Abstract
A key problem in understanding deuterostome evolution has been the origin of the chordate body plan. A biphasic life cycle with a sessile adult and a free-swimming larva is traditionally considered ancestral in chordates with subsequent neotenic loss of the sessile adult stage. Molecular phylogenies challenged this view, suggesting that the primitive life cycle in chordates was entirely free-living as in modern day larvaceans. Here, we report the precise cell lineage and fate map in the normal embryo of the larvacean Oikopleura dioica, using 4D microscopy technique and transmission electron microscopy. We document the extraordinary rapidity of cleavage and morphogenetic events until hatching and demonstrate that—compared with ascidians—fate restriction occurs considerably earlier in O. dioica and that clonal organization of the cell lineage is more tightly coupled to tissue fate. We show that epidermal cells in the trunk migrate through 90°, reminiscent of events in ascidian metamorphosis and that the axis of bilateral symmetry in the tail rotates in relation to the trunk. We argue that part of the tail muscle cells are ectomesodermal, because they are more closely associated with prospective epidermis than with other tissues in the cell lineage. Cladistic comparison with other deuterostomes suggests that these traits are derived within tunicates strengthening the hypothesis that the last common ancestor of tunicates had a sessile adult and thus support traditional morphology-derived scenarios. Our results allow hypothesizing that molecular developmental mechanisms known from ascidian models are restricted to fewer, yet identifiable, cells in O. dioica.
Keywords: heterochrony, larvacea, Oikopleura dioica, ontogeny
A key problem in understanding deuterostome evolution has been the origin of the tadpole-like chordate body plan. Ascidians are sessile tunicates with larval stages that possess chordate features such as notochord or dorsal nerve tube. Larvaceans are also tunicates but display these typical chordate features throughout their entirely planktonic life. Traditionally, a biphasic, ascidian-like life cycle with a free-swimming larva inherited from a deuterostome ancestor had been considered ancestral in chordates with subsequent neotenic loss of the sessile adult stage (1–5). Molecular phylogenies challenged this view and suggested that the primitive life cycle in chordates was entirely free-living as exemplified by modern day larvaceans (6–8). Thus, larvaceans are pivotal for the understanding of chordate evolution and figured prominently in discussions about the role neoteny might have played in the evolution of chordates (9–12). However, whereas cell lineage and molecular aspects of tissue restriction have been intensely studied in ascidians, culminating in the deciphering of the first metazoan whole-embryo gene regulatory network (13, 14), lack of comparable knowledge in larvaceans has hindered a deeper understanding of chordate ontogeny and evolution (2, 4, 15, 16). Here, we report the precise cell lineage and fate map in the embryo of the larvacean Oikopleura dioica, using 4D microscopy technique and transmission electron microscopy. We document the extraordinary rapidity of cleavage and morphogenetic events and demonstrate that—compared with ascidians—fate restriction occurs considerably earlier in O. dioica and that clonal organization of the cell lineage is more tightly coupled to tissue fate. We show that epidermal cells in the trunk migrate through 90°, reminiscent of events in ascidian metamorphosis, and that the tail rotates through ontogeny in relation to the trunk. Comparison with other deuterostomes shows that cell lineage characteristics are derived within tunicates supporting the hypothesis that the last common ancestor of tunicates had a sessile adult stage. Our results allow us to hypothesize that molecular developmental mechanisms known from ascidians are restricted to fewer, yet identifiable, cells in O. dioica. The present study reveals the simplest cell lineage (in the sense of ref. 17) known from metazoans and amounts to a major step forward in tracing evolutionary changes in cellular mechanisms that will ultimately facilitate understanding molecular mechanisms correlated with drastic changes in life history strategies.
Results and Discussion
To facilitate comparisons among animals, we adopted the nomenclature established by Conklin (18) for tunicate ascidians instead of the one introduced later by Delsman (15) for appendicularians. See Materials and Methods for explanation.
Early Cleavage.
