Abstract
Leaves of flowering plants are produced from the shoot apical meristem at regular intervals, with the time that elapses between the formation of two successive leaf primordia defining the plastochron. We have identified two genetic axes affecting plastochron length in Arabidopsis thaliana. One involves microRNA156 (miR156), which targets a series of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes. In situ hybridization studies and misexpression experiments demonstrate that miR156 is a quantitative, rather than spatial, modulator of SPL expression in leaf primordia and that SPL activity nonautonomously inhibits initiation of new leaves at the shoot apical meristem. The second axis is exemplified by a redundantly acting pair of cytochrome P450 genes, CYP78A5/KLUH and CYP78A7, which are likely orthologs of PLASTOCHRON1 of rice (Oryza sativa). Inactivation of CYP78A5, which is expressed at the periphery of the shoot apical meristem, accelerates the leaf initiation rate, whereas cyp78a5 cyp78a7 double mutants often die as embryos with supernumerary cotyledon primordia. The effects of both miR156-targeted SPL genes and CYP78A5 on organ size are correlated with changes in plastochron length, suggesting a potential compensatory mechanism that links the rate at which leaves are produced to final leaf size.
INTRODUCTION
In the aerial part of flowering plants, all organs, including leaves, stems, and flowers, originate from a small population of stem cells. These cells are embedded in the shoot apical meristem, on the flanks of which new leaf and flower primordia are produced in a regular spatial and temporal pattern. The spatial pattern is known as phyllotaxis and can be, for example, spiral, alternating (distichous), or decussate (forming 180° angles). Most species show a single phyllotactic pattern, but in some cases, a plant can switch from one pattern to another in response to developmental or external cues. The temporal pattern is characterized by the time that elapses between the formation of primordia, called the plastochron, which is the inverse of the leaf initiation rate.
Classical surgical experiments have indicated that existing leaf primordia produce a diffusible substance that inhibits the formation of new primordia, with new primordia being formed at the position with a minimal inhibitor concentration (Snow, 1929). Recent analyses have suggested an alternative phyllotactic model that is based on positive effects of the growth hormone auxin (Reinhardt et al., 2000, 2003; Vernoux et al., 2000; Jönsson et al., 2006; Smith et al., 2006). According to this model, auxin is imported by the shoot apical meristem and redistributed primarily through the epidermal cell layer (L1), with existing primordia acting as auxin sinks. Additional auxin maxima can thus only form between existing primordia, once these are sufficiently spaced apart. New primordia are then initiated at these sites.
While there has been considerable progress in knowledge about how phyllotaxis is governed and what role auxin plays in this process, a coherent understanding of the regulation of plastochron length is lacking. Inactivation of several genes, including rice (Oryza sativa) PLASTOCHRON1 (PLA1), maize (Zea mays) TERMINAL EAR1 (TE1) and its rice ortholog PLA2, and ALTERED MERISTEM PROGRAM1 (AMP1) in Arabidopsis thaliana, causes a shortening of the plastochron (Conway and Poethig, 1997; Itoh et al., 1998; Veit et al., 1998; Helliwell et al., 2001; Miyoshi et al., 2004; Kawakatsu et al., 2006). By contrast, phytochrome b (phyb) and serrate (se) mutants in Arabidopsis suffer from delayed initiation of new leaves (Reed et al., 1993; Clarke et al., 1999; Prigge and Wagner, 2001). All of these genes have pleiotropic effects, and the diverse nature of the encoded products, including a photoreceptor (PHYB), a cytochrome P450 enzyme (PLA1), and an RNA binding protein (TE1/PLA2), suggests a complex network for plastochron regulation in many species.
Several lines of evidence point to cytokinin as a regulator of leaf initiation. For example, amp1 mutants contain elevated levels of cytokinin (Chaudhury et al., 1993), and overexpressing the CYTOKININ OXIDASE1 (CKX1) gene increases plastochron length in Arabidopsis (Werner et al., 2003). Furthermore, maize plants lacking the ABPHYL1 (ABPH1) gene have, like amp1 mutants in Arabidopsis, a larger shoot apical meristem and altered phyllotaxy (Jackson and Hake, 1999; Nogué et al., 2000; Giulini et al., 2004). The ABPH1 gene product belongs to a group of ARABIDOPSIS RESPONSE REGULATOR proteins, which are important modulators of the cytokinin response (Ferreira and Kieber, 2005). Related genes in Arabidopsis function directly downstream of the meristem regulator WUSCHEL (Leibfried et al., 2005). Nevertheless, how meristem size and cytokinin affect primordium formation or how this is regulated by crosstalk between auxin and cytokinin is unclear.
Here, we describe two genetic axes affecting plastochron length in Arabidopsis. One includes microRNA156 (miR156), which targets several SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) transcription factor genes that are predominantly expressed at the shoot apex (Cardon et al., 1999; Rhoades et al., 2002; Schmid et al., 2003; Schwarz et al., 2008). By comparing endogenous expression domains of SPL genes with the results of misexpression studies, we demonstrate that miR156 limits plastochron length in developing leaf primordia. The effects of miR156/SPLs on leaf initiation rate appear to be independent of CYP78A5/KLUH, a likely ortholog of rice PLA1 (Itoh et al., 1998; Zondlo and Irish, 1999; Miyoshi et al., 2004; Anastasiou et al., 2007). Interestingly, organ size and plastochron length are coordinately affected by both miR156/SPL and CYP78A5, suggesting either that there are common regulators of organ size and plastochron length or that these two developmental parameters reciprocally influence each other.
RESULTS
Role of miR156-Targeted SPL Genes in Determining Plastochron Length
Among previously described mutants with increased plastochron lengths is se-1 (Clarke et al., 1999), which is defective in the processing of several miRNAs, including miR156 (Lobbes et al., 2006; Yang et al., 2006; Laubinger et al., 2008; see Supplemental Figures 1A and 1B online). We found that leaf initiation was similarly affected in ago1-27 plants, which carry a weak mutant allele of ARGONAUTE1 (AGO), the gene encoding the main slicer responsible for miRNA-directed transcript cleavage in Arabidopsis (Morel et al., 2002; Vaucheret et al., 2004; Baumberger and Baulcombe, 2005; see Supplemental Figure 1A online). Together, these observations indicate that an miRNA is limiting for the delay of the initiation of new leaf primordia. At least two lines of evidence suggest miR156 as the most promising candidate. First, overexpression of miR156 plants causes shortening of the plastochron (Schwab et al., 2005; see Supplemental Figure 1C online), a phenotype opposite to that of se-1 and ago1-27. Second, while Pro35S:MIR156f se-1 double mutants retained the serrated leaf phenotype typical of se-1 mutants, the plastochron defect caused by miR156 overexpression was suppressed in these plants (see Supplemental Figure 1C online).
In Arabidopsis, 11 of the 17 SPL genes are targeted by miR156 (Rhoades et al., 2002; Schwab et al., 2005; Gandikota et al., 2007). In microarray analyses, most miR156-targeted SPLs show similar patterns of RNA accumulation, with low levels in vegetative tissues and increased expression at the shoot apex upon flowering (Schmid et al., 2003; Schwab et al., 2005). However, the SPL proteins differ substantially in size. SPL3, SPL4, and SPL5 are much smaller than the other gene products, with the DNA binding domain making up most of the protein (Cardon et al., 1999). We identified several homozygous T-DNA insertion lines of miR156-targeted SPLs, including spl2, spl3, spl9, spl10, spl11, spl13, and spl15 (Figure 1A; see Supplemental Figure 2A online). Similar to spl3 mutants (Wu and Poethig, 2006), the other single-insertion mutants were phenotypically normal in terms of leaf initiation rate and flowering time (data not shown), indicating a high degree of functional redundancy.
Figure 1.
Modulation of Plastochron Length by SPL Genes.
(A) Diagram of SPL9 and SPL15 transcribed regions, with thin lines indicating introns. Arrowheads mark T-DNA insertion sites.
(B) Rosettes of 30-d-old short-day-grown plants.
(C) Appearance of visible leaves in wild-type, Pro35S:MIR156f, and spl9 spl15 plants grown in short days (n ≥ 10). Bars indicate sd.
