Abstract
The aggregation of proteins into amyloid fibrils is associated with several neurodegenerative diseases. In Parkinson's disease it is believed that the aggregation of α-synuclein (α-syn) from monomers by intermediates into amyloid fibrils is the toxic disease-causative mechanism. Here, we studied the structure of α-syn in its amyloid state by using various biophysical approaches. Quenched hydrogen/deuterium exchange NMR spectroscopy identified five β-strands within the fibril core comprising residues 35–96 and solid-state NMR data from amyloid fibrils comprising the fibril core residues 30–110 confirmed the presence of β-sheet secondary structure. The data suggest that β1-strand interacts with β2, β2 with β3, β3 with β4, and β4 with β5. High-resolution cryoelectron microscopy revealed the protofilament boundaries of ≈2 × 3.5 nm. Based on the combination of these data and published structural studies, a fold of α-syn in the fibrils is proposed and discussed.
Keywords: amyloid, NMR, Parkinson's disease, structure, aggregation
Parkinson's disease (PD) is the second most common neurodegenerative disease in the United States, currently affecting as many as 1.5 million individuals. The two pathological hallmarks of PD are the loss of dopaminergic neurons in the substantia nigra region of the brain, and the presence of intracellular inclusions: Lewy bodies and Lewy neurites. Recent observations indicate that PD is an amyloid disease involving the aggregation of α-synuclein [α-syn (1–4)]. The evidence includes the finding that α-syn is the major fibrillar protein in Lewy bodies, and the discovery that point mutations in the α-syn gene (i.e., A53T, A30P, and E46K) are observed in rare familial forms of early-onset PD, inherited as an autosomal dominant form. Furthermore, fly and other animal models have suggested an important role for α-syn in the etiology of PD (5, 6). On the one hand, cytotoxicity of α-syn has been correlated to a dosage effect by protein overexpression resulting from supernumerary copies of the α-syn gene (7, 8). On the other hand, cellular dysfunction might be caused by a change in the conformation of α-syn, because a few studies have pointed to the direct correlation between α-syn fibrillar inclusion bodies and neurodegeneration (9, 10), whereas others have suggested that α-syn oligomers are the major cause of cellular toxicity (11).
α-Syn is a 140-aa cytoplasmic or/and membrane-attached protein found in presynaptic terminals of neuronal cells (1). Although the precise physiological role of this protein is not fully understood, it has been suggested that α-syn is involved in the modulation of neurotransmitter release (12, 13), involved in ER/Golgi trafficking (8), or loosely associated with synaptic vesicles (2). The 3D NMR structure of α-syn in the presence of the membrane-mimetic SDS comprises two helices of residues 3–37 and 45–92 and a highly mobile C-terminal tail comprising residues 98–140 (14). In aqueous solution, α-syn is natively unfolded and hence highly dynamic (15–17). On aggregation, α-syn undergoes a conformational change into amyloid fibrils by intermediates, that is, oligomers, ring-like oligomers, and protofibrils, which are a kind of premature fibril (11). The amyloid fibrils composed of several protofilaments contain a cross β-structure in which individual β-strands run perpendicular to the fiber axis (18). EPR measurements (19, 20) and limited proteolysis experiments (21) indicate that residues 31–109 are folded in the fibrils, whereas the N-terminal ≈30 residues are heterogeneous and the C-terminal ≈30 residues are flexible. The solid-state NMR studies of α-syn fibrils support these findings and furthermore elucidate the presence of two structurally distinct isoforms, both composed of several β-strands (22). To obtain further structural insights, in particular, to elucidate the sequence-specific positions of amyloid fibril secondary structure elements, we studied the amyloid fibrils of α-syn and/or the fragment α-syn (30–110), which shows faster aggregation properties than wild-type α-syn [supporting information (SI) Fig. S1], by quenched hydrogen/deuterium exchange NMR, solid-state NMR, high-resolution cryoelectron microscopy, and site-directed mutagenesis.
Results
H/D Exchange of α-Synuclein Fibrils by NMR Reveals a Solvent-Protected Core.
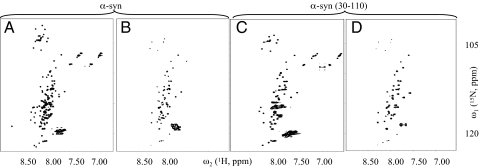
Toward the elucidation of the fold of the amyloid fibrils of recombinant α-syn grown in PBS (pH 7.4), we aimed to identify regular secondary structural elements by using quenched hydrogen/deuterium (H/D) exchange measured by solution-state NMR (23–25). This technique allows the identification of solvent-protected backbone amide protons indicative of hydrogen bond formation. Fig. 1A shows the reference [15N,1H] correlation NMR spectrum of α-syn in DMSO corresponding to fully protonated α-syn fibrils. The spectrum contains one cross-peak for each backbone 15N-1H moiety of α-syn, enabling a residue-specific determination of the hydrogen exchange rates. After 1 h of H/D exchange with the α-syn fibrils (PBS, pH 7.4) approximately half of the resonances are significantly reduced or absent from the spectrum (Fig. 1B). The corresponding amides have exchanged with solvent deuterons that are not observable in the [15N,1H] correlation NMR experiment. The close resemblance of this spectrum with a reference spectrum of the fragment α-syn (30–110) comprising residues 30–110 (Fig. 1C and Fig. S2), indicates that for full-length α-syn, most of the amides of residues 30–110 are solvent-protected, whereas most of the amides of the N- and C-terminal 30 residues are solvent-exposed and possibly not involved in a regular secondary structure. Most of the amides of residues 30–110 are also protected in α-syn (30–110) fibrils (Fig. 1D) suggesting a similar conformation for α-syn (30–110) and α-syn fibrils. The presence of a solvent-protected core in α-syn fibrils comprising amino acid residues ≈30–110 is in agreement with proteolysis-resistant experiments (21), EPR (19), and solid-state NMR studies (22).