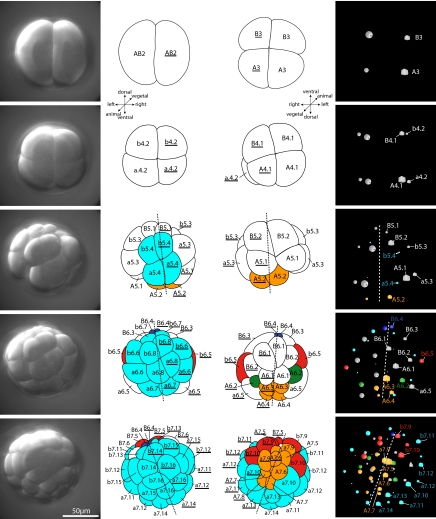
Cleavage in O. dioica is stereotyped and characterized by rapid divisions, early fate determinations, and overall bilateral symmetry [Fig. 1, supporting information (SI) Fig. S1, and Movies S1 and S2]. The first cleavage, 15 min after fertilization, in principle separates left from right (Figs. 1 and 2; see next paragraph). The second cleavages, 5 min after the first, occur longitudinally at right angles to the first one (Fig. 2). The third round of cell divisions is at right angles to both previous planes and separates animal from vegetal (Figs. 2 and 3). After the initial three cleavages, a stereoblastula is formed. The blastula stage lasts until after the fifth round of cell divisions, when most adult tissue types can be assigned to specific fate-restricted cells (Fig. 4). At the ventral vegetal side of the embryo, an exceptionally asymmetric cell division from B5.2 (B5.2) to B6.3 and B6.4 (B6.3, B6.4) takes place, B6.3 (B6.3) being bigger than B6.4 (B6.4). At this stage embryos can easily be oriented.
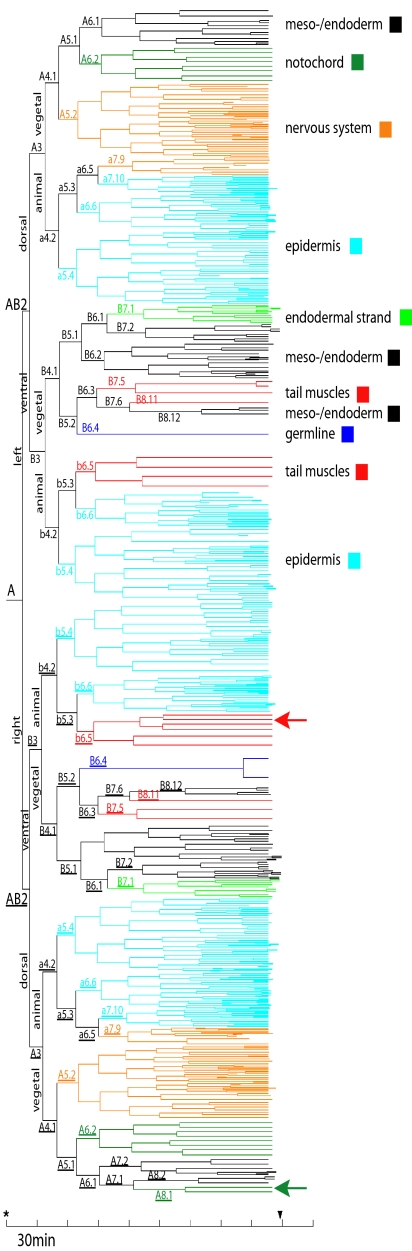
Fig. 1.
Tree representation and fate map of the tunicate Oikopleura dioica. Colors indicate different tissues identifiable at hatching. The nomenclature is according to Conklin (18) and Nishida (19) [Table S2 can be consulted to translate this nomenclature to the one used by Delsman (15)]. Note overall correspondences between tissue fates of left and right sides. Arrows indicate obvious deviations from bilateral symmetry in the muscle and notochord lineages. The asterisk indicates the time of fertilization; the arrowhead marks the time of hatching.
Fig. 2.
