Abstract
Numerous, varied, and widespread taxa have an internal circadian clock that allows anticipation of rhythmic changes in the environment. We have identified XAP5 CIRCADIAN TIMEKEEPER (XCT), an Arabidopsis thaliana gene important for light regulation of the circadian clock and photomorphogenesis. XCT is essential for proper clock function: xct mutants display a shortened circadian period in all conditions tested. Interestingly, XCT plays opposite roles in plant responses to light depending both on trait and wavelength. The clock in xct plants is hypersensitive to red but shows normal responses to blue light. By contrast, inhibition of hypocotyl elongation in xct is hyposensitive to red light but hypersensitive to blue light. Finally, XCT is important for ribulose-1,5-bisphosphate carboxylase/oxygenase production and plant greening in response to light. This novel combination of phenotypes suggests XCT may play a global role in coordinating growth in response to the light environment. XCT contains a XAP5 domain and is well conserved across diverse taxa, suggesting it has a common function in higher eukaryotes. Downregulation of the XCT ortholog in Caenorhabditis elegans is lethal, suggesting that studies in Arabidopsis may be instrumental to understanding the biochemical activity of XCT.
INTRODUCTION
As a consequence of the rhythmic, predictable cycles in light and temperature conferred by the Earth's rotation, organisms have evolved an internal mechanism to anticipate these changes. First reports of an endogenous oscillator in plants date back to 1729, when de Mairan observed leaf movement rhythms persisted in constant darkness (de Mairan, 1729). Since then, evidence for an internal circadian clock has been observed in nearly all eukaryotes and some prokaryotes. This suggests an evolutionary advantage in the capacity to predict environmental changes; indeed, a functional circadian clock has been shown to provide a physiological advantage when matched in periodicity to the external environment (Dodd et al., 2005; Johnson, 2005). However, individual components of the core oscillator are not related across higher taxa (Young and Kay, 2001).
In Arabidopsis thaliana, the core clock has been suggested to consist of a negative feedback loop involving the morning-phased genes CIRCADIAN CLOCK-ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) and the evening-phased gene TIMING OF CAB EXPRESSION1/PSEUDO RESPONSE REGULATOR1 (TOC1/PRR1) (Alabadí et al., 2001). CCA1 and LHY inhibit TOC1 expression by binding to the evening element motif found in its promoter and are thought to regulate many other evening-phased genes through the same mechanism (Alabadí et al., 2001; Harmer and Kay, 2005). TOC1 in turn has a positive effect on expression of CCA1 and LHY through an unknown mechanism. However, this simple model fails to explain all published data (Salomé and McClung, 2004), and the plant clock likely consists of multiple, interlocking feedback loops, similar to other systems (Emery and Reppert, 2004). Recently, genes such as GIGANTEA (GI) (Locke et al., 2005; Mizoguchi et al., 2005; Gould et al., 2006; Martin-Tryon et al., 2007), LUX ARRYTHMO/PHYTOCLOCK1 (LUX/PCL1) (Hazen et al., 2005; Onai and Ishiura, 2005), and EARLY FLOWERING4 (ELF4) (Kikis et al., 2005) have been suggested to form parallel evening loops with TOC1. Each of these components promotes expression of CCA1 and LHY, perhaps by acting together as a protein complex. Additionally, morning-phased loops comprised of PRR9, PRR7, CCA1, and LHY probably further act to ensure correct clock period and resetting to light (Farré et al., 2005; Locke et al., 2006; Zeilinger et al., 2006). Other components, such as ELF3 (McWatters et al., 2000; Covington et al., 2001; Liu et al., 2001), ZEITLUPE (ZTL) (Más et al., 2003b), and LIGHT INSENSITIVE PERIOD1 (LIP1) (Kevei et al., 2007), appear to function in entrainment of the clock to light, while the site of action of genes such as SENSITIVITY TO RED LIGHT REDUCED1 (SRR1) (Staiger et al., 2003) and TIME FOR COFFEE (TIC) (Hall et al., 2003; Ding et al., 2007), remains undefined. Inconsistencies between published data and current models suggest that additional components of the clock remain to be discovered.
Light input to the clock is mediated through multiple photoreceptors, including the F-box protein ZTL, phytochromes (PHYs) and cryptochromes (CRYs) (Somers et al., 1998a; Devlin and Kay, 2000; Kim et al., 2007). Generally speaking, PHYs perceive and transmit red and far-red light signals, while CRYs and ZTL mediate blue light responses. CRYs are conserved in both structure and function in many taxa, serving an important role in clock entrainment in plants and insects and in addition acting within the core oscillator in some animals (Cashmore, 2003). PHYs are photoreceptors that mediate light regulation of many processes, including the shade avoidance response and seed germination, as well as entrainment of the plant circadian clock to light (Millar, 2004). Interactions between PHY and CRY signals play an important role in directing photomorphogenic responses, such as the inhibition of hypocotyl elongation, through regulation of both repressors and promoters of photomorphogenesis.
To uncover components of the circadian system, we mutagenized seeds with ethyl methanesulfonate (EMS) and identified plants with altered free-running rhythms (Martin-Tryon et al., 2007). One such short period mutant was characterized and its molecular defect localized to a gene encoding a protein of unknown function. This protein contains an X-CHROMOSOME ASSOCIATED PROTEIN5 (XAP5) domain, a region with uncertain biochemical function but that is highly conserved in many eukaryotes. Accordingly, we named this protein XAP5 CIRCADIAN TIMEKEEPER (XCT). We show that wild-type XCT inhibits red light input to the circadian clock and has additional functions in photomorphogenesis. Intriguingly, these functions are dependent upon wavelength, as XCT acts to inhibit growth in red light but promote growth in blue light. XCT therefore likely functions as a light quality integrator that coordinates the activities of blue and red light signaling pathways on plant growth.
