing is Close to 100%
The most common type of quality control in which clinical laboratories participate is proficiency testing, in which testing is performed along with routine laboratory work. Proficiency testing is a method of externally validating the accuracy of laboratory performance by testing samples and comparing results of all participating laboratories. To comply with Clinical Laboratory Improvement Amendments of 1988, laboratories performing moderate and high complexity tests must be enrolled in regulatory proficiency testing for their particular specialties/subspecialties of testing. A convenient way to accomplish this is by subscribing to a proficiency testing program that monitors those analytes. In the United States, proficiency testing for MSI is provided by the College of American Pathologists (CAP).
In the most recent CAP Proficiency Survey for microsatellite instability testing (2007 MSI-B), 71 laboratories were enrolled in this survey. The participant summary provided evidence that mononucleotide markers have higher specificity (ie, a lower false positive rate) for instability than dinucleotide markers. CAP also provided a detailed summary on several clinically important issues such as the number and types of markers used, methods used to perform the assay, and definition of MSI-H and MSI-L phenotypes. This information is valuable to clinical laboratories that are currently offering this test as well as to those that are planning to launch this test. Thirty-four of the 53 laboratories reported performing IHC together with MSI, whereas 19 of 53 laboratories did not perform IHC testing. With two recent CAP Proficiency Surveys combined (2007 MSI-A and MSI-B), 101 of 103 laboratories (98%) reported the same results, indicating that the reproducibility of MSI testing can be evaluated and, more importantly, that it is satisfactorily high.
In terms of quality control and interpretation of MSI testing in clinical settings, studies from a six-center consortium indicated that optimal PCR quality is essential to getting interpretable results. In most cases, the shifted PCR products from all five mononucleotide markers were smaller in size than the germline allele, as deletion in polyA sequences is much more common than insertions.27,28 It has been suggested that a larger allele size does not correlate with loss of mismatch repair gene expression by immunostaining and overall tumor phenotype. Forcing a call tended to bias toward increased numbers of MSI-L or MSI-H cases. Duplicate readings (scoring MSI results independently by two people) can help to reduce errors.29
MSI Analysis Could Potentially Identify a Tumor That Had Defective DNA MMR But Intact Staining, Perhaps Due to Non-Truncating Missense Alteration
Some disease-causing non-truncating mutations (missense mutations or in-frame insertion/deletion mutations) may not affect the protein translation, stability, and antigenicity; therefore, IHC staining is intact. A common scenario for IHC is to find focal and weak, or ambiguous, MLH1 staining along with absent PMS2 staining; this most likely represents a germline missense mutation in MLH1. However, when a missense mutation or an in-frame insertion/deletion mutation in MLH1 resides outside the MLH1/PMS2 interacting domain, IHC for both MLH1 and PMS2 may be present. Just as occurs with MLH1, some MSH6 missense mutations increase the risk for cancer, but MSH6 protein is present in the tumors by IHC. Under these situations, although MLH1 or MSH6 staining is present, MSI is able to identify a tumor that has MMR deficiency.
MSI Analysis Could Potentially Identify a Tumor That Had Defective DNA MMR Due to Defects in Genes Other than MSH2, MLH1, MSH6, or PMS2
MSI is a functional analysis in that mutations that disrupt MMR function lead to microsatellite instability in the tumor cells. Since MMR involves a set of genes including, but not limited to, MLH1, MSH2, MSH6, PMS2, MSH3, and PMS1, MMR deficiency may be caused by mutations in MMR genes not tested by IHC or in as yet unidentified MMR gene. MSI testing shows positive results in the presence of a mutation that disrupts the normal MMR function, no matter if the mutation resides in a known gene or in an uncharacterized MMR gene. IHC cannot detect such abnormalities since the current testing is limited to the four proteins with available antibodies.