Among the larger SPLs, two unlinked genes are particularly similar to each other, SPL9 and SPL15 (Cardon et al., 1999; Yang et al., 2008). We found that spl9 spl15 double mutants had short plastochrons (Figure 1; see Supplemental Table 1 online), in agreement with a recent independent report (Schwarz et al., 2008). Although the phenotype was milder than that of the miR156 overexpressers, in which expression of eight additional SPL genes is strongly reduced (Schwab et al., 2005), this finding indicated an important redundant effect of SPL9 and SPL15 on plastochron length.
Based on overexpression experiments, the SPL3 gene has been implicated primarily in the regulation of flowering time and phase change (Cardon et al., 1997; Wu and Poethig, 2006). Unfortunately, knockout alleles for the other two small SPL genes, SPL4 and SPL5, are not available, making it more difficult to test whether redundancy among the three small SPL genes masks their contribution to plastochron length. We therefore designed an artificial miRNA that targeted the mRNAs of both SPL4 and SPL5 (Schwab et al., 2006) (see Supplemental Figures 2B and 2C online) and introduced these into spl3 mutants. These plants did not show obvious differences in plastochron length (data not shown), suggesting that the three small SPL genes likely play minor roles, if any, in the regulation of leaf initiation rate.
Effects of miR156 Misexpression
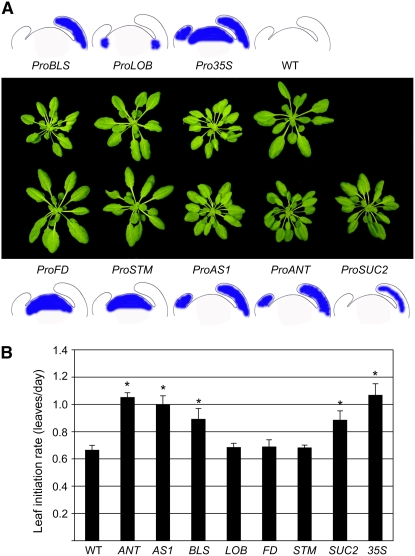
To understand how miR156 and its SPL targets modulate leaf initiation rate, we analyzed the effects of expressing miR156 in different domains of the shoot apex (Figure 2). We chose the promoter of SHOOT MERISTEMLESS (STM) for the shoot apical meristem (Kim et al., 2003), that of FLOWERING LOCUS D (FD) for the shoot apical meristem and leaf anlagen (Abe et al., 2005; Wigge et al., 2005), that of LATERAL ORGAN BOUNDARIES (LOB) for the periphery of the shoot apical meristem (Shuai et al., 2002), those of AINTEGUMENTA (ANT) and ASYMMETRIC LEAVES1 (AS1) for young leaf primordia (Schoof et al., 2000; Eshed et al., 2001), that of the SCARECROW homolog BLS for late leaf primordia (Lifschitz et al., 2006), and that of the SUCROSE-PROTON SYMPORTER2 (SUC2) for vascular tissue (Truernit and Sauer, 1995; Imlau et al., 1999). Specific activity of the promoters, as reported in the literature, was confirmed by promoter β-glucuronidase (GUS) fusions (see Supplemental Figure 3 online).
Figure 2.
Effects of MIR156f Misexpression.
(A) Thirty-day-old short-day-grown transgenic plants expressing MIR156f from different promoters, with expression domains indicated by blue color in the accompanying illustrations of shoot apices (see Supplemental Figure 3 online for expression data).
(B) Leaf initiation rates of transgenic plants, calculated from short-day-grown T1 individuals (n ≥ 10; see Supplemental Table 1 online). Bars indicate sd. Asterisks indicate a significant difference from the wild type (Student's t test with Bonferroni correction, P < 0.04).
Because one can imagine that the rate of primordium initiation is largely determined by the number of cells available in the periphery of the meristem, we suspected that miR156 targets would limit plastochron length in the meristem itself. To our surprise, transgenic miR156 expression in the meristem proper (ProFD:MIR156f and ProSTM:MIR156f) did not affect leaf initiation rate (Figure 2; see Supplemental Table 1 online). Similar to Pro35S:MIR156b plants (Schwab et al., 2005), directing miR156 expression to the periphery of the shoot apical meristem with the LOB promoter did not change plastochron length either (Figure 2; see Supplemental Table 1 online). By contrast, expression of MIR156f under control of the leaf primordium–specific ANT and AS1 promoters led to a similar plastochron phenotype to that seen in Pro35S:MIR156f plants. Thirty-day-old wild-type plants grown in short days had produced 20 leaves, whereas ProANT:MIR156f and ProAS1:MIR156f plants had produced 30 or half again as many leaves (Figure 2; see Supplemental Table 1 online). Finally, ProBLS:MIR156f and ProSUC2:MIR156f plants had an intermediate phenotype, indicating that misexpression of miR156 in older leaf primordia that already develop vasculature (or in the vasculature subtending the meristem proper) is less effective in altering plastochron length. Taken together, we conclude that miR156-targeted SPL genes contribute to nonautonomous effects of existing leaf primordia on the initiation of new leaf primordia at the shoot apical meristem.
Expression Pattern of SPL9
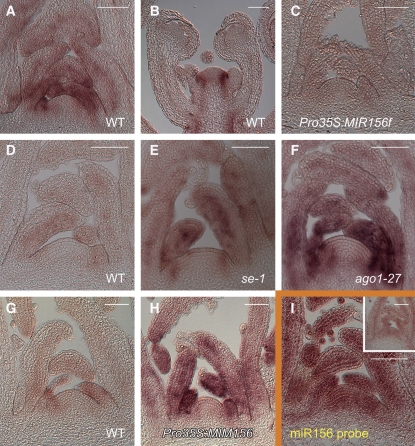
Consistent with other types of expression studies for SPL3 (Cardon et al., 1997; Wu and Poethig, 2006), microarray analyses have shown that most miR156-targeted SPL genes have a similar RNA expression pattern, with the highest levels in the shoot apex and an increase in expression during the transition to flowering (Schmid et al., 2003; Schwab et al., 2005). We used in situ hybridization to analyze in more detail the expression pattern of SPL9. During the vegetative phase, SPL9 mRNA was found in leaf anlagen and in developing leaf primordia (Figure 3A). As young leaves grew, SPL9 expression became gradually restricted to vascular tissues. Importantly, we did not detect any SPL9 transcripts in the central part of the meristem. In the inflorescence apex, the expression pattern of SPL9 was similar to that in the vegetative shoot apex. SPL9 mRNA was found only transiently in the youngest floral primordia (Figure 3B). In Pro35S:MIR156f plants, SPL9 transcripts were strongly reduced (Figure 3C), confirming that miR156 can target SPL9 mRNAs for degradation by miRNA-guided cleavage.
Figure 3.
In Situ Hybridizations Showing Expression Patterns of SPL9 and miR156.
(A) to (H) Expression of SPL9.
(A) and (B) Wild-type vegetative (A) and reproductive shoot apex (B).
(C) Pro35S:MIR156f vegetative shoot apex.
(D) to (F) Wild-type, se-1, and ago1-27 vegetative shoot apices. All three samples were stained for the same amount of time in the dark. The wild type was allowed to remain underdeveloped so that the stronger signals in the other two genotypes would not become saturated.
(G) and (H) Wild-type and Pro35S:MIM156 vegetative apices. Both samples were stained for the same amount of time in the dark. The wild type was allowed to remain underdeveloped so that the stronger signal in the Pro35S:MIM156 apex would not become saturated.
(I) Expression of miR156 in the wild type and se-1 (inset).
Shoot apices were dissected from 15-d-old plants grown in short days. Bars = 50 μm.