RESULTS
Multiple Circadian Outputs Are Disrupted in xct Mutants
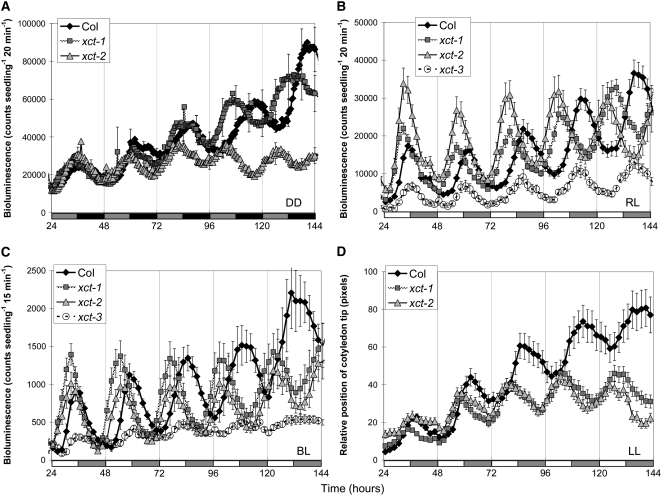
EMS-mutagenized Arabidopsis plants of the Columbia-0 (Col) ecotype transgenic for the COLD–CIRCADIAN RHYTHM–RNA BINDING2/GLYCINE-RICH RNA BINDING PROTEIN7 (CCR2/GRP7) promoter fused to the luciferase (LUC) reporter protein (CCR2pro:LUC) (Strayer et al., 2000) were entrained in light/dark (LD) cycles and then screened in constant darkness for alterations in circadian regulation of LUC activity. One mutant, later named xct-1, retains robust circadian rhythms but with a period ∼1.5 h shorter than the parental control when assayed in constant darkness (Figure 1A, Table 1). As some clock mutants display differential defects under specific conditions, we examined xct-1 in a variety of light conditions. A similar short-period phenotype for CCR2pro:LUC activity was also observed when plants were assayed in different colors of constant light (Figures 1B and 1C, Table 1). Furthermore, we examined the expression of 10 clock-associated genes using quantitative RT-PCR (qRT-PCR) and found all showed short-period rhythmic expression in xct-1 (see Supplemental Figure 1 online). We next determined whether this mutation affected a whole-plant output by monitoring leaf movement rhythms in constant white light. Similar to the transcriptional outputs, leaf movement rhythms in xct mutants were shortened ∼1.5 h relative to the wild type (Figure 1D, Table 1). Thus, multiple circadian outputs are altered in xct-1 under a variety of light conditions, suggesting XCT functions either within the core oscillator or in a clock input pathway.
Figure 1.
xct Mutants Display Short Period Rhythms for Multiple Circadian Clock Outputs.
After entrainment to 12:12 LD cycles for 6 d, CCR2pro:LUC displays rhythmic short-period expression in xct mutants held in constant darkness (A), constant red light (B), and constant blue light (C). Similarly, xct mutants demonstrate short period leaf movement rhythms in constant white light (D). Each data point is the average of 8 to 36 seedlings for CCR2pro:LUC and 42 to 80 cotyledons for leaf movement analysis, and error bars represent se. Error bars are presented only on every 12th data point in (A) for clarity. Shaded and closed or open and shaded bars on the x axis represent subjective day and subjective night, respectively.
Table 1.
Multiple Clock Outputs Are Disrupted in xct Mutants
| Period Estimate (Variance Weighted Mean ± se [n]) in Hours
|
|||||
|---|---|---|---|---|---|
| Genotype | DD | cRL (40 μE) | cBL (20 μE) | cR+BL (55 μE) | LMRA (20 μE) |
| Col | 25.64 ± 0.36 (6) | 25.60 ± 0.07 (35) | 24.37 ± 0.09 (17) | 24.81 ± 0.08 (16) | 24.97 ± 0.17 (31) |
| xct-1 | 24.17 ± 0.24 (8) | 23.01 ± 0.09 (33) | 22.61 ± 0.07 (18) | 22.93 ± 0.08 (16) | 23.59 ± 0.07 (49) |
| xct-2 | 22.71 ± 0.23 (8) | 22.47 ± 0.09 (27) | 22.52 ± 0.06 (13) | 22.27 ± 0.18 (16) | 22.23 ± 0.2 (49) |
| xct-3 | 25.28 ± 0.44 (7) | 25.29 ± 0.08 (33) | 24.02 ± 0.13 (16) | 24.49 ± 0.12 (16) | ND |
| xct-1 XCTpro:XCT | ND | 25.28 ± 0.03 (73) | ND | ND | ND |
| xct-2 XCTpro:XCT | ND | 25.16 ± 0.11 (18) | ND | ND | ND |
| xct-2 35Spro:XCT | ND | 24.92 ± 0.24 (15) | ND | ND | ND |
Intensities of constant light conditions are presented in μE (μmol m−2 s−1). CCR2pro:LUC activity was determined in constant darkness (DD), constant red light (cRL), constant blue light (cBL), or the combination of constant red and blue light (cR+BL). Cotyledon movement rhythms (LMRA) were monitored in constant white light. xct mutants transformed with a genomic copy of XCT under the control of the native XCT promoter (XCTpro:XCT) or the globally expressed cauliflower mosaic virus 35S promoter (35Spro:XCT) were assayed in the T1 generation, with similar rescued periods observed in resulting T2 and T3 families. ND represents data not determined; se is the standard error for (n) individuals.
XCT Is Well Conserved in Many Eukaryotes and Is Nuclear Localized
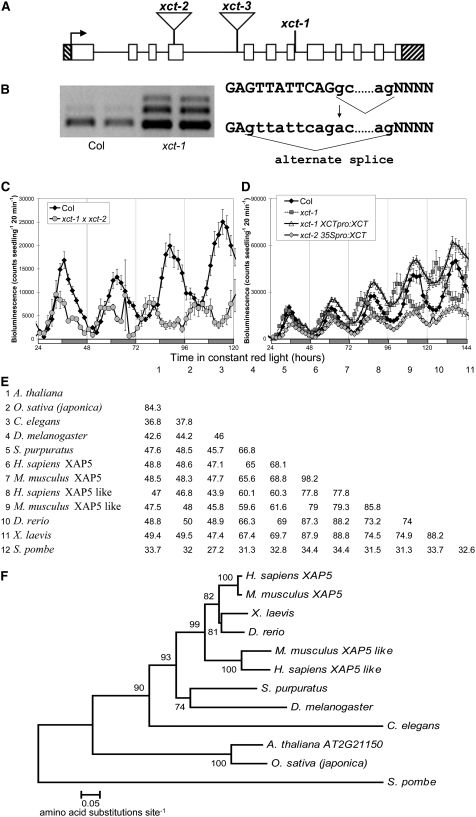
We initially localized xct-1 to the upper arm of chromosome 2 through bulked segregant analysis (Michelmore et al., 1991; Lukowitz et al., 2000) and then further mapped it to a 92-kb region containing 33 genes. Sequencing of candidate genes in this region revealed a G-to-A transition within At2g21150 (Figures 2A and 2B). This mutation occurred at the 5′ splice site of intron 7, changing the sequence from GC to AC, a nonconsensus splice site. Sequencing of xct-1 cDNA products indicates that the mutated splice site is not recognized by RNA splicing machinery, and a GT nine nucleotides upstream of the normal splice junction is used instead (Figure 2B), generating mRNA encoding a protein with an internal deletion of three amino acids.