First cleavages in the ontogeny of the tunicate Oikopleura dioica. (Left) Projections of the upper (animal) half of DIC-stacks recorded for 4D microscopy. (Line drawings in Left) View from animal pole. (Line drawings in Right) View from vegetal pole. Asterisk marks the position of the blastopore. (Right) 3D representation of the positions of the nuclei of each cell in the 4D microscopy software SIMI°BioCell as seen from the vegetal pole. Cubes derive from the left side (AB2), spheres from the right (AB2). Nomenclature and color code for tissues that are already restricted as in Fig. 1. Dashed line indicates axis of bilateral symmetry. Note bilateral symmetry in these early embryonic stages.
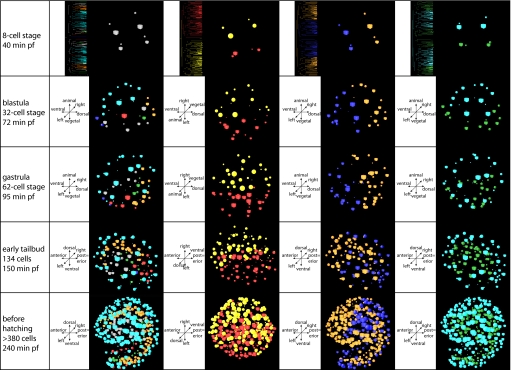
Fig. 3.
Virtual cell tracing experiments, using the 3D representation of the positions of the nuclei of each cell implemented in the 4D microscopy software SIMI°BioCell. Cubes derive from the left side (AB2); spheres derive from the right side (AB2). (Second column) Colors indicate tissues as in Fig. 1. (Third column) Red cubes derive from the left side (AB2); yellow spheres derive from the right side (AB2). Note that the orientation in this panel differs from the remaining ones to give a better view of the dorsal midline in the prehatchling state. (Fourth column) Orange shapes derive from dorsal cells A3 and A3, traditionally labeled “anterior” (see text for details); blue shapes derive from ventral cells B3 and B3, traditionally labeled “posterior.” Note that the center of orange colored nuclei shifts from dorsal to anterior between gastrula and early tailbud stage. (Far right column) Blue shapes derive from the animal cells a4.2, b4.2, a4.2, and b4.2; green shapes derive from the vegetal cells A4.2, B4.2, A4.2, and B4.2.
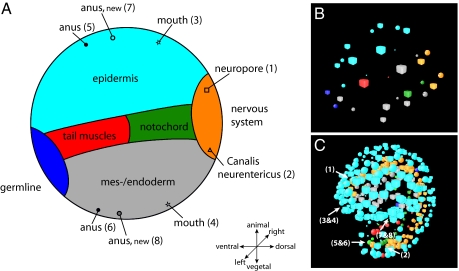
Fig. 4.
Virtual tracing of chordate landmarks. (A) Schematic projection of the prospective fate in Oikopleura dioica as derived from the blastula (B). Positions of anatomic landmarks that are indicated in the larva (C) are also indicated in the schematic projection. (B) Representation of nuclei with colors coding for prospective tissue fates (see Fig. 1). (C) Representation of nuclei in the larva with positions of anatomic landmarks indicated that are depicted in the schematic fate map in A.
Cell Divisions and Body Axes.
The planes of the first three cleavages in O. dioica coincide with embryonic axes at least to late neurula stages. The first cleavage separates left from right (Figs. 1 and 2), but, during later stages, cells cross the bilateral symmetry axis (Fig. 3, third column from left; Fig. S2; and Movie S2). This is most obvious in muscle cells in the b6.5 and b6.5 lines. Two other exceptions from bilateral symmetry are the additional divisions in the prospective muscle cell lineage b7.9 and the occurrence of two notochord cells originating in A8.1 on the right side (Fig. 1; Fig. S1). In addition, the symmetry axis of the tail rotates anti-clockwise as seen from posterior during later stages (Fig. S3). The second cleavage is traditionally thought to establish the anterior–posterior axis in tunicates (18–21). Our results, however, demonstrate that what traditionally was labeled “anterior” coincides with the prospective nervous system in early stages in O. dioica (Figs. 2–4), the very structure defining dorsal in chordates (22, 23). Therefore, cells A3 and A3 are considered dorsal in the present study, a usage consistent with practice in cephalochordates (24) and craniates (25). A remarkable pattern of cell movement occurs during late neurula/early tailbud stages. Many of the originally dorsally located epidermal cells migrate to occupy anterior positions in the hatchling (Fig. 3, fourth column; Movie S3). This migration can be traced back to the fate map in the early blastula (32-cell stage), where the anterior–posterior axes specified by prospective mouth and anus and by the prospective neuropore and Canalis neurentericus respectively are at an angle of 90° (Fig. 4). This means that, according to our interpretation, at this early stage, the prospective mouth is situated dorsally, the prospective anus is situated ventrally, and the observed cell movement secondarily brings epidermis cells surrounding the prospective mouth to the anterior end. The third division separates animal from vegetal (Figs. 2 and 3 and Movie S4). Epidermis is strictly animal in derivation, whereas notochord, germ-line, endodermal strand, and mesoderm and endoderm are purely vegetal. In contrast, nervous system and musculature derive partly from both halves of the embryo (Figs. 1 and 4).