Fig. 1.
A qualitative interpretation of quenched H/D exchange of α-syn and α-syn (30–110) fibrils based on NMR spectra. Fast [15N,1H]-HMQC-spectra of solubilized 15N-labeled α-syn in d6-DMSO containing 0.1% d1-TFA, corresponding to fully protonated (A) and 1 h exchanged amyloid fibrils of α-syn (B). Many cross-peaks are lost on H/D exchange for 1 h. In C and D fast [15N,1H]-HMQC-spectra of 15N-labeled α-syn (30–110) solubilized in d6-DMSO containing 0.1% d1-TFA, corresponding to fully protonated, and 1 h exchanged amyloid fibrils of α-syn (30–110), respectively. The amino acid assignment of the cross-peaks of the spectrum shown in C are shown in Fig. S2.
The H/D Exchange of α-Synuclein Fibrils Is Biphasic Suggesting the Presence of Two Distinct Structural Entities.
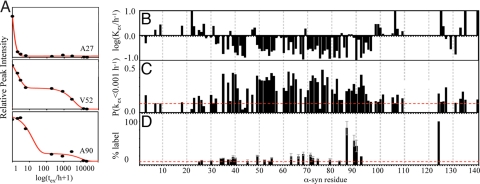
For a quantitative analysis of the H/D exchange measurements of α-syn fibrils, a residue-specific exchange was monitored for a period of 30 months as shown for three examples in Fig. 2A. In contrast to Ala-27, for which a fast monoexponential decay of the signal was observed (>10 h−1), the decay of the signals for Val-52 and Ala-90 have two distinct phases comprising a slow exchange (0.1–1 h−1) and an extremely slow exchange rate (<10−3 h−1). Overall, for the fast-exchanging residues only a single-exponential decay was observed, whereas for the slow-exchanging residues of the fibril core comprising residues ≈30–103 a biphasic decay was observed. Approximately one-half to two-thirds of the population of each backbone amide of the fibril core exhibited slow exchange, whereas the remainder of the population exhibited extremely slow exchange (Fig. 2 B and C). (Note: the biphasic decay is detected easily because the slow and extremely slow exchange rates differ by two orders of magnitude.) This heterogeneity is also observed in another H/D exchange experiment of a sample of α-syn fibrils grown in conventional Eppendorf tubes (data not shown) indicating the prevalence of this property for α-syn fibrils. Furthermore, α-syn (30–110) fibrils also have a biphasic exchange as shown in Fig. S3. However, for two-week-old α-syn (30–110) fibrils, only ≈20% of the population is in the extremely slow exchange regime and the average exchange rate for the population in the slow exchange regime is somewhat faster than in wild-type α-syn fibrils. When the H/D exchange of the same fibril sample of α-syn (30–110) was measured six months later, the subpopulation of extremely slow exchange increased to 30–50% (Fig. S3). This indicates that the extremely slow-exchanging population increases with time.
Fig. 2.
Detailed H/D exchange analysis and side-chain solvent accessibility of α-syn fibrils. (A) H/D exchange curves for several amide moieties as indicated. The peak volumes versus logarithm of the exchange time are shown. Data points are shown as spheres. Smooth red lines represent a superposition of two monoexponential fits of the raw data. For most residues a biphasic exchange is observed including a slow decay (≈1–10 h) and an extremely slow decay (>1,000 h). (B) Plot of the H/D exchange rates of the fast- and slow-exchanging population against the amino acid sequence. (C) Plot of the fraction of each amide moiety that exhibit extremely slow H/D exchange at rates <0.001 h−1. This representation enables us to highlight the solvent protection of amides of the population defined as extremely slow-exchanging by taking into consideration that this conformation may comprise fast-exchanging amides in loops and turns. Only if the fraction is >0.15 is it considered that the amide undergoes a very slow exchange in the sample, because an accuracy of ≈10% is obtained by fitting the data with a superposition of two monoexponential decays. The amide moieties for which H/D exchange data were obtained are labeled with a single-letter amino acid code. (D) Side-chain solvent accessibility of Cys in fibrils of α-syn variants. The fluorescence of Alexa Fluor 488 cross-linked to the Cys side chain is given for various α-syn variants relative to the A124C variant. The side chain of residue 124 is, in accordance to H/D exchange, EPR, solid-state NMR, and limited proteolysis studies, highly solvent exposed. A large fluorescence value is indicative of a high degree of solvent accessibility.
The biphasic behavior of the H/D exchange suggests the presence of two distinct conformations in the α-syn fibril sample. The exchange rates between the two conformations differ by >2 orders of magnitude and the population of the more solvent-protected conformation increases with time. Hence, the conformation with moderate solvent protection appears to be maturing into a highly solvent protected (fibrillar) conformation with exchange rates comparable to other amyloid fibrils (23, 24).
Secondary Structure Analysis of α-Synuclein Fibrils.