Summary
MSI offers the following advantages that IHC does not: i) unlike IHC, which requires well experienced pathologists, molecular laboratory directors can be easily trained to read MSI results; ii) compared with the IHC staining pattern, which may vary and result in uncertainty in interpretation, the MSI unstable pattern can be easily recognized and highly reproducible; iii) unlike IHC, the sensitivity of which is dependent on its antibody panel, with its satisfactorily high reproducibility and the availability of proficiency testing offered by CAP, the quality control of MSI testing can be easily performed in a clinical laboratory; iv) MSI analysis could potentially identify a tumor that had defective DNA MMR but intact staining possibly due to non-truncating missense alterations or mutations in other MMR proteins not included in the current IHC panel; v) when the availability of representative tumor sections is limited, unlike IHC, which requires at least four tumor sections, only one tumor section with reasonable size of tumor is sufficient for MSI testing.
Limitations of MSI
MSI Is Not Specific for HNPCC
Although microsatellite instability is a hallmark for HNPCC, it is not specific for HNPCC. Microsatellite instability has been demonstrated in 10 to 15% of sporadic colorectal cancers. Deficient mismatch repair is largely attributed to hypermethylation of the 5′ CpG island in the MLH1 promoter and its consequent transcriptional silencing. The methylation is often, but not invariably, associated with loss of MLH1 protein.15 This is a so-called epigenetic change that affects gene function by aberrant methylation of DNA without genetic changes at the DNA sequence level. Epigenetic silencing has been recognized as another pathway that inactivates tumor suppressor genes in cancer.30
MLH1 hypermethylation is the cause of the microsatellite instability phenotype in sporadic colorectal cancers. Although the frequency of MLH1 hypermethylation in tumors with well characterized germline genetic defects in MMR genes was significantly reduced relative to sporadic microsatellite instability tumors, the presence of MLH1 hypermethylation did not exclude the possibility of germline mutations in mismatch repair genes.15 Actually, MLH1 hypermethylation may act as the second hit that inactivates the wild-type MLH1 allele in HNPCC tumors in addition to somatic mutations and deletions.31,32
Recently, the V600E mutation in BRAF was reported to be associated with microsatellite-unstable colorectal tumors.33,34,35 BRAF (V600E) mutation is frequently present in sporadic colorectal cancers with MLH1 hypermethylation, but not present in HNPCC. BRAF (V600E) mutation was not present in 4/4 MSI-H cell lines with mutated MLH1 or in 20/20 MSI-H HNPCC tumors.31 In other studies, BRAF (V600E) mutation was absent in 18/18 HNPCC tumors,36 in 37/37 HNPCC tumors,37 in 111/111 HNPCC tumors, and in 45/45 cases with abnormal MSH2 immunostaining.38 Based on these recent studies, it was concluded that the presence of BRAF (V600E) mutation argues against the presence of a germline mutation in either the MLH1 or MSH2 gene in HNPCC-associated colorectal cancers (specificity ∼100%).31,36,37,38 Tumors that have the BRAF V600E mutation and demonstrate MLH1 promoter hypermethylation are almost certainly sporadic, whereas tumors that show neither are most likely inherited. This will avoid the fruitless germline sequencing and rearrangement analysis of MLH1, which is still costly and labor consuming.
Tumors with Germline Mutations in MSH6 Tend to Show Lower Levels of MSI
Although the MSH6 gene is a component of the DNA MMR machinery, tumors with germline mutations in MSH6 may not show MSI-H. Instead, they tend to show a lower level of MSI.39,40,41 This is the consequence of the partial redundancy of the function of MSH6 and MSH3 proteins.42,43 The MSH2/MSH6 heterodimer (MutSα) recognizes both base-base and insertion-deletion loops, whereas the MSH2/MSH3 heterodimer (MutSβ) mainly recognizes insertion-deletion mismatches larger than one nucleotide.42,44 This explains the fact that when MSH6 is mutated, the MSH2/MSH3 dimer is still functioning, so that MSI can be limited to mononucleotide tandem repeats. As a matter of fact, addition of the BAT-40 mononucleotide marker reclassified some MSI-L tumors as MSI-H and some MSS tumors to MSI-L.41 Since MSI in MSH6 carriers has been mainly observed at mononucleotide markers,45 we speculate that when a sufficient number of mononucleotide markers are used for MSI testing, more MSH6 tumors will be considered as MSI-H rather than MSI-L or MSS. This is an area that awaits further investigation.