Figure 2.
Identification of the XCT Locus.
(A) Gene structure of XCT: white blocks indicate exons and black lines introns; hatched bars indicate the 5′ and 3′ UTRs. Locations of mutations are indicated: xct-1 G1712A, xct-2 (SALK_134513) insertion after 771 nucleotides, and xct-3 (SALK_108639) insertion after 1287 nucleotides.
(B) xct-1 contains a G→A transition 1712 nucleotides after the translational start, which alters a 5′ intronic splice site and leads to missplicing of the XCT message.
(C) xct-2 fails to complement xct-1, with the F1 demonstrating a short period similar to both parents.
(D) xct-1 was rescued by transformation with XCT under the control of its own promoter (xct-1 XCTpro:XCT), and overexpression of XCT by the 35S promoter also produced a near normal free-running period in xct-2 (xct-2 35Spro:XCT, P > 0.1). For (C) and (D), plants were entrained and assayed as in Figure 1B, and error bars represent se.
(E) Percentage of identical amino acids between species across the entire XAP5 protein is presented in a similarity matrix.
(F) Neighbor-joining phylogenetic tree showing the relationships between XAP5 proteins from various species. Bootstrap values (10,000 replicates) are presented near each branch, and branch length represents the number of amino acid changes per site in accordance with the scale bar. Species of XAP5 proteins in (E) and (F) are Arabidopsis thaliana, Oryza sativa, Mus musculus, Strongylocentrotus purpuratus, Homo sapiens, Xenopus laevis, Danio rerio, Drosophila melanogaster, Caenorhabditis elegans, and Schizosaccharomyces pombe.
We also noted multiple XCT splice isoforms in xct-1 and Col (Figure 2B; see Supplemental Figure 2 online). A minor splice variant is observed in Col; sequencing of this band revealed that intron 5 had not been removed, and translation of this message would result in a truncated protein. In xct-1, this alternate form is not observed, but two other variants are generated. Sequencing reveals retention of intron 7 in one and both introns 6 and 7 in the other. Both of these forms contain premature stop codons and if translated would generate truncated proteins. The xct-1 period phenotype is recessive, suggesting that the alternatively spliced forms are nonfunctional. It is likely that the xct-1 phenotype is caused by the in-frame deletion in the predominant message rather than generation or loss of the minor alternatively spliced forms.
To confirm that the xct-1 phenotypes were due to the mutation in At2g21150, we obtained two lines from the Salk collection (Alonso et al., 2003), each with a T-DNA insertion within this gene, which we confirmed by sequencing. SALK_134513 (xct-2) contains an insertion in exon 4, and SALK_108639 (xct-3) contains an insertion in intron 4 (Figure 2A). No normal message is observed in xct-2, suggesting that it is a null allele. By contrast, xct-3 expresses a small amount (<10% of the wild type) of correctly spliced message (see Supplemental Figure 2 online). xct-2 exhibits a short period phenotype in constant light similar to that of xct-1 (Figure 1, Table 1). However, in constant darkness, the short period phenotype of xct-2 is more severe than xct-1 (Figure 1A, Table 1), indicating that xct-1 may be a partial loss-of-function allele. Although a change in amplitude of CCR2pro:LUC activity is observed in xct-3 (Figures 1B and 1C), levels of endogenous clock gene expression are not altered in xct-3 in constant light (see Supplemental Figure 1 online), and xct-3 shows no change in period for any output tested (Figure 1, Table 1; see Supplemental Figure 1 online). The reduction in luciferase expression in xct-3 is likely due to silencing of the LUC transgene, as we have observed previously in other Salk insertion lines (Martin-Tryon et al., 2007), and is consistent with the loss of kanamycin resistance (presumably due to silencing of the knockout T-DNA) observed in this line.
To verify that the xct-1 and xct-2 phenotypes are caused by alterations in At2g21150, we crossed these recessive mutants to each other. The resulting F1 has a short period phenotype similar to both homozygous parents, demonstrating a failure of complementation (Figure 2C). Furthermore, the short period phenotypes of both xct-1 and xct-2 can be rescued by transformation with a genomic copy of XCT (Table 1, Figure 2D). Notably, xct-2 mutants transformed with XCT driven by a strong viral promoter (xct-2 35Spro:XCT) also show a near wild-type period (P > 0.1, Student's t test) (Figure 2D, Table 1). In combination with the wild-type period seen in xct-3 plants, this indicates that the plant clock is relatively insensitive to levels of XCT expression but that loss of XCT function causes a short-period phenotype.
XCT encodes a novel protein of 337 amino acids that is highly conserved with proteins found in the genomes of many eukaryotes, including rice (Oryza sativa), human, zebra fish, sea urchin, and fission yeast (Schizosaccharomyces pombe). Alignment of these protein sequences reveals conservation of amino acids across the entire protein but most notably in the C-terminal 225 amino acids, which comprises a XAP5 domain. The amino acids surrounding the xct-1 deletion lie within the XAP5 domain and are widely conserved (see Supplemental Figure 3 online). Comparison of amino acid sequences between the entire Arabidopsis protein and its orthologs reveals 34 to 84% amino acid identity (Figure 2E), a remarkable degree of conservation between these distantly related taxa (Figure 2F). Amino acid identity when just the XAP5 domains are considered is even more impressive: these regions are 94% identical between Arabidopsis and rice, 64% identical between Arabidopsis and human, 47% identical between Arabidopsis and Caenorhabditis elegans, and 39% identical between Arabidopsis and S. pombe. In Arabidopsis, similar to the genomes of rice and Medicago truncatula, XCT is the only XAP5 domain-containing protein. To reflect this conserved domain and the role of At2g21150 in the clock, we have named it XAP5 CIRCADIAN TIMEKEEPER (XCT).