In favorable cases, residue-resolved quenched H/D exchange allows the determination of the positions of secondary structural elements of fibrils (23, 24). Here, successful H/D exchange analysis was obtained for ≈75% of all backbone amides including almost all backbone amides of the fibril core comprising residues 30–110 (Fig. 2). Furthermore, most of the nonassigned residues can be categorized to be in a fast-exchange regime, because most of the nonassigned cross-peaks in the [15N,1H] correlation spectrum are significantly reduced within 1 h of H/D exchange (Fig. 1). In agreement with the qualitative interpretation of the H/D exchange given above, sequence-specific analysis of the H/D exchange reveals the presence of three regions. The N-terminal residues 1–27 and the C-terminal residues 104–140 are in fast exchange, which indicates structural flexibility or absence of secondary structural elements. Conversely, in the segment comprising residues 30–103 most of the amides are solvent-protected (Fig. 2 B and C). Within this core region, five segments can be identified either by the grouping of residues that display slow exchange and are interrupted by residues with faster H/D exchange, or by grouping of the residues that have subpopulations of extremely slow exchangers. For the species with extremely slow H/D exchange, these five segments comprise residues ≈35–41, ≈49–56, ≈60–67, ≈69–82 (possibly interrupted at residues 75 and 79), and ≈86–94 (Fig. 2C). For the species with slow H/D exchange, the five segments are similar with residues ≈39–44, ≈49–56, ≈62–68, ≈70–79 (or ≈70–83), and ≈90–96 (Fig. 2B). Because α-syn fibrils are composed of β-sheets (Fig. S1) (18, 19, 22, 26, 27), the five protected segments for both species are presumed to be five distinct β-strands. These β-strands are thereby in good agreement with the solid-state NMR analysis of α-syn (30–110) fibrils (see below). In this context it is noteworthy to mention that both an algorithm for the prediction of secondary structural elements [GOR-IV (28)] as well as an algorithm for the prediction of aggregation-prone segments (29) highlight with astonishing accuracy the identified β-strands (SI Text).
Solid-State NMR of α-syn (30–110) Fibrils.
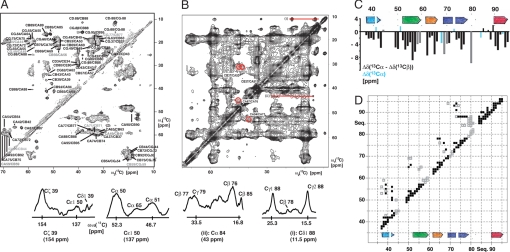
To collect distance and angular restraints for α-syn fibrils, solid-state NMR experiments with 10-month-old sample of 15N,13C-labeled α-syn (30–110) were measured, exploiting the unique sequence properties of the α-syn variant. Although the quality of the spectra (Fig. 3) with respect to line width and resolution are not comparable with the fibrils of the HET-s prion (23), the sequence of α-syn (30–110) comprising 1 Leu (Leu-38), 1 Tyr (Tyr 39), 2 Ser (Ser-42, Ser-87), 1 His (His-50), 1 Asn (Asn-65), 1 Gly–Gly pair (Gly-67–Gly-68), 1 Thr–Ala pair (Thr-75–Ala-76), 1 Ala–Gly pair (Ala-85–Gly-86), 1 Ile (Ile-88), 1 Phe (Phe-94) provided 14 starting points for the sequence-specific chemical shift assignment. Side-chain assignment was obtained by using the DREAM (30) spectra (Fig. 3A), in combination with the 13C–13C proton-driven spin diffusion (PDSD) spectra (Fig. 3B). Sequential assignment was obtained by analysis of the 13C–13C PDSD (31) spectra and confirmed by the sequential backbone spectra from the experiments NCA (Fig. S4) and N(CO)CA (32, 33). The assignment of the 14 unique residues and residue pairs and their neighboring residues yielded almost 50% of the sequential assignment of the fibrillar core comprising residues 35–95. With a further analysis of the solid-state NMR spectra, assignments were obtained for 70% of the core residues 35–95 (Fig. 3 A and C). In contrast to the H/D exchange data, and the solid-state NMR studies performed by Heise et al. (22), minimal peak doubling due to sample heterogeneity was observed.
Fig. 3.
Solid-state NMR of α-syn (30–110) fibrils. (A) 13C–13C DREAM spectrum for the intraresidual assignment. (B) 13C–13C proton-driven spin diffusion (PDSD) spectrum with a mixing time of 250 ms. Long-range cross-peaks are indicated by red circles and labeled accordingly. In addition, several cross-sections with long-range cross-peaks are shown and labeled accordingly. (C) Secondary chemical shift plot of the difference of Δδ(13Cα) − Δδ(13Cβ) with Δδ(13Cα) and Δδ(13Cβ), which are the differences between experimental 13Cα or 13Cβ chemical shifts and their corresponding “random-coil” chemical shifts. Highlighted in cyan are Gly residues for which only the Δδ(13Cα) could be calculated. Highlighted in gray are ambiguous assignments. This secondary chemical shift plot indicates the presence of β-strand motifs as indicated. (D) 2D distance plot of α-syn (30–110) fibrils showing distance restraints < ≈6 Å between residues, which have been extracted from the 13C–13C proton-driven spin diffusion spectra. Black squares indicate unambiguous distance restraints between residues. Gray squares indicate distances, which are only unambiguous under the assumption that α-synuclein fibrils form a five-layered β-sandwich. In addition, the β-strand motifs extracted from C are shown.