The fact that few or no differences in expressivity have been detected between MLH1 and MSH2 mutations indicates that both are equally important for MMR. In this regard, mutations in MSH6 behave quite differently. The so-called “attenuated” type of HNPCC caused by mutations in MSH6 is characterized by lower penetrance, higher age at onset, and more frequently distally localized colon cancers.39,46 It is reasonable to speculate that some MSS colon tumors in MSH6 patients are likely to have been sporadic tumors that did not develop because of MMR deficiency.41 Therefore, HNPCCs with MSH6 mutations may not be easily recognized because these features differ from those with mutations in MLH1 or MSH2.
Summary
MSI testing can be seen in both HNPCC tumors with germline mutations in MLH1 and sporadic colorectal cancers with MLH1 hypermethylation. However, IHC cannot distinguish these two types of colorectal cancers either. Since mutation in BRAF is present in the majority of tumors with hypermethylation of the MLH1 promoter but, importantly, not in cases with germline MLH1 mutations, the combination of microsatellite instability testing, MLH1 hypermethylation, and BRAF (V600E) mutation analysis can help distinguish a sporadic colorectal cancer from one associated with HNPCC. As for the MSH6 mutations, since the frequency of missense mutations is similar to that of truncating mutations and missense mutations in MSH6 may not alter its antigenicity, some mutations in MSH6 will be equally missed by IHC. In this regard, MSH6 mutation analysis should be considered in all patients suspected to have HNPCC. Neither MSI nor IHC should be a definitive selection criterion for MSH6 mutation analysis.39
Conclusion and Perspectives
MSI analysis has been used as a screening test for HNPCC in the original and revised Bethesda guidelines for over 10 years, and we have accumulated a wealth of experience with this test. The recent development of the five-mononucleotide marker system (the Promega MSI Analysis System) has greatly enhanced the accuracy and reduced the cost of MSI testing. Since the Promega system can help resolve cases of MSI-L into either MSI-H or MSS, and MSI-L is a major issue of MSH6 tumors, we speculate that, with the utility of this new MSI system, MSI testing will be better able to characterize the MSH6 tumors and assist in the search for germline mutations in MSH6. With its satisfactorily high sensitivity, specificity, and reproducibility, MSI offers myriad advantages for HNPCC screening. It is worthwhile to mention that CAP proficiency testing on MSI makes quality control as easy as possible to implement in clinical settings. Therefore, it is recommended that for patients who meet the Bethesda guidelines, the first step would be MSI analysis followed by IHC of all tumors classified as MSI-H.47
It is a fact that about half of all families clinically defined as HNPCC do not have mutations in any of the known MMR genes. This is an active research area in which new genes involved in MMR await to be identified and characterized. In this regard, MSI analysis could potentially identify mutations in other MMR genes not included in the current IHC panel. That is, if no mutations are present in the currently well studied MMR genes, a deleterious mutation might be present in hitherto unidentified genes. It is also possible that a mutation is present in the currently known MMR genes, but leads to HNPCC via completely different molecular mechanisms. The recent reports on germline methylation of the MLH1 or MSH2 promoter leading to HNPCC provide examples of these novel mechanisms.48,49,50,51
To some extent, MSI and IHC are complementary to each other in identifying HNPCC. The issue is that, currently, molecular testing for HNPCC screening is available at academic hospitals and major cancer centers as well as some private diagnostic companies; it is important that we make efforts to extend the current small-scale use of these molecular diagnostic tools and make them available to all clinicians. In order for this effort to be successful, clinicians must become aware of the advantages and disadvantages of these tests.