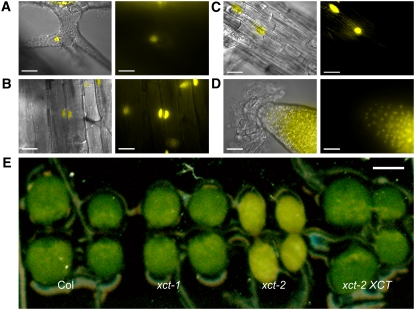
The S. pombe XCT homolog was found to be nuclear localized in a genome-wide study (Matsuyama et al., 2006), and XCT contains a predicted nuclear localization sequence (KKRK) at amino acids 112 to 115 (Horton et al., 2007). To determine the intracellular localization of XCT, we generated plants expressing XCT fused to yellow fluorescent protein (YFP) and the hemagglutinin epitope tag (HA) under control of the native XCT promoter, which rescues the xct phenotype (Figures 2D and 3E, Table 1). We found that XCT protein is restricted to and present in the nuclei of all cell types examined, including trichome, hypocotyl, and root cells of light-grown seedlings (Figures 3A to 3D). XCT-YFP-HA is also found in the nuclei of etiolated seedlings, juvenile leaf mesophyll, and adult leaves, stems, and flowers (see Supplemental Figure 4 online; data not shown), indicating that light is not required for XCT expression and that XCT protein is present in the nucleus throughout development. XCT translationally fused to YFP at the N terminus was also present in nuclei (see Supplemental Figure 4 online), indicating that this localization pattern is not an artifact of the fusion protein.
Figure 3.
XCT Is a Widely Expressed, Nuclear-Localized Protein.
(A) to (D) XCT-YFP-HA expressed under the control of the native XCT promoter is found in the nuclei of cells throughout the plant, including trichome (A), hypocotyl (B), root (C), and root meristem (D). Right panels show YFP signal alone (imaged by fluorescence microscopy), and an overlay of the YFP signal with bright field image is presented in the left panels. Bars = 25 μm.
(E) Six-day-old xct-2 seedlings display a delayed greening of cotyledons and young tissues, which is rescued by transformation with genomic XCT under the control of the native promoter. Bar = 1 mm.
Regulation of XCT Protein Levels Is Complex
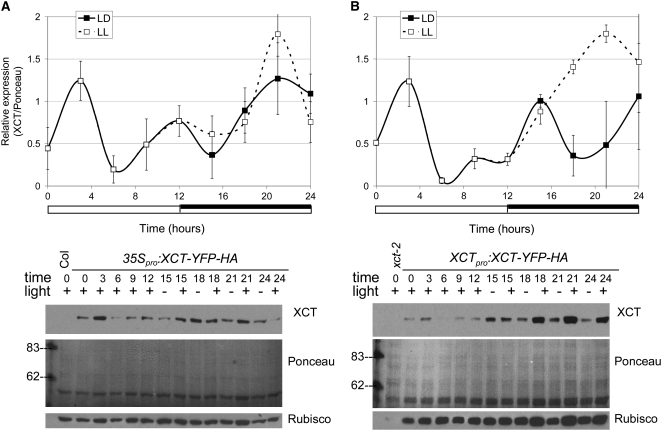
Many, but not all, clock-associated genes show rhythmic regulation of mRNA levels. To examine whether XCT expression is under clock control, we entrained wild-type seedlings in LD cycles and then released them to constant light (LL) and collected time-course samples. XCT mRNA levels do not show 24-h rhythms either in LL or LD (data not shown). However, using an HA antibody to detect tagged XCT expressed under the control of its own promoter or the constitutively expressed 35S promoter, we found that XCT protein levels fluctuate in 6-d-old entrained seedlings. When plants are maintained in LD, XCT protein levels are low during the middle of the day and early evening (ZT6 to ZT12) and highest shortly after dawn (ZT3), regardless of whether expression is driven by the 35S or native promoter (Figure 4). Thus, a posttranscriptional mechanism regulates XCT abundance during the day.
Figure 4.
XCT Protein Levels Undergo Complex Regulation.
Levels of XCT-YFP-HA fusion protein expressed under the control of the 35S (A) or native promoter (B) were determined with an anti-HA antibody and normalized to Ponceau staining. Plants were maintained in 12:12 LD cycles or constant light (LL). Data in the top panels are the mean ± se of three to four independent experiments; white and black bars below traces represent light and dark periods, respectively. Representative immunoblots are shown in the bottom panels. Rubisco expression was determined using a specific antibody and is shown in the bottom panels. Rubisco levels are impaired in xct-2 mutants (right panel, first lane) and rescued by expression of XCT-YFP-HA; however, overexpression of XCT does not cause an increase in Rubisco (left panel).
To distinguish between effects caused by light versus the circadian clock, we examined XCT levels in plants maintained in LL or in LD conditions. LL samples 15 to 24 were harvested during the subjective night, corresponding to times when the plants were in the dark during entrainment but were exposed to light during the collection period. There is no obvious difference in XCT protein levels during the true and subjective nights (LD and LL samples, respectively) when XCT-YFP-HA is expressed under control of the 35S promoter (Figure 4A). By contrast, more XCT protein accumulates during the subjective night than during the true night when XCT-YFP-HA is expressed under the control of the native XCT promoter (Figure 4B). No consistent difference in XCT mRNA level is observed when entrained plants are held in constant light versus diurnal cycles (data not shown), suggesting that this light-induced increase in XCT protein at night might also be due to posttranscriptional regulation.
XCT Inhibits Clock Gene Expression in Diurnal Cycles
The plant circadian clock is thought to consist of multiple interlocked transcription/translation feedback loops (Salomé and McClung, 2004). We therefore examined the effect of XCT mutations on the expression levels of many known clock-associated genes. As expected, rhythmic expression of all genes assayed in LL showed a short-period phenotype in xct-1 and xct-2 but a near wild-type period in xct-3 (see Supplemental Figure 1 online). Immediately after release to constant environmental conditions, TOC1, ELF4, PRR3, and PRR5 showed significantly higher peak expression levels in xct-1 and xct-2 compared with Col (see Supplemental Figures 1A to 1D online, P < 0.05 for Student's t test comparison of peaks), with these differences generally decreasing by the second and third days in LL. Although there was no consistent significant difference in peak expression for PRR7, PRR9, LHY, CCA1, GI, or LUX (see Supplemental Figures 1E to 1J online), some of these genes appeared to also have elevated peak expression early in this time course.