Deviations of the combined 13Cα/13Cβ chemical shifts from random coil values were then used to identify the type of secondary structure present in α-syn (30–110) fibrils. Negative deviations (Fig. 3C) indicate β-strand secondary structure for residues 37–43, 52–59, 62–66, 68–77, and 90–95. The strong correlation between the exchange data (Fig. 2 and Fig. S4) and the chemical shift data (Fig. 3C) allows for the confident identification of five β-strands in the secondary structure of α-syn in its fibril form.
To determine the topology of the β-strands, through-space distance restraints with a distance smaller than ≈6 Å were collected from the 13C–13C PDSD experiment with a mixing time of 250 ms (Fig. 3B). Not surprisingly this approach resulted in many distance restraints between residues close in sequence (squares along the diagonal in Fig. 3D). To obtain unambiguous long-range distance restraints, which are defined as distances between residues that are >4 residues apart from each other in sequence, we again made use of the unique sequence properties of α-syn (30–110) (i.e., resonances from the aromatic side chains of Tyr-39, His-50, and Phe-94, and the resonances from Leu-38, Ser-42, Ser-87, Ile-88, and the Gly pair Gly-67–Gly-68). By using this procedure long-range distance restraints were obtained for each β-strand (indicated as black squares in Fig. 3D). These distance restraints indicate that β1 is close in space to β2, β2 is close to β3, β3 is possibly close to β4, and β4 is close to β5. With this information in mind, many more distance restraints were collected (indicated as gray squares in Fig. 3D), but they are only unambiguous considering the tertiary structural information gathered by the unambiguous distance restraints. This procedure yielded a total of ≈100 long-range distance restraints listed in Fig. 3D. Although these data must be interpreted with caution, because several residues were not assigned and many chemical shifts are overlapping, the data obtained suggest that the fold of α-syn fibrils is a β-sandwich composed of five β-strands (see below).
High-Resolution Cryoelectron Microscopy of α-Synuclein Fibrils.
To obtain more information about the morphology, the filament composition, the conformational heterogeneity of α-syn fibrils, and the protofilament size, we carried out high-resolution cryoelectron microscopy experiments, with both full-length α-syn and α-syn (30–110) fibrils. Both synuclein variants showed twisted and straight filament strands, as seen in cryo-negative stained preparations in Fig. 4 A and B. These features were confirmed by imaging of nonstained cryo-EM preparations (data not shown). This structural heterogeneity is in good agreement with the presented heterogeneity on the level of H/D exchange as well as the documented heterogeneity in the solid-state NMR spectra and the presence of twisted and straight fibrils in the solid-state NMR samples of the Baldus group (22).
Fig. 4.
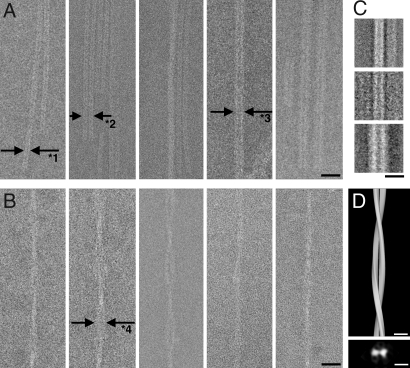
Cryo-negative stain transmission electron microscopy of straight wild-type α-syn and twisted α-syn (30–110) fibrils. In both cases, different bundling states can be observed. The straight filament is composed of a basic strand formed of two denser lines of 2.0-nm width separated by a gap of 1.5 nm (A, arrows labeled with “*1”) and tends to dimerize (A, arrows labeled with “*2”). Thicker filaments can be interpreted as tilted views of dimeric strands (A Right Center and Right), or as a different aggregation state. Straight filaments allowed the calculation of particle class averages, reproduced in C from 146, 39, and 71 particles (Top to Bottom). Twisted filaments also show variable thicknesses or density (B) and allowed a helical 3D reconstruction in D. D Lower shows the cross-section of the helical filament. (Scale bars: A and B, 20 nm; C and D, 10 nm.)
In the projection images the straight fibrils showed a width of 5.5 ± 0.5 nm (average from 62 filaments) with a region of lower density of ≈1.5 nm as a central darker line in each filament, suggesting that these filaments are composed of two protofilaments of ≈2.0-nm width each. One such straight fibrillar filament is indicated by the label “*1” in Fig. 4A Left. Two such fibrillar filaments were often found in a lateral arrangement, separated by a gap of ≈1.4 nm or more, giving the dimeric filament a total width of 13 ± 1 nm (Fig. 4A Left Center, arrows labeled “*2”). Occasionally, filamentous structures with stronger contrast and a total width of 8–12 nm were observed (e.g., Fig. 4A Right Center and Right, arrows labeled “*3”). From these filaments, 256 square particles of 128 × 128 pixels were windowed and classified and class averages were calculated (Fig. 4C), showing the tendency of the filaments to dimerize or form wider, stronger contrasted structures, which could be interpreted as side views of the tilted dimeric filament bundle, or as a different aggregation state. In line with these data, a straight fibril type of α-syn with a filament structure of ≈10 nm and a 5-nm substructure has been documented to be present in deceased PD patients (34).