References
- 1.Aaltonen LA, Salovaara R, Kristo P, Canzian F, Hemminki A, Peltomaki P, Chadwick RB, Kaariainen H, Eskelinen M, Jarvinen H, Mecklin JP, de la Chapelle A. Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med. 1998;338:1481–1487. doi: 10.1056/NEJM199805213382101. [DOI] [PubMed] [Google Scholar]
- 2.Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, Nakagawa H, Sotamaa K, Prior TW, Westman J, Panescu J, Fix D, Lockman J, Comeras I, de la Chapelle A. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer) N Engl J Med. 2005;352:1851–1860. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 3.Salovaara R, Loukola A, Kristo P, Kaariainen H, Ahtola H, Eskelinen M, Harkonen N, Julkunen R, Kangas E, Ojala S, Tulikoura J, Valkamo E, Jarvinen H, Mecklin JP, Aaltonen LA, de la Chapelle A. Population-based molecular detection of hereditary nonpolyposis colorectal cancer. J Clin Oncol. 2000;18:2193–2200. doi: 10.1200/JCO.2000.18.11.2193. [DOI] [PubMed] [Google Scholar]
- 4.Schmeler KM, Lynch HT, Chen LM, Munsell MF, Soliman PT, Clark MB, Daniels MS, White KG, Boyd-Rogers SG, Conrad PG, Yang KY, Rubin MM, Sun CC, Slomovitz BM, Gershenson DM, Lu KH. Prophylactic surgery to reduce the risk of gynecologic cancers in the Lynch syndrome. N Engl J Med. 2006;354:261–269. doi: 10.1056/NEJMoa052627. [DOI] [PubMed] [Google Scholar]
- 5.Aaltonen LA, Peltomaki P, Leach FS, Sistonen P, Pylkkanen L, Mecklin JP, Jarvinen H, Powell SM, Jen J, Hamilton SR. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 6.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 7.Peltomaki P, Aaltonen LA, Sistonen P, Pylkkanen L, Mecklin JP, Jarvinen H, Green JS, Jass JR, Weber JL, Leach FS. Genetic mapping of a locus predisposing to human colorectal cancer. Science. 1993;260:810–812. doi: 10.1126/science.8484120. [DOI] [PubMed] [Google Scholar]
- 8.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 9.Pinol V, Castells A, Andreu M, Castellvi-Bel S, Alenda C, Llor X, Xicola RM, Rodriguez-Moranta F, Paya A, Jover R, Bessa X. Accuracy of revised Bethesda guidelines, microsatellite instability, and immunohistochemistry for the identification of patients with hereditary nonpolyposis colorectal cancer. JAMA. 2005;293:1986–1994. doi: 10.1001/jama.293.16.1986. [DOI] [PubMed] [Google Scholar]
- 10.Tautz D. Hypervariability of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res. 1989;17:6463–6471. doi: 10.1093/nar/17.16.6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weber JL, May PE. Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am J Hum Genet. 1989;44:388–396. [PMC free article] [PubMed] [Google Scholar]
- 12.Peltomaki P, Lothe RA, Aaltonen LA, Pylkkanen L, Nystrom-Lahti M, Seruca R, David L, Holm R, Ryberg D, Haugen A. Microsatellite instability is associated with tumors that characterize the hereditary non-polyposis colorectal carcinoma syndrome. Cancer Res. 1993;53:5853–5855. [PubMed] [Google Scholar]
- 13.Vogelstein B, Kinzler KW. The Genetic Basis of Human Cancer. McGraw-Hill, Medical Pub Division; New York: 2002. [Google Scholar]
- 14.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 15.Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, Kane MF, Kolodner RD, Vogelstein B, Kunkel TA, Baylin SB. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Umar A, Risinger JI, Hawk ET, Barrett JC. Testing guidelines for hereditary non-polyposis colorectal cancer. Nat Rev Cancer. 2004;4:153–158. doi: 10.1038/nrc1278. [DOI] [PubMed] [Google Scholar]