The difference in expression levels primarily seen shortly after the transition to LL prompted us to examine expression levels of these genes in short-day (8:16) LD cycles. ELF4, PRR5, PRR7, and PRR9 showed an increase in peak expression levels in short-day conditions consistent across 2 d of collection (see Supplemental Figures 5A to 5D online, P < 0.05 for Student's t test comparison of peaks). These data suggest that XCT does not strongly affect expression levels of known clock genes in constant conditions but can modulate the expression level of a subset of these genes under entrained conditions.
In the short-day LD samples, clear phase advances consistent with the short period phenotype of xct can be seen for ELF4, TOC1, LUX, and PRR3 but not for PRR5, PRR7, PRR9, or GI (see Supplemental Figure 5 online). PRR9 likely shows a normal dawn phase because it is directly light inducible (Tepperman et al., 2001), and increased expression and duration of PRR5 and PRR7 may mask a slightly earlier phase of expression of these genes. However, the lack of an early phase of GI expression in xct-1 and xct-2 (see Supplemental Figure 5F online) motivated us to examine the photoperiodic pathway in these plants. Like GI, we found no change in the phase of CONSTANS expression in xct plants grown in short days (see Supplemental Figure 6 online). Consistent with the normal phases of expression of these genes, xct mutants grown in short days do not flower early, unlike other short-period mutants, such as gi-200, toc1-2, and lhy-100 (see Supplemental Figure 7 online). xct mutants also flower with the same number of leaves as Col when grown in long days (see Supplemental Figure 7 online), indicating that XCT does not play an important role in the control of flowering time.
XCT Coordinates Plant Developmental Responses to Light
Although xct-1 and xct-2 plants assayed in constant light have very similar period phenotypes (Figure 1, Table 1), xct-2 demonstrates a light-dependent phenotype not seen in xct-1. When grown in any fluence or wavelength of light tested, xct-2 cotyledons initially appear yellow and only turn green 5 to 6 d after germination (Figure 3E). A similar phenotype is apparent as postembryonic tissues emerge; the shoot apical meristems of xct-2 plants maintain a yellow appearance throughout development, and floral organs, such as sepals, also demonstrate delayed greening (data not shown). This suggests that chloroplast development is impaired in xct mutants, so we examined levels of the abundant chloroplast protein ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco). Indeed, we found Rubisco levels are greatly decreased in light-grown xct-2 plants (Figure 4B) and that both the delayed greening and low Rubisco phenotypes are rescued by transformation with genomic XCT (Figures 3E and 4B). However, overexpression of XCT in the Col background does not lead to upregulation of Rubisco (Figure 4A) or enhanced greening of tissues (data not shown).
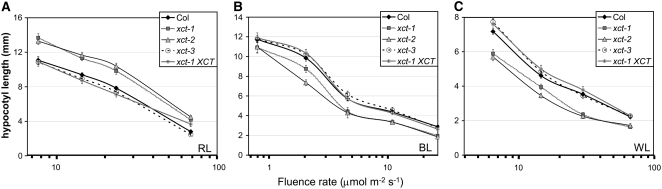
Many mutants with alterations in circadian function also display defects in photomorphogenesis (Nozue and Maloof, 2006). We therefore examined inhibition of hypocotyl elongation in xct plants in a variety of light conditions. Hypocotyl length of etiolated xct mutants was virtually identical to that of the wild type (see Supplemental Figure 8 online). Like many other clock mutants, xct plants are tall in constant red light (Figure 5A; P < 0.01 at all fluence rates). Unlike other clock mutants, however, xct mutants show the opposite phenotype of a shortened hypocotyl when grown in constant blue light (Figure 5B; P < 0.01 at all fluence rates), constant white light (Figure 5C; P < 0.01 at all fluence rates), or white LD cycles (data not shown). All of these phenotypes are rescued by transformation with a genomic copy of XCT expressed under control of the native promoter (Figure 5). Thus, XCT promotes the inhibition of hypocotyl elongation in response to red light but negatively regulates this response in blue or white light (Figure 8).
Figure 5.
XCT Functions in Light Signaling, with Distinct Phenotypes in Red Compared with Blue and White Light.
In constant red light (A), xct mutants have elongated hypocotyls across multiple fluence rates (P < 0.01, Student's t test), whereas in constant blue (B) or constant white light (C), xct mutant hypocotyls are shortened compared with Col (P < 0.01, Student's t test). In each case, the hypocotyl phenotype was rescued by transformation with genomic XCT under the control of the native promoter; xct-1 XCT, a representative T3 single insertion homozygous line, is shown. Error bars represent se; n = 15 to 20.
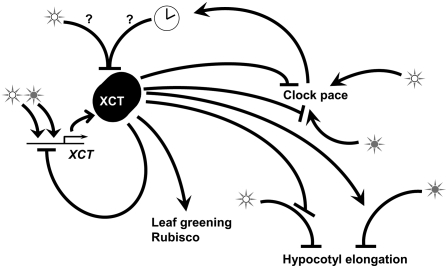
Figure 8.
XCT Has Variable Functions in Inhibition and Activation of Downstream Responses, such as Clock Pace, Hypocotyl Elongation, and Leaf Greening.
Open stars represent white light, and closed stars represent red light. XCT protein levels are reduced via a posttranscriptional mechanism 6 h after dawn, either due to direct effects of light (open star) or regulation by the circadian clock (clock symbol). In addition, XCT acts to repress induction of XCT expression in response to light, particularly red light. See text for further details.