Also the twisted fibril type (Fig. 4B) has been documented earlier both in vitro (35) and in deceased PD patients (34). We observed twisted filament bundles of various thicknesses, which could indicate a variable number of twisted filaments contributing to one strand. The twisted filaments showed a repeat of 120 ± 10 nm for a full 360° rotation, with most filaments showing a width of the zone of widest diameter of 11 ± 1 nm with a ≈3 nm low electron density in the center (Fig. 4B Left Center, arrows labeled “*4”). The diameter of the narrowest knot in most twisted filaments is 5 ± 1 nm. These dimensions are compatible with the twisted filament type being a coiled form of the double-filament strands shown in Fig. 4A (label “*2”). These data are in approximate agreement with atomic force microscopy data, which measured single protofilaments of α-syn in the presence of heparin with a diameter of 3.8 ± 0.5 nm, a diameter of 6.5 ± 0.6 nm for twisted filaments, and a fibril diameter of 9.8 ± 1.1 nm (35). The twisted filament shown in Fig. 4B Center was windowed into 492 particles and subjected to helical image processing. The 3D reconstruction at 3.3-nm resolution shows the two coiling strands (Fig. 4D), confirming the above dimensions.
The observations with the twisted fibrils and the straight-dimeric fibrils imply that the fibrils share a similar protofilament structure with a ≈5-nm diameter, which is composed of two subfilaments with size 2 × 4 nm. However, the two fibril types differ in their protofilament interactions.
Discussion
A Possible Fold of α-Synuclein Fibrils.
X-ray fiber diffraction of α-syn fibrils suggested the presence of a β-sheet aligned along the fibril axis (36). EPR studies and solid-state NMR studies†† indicated that these β-sheets are parallel and in-register (19, 20). The H/D exchange data together with the solid-state NMR analysis of α-syn fibrils identified five possible β-strands in the core of the fibrils with adjacent heterogeneous N- and flexible C-terminal segments. The first three β-strands have a length of ≈6–8 residues, whereas the latter two have a length of ≈9–13 residues (Fig. 2). In addition, the solid-state NMR data (Fig. 3) suggest that β1 is close in space to β2, β2 is adjacent to β3, β3 is probably close to β4, and β4 is adjacent to β5, reminiscent of a five-layered β-sandwich. Furthermore, side-chain solvent accessibility data suggest that β1 and β5 are more solvent-exposed than β2–β4 (Fig. 2 D and SI Text). Negative-stain cryo-EM set the boundaries of the filament core to ≈4 × 2 nm.
In Fig. 5, we propose a possible fold of the core of α-syn fibrils, which is consistent with all of the experimental data listed above including published solid-state NMR and EPR studies (19, 22). The proposed fold comprises a five-layered β1–loop–β2–loop–β3–loop–β4–loop–β5 β-sandwich, which, when incorporated into a protofilament of a fibril, generates five layers of parallel, in-register β-sheets. In the straight fibril type, two protofilaments align with each other to form a fibril, which can align again itself. In the twisted fibrils two protofilaments twist around each other, and such a twisted filament twists again with another one. It is evident that various protofilament packings are possible. In Fig. S5 all of the possible packings of our model are shown and discussed. However, with the assumption that the protofilaments are unidirectional oriented, and have an identical conformation, the possible packing is limited to the two shown in Fig. 5.
Fig. 5.
Proposed fold of α-syn fibrils. The proposed fold of a monomeric α-syn within a protofilament is shown in Center. The incorporation of a protofilament into the straight (Left) and twisted (Right) fibril type is indicated by a schematic drawing.
The comparison of the two packings of straight and twisted fibrils also requests different filament interactions. In the models proposed, the straight filaments composed of two protofilaments interact through strands β4 and β5, for which the largest heterogeneity both in the H/D exchange data and solid-state NMR data have been observed (22). In the twisted fibrils the filaments are ≈3 nm apart from each other suggesting that they interact through residues ≈20–30, which are not part of the core of the fibril, but are partially solvent-protected (Fig. 2 C and D), static heterogeneous, and have been suggested to be involved in protofilament interactions (36).
The Fold of α-Synuclein Fibrils in Context of Its Amino Acid Sequence.
Because the amyloid fibrils of α-syn are associated with and are a pathological hallmark of the age-related disease PD, it is tempting to ask whether the amino acid sequence of α-syn evolved to avoid the formation of amyloid fibrils. The protein sequence comparison of synucleins (including α-, β-, and γ-syn) throughout the animal kingdom shown in Fig. S6 elucidates a high level of conservation, in particular, within the first 100 aa residues, which includes the residues involved in fibril formation. A comparison between the β-sheet secondary structural elements of the fibrils and the protein sequences reveals, for instance, that the deletions of residues 74–84 in mammalian β-synuclein interferes with the formation of amyloid fibrils (37, 38), because these residues constitute β-strand β4 in the fibrils. Similarly, the deletion of residues 61–67 and 72–86 in fish β-synuclein should reduce the aggregation propensity when compared with fish α-syn, because these residues constitute β-strands β3 and β4 in the fibrils. Indeed, both mammalian and fish β-synucleins have a much lower aggregation tendency than their α-syn counterpart (37–42). The puzzling size of the deletions (i.e., 11 and 21 residues) in β-synucleins suggested to inhibit fibril formation can be explained by the proposed fold of the synuclein fibrils (Fig. 5): β3- and β4-strands are in the center of the fibril fold and their deletion is deleterious for the formation of fibrils. In contrast, deletions in either strand β1, β2, or β5 might not be sufficient to stop fibril formation, because they are at the periphery of the syn fold in the fibril.