Since transgene-directed expression of miR156 in the meristem itself had little effect on plastochron length, one might conclude that this is the primary site of endogenous miR156 activity and that the main function of miR156 is to prevent accumulation of SPL transcripts in the meristem proper. We therefore analyzed SPL9 expression in two mutants with reduced miR156 levels, se-1 and ago1-27. Compared with the wild type, SPL9 RNA levels were substantially elevated in both mutants. However, the overall pattern of expression was similar to that seen in the wild type (Figures 3D to 3F). To exclude that this was an indirect effect of other miRNAs, we employed transgenic plants in which miR156 activity is specifically reduced by constitutive expression of a target mimic (Franco-Zorrilla et al., 2007). SPL9 mRNA expression was changed in these Pro35S:MIM156 plants in a similar manner as in se-1 and ago1-27 plants (Figures 3G and 3H).
The analysis of SPL9 expression in mutants indicates that miR156 primarily affects SPL9 expression in a quantitative, rather than spatial manner. That miR156 is not an important regulator of the domain of SPL9 expression was also supported by a direct analysis of miR156 expression by in situ hybridization, which revealed that miR156 accumulated to similar levels in both the shoot apical meristem proper and leaf primordia (Figure 3I). The specificity of the hybridization signal was confirmed using se-1 plants, in which the level of miR156 was greatly reduced (Figure 3I, inset).
In summary, the expression studies confirm that SPL9, and likely other SPL genes, either mediate or trigger nonautonomous effects of existing leaf primordia on the initiation of new leaf primordia by the meristem proper and that the primary role of miR156 is to dampen overall levels of SPL RNA (and probably also protein; Gandikota et al., 2007) in leaf primordia, rather than shaping its spatial expression pattern.
Increased Plastochron Length in Response to Elevated SPL9 and SPL10 Activity
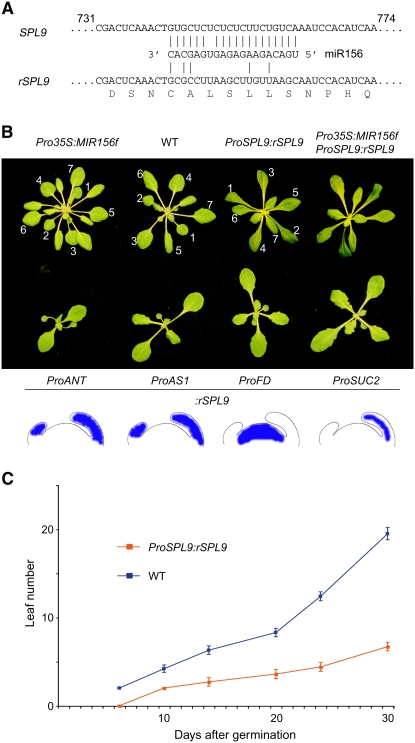
To determine whether SPL genes are not only required to prevent shortening of plastochron length but are also sufficient to increase plastochron length, we prepared forms of SPL genes that can no longer be targeted by miR156 according to known rules for effective miRNA targeting in plants (Schwab et al., 2005). We refer to these mutants as nontargeted or resistant SPLs (rSPLs) (Figure 4A).
Figure 4.
Effects of a miR156-Insensitive Form of SPL9.
(A) Diagram of the miR156 target sites of the wild-type and modified version of SPL9. Capital letters below indicate encoded amino acids.
(B) Forty-day-old (ProSPL9:rSPL9 genotypes) and 20-d-old plants (all other genotypes) grown in short days. Misexpression of rSPL9 with different promoters causes a similar delay in leaf initiation. Numbers indicate the order of leaves, with 1 referring to the oldest leaf. Note that there is no apparent disruption of normal phyllotaxis in plants with decreased or increased SPL activity.
(C) Appearance of visible leaves in short-day-grown plants (n = 10). Bars indicate sd.
Overexpression of rSPL3 caused early flowering as reported (Wu and Poethig, 2006; Gandikota et al., 2007) but caused only a minor decrease in leaf initiation rate (see Supplemental Table 1 online), suggesting limited crosstalk between miR156 targets of the SPL3/4/5 and SPL9/15 groups.
It was difficult to recover Pro35S:rSPL9 plants, suggesting that very high levels of SPL9 protein cause embryonic lethality. Plants that expressed rSPL9 under the control of the native promoter had a very strong plastochron phenotype, with the leaf initiation rate reduced to one-third of that of the wild type (Figures 4B and 4C; see Supplemental Table 1 online). We observed a similar, but weaker, phenotype in a few plants expressing the nonmutated form of SPL9, in line with the hypothesis of a quantitative interaction between miR156 and its targets. We confirmed that rSPL9 is insensitive to miR156 action by introducing the Pro35S:MIR156f transgene into these plants. The doubly transgenic plants were indistinguishable from ProSPL9:rSPL9 plants (Figure 4B).
The results presented in the previous two sections had suggested that SPL9 functions nonautonomously in existing leaf primordia to time the emergence of new primordia from the shoot apical meristem. To assess whether SPL9 can also act directly in the meristem, we misexpressed rSPL9 from different promoters. Ectopic expression from the primordium-specific ANT and AS1 promoters reduced the number of leaves to less than half of that of the wild type after 30 d in short-day conditions. A similar phenotype was observed in ProFD:rSPL9 plants (Figure 4B; see Supplemental Table 1 online), in which SPL9 was ectopically expressed in the shoot meristem. Misexpression of rSPL10 had similar, though less dramatic, effects than rSPL9 (see Supplemental Table 1 online), consistent with overlapping roles of SPL9 and SPL10, which both belong to the group of large SPL genes.
In summary, elevated activity of SPL genes can increase plastochron length, indicating an instructive role of the miR156/SPL axis in timing the emergence of new leaf primordia.
Effects of CYP78A5/KLUH and CYP78A7 on Plastochron Length
A short plastochron phenotype has also been described for rice plants with mutations in PLA1, which is likely an ortholog of CYP78A5/KLUH in Arabidopsis. Both genes are expressed at the periphery of the meristem proper and at the base of new leaf primordia (Zondlo and Irish, 1999; Miyoshi et al., 2004; Anastasiou et al., 2007). We found that CYP78A5 loss-of-function mutants had shortened plastochrons in both long and short days. In addition, the first flowers formed a few days earlier than in the wild type (Figures 5A to 5C; see Supplemental Table 2 online).
Figure 5.
cyp78a5 and cyp78a7 Mutant Phenotypes.
(A) Diagram of CYP78A5 and CYP78A7 transcribed regions, with thin lines indicating introns. Arrowheads mark T-DNA insertion sites. The cyp78a5 allele has also been described as klu-4 (Anastasiou et al., 2007).
(B) Rosettes of 30-d-old plants grown in short days.
(C) Appearance of visible leaves in short-day-grown plants (n = 10). Bars indicate sd.
(D) and (E) Wild-type embryos at bent-cotyledon stage (D) and at maturity (E).
(F) and (G) cyp78a5 cyp78a7 embryos at bent-cotyledon stage (F) and at maturity (G). The latter was dissected from the seed coat. More than 10 siliques were examined, and representative embryos are shown. Note the enlarged shoot apical meristem in (F) and multiple cotyledons in (G).
Bars = 150 μm.
CYP78A5, located on chromosome 5, has a homolog on chromosome 1, CYP78A7. Unlike cyp78a5, the cyp78a7 single mutant appeared phenotypically normal (see Supplemental Figure 4 online). To reveal functional redundancy between these two members, we crossed the cyp78a5 and cyp78a7 mutant lines. Because we did not readily identify double mutants in the F2 generation, we examined developing embryos in siliques of cyp78a5/cyp78a5 cyp78a7/+ plants. Little phenotypic change was observed among embryos in cyp78a5/cyp78a5 cyp78a7/+ siliques until the torpedo stage. From that stage on, almost a quarter of embryos did not increase much in size. The shoot apical meristem, however, continued to enlarge and became much bigger than the arrested cotyledons. The abnormal shoot apical meristem initiated supernumerary cotyledons (Figures 5D to 5G). The double mutants could sometimes survive, and we recovered a few cyp78a5 cyp78a7 seedlings from a cyp78a5/cyp78a5 cyp78a7/+ parent (five double mutants among 87 progeny tested). These plants, which produced no seeds, were small with compacted rosette leaves and an increased leaf initiation rate. The seedlings had three or four cotyledons (see Supplemental Figure 4 online), similar to amp1 mutants, which also have an embryonic phenotype reminiscent of cyp78a5 cyp78a7 double mutants (Chaudhury et al., 1993; Conway and Poethig, 1997). In summary, these results indicate that CYP78A5 and CYP78A7 play redundant roles in regulating relative growth of the shoot apical meristem and the rest of the plant.