XCT Mediates Red Light Input to the Clock
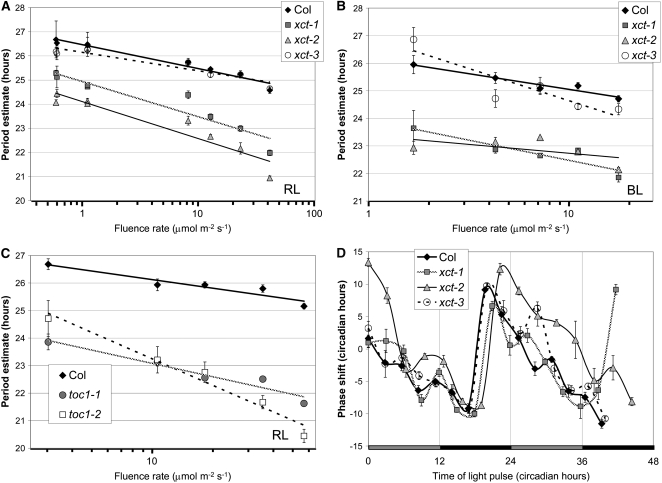
Since XCT plays a positive role in red light and a negative role in blue light regulation of photomorphogenesis, we next investigated its involvement in light input to the clock. Increased light intensity causes the internal circadian clocks of diurnal organisms, such as Arabidopsis, to run at a faster pace (Aschoff's rule; Aschoff, 1979; Millar, 2004). We therefore examined the relationship between fluence rate and free-running period in xct plants maintained in either monochromatic red or blue light. xct-1 and xct-2 mutants are hypersensitive to red light, exhibiting a steeper slope of the period response to fluence rate compared with Col (Figure 6A; P < 0.01). In contrast, xct and Col plants show a very similar response to increasing fluence rates of constant blue light (Figure 6B, P > 0.1). Therefore XCT plays a role in red light, but not blue light, input to the circadian clock.
Figure 6.
XCT Inhibits Red Light Input to the Clock.
(A) and (B) Period length of CCR2pro:LUC expression in xct mutants assayed in constant red (A) and constant blue (B) light of various fluence rates (n = 5 to 12). Mutants are more responsive than Col to increased fluence rates of red light (A) (P < 0.01 by analysis of variance [ANOVA]) but not blue light (B) (P > 0.1 by ANOVA).
(C) Period fluence rate response curve for toc1 mutants in constant red light (n = 12). The pace of the clock in toc1-1 responds to red light at the same rate as Col (P > 0.1 by ANOVA), whereas toc1-2 demonstrates an increased rate of period change from Col with increasing red light (P < 0.01 by ANOVA).
(D) Phase response curve of CCR2pro:LUC expression. Red light pulses were given at the indicated circadian time, and the magnitude of the resulting circadian phase shift in the peak of CCR2pro:LUC expression is shown on the y axis. Error bars represent the se of response. Shaded and filled bars represent subjective day and night, respectively.
The short-period and tall hypocotyl in red light phenotypes of xct mutants are also seen in the recessive TOC1 allele toc1-2 (Más et al., 2003a). We therefore investigated whether toc1-2 plants are hypersensitive to red light input to the clock. Just as for xct-1 and xct-2, we found the clock to be hypersensitive to increasing fluence rates of constant red light in toc1-2 (Figure 6C; P < 0.01). By contrast, the short-period semidominant toc1-1 allele and Col showed similar responsiveness to increasing fluence rates, as previously reported (Somers et al., 1998b; Figure 6C). This indicates that red light input to the clock is altered in toc1-2 but not toc1-1, consistent with the toc1-1 allele retaining some function (Strayer et al., 2000; Alabadí et al., 2001).
A further means of examining light signal transduction to the clock is by determining the change in phase of a dark-adapted circadian rhythm in response to a pulse of light. The clock modulates the magnitude of response, such that depending on circadian time of the light pulse, the clock will advance or delay to varying degrees. Dark-adapted xct mutants were tested over two consecutive days for a change in phase of CCR2pro:LUC activity in response to a pulse of red light. xct-1 plants show nearly wild-type responses, whereas xct-2 shows stronger phase advances than the Col control, particularly during the subjective day (Figure 6D). This confirms that the clock in xct-2 plants is hypersensitive to red light and suggests that XCT may play an important role in inhibiting light input to the clock during the subjective day.
XCT Acts as a Light Quality Sensor Directing Both Negative and Positive Transcriptional Regulation
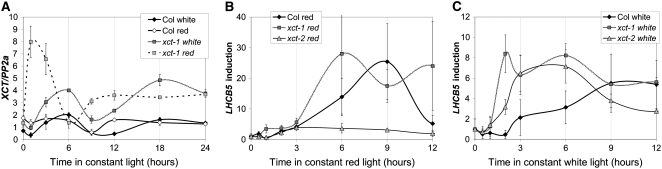
Since many light signaling and some clock genes are acutely induced by light (Tepperman et al., 2001), we examined the effect of light on XCT expression in etiolated plants. In Col, treatment of etiolated plants with white light causes a slow and modest increase in XCT levels, with peak expression ∼6 h after light onset immediately followed by a reduction; by contrast, red light has little effect on message levels (Figure 7A). White light treatment of xct-1 causes an increase in XCT message with similar kinetics to Col but greater fold induction; however, in xct-1, red light exposure leads to a rapid and pronounced increase in XCT expression (Figure 7A). XCT message levels remain elevated in xct-1 relative to Col many hours after transfer from darkness to either red or white light (Figure 7A). Thus, the xct-1 mutation causes enhanced induction of XCT mRNA in response to light, suggesting that XCT acts to inhibit its own expression (Figure 8).
Figure 7.
Etiolated xct Seedlings Show Altered Transcriptional Responses after Transfer to Light.
(A) Etiolated xct-1 seedlings show a marked upregulation of XCT message in response to red light, which is not apparent in Col or in response to white light.
(B) and (C) In red light (B), xct-2 fails to upregulate LHCB5, whereas white light (C) can overcome this and leads to elevated, early expression of LHCB5 in both xct-1 and xct-2 relative to the wild type. In (B) and (C), LHCB5 expression is normalized to time 0 to facilitate comparison of the induction kinetics. Data are the mean of three technical replicates; se is shown. Results are representative of two biological replicates.