Conclusion
Recently, an increase in the knowledge of 3D structures of amyloid fibrils has been reported. The folding of these macromolecular aggregates shows a similar repertoire of secondary structural elements, mainly in-register parallel β-sheets linked by flexible loops (23, 24, 43–48). Although many more restraints must be collected to determine the 3D structure of α-syn fibrils, the presented study supports a similar in-register parallel β-sheet conformation for α-syn fibrils, establishing further ties between the diverse members of amyloid proteins. The proposed fold of α-syn may also serve as a starting point toward the establishment of a structural understanding of α-syn aggregation and the elucidation of a structure–toxicity relationship in PD.
Materials and Methods
Electron Microscopy (EM).
For cryo and cryo-negative stain transmission electron microscopy amyloid fibrils of wild-type α-syn and α-syn (30–110) were applied to EM grids coated with holey carbon film (Quantifoil Micro Tools GmbH). The grids were blotted and quick-frozen by plunging into liquid ethane to obtain an unstained vitrified cryoelectron microscopy specimen. These samples were imaged by using a JEOL JEM-2100F transmission electron microscope. Further details are described in SI Text.
H/D Exchange.
For the quenched H/D-exchange studies, amyloid fibrils of recombinant 15N-labeled α-syn or α-syn (30–110) were used. The samples were either 2 weeks old or 6 months old. To start the H/D exchange, the fibrils were sedimented at 3,600 × g for 10 min, washed with PBS pH 7.4, and resuspended in D2O. At suitable intervals between 0 h and 30 months, aliquots were sedimented at 24,000 × g for 2 min, and frozen in liquid nitrogen to quench hydrogen exchange. For the NMR measurements, the fibrils were dissociated in perdeuterated dimethyl sulfoxide (d6-DMSO) containing 0.1% deuterated trifluoroacetic acid (d1-TFA). Further details are described in SI Text.
Solid-State NMR.
Approximately 30 mg of a 10-month-old sample of 15N,13C-labeled α-syn (30–110) was washed in H2O, centrifuged, and partially dried under reduced pressure in a desiccator. The resulting material, which had a gel-like consistency, was packed into ZrO Magic Angle Spinning (MAS) rotors and sealed with two-component epoxy adhesive to prevent further dehydration. Solid-state NMR spectra were recorded on a Varian/Chemagnetics Infinity + 500 spectrometer by using 4-mm double-resonance MAS and 2.5-mm triple-resonance MAS probe heads and on a Bruker AV8 50-MHz spectrometer by using a triple-resonance 3.2-mm MAS probe head in double-resonance mode. The 13C-13C DREAM (30) spectra together with the 13C-13C PDSD (49) and DARR experiments (50) were used for the identification of spin systems and 13C side chain assignment. The sequential assignment and the collection of distance restraints were extracted from 13C-13C PDSD (49) and DARR (50) spectra. The assignment was confirmed by the N(CO)CA (33, 51) and NCA (52) correlation spectra by using the CARA program (http://www.nmr.ch).
Further methods and materials used are described in SI Text.
Supplementary Material
Acknowledgments.
We thank D. Zellmer for the diagram drawings and Dr. J. Greenwald for careful reading. This work was supported in part by the National Institutes of Health and the Swiss National Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Heise H, et al. (2006) Investigating the 3D structural organization of full-length alpha synuclein fibrils by solid state NMR. Poster presented at International Conference on Magnetic Resonance in Biological Systems, August 20–25, Göttingen, Germany.
References
- 1.Goedert M. Alpha-synuclein and neurodegenerative diseases. Nat Rev Neurosci. 2001;2:492–501. doi: 10.1038/35081564. [DOI] [PubMed] [Google Scholar]
- 2.Cookson MR. The biochemistry of Parkinson's disease. Annu Rev Biochem. 2005;74:29–52. doi: 10.1146/annurev.biochem.74.082803.133400. [DOI] [PubMed] [Google Scholar]
- 3.Recchia A, et al. Alpha-synuclein and Parkinson's disease. FASEB J. 2004;18:617–626. doi: 10.1096/fj.03-0338rev. [DOI] [PubMed] [Google Scholar]
- 4.Lee VM, Trojanowski JQ. Mechanisms of Parkinson's disease linked to pathological alpha-synuclein: New targets for drug discovery. Neuron. 2006;52:33–38. doi: 10.1016/j.neuron.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 5.Hashimoto M, Rockenstein E, Masliah E. Transgenic models of alpha-synuclein pathology: Past, present, and future. Ann NY Acad Sci. 2003;991:171–188. [PubMed] [Google Scholar]
- 6.Feany MB, Bender WW. A Drosophila model of Parkinson's disease. Nature. 2000;404:394–398. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- 7.Ibanez P, et al. Causal relation between alpha-synuclein gene duplication and familial Parkinson's disease. Lancet. 2004;364:1169–1171. doi: 10.1016/S0140-6736(04)17104-3. [DOI] [PubMed] [Google Scholar]