Gastrulation.
Gastrulation begins after the fifth round of cell divisions and is completed when the last of the prospective muscle cells, the animal-derived b7.9, b7.10, b7.9, and b7.10, are internalized and prospective epidermis cells close over the blastopore after the seventh round of cell divisions. Although there is no archenteron formed, the blastopore can be recognized as the site where cells ingress into the embryo (star in Fig. 2). Prospective muscle cells invaginate over its ventral lip and lateral sides, whereas prospective nervous system cells border the dorsal side of the blastopore at this stage (Movie S1).
Neurulation.
Neurulation overlaps with gastrulation, because the prospective nervous system cells arrange along the dorsal midline after the fifth round of cell divisions, bordering the dorsal side of the blastopore (Movie S1). From the border of the blastopore in animal direction, the identity of prospective neural cells is a7.9, A7.5, A7.6, A7.7, and A7.8 on the left and their counterparts on the right. The cells sink beneath the epidermis, starting with the animal-most and vegetal-most of the vegetal cells (A7.5, A7.5, A7.8, and A7.8). All prospective nervous system cells are covered by epidermis after the seventh round of cell divisions, with animal cells (a7.9 and a7.9) being the last to sink beneath the epidermis. Although posterior nuclei of prospective nervous system cells arrange themselves in double layer as early as 2 h 10 min postfertilization (pf), anterior ones remain in single file at least half an hour longer and a dilated anterior brain is only recognizable at 3 h and 30 min pf.
Clonal Tissue Restriction.
We used transmission electron microscopy (TEM) of a complete series of sections of a newly hatched individual to reconstruct its 3D anatomy (Figs. S5–S15). In the tail, a central notochord, delimited by an extracellular matrix (ecm), is surrounded by muscle-cells equipped with myofilaments, the neural tube with a central canal, and endodermal strand cells. In the trunk, the nervous system can be recognized, based on its dorsal position, smaller cell size, and connections with the neural tube in the tail. Although in the anterior half of the trunk the ventral endodermal cells are epithelially organized surrounding a primordial intestinal cavity, in the posterior half of the trunk, cells are less differentiated. Because in this area the mesodermal pericard develops, we refrained from distinguishing between endoderm and mesoderm in this region and used the term “meso-/endoderm.” Based on the detailed anatomical knowledge derived from the TEM investigation, we could assign almost every cell of the hatchling to a tissue fate that was then traced back by using 4D microscopy to result in the cell lineage fate map (Fig. 1, Fig. S1, and Movie S1) and 3D representation of nuclei in time (Figs. 2–4).