Since the acute induction of XCT message in response to light is altered in xct-1, we next examined regulation of LIGHT HARVESTING COMPLEX B5 (LHCB5), which is induced by both red and white light (Zimmermann et al., 2004). LHCB5 induction is severely impaired in etiolated xct-2 mutants transferred to red light (Figure 7B), suggesting that XCT is necessary for induction of LHCB5. However, induction of LHCB5 is very similar in xct-1 and Col in response to red light (Figure 7B), suggesting that the three–amino acid deletion present in xct-1 does not affect its function in this process. Upon transfer to white light, both xct-1 and xct-2 show similar upregulation of LHCB5, with more rapid kinetics than seen in Col (Figure 7C); however, LHCB levels are lower in xct-2 than the wild type by the end of this time course. Therefore, XCT appears to play a positive role in the induction of LHCB5 in response to red light, but in the acute response to white light, XCT acts to inhibit and delay its expression to the appropriate phase. Sustained upregulation of LHCB5 in response to either red or white light requires XCT function that can be provided by the xct-1 but not the xct-2 allele. Considered along with the variable phenotypes of xct mutants grown in red or blue light, these differential effects on gene expression in response to red or white light indicate that XCT plays different roles in different light qualities. XCT may therefore play an important role in coordinating plant responses to the light environment.
DISCUSSION
We have cloned and characterized Arabidopsis XCT, a nuclear protein containing a XAP5 domain that negatively regulates red light input to the clock. XCT functions beyond the circadian clock as well, coordinating growth and gene expression in response to light signals and playing a role in chloroplast development. As the XCT protein is highly conserved across varied taxa, it is likely that XAP5 proteins in diverse organisms have similar biochemical functions.
XCT Is Posttranscriptionally Regulated
We did not observe rhythmic expression of XCT mRNA either when plants were grown in LL or LD (data not shown). However, XCT protein levels show a consistent increase ∼3 h after dawn followed by a decrease during the rest of the day, with a trough of expression at ZT6. Consistent with this being a posttranscriptional effect, a similar pattern was observed whether the XCT or 35S promoter was used to drive XCT expression (Figure 4). One possible explanation for this pattern would be destabilization of XCT protein, either in response to light or by a clock-regulated protein with peak activity between CT4 and CT8. Regulation of protein degradation is known to be an important regulatory mechanism in both light and clock signaling (Lechner et al., 2006).
Interestingly, light exposure during the subjective night leads to an increase in XCT protein levels in plants expressing the XCTpro:XCT-YFP-HA construct (Figure 4B) but not the 35Spro:XCT-YFP-HA construct (Figure 4A). Despite the marked increase in XCT protein in LL, no similar change in XCT message was observed under these conditions (data not shown), again suggesting that a posttranscriptional mechanism might be at work. One important difference between these constructs is that the 35S-driven XCT contains the viral 5′ untranslated region (UTR), while the XCT-driven construct contains the native 5′ UTR. Some 5′ UTR sequences are known to control translation (Kawaguchi and Bailey-Serres, 2002). Of particular interest, translation of both LHY (Kim et al., 2003) and many genes implicated in photosynthesis (Berry et al., 1988, 1990; Hansen et al., 2001; Tang et al., 2003) is light regulated. Although we cannot unequivocally conclude that translation of XCT message and degradation of XCT protein are light and clock regulated, these are interesting possibilities that merit further study. Since a low level of XCT expression is sufficient to maintain wild-type period and gene expression profiles in xct-3 (Table 1, Figure 1; see Supplemental Figure 1 online) and overexpression of XCT does not lead to a lengthened period (Table 1), it seems likely that posttranscriptional regulation of XCT is more important than control of its transcript level.
XCT Regulates Red Light Input to the Clock
We found that xct mutants are hypersensitive to both continuous red light (Figure 6A) and pulses of red light (Figure 6D). However, the free-running period of xct mutants shows normal responsiveness to increasing fluence rates of blue light (Figure 6B), indicating that XCT specifically regulates red light input to the clock. Similarly, we found the clock in toc1-2 mutants to be hypersensitive to red light but show normal blue light responsiveness (Figure 6C; data not shown). xct and toc1-2 mutants share other phenotypes as well; they both have tall hypocotyls in red light and short free-running periods. It is therefore possible that XCT and TOC1 have related functions in red light signaling and clock pathways. The localization of XCT to the nucleus (Figures 3A to 3D) places it in the same compartment of the cell as TOC1 and other clock-associated proteins, such as ELF3, (Liu et al., 2001), GI (Kim et al., 2007), LIP1 (Kevei et al., 2007), TIC (Ding et al., 2007), and SRR1 (Staiger et al., 2003).
However, unlike toc1-2 mutants, xct plants don't show reduced expression of clock genes such as CCA1 and LHY (see Supplemental Figure 1 online; Alabadí et al., 2001); instead, some clock-associated genes show transiently enhanced expression soon after the transition from entrained to free-running conditions (see Supplemental Figure 1 online). This enhancement can be seen more strongly in plants harvested in LD cycles; we observed consistent upregulation of ELF4, PRR5, PRR7, and PRR9 near the expected peak of expression in diurnal cycles in xct plants (see Supplemental Figure 5 online). Therefore, a function of XCT is to inhibit expression of these genes, either directly or indirectly, in diurnal conditions. Enhanced expression of these genes is consistent with the increased sensitivity of xct plants to red light–induced clock resetting.
Interestingly, PRR5, PRR7, and ELF4 are phased with peak expression coinciding with the trough of XCT protein levels in diurnal cycles (Figure 4), suggesting that XCT could act directly to inhibit the expression of these genes. Alternatively, since many clock genes appear to be upregulated to a certain extent in xct mutants in diurnal cycles (see Supplemental Figure 1, times 0 to 12, and Supplemental Figure 4 online), this may be a general consequence of the clock being more sensitive to light input in the absence of XCT. A similar environment-dependent phenotype was recently reported for another clock input mutant; levels of TOC1 expression in lip1 plants are similar to the wild type in constant light but reduced in LD cycles (Kevei et al., 2007).
xct mutants display a shortened circadian period for multiple clock outputs in all light conditions tested, including constant darkness (Figure 1, Table 1; see Supplemental Figure 1 online). Other components involved in light entrainment of the circadian system also have phenotypes in both constant darkness and constant light. For example, ZTL regulates light input to the clock, but ztl mutants have long-period phenotypes in all light conditions tested, including constant darkness (Somers et al. 2004). ZTL interacts with TOC1 and targets it for degradation; this interaction is light independent, and ZTL destabilizes TOC1 even in the dark (Más et al., 2003b; Kim et al., 2007). Thus, ZTL has both light-dependent and light-independent functions within the circadian system. Similarly, TOC1 also plays separable roles in clock input and core clock function, since toc1-2, but not toc1-1, is hypersensitive to red light input to the clock (Figure 6C) despite the similar short-period phenotypes of these alleles. Thus, it is possible that XCT acts near or within the core oscillator in both light-dependent and light-independent ways.