- 8.Cooper AA, et al. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson's models. Science. 2006;313:324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee HJ, Lee SJ. Characterization of cytoplasmic alpha-synuclein aggregates. Fibril formation is tightly linked to the inclusion-forming process in cells. J Biol Chem. 2002;277:48976–48983. doi: 10.1074/jbc.M208192200. [DOI] [PubMed] [Google Scholar]
- 10.Dawson T, Mandir A, Lee M. Animal models of PD: Pieces of the same puzzle? Neuron. 2002;35:219–222. doi: 10.1016/s0896-6273(02)00780-8. [DOI] [PubMed] [Google Scholar]
- 11.Caughey B, Lansbury PT. Protofibrils, pores, fibrils, and neurodegeneration: Separating the responsible protein aggregates from the innocent bystanders. Annu Rev Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 12.Lotharius J, Brundin P. Pathogenesis of Parkinson's disease: Dopamine, vesicles and alpha-synuclein. Nat Rev Neurosci. 2002;3:932–942. doi: 10.1038/nrn983. [DOI] [PubMed] [Google Scholar]
- 13.Norris EH, Giasson BI, Lee VM. Alpha-synuclein: Normal function and role in neurodegenerative diseases. Curr Top Dev Biol. 2004;60:17–54. doi: 10.1016/S0070-2153(04)60002-0. [DOI] [PubMed] [Google Scholar]
- 14.Ulmer TS, Bax A, Cole NB, Nussbaum RL. Structure and dynamics of micelle-bound human alpha-synuclein. J Biol Chem. 2005;280:9595–9603. doi: 10.1074/jbc.M411805200. [DOI] [PubMed] [Google Scholar]
- 15.Eliezer D, Kutluay E, Bussell R, Jr, Browne G. Conformational properties of alpha-synuclein in its free and lipid-associated states. J Mol Biol. 2001;307:1061–1073. doi: 10.1006/jmbi.2001.4538. [DOI] [PubMed] [Google Scholar]
- 16.Dedmon MM, Lindorff-Larsen K, Christodoulou J, Vendruscolo M, Dobson CM. Mapping long-range interactions in alpha-synuclein using spin-label NMR and ensemble molecular dynamics simulations. J Am Chem Soc. 2005;127:476–477. doi: 10.1021/ja044834j. [DOI] [PubMed] [Google Scholar]
- 17.Bernado P, Bertoncini CW, Griesinger C, Zweckstetter M, Blackledge M. Defining long-range order and local disorder in native alpha-synuclein using residual dipolar couplings. J Am Chem Soc. 2005;127:17968–17969. doi: 10.1021/ja055538p. [DOI] [PubMed] [Google Scholar]
- 18.Serpell LC, Berriman J, Jakes R, Goedert M, Crowther RA. Fiber diffraction of synthetic alpha-synuclein filaments shows amyloid-like cross-beta conformation. Proc Natl Acad Sci USA. 2000;97:4897–4902. doi: 10.1073/pnas.97.9.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Der-Sarkissian A, Jao CC, Chen J, Langen R. Structural organization of alpha-synuclein fibrils studied by site-directed spin labeling. J Biol Chem. 2003;278:37530–37535. doi: 10.1074/jbc.M305266200. [DOI] [PubMed] [Google Scholar]
- 20.Chen M, Margittai M, Chen J, Langen R. Investigation of alpha-synuclein fibril structure by site-directed spin labeling. J Biol Chem. 2007;282:24970–24979. doi: 10.1074/jbc.M700368200. [DOI] [PubMed] [Google Scholar]
- 21.Miake H, Mizusawa H, Iwatsubo T, Hasegawa M. Biochemical characterization of the core structure of alpha-synuclein filaments. J Biol Chem. 2002;277:19213–19219. doi: 10.1074/jbc.M110551200. [DOI] [PubMed] [Google Scholar]
- 22.Heise H, et al. Molecular-level secondary structure, polymorphism, and dynamics of full-length α-synuclein fibrils studied by solid-state NMR. Proc Natl Acad Sci USA. 2005;102:15871–15876. doi: 10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ritter C, et al. Correlation of structural elements and infectivity of the HET-s prion. Nature. 2005;435:844–848. doi: 10.1038/nature03793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luhrs T, et al. 3D structure of Alzheimer's amyloid-beta(1–42) fibrils. Proc Natl Acad Sci USA. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoshino M, et al. Mapping the core of the beta(2)-microglobulin amyloid fibril by H/D exchange. Nat Struct Biol. 2002;9:332–336. doi: 10.1038/nsb792. [DOI] [PubMed] [Google Scholar]
- 26.Conway KA, et al. Accelerated oligomerization by Parkinson's disease linked alpha-synuclein mutants. Ann NY Acad Sci. 2000;920:42–45. doi: 10.1111/j.1749-6632.2000.tb06903.x. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Uversky VN, Fink AL. Effect of familial Parkinson's disease point mutations A30P and A53T on the structural properties, aggregation, and fibrillation of human alpha-synuclein. Biochemistry. 2001;40:11604–11613. doi: 10.1021/bi010616g. [DOI] [PubMed] [Google Scholar]
- 28.Garnier J, Gibrat JF, Robson B. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 1996;266:540–543. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- 29.Pawar AP, et al. Prediction of “aggregation-prone” and “aggregation-susceptible” regions in proteins associated with neurodegenerative diseases. J Mol Biol. 2005;350:379–392. doi: 10.1016/j.jmb.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 30.Verel R, Ernst M, Meier BH. Adiabatic dipolar recoupling in solid-state NMR: The DREAM scheme. J Magn Reson. 2001;150:81–99. doi: 10.1006/jmre.2001.2310. [DOI] [PubMed] [Google Scholar]