Clonal organization of the tissues is essentially invariant among individuals (Fig. S1), and fate restriction occurs as early as the 16-cell stage, where a5.4, b5.4 and their counterparts a5.4, b5.4 are destined to become epidermis, and A5.2 and A5.2 are restricted to nervous fate. Most lineages achieve clonal restriction at the 62-cell stage, many already at the 32-cell stage. Epidermis, albeit being entirely animal in derivation, has a variety of clonal precursors (a5.4, a6.6, a7.10, b5.4, b6.6, and their counterparts). The majority of cells in the nervous system derive from vegetal cells A5.2 and A5.2; only a few derive from the animal a7.9 and a7.9. Similarly, tail muscle cells are in part vegetal (B7.5, B8.11, B7.5, and B8.11) and part animal (b6.5 and b6.5) in origin. Muscle cells and notochord cells display a stereotyped pattern of complex morphogenetic movements ending up in precise and invariant positions in the hatchling (Fig. S4). Vegetal muscle cells end up anteriorly in the tail and remain in their respective bilateral sides. Animal muscle cells become more posteriorly situated and—like many epidermal cells—partly cross the bilateral symmetry axis. What we termed meso-/endoderm is entirely vegetal in origin, but we were unable to assign a more specific fate to these cells. Part of the vegetal progeny, the lines originating in B7.6 and B7.6 can be assigned to the endodermal strand in the tail. B6.4 and B6.4 derive via a strongly asymmetric cleavage from B5.2 and B5.2 and arrest cleavage until hatching, characteristic for germ line cells (in one individual one division in the B6.4 line was seen 20 min before hatching). These conspicuous small cells are situated in the ventral midline in early stages and invaginate during gastrulation via the ventral blastopore lip. Delsman, in the classic description of the ontogeny of Oikopleura dioica, was unsure whether these cells were not merely plasmaprotrusions (15). Later, these cells become situated in the posterior trunk and are impacted by the tail rotation, because the right cell is more dorsally located than the left. On the dorsal vegetal side, eight notochord cells derive from A6.2 and A6.2, and, on the right side, an additional pair derive from A8. Thus, using 4D microscopy, we identified 18 notochord cells in the hatchling, 19 using TEM, and 20 that were described later as juveniles (15, 16, 26, 27). Notochord cells position themselves along the embryonic midline and start intercalating ≈2.5 h pf. Intercalation proceeds from anterior to posterior and is completed ≈3.5 h pf. It remains unclear where the two remaining cells needed to complete juvenile cell number originate.
The cell lineage and fate map of O. dioica share similarities with those of ascidians, such as rapid and precise determinative cleavage, general bilateral symmetry of cleavage pattern, and the derivation of nervous system and musculature from both vegetal and animal lineages (18–20). However, closer inspection reveals also major differences between larvaceans and ascidians. Fate restrictions occur considerably earlier, and clonal organization of the cell lineage is even more tightly coupled to tissue fate in O. dioica. For example, tail muscle cells derive from four distinct lineages in the ascidian embryo (A8.16, B7.4, B7.5, B7.8, and b8.17) (13, 19), whereas, in O. dioica, no A-cell line gives rise to musculature. Similarly, whereas the nervous system derives from A-, a-, and b-lines in ascidians (19, 28, 29), in O. dioica, nervous tissue originates in the lineages A5.2 and a7.9, but no b-line contribution to the nervous tissue was found. Another example is seen in the development of the notochord; in ascidian embryos, the anterior 32 primary notochord cells derive from A7.3, A7.7, and the posterior eight secondary notochord cells derive from B8.6 (13, 30). In O. dioica notochord originates only in A-cell lines, indicating the absence of secondary notochordal cells. In Ciona intestinalis, notochordal cells in the A-line depend on activation of brachyury, which depends on ZicL and Fgf9/16/20 activation. ZicL is activated by FoxA-a and FoxD, and Fgf9/16/20 is mediated by phosphorylation of the ETS-containing transcription factor ets/pointed2 (13). Although we would predict that a similar network is responsible for the determination of A6.2, A6.2, and A8.1 in O. dioica, it will be interesting to see whether the slightly different circuit that results in secondary notochordal cells in C. intestinalis (13, 30) is missing or silenced in O. dioica like genes in the case of the tailless ascidian larva of Molgula occulta that is evolutionarily derived from a tailed ancestor (31, 32). Based on our results, it is now possible to target molecular networks for comparative analysis elucidating ontogenetic mechanisms in the evolution of drastically divergent life cycles.