- 17.Kuismanen SA, Moisio AL, Schweizer P, Truninger K, Salovaara R, Arola J, Butzow R, Jiricny J, Nystrom-Lahti M, Peltomaki P. Endometrial and colorectal tumors from patients with hereditary nonpolyposis colon cancer display different patterns of microsatellite instability. Am J Pathol. 2002;160:1953–1958. doi: 10.1016/S0002-9440(10)61144-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myeroff LL, Parsons R, Kim SJ, Hedrick L, Cho KR, Orth K, Mathis M, Kinzler KW, Lutterbaugh J, Park K. A transforming growth factor beta receptor type II gene mutation common in colon and gastric but rare in endometrial cancers with microsatellite instability. Cancer Res. 1995;55:5545–5547. [PubMed] [Google Scholar]
- 19.Parsons R, Myeroff LL, Liu B, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Microsatellite instability and mutations of the transforming growth factor beta type II receptor gene in colorectal cancer. Cancer Res. 1995;55:5548–5550. [PubMed] [Google Scholar]
- 20.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 21.Rodriguez-Bigas MA, Boland CR, Hamilton SR, Henson DE, Jass JR, Khan PM, Lynch H, Perucho M, Smyrk T, Sobin L, Srivastava S. A National Cancer Institute Workshop on Hereditary Nonpolyposis Colorectal Cancer Syndrome: meeting highlights and Bethesda guidelines. J Natl Cancer Inst. 1997;89:1758–1762. doi: 10.1093/jnci/89.23.1758. [DOI] [PubMed] [Google Scholar]
- 22.Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Ruschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R, Hamilton SR, Hiatt RA, Jass J, Lindblom A, Lynch HT, Peltomaki P, Ramsey SD, Rodriguez-Bigas MA, Vasen HF, Hawk ET, Barrett JC, Freedman AN, Srivastava S. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bacher JW, Flanagan LA, Smalley RL, Nassif NA, Burgart LJ, Halberg RB, Megid WM, Thibodeau SN. Development of a fluorescent multiplex assay for detection of MSI-High tumors. Dis Markers. 2004;20:237–250. doi: 10.1155/2004/136734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suraweera N, Duval A, Reperant M, Vaury C, Furlan D, Leroy K, Seruca R, Iacopetta B, Hamelin R. Evaluation of tumor microsatellite instability using five quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology. 2002;123:1804–1811. doi: 10.1053/gast.2002.37070. [DOI] [PubMed] [Google Scholar]
- 25.Murphy KM, Zhang S, Geiger T, Hafez MJ, Bacher J, Berg KD, Eshleman JR. Comparison of the microsatellite instability analysis system and the Bethesda panel for the determination of microsatellite instability in colorectal cancers. J Mol Diagn. 2006;8:305–311. doi: 10.2353/jmoldx.2006.050092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baker K, Raut P, Jass JR. Microsatellite unstable colorectal cancer cell lines with truncating TGFbetaRII mutations remain sensitive to endogenous TGFbeta. J Pathol. 2007;213:257–265. doi: 10.1002/path.2235. [DOI] [PubMed] [Google Scholar]
- 27.Blake C, Tsao JL, Wu A, Shibata D. Stepwise deletions of polyA sequences in mismatch repair-deficient colorectal cancers. Am J Pathol. 2001;158:1867–1870. doi: 10.1016/S0002-9440(10)64143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou XP, Hoang JM, Cottu P, Thomas G, Hamelin R. Allelic profiles of mononucleotide repeat microsatellites in control individuals and in colorectal tumors with and without replication errors. Oncogene. 1997;15:1713–1718. doi: 10.1038/sj.onc.1201337. [DOI] [PubMed] [Google Scholar]