Genetic Interaction between miR156/SPL and CYP78A5
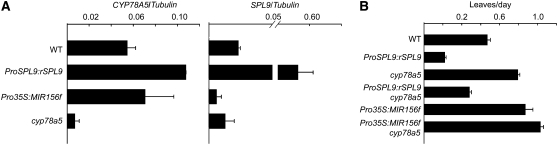
Our miR156 misexpression experiments indicated that miR156-targeted SPLs act predominantly in leaf primordia, while CYP78A5 is expressed in the periphery of the shoot apical meristem (Zondlo and Irish, 1999). Nevertheless, given the phenotypic similarity of Pro35S:MIR156f and cyp78a5 mutants, we wanted to test whether miR156/SPL and CYP78A5 operate in the same genetic pathway. Since SPL genes encode transcription factors, we first assessed RNA expression levels. There was no significant change of CYP78A5 transcript levels in either Pro35S:MIR156f or ProSPL9:rSPL9 plants (Figure 6A). Similarly, SPL9 levels were unaffected by cyp78a5 (Figure 6A).
Figure 6.
Genetic Interaction between miR156/SPL and CYP78A5.
(A) Expression of CYP78A5 and SPL9 in wild-type, mutant, and transgenic plants. Total RNA was extracted from 7-d-old long-day-grown plants and analyzed by real-time RT-PCR with three technical replicates. Expression was normalized relative to that of β-TUBULIN2. Two biological replicates were performed, both with similar results.
(B) Leaf initiation rate in wild-type, mutant, and transgenic plants, calculated from 10 short-day-grown individuals (see Supplemental Table 1 online).
Bars indicate sd.
To further clarify the relationship between CYP78A5 and the miR156/SPL pathway, we examined genetic interactions. The plastochron length of ProSPL9:rSPL9 cyp78a5 plants was intermediate between that of the parental lines, which have opposite phenotypes, suggesting parallel effects of the two genetic systems. This was supported by the Pro35S:MIR156f cyp78a5 combination, which had an even faster leaf initiation rate than either single mutant (Figure 6B).
CYP78A5, also known as KLUH, has recently been implicated in growth regulation, with small leaves and floral organs in cyp78a5 mutants because of prematurely arrested growth (Anastasiou et al., 2007), similar to what was shown for the apparent rice ortholog, PLA1 (Itoh et al., 1998). We found that Pro35S:MIR156f plants, which have a similar plastochron phenotype to cyp78a5 mutants, also exhibited smaller leaf size (Table 1; see Supplemental Figure 5 online). Similar effects on organ and cell size were observed in petals (see Supplemental Table 3 online). Thus, the effects of decreasing SPL activity on organ growth parallel the changes caused by inactivation of PLA1 or CYP78A5/KLUH (Itoh et al., 1998; Anastasiou et al., 2007). Together, these observations support a close relationship between leaf growth and plastochron length.
Table 1.
Leaf and Leaf Cell Sizes in Different Genotypes
| Wild Type | cyp78a5 | Pro35S:MIR156f | |
|---|---|---|---|
| First leaf length (mm) | 8.20 ± 0.50 | 7.38 ± 0.23* | 8.00 ± 0.21 |
| First leaf width (mm) | 7.58 ± 0.29 | 5.38 ± 0.23* | 7.45 ± 0.33 |
| Third leaf length (mm) | 17.58 ± 0.36 | 12.54 ± 0.33* | 13.13 ± 0.43* |
| Third leaf width (mm) | 9.71 ± 0.58 | 8.04 ± 0.26* | 9.21 ± 0.26 |
| Largest leaf length (mm) | 21.79 ± 0.81 | 17.42 ± 0.36* | 17.38 ± 0.38* |
| Largest leaf width (mm) | 12.79 ± 0.50 | 8.92 ± 0.42* | 10.79 ± 0.45* |
| Cell size (μm2)a | 7446 ± 319 | 7436 ± 364 | 7314 ± 367 |
For each genotype, 12 20-d-old long-day-grown plants were scored. sd is given. Asterisks indicate significant difference from the wild type (Student's t test with Bonferroni correction, P < 0.01).
Subepidermal palisade cells of the largest leaf were measured.
Correlation between Plastochron Length, Cell Division Rate, and Meristem Size
From first principles, the leaf initiation rate could be affected either by the size of the meristem or by the rate of cell division in the meristem. Indeed, increased shoot apical meristem size in clv1 mutants is paralleled by shortened plastochron length (Kwon et al., 2005). We therefore examined vegetative shoot apices by scanning electron microscopy. Meristem shape and phyllotaxis of cyp78a5, and Pro35S:MIR156f and ProSPL9:rSPL9 plants were similar to those of the wild type, even though they had either shorter or longer plastochrons than the wild type, suggesting that the plastochron changes were not caused by defects in leaf positioning (Figures 7A to 7D). The two lines with shortened plastochrons, cyp78a5 and Pro35S:MIR156f, had meristems of similar sizes to the wild type, while meristems were smaller in ProSPL9:rSPL9 plants (Figure 7, Table 2).
Figure 7.
Morphology of Shoot Apical Meristem and Dividing Cells.
(A) to (D) Scanning electron micrographs of shoot apices from 30-d-old short-day-grown plants. At least 10 individuals were examined for each genotype, and representative images are shown.
(E) to (H) Histone H4 expression in shoot apices of 15-d-old short-day-grown plants.
The ProSPL9:rSPL9 plants were grown at a later time point, but the wild-type controls were similar as for the other two genotypes. Bars = 50 μm.
Table 2.
Shoot Apical Meristem Characteristics of Different Genotypes Grown for 30 d in Short Photoperiods
| Genotype | Leaves/Day | Width (μm) | Height (μm) | H4 Positive Cellsa |
|---|---|---|---|---|
| Wild type | 0.66 ± 0.03 | 125.2 ± 0.8 | 40.3 ± 0.8 | 16.0 ± 0.81 |
| cyp78a5 | 0.98 ± 0.05* | 125.8 ± 0.9 | 39.6 ± 0.8 | 19.1 ± 1.3* |
| Pro35S:MIR156f | 1.07 ± 0.08* | 125.7 ± 1.1 | 38.8 ± 0.8 | 17.1 ± 0.89 |
| ProSPL9:rSPL9 | 0.23 ± 0.01* | 96.9 ± 2.1* | 32.2 ± 1.1* | 11.4 ± 0.78* |
n = 10 for all genotypes. sd is given. Asterisks indicate significant difference from the wild type (Student's t test with Bonferroni correction, P < 0.015).
Number of Histone H4 positive cells per longitudinal section.
Histone H4 is highly transcribed in G1-S cells and is a useful marker for cell division (Krizek, 1999; Gaudin et al., 2000). We performed in situ hybridization with different genotypes and counted the number of Histone H4 positive cells per median longitudinal section (Table 2). We found that there were substantially fewer Histone H4–expressing cells in the ProSPL9:rSPL9 shoot apical meristem (Figure 7F), suggesting that the increased plastochron length in rSPL9 plants correlates with both fewer dividing cells and a smaller overall meristem. The picture was less clear for cyp78a5 and Pro35S:MIR156f plants, which have normal-sized shoot apical meristems. There were more Histone H4–expressing cells in cyp78a5 and Pro35S:MIR156f plants (Figure 7G), but the difference from the wild type was statistically significant only in cyp78a5 (Table 2, Figures 7E and 7G). Even so, together with the normal-sized shoot apical meristem, we conclude that more rapid cell division in the shoot apical meristems of both miR156-overexpressing plants and in cyp78a5 mutants compensates for an increase in leaf initiation rate.
DISCUSSION
miR156 and its SPL targets have previously been shown to affect flowering time and phase change (Cardon et al., 1997; Schwab et al., 2005; Wu and Poethig, 2006). Here, we have described a critical role of miR156/SPL in regulating plastochron length. Although the miR156/SPL axis appears to act largely in parallel with CYP78A5/KLUH, the likely ortholog of rice PLA1, the two genetic systems have similar effects on both organ size and plastochron length, suggesting a regulatory link between these traits, which are important determinants of overall plant growth.