- 31.Detken A, et al. Methods for sequential resonance assignment in solid, uniformly 13C, 15N labelled peptides: Quantification and application to antamanide. J Biomol NMR. 2001;20:203–221. doi: 10.1023/a:1011212100630. [DOI] [PubMed] [Google Scholar]
- 32.Hediger S, Meier B, Ernst RR. Adiabatic passage Hartmann-Hahn cross polarization in NMR under magic angle sample spinning. Chem Phys Lett. 1995;240:449–456. [Google Scholar]
- 33.Verel R, Meier BH. Polarization-transfer methods in solid-state magic-angle-spinning NMR: Adiabatic CN pulse sequences. ChemPhysChem. 2004;5:851–862. doi: 10.1002/cphc.200301074. [DOI] [PubMed] [Google Scholar]
- 34.Crowther RA, Daniel SE, Goedert M. Characterisation of isolated alpha-synuclein filaments from substantia nigra of Parkinson's disease brain. Neurosci Lett. 2000;292:128–130. doi: 10.1016/s0304-3940(00)01440-3. [DOI] [PubMed] [Google Scholar]
- 35.Khurana R, et al. A general model for amyloid fibril assembly based on morphological studies using atomic force microscopy. Biophys J. 2003;85:1135–1144. doi: 10.1016/S0006-3495(03)74550-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conway KA, Harper JD, Lansbury PT., Jr Fibrils formed in vitro from alpha-synuclein and two mutant forms linked to Parkinson's disease are typical amyloid. Biochemistry. 2000;39:2552–2563. doi: 10.1021/bi991447r. [DOI] [PubMed] [Google Scholar]
- 37.Giasson BI, Murray IV, Trojanowski JQ, Lee VM. A hydrophobic stretch of 12 amino acid residues in the middle of alpha-synuclein is essential for filament assembly. J Biol Chem. 2001;276:2380–2386. doi: 10.1074/jbc.M008919200. [DOI] [PubMed] [Google Scholar]
- 38.George JM. The synucleins. Genome Biol. 2002;3:REVIEWS3002. doi: 10.1186/gb-2001-3-1-reviews3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshida H, et al. Synuclein proteins of the pufferfish Fugu rubripes: Sequences and functional characterization. Biochemistry. 2006;45:2599–2607. doi: 10.1021/bi051993m. [DOI] [PubMed] [Google Scholar]
- 40.Park JY, Lansbury PT., Jr Beta-synuclein inhibits formation of alpha-synuclein protofibrils: A possible therapeutic strategy against Parkinson's disease. Biochemistry. 2003;42:3696–3700. doi: 10.1021/bi020604a. [DOI] [PubMed] [Google Scholar]
- 41.Uversky VN, et al. Biophysical properties of the synucleins and their propensities to fibrillate: Inhibition of alpha-synuclein assembly by beta- and gamma-synucleins. J Biol Chem. 2002;277:11970–11978. doi: 10.1074/jbc.M109541200. [DOI] [PubMed] [Google Scholar]
- 42.Hashimoto M, Rockenstein E, Mante M, Mallory M, Masliah E. beta-Synuclein inhibits alpha-synuclein aggregation: A possible role as an anti-parkinsonian factor. Neuron. 2001;32:213–223. doi: 10.1016/s0896-6273(01)00462-7. [DOI] [PubMed] [Google Scholar]
- 43.Ferguson N, et al. General structural motifs of amyloid protofilaments. Proc Natl Acad Sci USA. 2006;103:16248–16253. doi: 10.1073/pnas.0607815103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iwata K, et al. 3D structure of amyloid protofilaments of beta2-microglobulin fragment probed by solid-state NMR. Proc Natl Acad Sci USA. 2006;103:18119–18124. doi: 10.1073/pnas.0607180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nelson R, et al. Structure of the cross-β spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelson R, Eisenberg D. Recent atomic models of amyloid fibril structure. Curr Opin Struct Biol. 2006;16:260–265. doi: 10.1016/j.sbi.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 47.Nelson R, Eisenberg D. Structural models of amyloid-like fibrils. Adv Protein Chem. 2006;73:235–282. doi: 10.1016/S0065-3233(06)73008-X. [DOI] [PubMed] [Google Scholar]
- 48.Petkova AT, et al. Solid state NMR reveals a pH-dependent antiparallel beta-sheet registry in fibrils formed by a beta-amyloid peptide. J Mol Biol. 2004;335:247–260. doi: 10.1016/j.jmb.2003.10.044. [DOI] [PubMed] [Google Scholar]
- 49.Suter D, Ernst RR. Spin diffusion in resolved solid-state NMR spectra. Phys Rev B Condens Matter. 1985;32:5608–5627. doi: 10.1103/physrevb.32.5608. [DOI] [PubMed] [Google Scholar]
- 50.Takegoshi K, Yano T, Takeda K, Terao T. Indirect high-resolution observation of 14N NMR in rotating solids. J Am Chem Soc. 2001;123:10786–10787. doi: 10.1021/ja016789a. [DOI] [PubMed] [Google Scholar]
- 51.Verel R, van Beek JD, Meier B. Inadequate-CR experiments in the solid state. J Magn Reson. 1999;140:300–303. doi: 10.1006/jmre.1999.1880. [DOI] [PubMed] [Google Scholar]
- 52.Baldus M, Geurts DG, Hediger S, Meier BH. Efficient 15N–13C Polarization transfer by adiabatic-passage Hartmann-Hahn cross polarization. J Magn Reson. 1996;118:140–144. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.