- 29.Lindor NM, Smalley R, Barker M, Bigler J, Krumroy LM, Lum-Jones A, Plummer SJ, Selander T, Thomas S, Youash M, Seminara D, Casey G, Bapat B, Thibodeau SN. Ascending the learning curve—MSI testing experience of a six-laboratory consortium. Cancer Biomark. 2006;2:5–9. doi: 10.3233/cbm-2006-21-202. [DOI] [PubMed] [Google Scholar]
- 30.Kondo Y, Issa JP. Epigenetic changes in colorectal cancer. Cancer Metastasis Rev. 2004;23:29–39. doi: 10.1023/a:1025806911782. [DOI] [PubMed] [Google Scholar]
- 31.Deng G, Bell I, Crawley S, Gum J, Terdiman JP, Allen BA, Truta B, Sleisenger MH, Kim YS. BRAF mutation is frequently present in sporadic colorectal cancer with methylated hMLH1, but not in hereditary nonpolyposis colorectal cancer. Clin Cancer Res. 2004;10:191–195. doi: 10.1158/1078-0432.ccr-1118-3. [DOI] [PubMed] [Google Scholar]
- 32.Young J, Simms LA, Biden KG, Wynter C, Whitehall V, Karamatic R, George J, Goldblatt J, Walpole I, Robin SA, Borten MM, Stitz R, Searle J, McKeone D, Fraser L, Purdie DR, Podger K, Price R, Buttenshaw R, Walsh MD, Barker M, Leggett BA, Jass JR. Features of colorectal cancers with high-level microsatellite instability occurring in familial and sporadic settings: parallel pathways of tumorigenesis. Am J Pathol. 2001;159:2107–2116. doi: 10.1016/S0002-9440(10)63062-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 34.Oliveira C, Pinto M, Duval A, Brennetot C, Domingo E, Espin E, Armengol M, Yamamoto H, Hamelin R, Seruca R, Schwartz S., Jr BRAF mutations characterize colon but not gastric cancer with mismatch repair deficiency. Oncogene. 2003;22:9192–9196. doi: 10.1038/sj.onc.1207061. [DOI] [PubMed] [Google Scholar]
- 35.Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 36.Kambara T, Simms LA, Whitehall VL, Spring KJ, Wynter CV, Walsh MD, Barker MA, Arnold S, McGivern A, Matsubara N, Tanaka N, Higuchi T, Young J, Jass JR, Leggett BA. BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut. 2004;53:1137–1144. doi: 10.1136/gut.2003.037671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Domingo E, Niessen RC, Oliveira C, Alhopuro P, Moutinho C, Espin E, Armengol M, Sijmons RH, Kleibeuker JH, Seruca R, Aaltonen LA, Imai K, Yamamoto H, Schwartz S, Jr, Hofstra RM. BRAF-V600E is not involved in the colorectal tumorigenesis of HNPCC in patients with functional MLH1 and MSH2 genes. Oncogene. 2005;24:3995–3998. doi: 10.1038/sj.onc.1208569. [DOI] [PubMed] [Google Scholar]
- 38.Domingo E, Laiho P, Ollikainen M, Pinto M, Wang L, French AJ, Westra J, Frebourg T, Espin E, Armengol M, Hamelin R, Yamamoto H, Hofstra RM, Seruca R, Lindblom A, Peltomaki P, Thibodeau SN, Aaltonen LA, Schwartz S., Jr BRAF screening as a low-cost effective strategy for simplifying HNPCC genetic testing. J Med Genet. 2004;41:664–668. doi: 10.1136/jmg.2004.020651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berends MJ, Wu Y, Sijmons RH, Mensink RG, van der Sluis T, Hordijk-Hos JM, de Vries EG, Hollema H, Karrenbeld A, Buys CH, van der Zee AG, Hofstra RM, Kleibeuker JH. Molecular and clinical characteristics of MSH6 variants: an analysis of 25 index carriers of a germline variant. Am J Hum Genet. 