Regulation of SPL Expression by miR156
It had been initially proposed that plant miRNAs mostly control the spatial expression pattern of their targets (Juarez et al., 2004; Parizotto et al., 2004). However, in other cases, miRNAs seem to be important for both spatially confining target mRNAs and for dampening their levels (McConnell et al., 2001; Kidner and Martienssen, 2004; Sieber et al., 2007). In se-1 and ago1-27 mutants as well as Pro35S:MIM156 plants, all of which have reduced miR156 activity, we observed a clear increase in the levels of the SPL9 target mRNA but no obvious expansion of its expression domain. Thus, miR156 appears to be at the other end of possibilities for miRNA target interactions, with its main role being quantitative control of target mRNA levels, similar to what has been reported for miR169 and its targets in snapdragon (Antirrhinum majus) and petunia (Petunia hybrida; Cartolano et al., 2007). This conclusion is supported by a comparison of SPL9 and miR156 expression patterns, which strongly overlap (Figure 3).
Nonautonomous Effects on Plastochron Length
Classic experiments have shown that removal of young leaf primordia accelerates the rate of leaf initiation at the shoot apex (Snow, 1929). Based on this finding, it has been proposed that young leaf primordia synthesize an inhibitory, mobile factor that acts in the meristem proper. Two lines of evidence indicate that the miR156/SPL axis fits the tenets of this hypothesis. First, overexpression of miR156 in leaves or leaf primordia shortens plastochron length, whereas increased SPL activity in the leaf primordium has an opposite effect. Second, at least one of the SPL genes, SPL9, is predominantly expressed in leaf primordia.
Auxin is required for the initiation of leaf primordia, and blocking auxin transport or signaling inhibits primordium formation (Reinhardt et al., 2000, 2003; Benkova et al., 2003). Since existing leaf primordia, as auxin sinks, have an important effect on the generation of new auxin maxima, which then direct the initiation of new primordia, it is formally possible that SPLs affect plastochron length by modulating auxin accumulation or sensitivity. A similar formal possibility is that SPL proteins themselves are the mobile signal, which would be consistent with our observation that overexpressing SPLs in either leaf primordia or the meristem proper had similar effects on plastochron length. The latter observations also conform to a model in which a secondary inhibitor is directly regulated by SPLs. Some of these alternatives could be resolved by identifying direct targets of SPL transcription factors.
Several other genes have similar effects on plastochron length to those of the miR156/SPL axis. Mutations in both TE1 and in its rice ortholog PLA2 cause shortened plastochrons (Itoh et al., 1998; Veit et al., 1998; Miyoshi et al., 2004). These genes encode RNA binding proteins related to yeast MEI2, a master regulator of meiosis (Ohno and Mattaj, 1999). Both are preferentially expressed in young leaf primordia, and they could potentially act in a pathway dependent on miR156 and its SPL targets, which are conserved in monocotyledonous plants (Xie et al., 2006; Chuck et al., 2007). Consistent with this hypothesis is the finding that PLA2 appears to function independently of PLA1 in rice (Kawakatsu et al., 2006), similar to the relationship between miR156/SPLs and CYP78A5 in Arabidopsis. Unfortunately, it is unclear whether TE1/PLA2 homologs fulfill related roles in Arabidopsis, especially since the most similar genes, TEL1 and TEL2, are expressed in the shoot apical meristem only, different from TE1 and PLA2 (Anderson et al., 2004).
In contrast with the case for PLA2, a clear candidate for a PLA1 ortholog exists in Arabidopsis, CYP78A5/KLUH, which delays the initiation of new leaves, just like PLA1 (Itoh et al., 1998; Miyoshi et al., 2004; this work). The phenotype of embryos that lack activity of both CYP78A5 and its most similar Arabidopsis homolog, CYP78A7, is reminiscent of amp1 mutants (Chaudhury et al., 1993; Conway and Poethig, 1997). The cyp78a double mutant phenotype appears to be stronger than that of amp1 mutants, being often embryonic lethal and always causing several cotyledons to develop in surviving seedlings. AMP1 encodes an enzyme with unknown substrate (Helliwell et al., 2001; Vidaurre et al., 2007), and it remains to be seen whether the CYP78A5/7 and AMP1 proteins operate in the same metabolic pathway.
Plastochron Length and Shoot Apical Meristem Size
Several reports have suggested that plastochron length and shoot apical meristem size are inversely correlated. Increased meristem size and shortened plastochron characterize clv1 mutants (Kwon et al., 2005), while the opposite is true of plants that overexpress CKX genes (Werner et al., 2003). A longer plastochron is also seen in plants with mutations in multiple cytokinin receptor genes (Riefler et al., 2006). These observations are consistent with our finding that ProSPL9:rSPL9 plants have both a reduced meristem and increased plastochron length.
There is, however, no simple correlation between shoot apical meristem size and plastochron length, as neither miR156-overexpressing plants nor cyp78a5 mutants have strongly enlarged shoot apical meristems, despite a substantial shortening of plastochron length. In these plants, it appears that the increase in plastochron length is mostly due to increased cell division rates, which is also the case in several rice mutants, including pla1 and pla2 (Itoh et al., 1998; Ikeda et al., 2005; Kawakatsu et al., 2006). A direct link between cell division and plastochron length has been made with plants that overexpress Cyclin D2, which leads to faster leaf initiation correlated with higher cell division rates. At least early on, size and organization of the shoot apical meristem are normal in these plants (Cockcroft et al., 2000; Boucheron et al., 2005). However, while cell division can under certain circumstances be limiting for shortening plastochron length, slowing down cell division with a dominant-negative version of CDC2a, a cyclin-dependent kinase that interacts with Cyclin D2, did not affect leaf initiation rate, apparently because increased cell size compensated for lower cell numbers (Hemerly et al., 1995).
Plastochron Length and Phase Change
Another defect of plants with reduced SPL activity due to miR156 overexpression is a delay in phase change, with the opposite effect caused by constitutive overexpression of an miRNA-insensitive version of SPL3 (Wu and Poethig, 2006). An even stronger phenotype is seen in ProSPL9:rSPL9 plants, in which the juvenile phase is lost (this work). Clonal analyses with maize have indicated that vegetative phase change is not conferred by the shoot apical meristem, but rather that phase identity is determined autonomously in each leaf primordium (Orkwiszewski and Poethig, 2000). Our results are consistent with this observation, as SPL9 is normally expressed in leaf primordia, and increasing SPL9 levels in leaf primordia by expression from the ANT promoter is sufficient to accelerate phase change.
A similar correlation between reduced plastochron length and delayed phase change is seen in rice pla1 and pla2 mutants (Itoh et al., 1998; Kawakatsu et al., 2006). A net outcome of this would be that phase change is more normal in these mutants when measured in absolute time. However, there does not seem to be an inevitable link between plastochron length and phase change. For example, Teopod mutants in maize suffer from delayed phase change, but leaf initiation rates are normal (Poethig, 1988). Conversely, inactivation of the TAS3 pathway causes an acceleration of phase change. However, leaf initiation rates are not altered in RNA-dependent RNA polymerase6, dicer-like4, or zippy/ago7 mutants (J.-W. Wang and D. Weigel, unpublished results), all of which are affected in TAS3-dependent phase change (Hunter et al., 2003, 2006; Xie et al., 2005; Adenot et al., 2006; Fahlgren et al., 2006; Garcia et al., 2006). As with meristem size, the relationship between phase change and plastochron length is complex.
A Compensatory Mechanism Linking Plastochron Length and Organ Size?