Larvaceans have been used as proxies for the suggested role heterochrony might have played in the evolution of the chordate body plan (2, 4, 33), but Ruppert (34) used a cladistic approach to argue that heterochrony occurred in the stem lineage leading to tunicates and that developmental speed was even more derived in larvaceans. Comparison of our data to available data from other deuterostomes (Table S1 and Fig. S16) supports Ruppert's conclusions and strengthens the hypothesis that larvaceans are derived within tunicates. The observed rotation of the tail can be used as an informative phylogenetic character to place larvaceans more precisely as the sister group to aplousobranch ascidians (35, 36).
The observed acceleration in the development of larvaceans is not only achieved by shortening cell cycles or lowering number of cleavages at which specific events occur, but the lineage tree is altered. This finding is consistent with the hypothesis that selection for decreased cell lineage complexity played a major role in the evolution of metazoan ontogenies (17, 37). Although Caenorhabditis elegans had been considered a prime example of mosaic development (38), modern investigations have demonstrated that its ontogeny is based on principles of regional specification rather than clonal organization of cell lineage (39, 40). Thus, although tunicates also show regulative cell interactions during ontogeny, their fate map is tightly coupled to cell lineage and in that sense remain extreme examples of determinative (mosaic) development, with Oikopleura dioica as the most extreme example.
Materials and Methods
For recordings, two- or four-cell embryos were mounted on a microscope slide coated with 0.01% polylysine. Fundamentals of 4D microscopy are described by Schnabel et al. (40). A Zeiss Axioplan Imaging 2 microscope with internal focus drive was used to move the temperature-controlled stage to record a z series with a Hamamatsu Newvicon camera. Images were digitized with an Inspecta 3 frame grabber (Mikroton) and compressed with a wavelet function (Lurawave). The microscope is controlled with a software programmed by K. Schulz and R. Schnabel. Embryos were recorded at 15°C (measured on stage) until hatching. Recordings of three different embryos—all of which were developing normally and, after hatching, actively swimming—stemming from three different parental pairs were analyzed by using SIMI°BioCell software (SIMI). Z projections of the upper half of the DIC-stacks recorded at specific times were generated in ImageJ software (http://rsb.info.nih.gov/ij).
The anatomy of a hatched larva was analyzed by using TEM. Fixation was in 1.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) adjusted to 950 mosM with NaCl followed by a postfixation in 1% OsO4. Araldite was used as embedding resin, and a complete series of ultrathin sections prepared. Analysis was on a Philips Biotwin CM 120 (FEI) and a virtual 3D reconstruction was prepared by using 3Ds Studio Max (Autodesk).
In this article, we used the standard nomenclature established for tunicate ascidians by Conklin (18). In a study of Oikopleura dioica, Delsman (15) introduced a different nomenclature. We provide a translation table for the two systems in Table S2 and compare the two systems in Fig. S1. We preferred the Conklin system, because by adopting Conklin's nomenclature the similarity of the fate maps becomes immediately obvious and allows detailed evolutionary comparisons.
Supplementary Material
Acknowledgments.
T.S. thanks Prof. W. Dohle (Freie Universität, Berlin) for many fruitful discussions and Prof. H. Nishida for critical remarks on earlier versions of the manuscript. This work was supported by Deutsche Forschungsgemeinschaft Grant Sta651-1/2.
Note.
While this article was under review, a description of the early development up to the 64-cell stage of O. dioica was published by Fuji et al. (41). Note that these authors use Delsman's nomenclature as explained here and in Table S2.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710196105/DCSupplemental.