2002;70:26–37. doi: 10.1086/337944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buttin BM, Powell MA, Mutch DG, Babb SA, Huettner PC, Edmonston TB, Herzog TJ, Rader JS, Gibb RK, Whelan AJ, Goodfellow PJ. Penetrance and expressivity of MSH6 germline mutations in seven kindreds not ascertained by family history. Am J Hum Genet. 2004;74:1262–1269. doi: 10.1086/421332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hendriks YM, Wagner A, Morreau H, Menko F, Stormorken A, Quehenberger F, Sandkuijl L, Moller P, Genuardi M, Van Houwelingen H, Tops C, Van Puijenbroek M, Verkuijlen P, Kenter G, Van Mil A, Meijers-Heijboer H, Tan GB, Breuning MH, Fodde R, Wijnen JT, Brocker-Vriends AH, Vasen H. Cancer risk in hereditary nonpolyposis colorectal cancer due to MSH6 mutations: impact on counseling and surveillance. Gastroenterology. 2004;127:17–25. doi: 10.1053/j.gastro.2004.03.068. [DOI] [PubMed] [Google Scholar]
- 42.Acharya S, Wilson T, Gradia S, Kane MF, Guerrette S, Marsischky GT, Kolodner R, Fishel R. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci USA. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marsischky GT, Filosi N, Kane MF, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 44.Edelmann W, Yang K, Umar A, Heyer J, Lau K, Fan K, Liedtke W, Cohen PE, Kane MF, Lipford JR, Yu N, Crouse GF, Pollard JW, Kunkel T, Lipkin M, Kolodner R, Kucherlapati R. Mutation in the mismatch repair gene Msh6 causes cancer susceptibility. Cell. 1997;91:467–477. doi: 10.1016/s0092-8674(00)80433-x. [DOI] [PubMed] [Google Scholar]
- 45.Verma L, Kane MF, Brassett C, Schmeits J, Evans DG, Kolodner RD, Maher ER. Mononucleotide microsatellite instability and germline MSH6 mutation analysis in early onset colorectal cancer. J Med Genet. 1999;36:678–682. [PMC free article] [PubMed] [Google Scholar]
- 46.de la Chapelle A. Genetic predisposition to colorectal cancer. Nat Rev Cancer. 2004;4:769–780. doi: 10.1038/nrc1453. [DOI] [PubMed] [Google Scholar]
- 47.Lynch HT, Boland CR, Gong G, Shaw TG, Lynch PM, Fodde R, Lynch JF, de la Chapelle A. Phenotypic and genotypic heterogeneity in the Lynch syndrome: diagnostic, surveillance and management implications. Eur J Hum Genet. 2006;14:390–402. doi: 10.1038/sj.ejhg.5201584. [DOI] [PubMed] [Google Scholar]
- 48.Chan TL, Yuen ST, Kong CK, Chan YW, Chan AS, Ng WF, Tsui WY, Lo MW, Tam WY, Li VS, Leung SY. Heritable germline epimutation of MSH2 in a family with hereditary nonpolyposis colorectal cancer. Nat Genet. 2006;38:1178–1183. doi: 10.1038/ng1866. [DOI] [PubMed] [Google Scholar]
- 49.Hitchins MP, Wong JJ, Suthers G, Suter CM, Martin DI, Hawkins NJ, Ward RL. Inheritance of a cancer-associated MLH1 germ-line epimutation. N Engl J Med. 2007;356:697–705. doi: 10.1056/NEJMoa064522. [DOI] [PubMed] [Google Scholar]
- 50.Suter CM, Martin DI, Ward RL. Germline epimutation of MLH1 in individuals with multiple cancers. Nat Genet. 2004;36:497–501. doi: 10.1038/ng1342. [DOI] [PubMed] [Google Scholar]
- 51.Valle L, Carbonell P, Fernandez V, Dotor AM, Sanz M, Benitez J, Urioste M. MLH1 germline epimutations in selected patients with early-onset non-polyposis colorectal cancer. Clin Genet. 2007;71:232–237. doi: 10.1111/j.1399-0004.2007.00751.x. [DOI] [PubMed] [Google Scholar]
- 52.Dietmaier W, Wallinger S, Bocker T, Kullmann F, Fishel R, Ruschoff J. Diagnostic microsatellite instability: definition and correlation with mismatch repair protein expression. Cancer Res. 1997;57:4749–4756. [PubMed] [Google Scholar]