A final phenotype that is shared between several mutants is the reduced size of leaves and floral organs (Itoh et al., 1998; Veit et al., 1998; Kawakatsu et al., 2006; Anastasiou et al., 2007; this work). An increase in plastochron length, at least during inflorescence development, has also been suggested for the big brother mutant, which has enlarged organs as well (Disch et al., 2006). Similar to how coordinated changes in plastochron length and phase change would maintain the absolute timing of phase change, an effect of organ size on plastochron length or vice versa would lead to overall biomass changes being kept to a minimum. There is precedence for a related phenomenon during leaf growth, namely the compensation of changes in cell size by total cell number (Horiguchi et al., 2006). Alternatively, rather than organ size and plastochron length reciprocally affecting each other, one can envision that coordinated behavior of the two traits is due to a common regulator. Nevertheless, the connection between organ size and plastochron length is not static. For example, manipulating leaf size by altering activity of the STRUBBELIG-RECEPTOR FAMILY4 gene (Eyüboglu et al., 2007) does not have the same effects on plastochron length as altering miR156/SPL activity (K. Schneitz, personal communication).
In summary, we have described how and where the miR156/SPL axis affects plastochron length and organ size and found that it acts largely independently of CYP78A5/KLUH, despite similar mutant phenotypes. Apart from the nature of the leaf-derived signal that affects plastochron length, an important challenge for future studies is the question of how ubiquitous coordinated changes in plastochron length and leaf size are.
METHODS
Oligonucleotide primers used in this work are given in Supplemental Table 4 online.
Plant Material
Arabidopsis thaliana plants, ecotype Columbia (Col-0), were grown at 23°C in long days (16 h light/8 h dark) or short days (8 h light/16 h dark). Individual spl and cyp78a T-DNA insertion lines (Tissier et al., 1999; Sessions et al., 2002; Alonso et al., 2003) were obtained from the European Arabidopsis Stock Centre (http://Arabidopsis.info). The cyp78a5 allele has also been described as klu-4 (Anastasiou et al., 2007). miR156 target mimic expressing Pro35S:MIM156 plants have been described (Franco-Zorrilla et al., 2007).
Transgenic Plants
For promoter fusions, AS1 and BLS promoters were amplified by PCR with AS1:LhG4 or BLS:LhG4 plasmid DNA as templates. All other promoters and SPL genes were amplified by PCR with genomic DNA from Col-0 as template, using Turbo Hotstart Pfu DNA polymerase (Stratagene). rSPL9 was made by two rounds of mutagenic PCR using Turbo Hot-start Pfu DNA polymerase (Stratagene). An artificial miRNA construct against SPL4 and SPL5 was generated as described (Schwab et al., 2006). The binary constructs were delivered into Agrobacterium tumefaciens strain GV3101 (pMP90RK) by the freeze-thaw method (Weigel and Glazebrook, 2002). Arabidopsis plants were transformed using the flower-dip method (Clough and Bent, 1998). Transgenic seedlings were selected with 50 μg/mL kanamycin on plates or 0.1% glufosinate (BASTA) on soil. At least 50 T1 seedlings were analyzed for each construct.
GUS Staining and Histology
One-week-old seedlings were fixed in 90% acetone for 20 min on ice. GUS staining was performed as described (Blázquez et al., 1997). Stained tissue was embedded, sectioned, and mounted in Clarion Mounting Medium (Sigma-Aldrich). Developing embryos were dissected from siliques and mounted in a mixture of chloralhydrate/glycerol/water (8:1:2) and photographed using differential interference contrast optics.
Expression Analysis
Total RNA was extracted from 1-week-old seedlings or vegetative shoot apices with the Plant RNeasy Mini kit (Qiagen). One microgram of total RNA was treated with DNase I and used for cDNA synthesis with oligo(dT) primer and Superscript reverse transcriptase (Invitrogen). Quantitative RT-PCR was performed with SYBR-Green PCR Mastermix (Invitrogen), and amplification was real-time monitored on an Opticon continuous fluorescence detection system (MJR). For small RNA gel blots, a mixed DNA/locked nucleic acid (LNA; Exiqon; Wahlestedt et al., 2000) oligonucleotide probe (gtgmCtcActmCtcTgtmCa, where uppercase letters indicate LNA bases and lowercase letters DNA bases) was used.
In Situ Hybridization
SPL9 and Histone H4 cDNAs were PCR amplified and cloned into pGEM-T easy (Promega). Digoxigenin-labeled sense and antisense probes were synthesized with T7 or SP6 RNA polymerase (Roche). For the miR156 probe, LNA oligonucleotides were end labeled with the DIG oligonucleotide 3′-end labeling kit (Roche). Shoot apices from 20-d-old short-day-grown plants were dissected and fixed in formalin/acetic acid/ethanol (1:1:18). Paraffin-embedded material was sectioned to 8 μm thickness. Hybridization and detection were performed as previously described (Palatnik et al., 2003).
Scanning Electron Microscopy and Shoot Apical Meristem and Organ Size Measurement
Vegetative shoot apices of 30-d-old short-day-grown plants were dissected, fixed in methanol, washed twice with 100% ethanol, critical point dried, and mounted. After gold coating, at least 10 apices per genotype were examined on a Hitachi S800 electron microscope. For size measurements, apices were embedded in paraffin and sectioned. The width and height of the meristem were scored for each section. To measure cell size, the leaves or petals were dissected, incubated in 80% ethanol for 2 h, mounted on slides with a drop of chloralhydrate/glycerol/water (8:1:2), and photographed using differential interference contrast optics. Cell size was calculated using AxioVision software (Zeiss).
Leaf Initiation Rate Measurement
The number of visible leaves (∼1 mm in width) was recorded daily. Average leaf initiation rate was calculated by dividing total leaf number by days after germination.
Accession Numbers
Arabidopsis Genome Initiative gene identifiers are as follows: SPL2 (At5g43270), SPL3 (At2g33810), SPL4 (At1g53160), SPL5 (At3g15270), SPL9 (At2g42200), SPL10 (At1g27370), SPL11 (At1g27360), SPL13 (At5g50670), SPL15 (At3g57920), CYP78A5 (At1g13710), CYP78A7 (At5g09970), LOB (At5g63090), SUC2 (At1g22710), FD (At4g35900), BLS (At3g49950), STM (At1g62360), ANT (At4g37750), AS1 (At2g37630), β-TUBULIN-2 (At5g62690), and Histone H4 (At5g59690).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. miRNA Dependence of Plastochron Length.
Supplemental Figure 2. T-DNA Insertion Mutants and amiRNA Targeting SPL4/5.
Supplemental Figure 3. Activity of Promoter Fusions.
Supplemental Figure 4. Phenotype of the cyp78a5 cyp78a7 Double Mutant.
Supplemental Figure 5. Leaf and Cell Size in Wild-Type, Mutant, and Transgenic Plants.
Supplemental Table 1. Leaf Initiation Rates of Different Genotypes.
Supplemental Table 2. Flowering Time of cyp78a5 Mutants.
Supplemental Table 3. Petal and Petal Cell Size of Different Genotypes.
Supplemental Table 4. Oligonucleotide Primer Sequences.