References
- 1.Garstang W. Preliminary note on a new theory of the phylogeny of the Chordata. Zool Anz. 1894;17:122–125. [Google Scholar]
- 2.Garstang W. The morphology of the Tunicata, and its bearings on the phylogeny of the Chordata. Q J Mic Sci. 1928;72:51–187. [Google Scholar]
- 3.Gee H. Before the backbone. Views on the origin of the vertebrates. London: Chapman & Hall; 1996. [Google Scholar]
- 4.Lacalli TC. Protochordate body plan and the evolutionary role of larvae: Old controversies resolved? Can J Zool. 2005;83:216–224. [Google Scholar]
- 5.Nielsen C. Origin of the chordate central nervous system—and the origin of the chordates. Dev Genes Evol. 1999;209:198–205. doi: 10.1007/s004270050244. [DOI] [PubMed] [Google Scholar]
- 6.Bourlat SJ, et al. Deuterostome phyology reveals monophyletic chordates and the new phylum Xenoturbellida. Nature. 2006;444:1–4. doi: 10.1038/nature05241. [DOI] [PubMed] [Google Scholar]
- 7.Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- 8.Wada H. Evolutionary history of free-swimming and sessile lifestyles in urochordates as deduced from 18S rDNA molecular phylogeny. Mol Biol Evol. 1998;15:1189–1194. doi: 10.1093/oxfordjournals.molbev.a026026. [DOI] [PubMed] [Google Scholar]
- 9.Nishino A, Satoh N. the simple tail of chordates: Phylogenetic significance of appendicularians. Genesis. 2001;29:36–45. doi: 10.1002/1526-968x(200101)29:1<36::aid-gene1003>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 10.Seo H-C, et al. Hox cluster disintegration with persistent anteroposterior order of expression in Oikopleura dioica. Nature. 2004;431:67–71. doi: 10.1038/nature02709. [DOI] [PubMed] [Google Scholar]
- 11.Ruppert EE. Key characters uniting hemichordates and chordates: Homologies or homoplasies? Can J Zool. 2005;83:8–23. [Google Scholar]
- 12.Canestro C, Bassham S, Postlethwait JH. Development of the central nervous system in the larvacean Oikipleura dioica and the evolution of the chordate brain. Dev Biol. 2005;285:298–315. doi: 10.1016/j.ydbio.2005.06.039. [DOI] [PubMed] [Google Scholar]
- 13.Imai KS, Levine M, Satoh N, Satou Y. Regulatory blueprint for a chordate embryo. Science. 2006;312:1183–1187. doi: 10.1126/science.1123404. [DOI] [PubMed] [Google Scholar]
- 14.Lemaire P. How many ways to make a chordate? Science. 2006;312:1145–1146. doi: 10.1126/science.1128784. [DOI] [PubMed] [Google Scholar]
- 15.Delsman HC. Beiträge zur Entwicklungsgeschichte von Oikopleura dioica. Verhandelingen uit het Rijksinstituut voor het Onderzoek der Zee. 1910;3:1–24. [Google Scholar]
- 16.Delsman HC. Weitere Beobachtungen über die Entwicklung von Oikopleura dioica. Tijdschr Ned Dierk Ver (Ser 2) 1912;12:197–215. [Google Scholar]
- 17.Azevedo RBR, et al. The simplicity of metazoan cell lineages. Nature. 2005;433:152–156. doi: 10.1038/nature03178. [DOI] [PubMed] [Google Scholar]
- 18.Conklin EG. Organization and cell lineage of the ascidian egg. J Acad Nat Sci Philadelphia. 1905;13:1–119. Series 2. plates I-XII. [Google Scholar]
- 19.Nishida H. Cell lineage analysis in ascidian embryos by intracellular injection of a tracer enzyme. Dev Biol. 1987;121:526–541. doi: 10.1016/0012-1606(87)90188-6. [DOI] [PubMed] [Google Scholar]
- 20.Zalokar M, Sardet C. Tracing of cell lineage in embryonic development of Phallusia mammillata (Ascidia) by vital staining of mitochondria. Dev Biol. 1984;102:195–205. doi: 10.1016/0012-1606(84)90184-2. [DOI] [PubMed] [Google Scholar]
- 21.Nishida H. Specification of embryonic axis and mosaic development in ascidians. Dev Dynam. 2005;233:1177–1193. doi: 10.1002/dvdy.20469. [DOI] [PubMed] [Google Scholar]
- 22.Arendt D, Nübler-Jung K. Inversion of dorsoventral axis? Nature. 1995;371:26. doi: 10.1038/371026a0. [DOI] [PubMed] [Google Scholar]