Supplementary Material
Acknowledgments
We thank the European Arabidopsis Stock Centre for seeds, Yuval Eshed for the AS1 and BLS promoter constructs, Marco Todesco for Pro35S:MIM156 plants, Jürgen Berger for scanning electron microscopy, Kay Schneitz for communicating unpublished data, and members of Team MiRNA for discussion. This work was supported by an EMBO Long-Term Fellowship to J.-W.W. (ALTF 274-2006), by a DFG-SFB 446 grant, and by European Community FP6 IPs SIROCCO (Contract LSHG-CT-2006-037900) and AGRON-OMICS (Contract LSHG-CT-2006-037704).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Detlef Weigel (weigel@weigelworld.org).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., Ichinoki, H., Notaguchi, M., Goto, K., and Araki, T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309 1052–1056. [DOI] [PubMed] [Google Scholar]
- Adenot, X., Elmayan, T., Lauressergues, D., Boutet, S., Bouche, N., Gasciolli, V., and Vaucheret, H. (2006). DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 16 927–932. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Anastasiou, E., Kenz, S., Gerstung, M., MacLean, D., Timmer, J., Fleck, C., and Lenhard, M. (2007). Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Dev. Cell 13 843–856. [DOI] [PubMed] [Google Scholar]
- Anderson, G.H., Alvarez, N.D.G., Gilman, C., Jeffares, D.C., Trainor, V.C.W., Hanson, M.R., and Veit, B. (2004). Diversification of genes encoding Mei2-Like RNA binding proteins in plants. Plant Mol. Biol. 54 653–670. [DOI] [PubMed] [Google Scholar]
- Baumberger, N., and Baulcombe, D.C. (2005). Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 102 11928–11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkova, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertova, D., Jürgens, G., and Friml, J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115 591–602. [DOI] [PubMed] [Google Scholar]
- Blázquez, M.A., Soowal, L., Lee, I., and Weigel, D. (1997). LEAFY expression and flower initiation in Arabidopsis. Development 124 3835–3844. [DOI] [PubMed] [Google Scholar]
- Boucheron, E., Healy, J.H., Bajon, C., Sauvanet, A., Rembur, J., Noin, M., Sekine, M., Riou-Khamlichi, C., Murray, J.A., Van Onckelen, H., and Chriqui, D. (2005). Ectopic expression of Arabidopsis CYCD2 and CYCD3 in tobacco has distinct effects on the structural organization of the shoot apical meristem. J. Exp. Bot. 56 123–134. [DOI] [PubMed] [Google Scholar]
- Cardon, G., Hohmann, S., Klein, J., Nettesheim, K., Saedler, H., and Huijser, P. (1999). Molecular characterisation of the Arabidopsis SBP-box genes. Gene 237 91–104. [DOI] [PubMed] [Google Scholar]
- Cardon, G., Hohmann, S., Nettesheim, K., Saedler, H., and Huijser, P. (1997). Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: A novel gene involved in the floral transition. Plant J. 12 367–377. [DOI] [PubMed] [Google Scholar]
- Cartolano, M., Castillo, R., Efremova, N., Kuckenberg, M., Zethof, J., Gerats, T., Schwarz-Sommer, Z., and Vandenbussche, M. (2007). A conserved microRNA module exerts homeotic control over Petunia hybrida and Antirrhinum majus floral organ identity. Nat. Genet. 39 901–905. [DOI] [PubMed] [Google Scholar]
- Chaudhury, A.M., Letham, S., Craig, S., and Dennis, E.S. (1993). amp1 — A mutant with high cytokinin levels and altered embryonic pattern, faster vegetative growth, constitutive photomorphogenesis and precocious flowering. Plant J. 4 907–916. [Google Scholar]
- Chuck, G., Cigan, A.M., Saeteurn, K., and Hake, S. (2007). The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat. Genet. 39 544–549. [DOI] [PubMed] [Google Scholar]
- Clarke, J.H., Tack, D., Findlay, K., Van Montagu, M., and Van Lijsebettens, M. (1999). The SERRATE locus controls the formation of the early juvenile leaves and phase length in Arabidopsis. Plant J. 20 493–501. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Cockcroft, C.E., den Boer, B.G.W., Healy, J.M.S., and Murray, J.A.H. (2000). Cyclin D control of growth rate in plants. Nature 405 575–579. [DOI] [PubMed] [Google Scholar]
- Conway, L.J., and Poethig, R.S. (1997). Mutations of Arabidopsis thaliana that transform leaves into cotyledons. Proc. Natl. Acad. Sci. USA 94 10209–10214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disch, S., Anastasiou, E., Sharma, V.K., Laux, T., Fletcher, J.C., and Lenhard, M. (2006). The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Curr. Biol. 16 272–279. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S.F., Perea, J.V., and Bowman, J.L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol. 11 1251–1260. [DOI] [PubMed] [Google Scholar]
- Eyüboglu, B., Pfister, K., Haberer, G., Chevalier, D., Fuchs, A., Mayer, K.F.X., and Schneitz, K. (2007). Molecular characterisation of the STRUBBELIG-RECEPTOR FAMILY of genes encoding putative leucine-rich repeat receptor-like kinases in Arabidopsis thaliana. BMC Plant Biol. 7 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren, N., Montgomery, T.A., Howell, M.D., Allen, E., Dvorak, S.K., Alexander, A.L., and Carrington, J.C. (2006). Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 16 939–944. [DOI] [PubMed] [Google Scholar]
- Ferreira, F.J., and Kieber, J.J. (2005). Cytokinin signaling. Curr. Opin. Plant Biol. 8 518–525. [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla, J.M., Valli, A., Todesco, M., Mateos, I., Puga, M.I., Rubio-Somoza, I., Leyva, A., Weigel, D., García, J.A., and Paz-Ares, J. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39 1033–1037. [DOI] [PubMed] [Google Scholar]
- Gandikota, M., Birkenbihl, R.P., Hohmann, S., Cardon, G.H., Saedler, H., and Huijser, P. (2007). The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 49 683–693. [DOI] [PubMed] [Google Scholar]
- Garcia, D., Collier, S.A., Byrne, M.E., and Martienssen, R.A. (2006). Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr. Biol. 16 933–938. [DOI] [PubMed] [Google Scholar]
- Gaudin, V., Lunness, P.A., Fobert, P.R., Towers, M., Riou-Khamlichi, C., Murray, J.A., Coen, E., and Doonan, J.H. (2000). The expression of D-cyclin genes defines distinct developmental zones in snapdragon apical meristems and is locally regulated by the CYCLOIDEA gene. Plant Physiol. 122 1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulini, A., Wang, J., and Jackson, D. (2004). Control of phyllotaxy by the cytokinin-inducible response regulator homologue ABPHYL1. Nature 430 1031–1034. [DOI] [PubMed] [Google Scholar]
- Helliwell, C.A., Chin-Atkins, A.N., Wilson, I.W., Chapple, R., Dennis, E.S., and Chaudhury, A. (2001). The Arabidopsis AMP1 gene encodes a putative glutamate carboxypeptidase. Plant Cell 13 2115–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly, A., Engler, J., Bergounioux, C., Van Montagu, M., Engler, G., Inzé, D., and Ferreira, P. (1995). Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO J. 14 3925–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi, G., Ferjani, A., Fujikura, U., and Tsukaya, H. (2006). Coordination of cell proliferation and cell expansion in the control of leaf size in Arabidopsis thaliana. J. Plant Res. 119 37–42. [DOI] [PubMed] [Google Scholar]
- Hunter, C., Sun, H., and Poethig, R.S. (2003). The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr. Biol. 13 1734–1739. [DOI] [PubMed] [Google Scholar]
- Hunter, C., Willmann, M.R., Wu, G., Yoshikawa, M., de la Luz Gutiérrez-Nava, M., and Poethig, S.R. (2006). Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133 2973–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda, K., Nagasawa, N., and Nagato, Y. (2005). ABERRANT PANICLE ORGANIZATION 1 temporally regulates meristem identity in rice. Dev. Biol. 282 349–360. [DOI] [PubMed] [Google Scholar]
- Imlau, A., Truernit, E., and Sauer, N. (1999). Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell 11 309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, J.I., Hasegawa, A., Kitano, H., and Nagato, Y. (1998). A recessive heterochronic mutation, plastochron1, shortens the plastochron and elongates the vegetative phase in rice. Plant Cell 10 1511–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, D., and Hake, S. (1999). Control of phyllotaxy in maize by the abphyl1 gene. Development 126 315–323. [DOI] [PubMed] [Google Scholar]
- Jönsson, H., Heisler, M.G., Shapiro, B.E., Meyerowitz, E.M., and Mjolsness, E. (2006). An auxin-driven polarized transport model for phyllotaxis. Proc. Natl. Acad. Sci. USA 103 1633–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez, M.T., Kui, J.S., Thomas, J., Heller, B.A., and Timmermans, M.C. (2004). microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428 84–88. [DOI] [PubMed] [Google Scholar]