- 23.Lowe CJ, et al. Dorsoventral patterning in hemichordates: Insights into early chordate evolution. PLoS Biol. 2006;4:e291. doi: 10.1371/journal.pbio.0040291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu J-K, et al. Axial patterning in cephalochordates and the evolution of the organizer. Nature. 2007;445:613–617. doi: 10.1038/nature05472. [DOI] [PubMed] [Google Scholar]
- 25.De Robertis EM, Kuroda H. Dorsal-ventral patterning and neural induction in Xenopus embryos. An Rev Cell Dev Biol. 2004;20:285–308. doi: 10.1146/annurev.cellbio.20.011403.154124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galt C. Seattle: University of Washington; 1972. Development of Oikopleura dioica (Utochordata: Larvacea) PhD dissertation. [Google Scholar]
- 27.Nishino A, Satou Y, Morisawa M, Satoh N. Brachyury (T) gene expression and notochord development in Oikopleura longicauda (Appendicularia, Urochordata) Dev Genes Evol. 2001;211:219–231. doi: 10.1007/s004270100141. [DOI] [PubMed] [Google Scholar]
- 28.Lemaire P, Bertrand V, Hudson C. Early steps in the formation of neural tissue in ascidian embryos. Dev Biol. 2002;252:151–169. doi: 10.1006/dbio.2002.0861. [DOI] [PubMed] [Google Scholar]
- 29.Taniguchi K, Nishida H. Tracing cell fate in brain formation during embryogenesis of the ascidian Halocynthia roretzi. Dev Growth Diff. 2004;46:163–180. doi: 10.1111/j.1440-169X.2004.00736.x. [DOI] [PubMed] [Google Scholar]
- 30.Hudson C, Yasuo H. A signalling relay involving nodal and delta ligands acts during secondary notochord induction in Ciona embryos. Development. 2006;133:2855–2864. doi: 10.1242/dev.02466. [DOI] [PubMed] [Google Scholar]
- 31.Swalla BJ, Jeffery WR. Requirement of the Manx gene for expression of chordate features in a tailless ascidian larva. Science. 1996;274:1205–1208. doi: 10.1126/science.274.5290.1205. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen C. Tail evolution. Science. 1997;277:1422. doi: 10.1126/science.277.5331.1421b. [DOI] [PubMed] [Google Scholar]
- 33.Berrill NJ. The Origin of Vertebrates. Oxford: Clarendon; 1955. [Google Scholar]
- 34.Ruppert EE. Hemichordata, Chaetognatha, and the invertebrate chordates. In: Harrison FW, Ruppert EE, editors. Microscopic Anatomy of Invertebrates. New York: Willey-Liss; 1997. pp. 1–13. [Google Scholar]
- 35.Stach T. Comparison of the serotonergic nervous system among Tunicata: Implications for its evolution within Chordata. Org Div Evol. 2005;5:15–24. [Google Scholar]
- 36.Stach T. Ontogeny of the appendicularian Oikopleura dioica (Tunicata, Chordata) reveals characters similar to ascidian larvae with sessile adults. Zoomorphology. 2007:203–214. [Google Scholar]
- 37.Houthoofd W, et al. Embryonic cell lineage of the marine nematode Pellioditis marina. Dev Biol. 2003;258:57–69. doi: 10.1016/s0012-1606(03)00101-5. [DOI] [PubMed] [Google Scholar]
- 38.Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 39.Schnabel R, et al. Global cell sorting in the C. elegans embryo defines a new mechanism for pattern formation. Dev Biol. 2006;294:418–431. doi: 10.1016/j.ydbio.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 40.Schnabel R, Hutter H, Moermann D, Schnabel H. Assessing normal embryogenesis in Caenorhabditis elegans using a 4D microscope: Variability of development and regional specification. Dev Biol. 1997;184:234–265. doi: 10.1006/dbio.1997.8509. [DOI] [PubMed] [Google Scholar]
- 41.Fuji S, Nishio T, Nishida H. Cleavage pattern, gastrulation, and neurulation in the appendicularian, Oikopleura dioica. Dev Genes Evol. 2008;218:69–79. doi: 10.1007/s00427-008-0205-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.