- Kawakatsu, T., Itoh, J.-I., Miyoshi, K., Kurata, N., Alvarez, N., Veit, B., and Nagato, Y. (2006). PLASTOCHRON2 regulates leaf initiation and maturation in rice. Plant Cell 18 612–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidner, C.A., and Martienssen, R.A. (2004). Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428 81–84. [DOI] [PubMed] [Google Scholar]
- Kim, J.Y., Yuan, Z., and Jackson, D. (2003). Developmental regulation and significance of KNOX protein trafficking in Arabidopsis. Development 130 4351–4362. [DOI] [PubMed] [Google Scholar]
- Krizek, B.A. (1999). Ectopic expression of AINTEGUMENTA in Arabidopsis plants results in increased growth of floral organs. Dev. Genet. 25 224–236. [DOI] [PubMed] [Google Scholar]
- Kwon, C.S., Chen, C., and Wagner, D. (2005). WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev. 19 992–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger, S., Sachsenberg, T., Zeller, G., Busch, W., Lohmann, J.U., Rätsch, G., and Weigel, D. (2008). Dual roles of the nuclear cap binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- Leibfried, A., To, J.P., Busch, W., Stehling, S., Kehle, A., Demar, M., Kieber, J.J., and Lohmann, J.U. (2005). WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438 1172–1175. [DOI] [PubMed] [Google Scholar]
- Lifschitz, E., Eviatar, T., Rozman, A., Shalit, A., Goldshmidt, A., Amsellem, Z., Alvarez, J.P., and Eshed, Y. (2006). The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. USA 103 6398–6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobbes, D., Rallapalli, G., Schmidt, D.D., Martin, C., and Clarke, J. (2006). SERRATE: A new player on the plant microRNA scene. EMBO Rep. 7 1052–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell, J.R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411 709–713. [DOI] [PubMed] [Google Scholar]
- Miyoshi, K., Ahn, B.-O., Kawakatsu, T., Ito, Y., Itoh, J.-I., Nagato, Y., and Kurata, N. (2004). PLASTOCHRON1, a timekeeper of leaf initiation in rice, encodes cytochrome P450. Proc. Natl. Acad. Sci. USA 101 875–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel, J.-B., Godon, C., Mourrain, P., Beclin, C., Boutet, S., Feuerbach, F., Proux, F., and Vaucheret, H. (2002). Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogué, F., Grandjean, O., Craig, S., Dennis, E., and Chaudhury, A. (2000). Higher levels of cell proliferation rate and cyclin CycD3 expression in the Arabidopsis amp1 mutant. Plant Growth Regul. 32 275–283. [Google Scholar]
- Ohno, M., and Mattaj, I.W. (1999). Meiosis: MeiRNA hits the spot. Curr. Biol. 9 R66–R69. [DOI] [PubMed] [Google Scholar]
- Orkwiszewski, J.A.J., and Poethig, R.S. (2000). Phase identity of the maize leaf is determined after leaf initiation. Proc. Natl. Acad. Sci. USA 97 10631–10636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik, J.F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J.C., and Weigel, D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425 257–263. [DOI] [PubMed] [Google Scholar]
- Parizotto, E.A., Dunoyer, P., Rahm, N., Himber, C., and Voinnet, O. (2004). In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 18 2237–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig, R.S. (1988). Heterochronic mutations affecting shoot development in maize. Genetics 119 959–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge, M.J., and Wagner, D.R. (2001). The Arabidopsis SERRATE gene encodes a zinc-finger protein required for normal shoot development. Plant Cell 13 1263–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, J.W., Nagpal, P., Poole, D.S., Furuya, M., and Chory, J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt, D., Mandel, T., and Kuhlemeier, C. (2000). Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt, D., Pesce, E.-R., Stieger, P., Mandel, T., Baltensperger, K., Bennett, M., Traas, J., Friml, J., and Kuhlemeier, C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426 255–260. [DOI] [PubMed] [Google Scholar]
- Rhoades, M.W., Reinhart, B.J., Lim, L.P., Burge, C.B., Bartel, B., and Bartel, D.P. (2002). Prediction of plant microRNA targets. Cell 110 513–520. [DOI] [PubMed] [Google Scholar]
- Riefler, M., Novak, O., Strnad, M., and Schmülling, T. (2006). Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18 40–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, M., Uhlenhaut, N.H., Godard, F., Demar, M., Bressan, R., Weigel, D., and Lohmann, J.U. (2003). Dissection of floral induction pathways using global expression analysis. Development 130 6001–6012. [DOI] [PubMed] [Google Scholar]
- Schoof, H., Lenhard, M., Haecker, A., Mayer, K.F.X., Jürgens, G., and Laux, T. (2000). The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100 635–644. [DOI] [PubMed] [Google Scholar]
- Schwab, R., Ossowski, S., Riester, M., Warthmann, N., and Weigel, D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab, R., Palatnik, J.F., Riester, M., Schommer, C., Schmid, M., and Weigel, D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8 517–527. [DOI] [PubMed] [Google Scholar]
- Schwarz, S., Grande, A.V., Bujdoso, N., Saedler, H., and Huijser, P. (2008). The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 67 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions, A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai, B., Reynaga-Pena, C.G., and Springer, P.S. (2002). The LATERAL ORGAN BOUNDARIES gene defines a novel, plant-specific gene family. Plant Physiol. 129 747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber, P., Wellmer, F., Gheyselinck, J., Riechmann, J.L., and Meyerowitz, E.M. (2007). Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development 134 1051–1060. [DOI] [PubMed] [Google Scholar]
- Smith, R.S., Guyomarc'h, S., Mandel, T., Reinhardt, D., Kuhlemeier, C., and Prusinkiewicz, P. (2006). A plausible model of phyllotaxis. Proc. Natl. Acad. Sci. USA 103 1301–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow, R. (1929). The young leaf as the inhibiting organ. New Phytol. 28 345–348. [Google Scholar]
- Tissier, A.F., Marillonnet, S., Klimyuk, V., Patel, K., Torres, M.A., Murphy, G., and Jones, J.D.G. (1999). Multiple independent defective suppressor-mutator transposon Insertions in Arabidopsis: A tool for functional genomics. Plant Cell 11 1841–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit, E., and Sauer, N. (1995). The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of β-glucuronidase to the phloem: Evidence for phloem loading and unloading by SUC2. Planta 196 564–570. [DOI] [PubMed] [Google Scholar]
- Vaucheret, H., Vazquez, F., Crété, P., and Bartel, D.P. (2004). The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18 1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit, B., Briggs, S.P., Schmidt, R.J., Yanofsky, M.F., and Hake, S. (1998). Regulation of leaf initiation by the terminal ear 1 gene of maize. Nature 393 166–168. [DOI] [PubMed] [Google Scholar]
- Vernoux, T., Kronenberger, J., Grandjean, O., Laufs, P., and Traas, J. (2000). PIN-FORMED 1 regulates cell fate at the periphery of the shoot apical meristem. Development 127 5157–5165. [DOI] [PubMed] [Google Scholar]
- Vidaurre, D.P., Ploense, S., Krogan, N.T., and Berleth, T. (2007). AMP1 and MP antagonistically regulate embryo and meristem development in Arabidopsis. Development 134 2561–2567. [DOI] [PubMed] [Google Scholar]
- Wahlestedt, C., et al. (2000). Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc. Natl. Acad. Sci. USA 97 5633–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, D., and Glazebrook, J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Werner, T., Motyka, V., Laucou, V., Smets, R., Van Onckelen, H., and Schmülling, T. (2003). Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15 2532–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge, P.A., Kim, M.C., Jaeger, K.E., Busch, W., Schmid, M., Lohmann, J.U., and Weigel, D. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309 1056–1059. [DOI] [PubMed] [Google Scholar]
- Wu, G., and Poethig, R.S. (2006). Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133 3539–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, K., Wu, C., and Xiong, L. (2006). Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 142 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Allen, E., Wilken, A., and Carrington, J.C. (2005). DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102 12984–12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L., Liu, Z., Lu, F., Dong, A., and Huang, H. (2006). SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 47 841–850. [DOI] [PubMed] [Google Scholar]
- Yang, Z., Wang, X., Gu, S., Hu, Z., Xu, H., and Xu, C. (2008). Comparative study of SBP-box gene family in Arabidopsis and rice. Gene 407 1–11. [DOI] [PubMed] [Google Scholar]
- Zondlo, S.C., and Irish, V.F. (1999). CYP78A5 encodes a cytochrome P450 that marks the shoot apical meristem boundary in Arabidopsis. Plant J. 19 259–268. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.