XCT Coordinates Environmental Signals to Direct Photomorphogenesis
The opposite hypocotyl phenotypes of xct mutants in red and blue light are very striking and suggest that XCT has different functions in PHY- and CRY-mediated regulation of photomorphogenesis (Figure 8). Plants with consistently opposite hypocotyl phenotypes across a broad range of red and blue light fluence rates have not been previously reported, either in circadian or classical photomorphogenic mutants. However, OBF4 BINDING PROTEIN3 has been suggested to play a positive role in PHYB-mediated inhibition of hypocotyl elongation and a negative role in CRY1-mediated cotyledon expansion (Ward et al., 2005). A number of signaling components are thought to act similarly downstream of both PHY and CRY photoreceptors (Osterlund et al., 2000; Pepper et al., 2001; Kang et al., 2005). It is possible that in xct mutants such shared signaling components are skewed away from the PHY pathway and toward the CRY pathway, leading to reduced red light and enhanced blue light responses. It seems likely in natural, white light conditions, that XCT coordinates the activity of blue and red light signaling pathways to control hypocotyl elongation (Figure 8).
Throughout development, xct-2 but not xct-1 plants display delayed greening of new tissues (Figure 3E), suggesting that xct-1 represents a partial loss-of-function allele. This delay in greening is also observed when seedlings are grown in monochromatic red or blue light (data not shown). Thus, one function for XCT is to direct the process of chlorophyll development (Figure 3E) and Rubisco expression (Figure 4) after exposure to light, potentially through transcriptional activation of key components in the process (Figure 8). Notably, xct-1 and xct-2 display similar short period phenotypes in constant light of various wavelengths (Table 1), indicating that the role of XCT in the accumulation of photosynthetic machinery is biochemically separable from its function within the clock.
It is interesting that the role of XCT in greening is wavelength independent but allele dependent (this phenotype is observed in xct-2 but not xct-1). By contrast, the xct-1 and xct-2 hypocotyl phenotypes are wavelength dependent but similar to each other (Figure 5). Acute light-mediated induction of gene expression cannot be neatly summarized in this manner. A stronger effect on the acute induction of both XCT and LHCB expression is seen in etiolated xct plants treated with red rather than with white light (Figure 7), suggesting wavelength dependence on immediate responses. However, by 12 h after transfer to either red or white light, xct-2 but not xct-1 seedlings have lower LHCB levels than the wild type (Figures 7B and 7C), suggesting a wavelength-independent but allele-dependent effect on this gene. It is likely that such longer-term, allele-specific but wavelength-independent effects on gene expression underlie the delayed greening seen in xct-2. Furthermore, these data indicate that XCT can have both positive and negative effects on gene expression depending upon the allele, the light quality, and the duration of treatment.
XCT May Act as a Link between the Clock and the Chloroplast
The circadian clock affects chloroplast development through coordination of pigment synthesis (Beator and Kloppstech, 1993), and chloroplasts may influence nuclear function through retrograde transcriptional signaling (Ruckle et al., 2007), suggesting that chloroplast status might influence the circadian clock. Anomalous greening in response to light occurs in some clock mutants; etiolated plants overexpressing TOC1 show delayed greening upon transfer to light, whereas plants overexpressing CCA1 green earlier than Col after transfer (Kato et al., 2007). However, no similar deficiency in greening has been reported in light-grown TOC1-overexpressing or CCA1-overexpressing plants, although overexpression of CCA1 does cause strong growth defects in very short days (Green et al., 2002). The loss of XCT more severely disrupts chloroplast development than overexpression of core clock components, suggesting that one function of XCT may be within a feedback loop between light signaling, the clock, and chloroplast pigment development.
XAP5 Domain Proteins in Other Organisms
The strong similarity between XCT orthologs in divergent species raises the possibility that XCT represents a link between clocks of diverse taxa. Interestingly, RNA interference–mediated downregulation of the XCT homolog in C. elegans causes embryonic lethality, suggesting an early and essential developmental requirement for this protein (Piano et al., 2002). Although the human XAP5 gene is ubiquitously expressed, levels are higher in fetal relative to adult tissues, suggesting that it may also play an important role in early mammalian development (Mazzarella et al., 1997). The human XAP5 gene is located in Xq28, a gene-dense region of the X chromosome. The presence of polymorphic numbers of trinucleotide repeats in the human 5′ UTR has led to the suggestion that XAP5 may be involved in genetic diseases (Mazzarella et al., 1997), and linkage analysis indicates that XAP5 is within the interval for many X-linked genes that cause mental retardation when altered (for review, see Chiurazzi et al., 2000). However, currently nothing is known about the molecular function of XAP5 domain-containing proteins in humans or other organisms. Since XCT function is required for viability in C. elegans (Piano et al., 2002) but not plants, Arabidopsis may be an excellent model system in which to investigate the biochemical activity of this intriguing protein.
METHODS
Plant Materials and Genotyping
Arabidopsis thaliana seeds of the Col ecotype were mutagenized as described in Martin-Tryon et al. (2007) and mutants mapped through bulked segregant analysis (Michelmore et al., 1991; Lukowitz et al., 2000) followed by fine scale mapping in a Landsberg erecta outcross population to isolate xct-1. The mutation was confirmed with a derived cleaved-amplified polymorphic sequence marker, F 5′-GAAACGAATTTCCTGCCTGA-3′ and R 5′-AAGGCTCAAGCAGTATGACAAAGGTGTAGTTTGTGCTAG-3′, followed by digestion with NheI (New England Biolabs) and electrophoresis to resolve bands of 163 and 39 bp for Col and 202 bp for xct-1. T-DNA insertion mutants xct-2 (SALK_134513) and xct-3 (SALK_108639) (Alonso et al., 2003) were obtained from the ABRC. xct-2 plants were genotyped with F 5′-TCTTGCTATTTGGGCTGTGA-3′ and R 5′-GACAGCATGTATCACACCTAATCC-3′ to give a 620-bp band in Col. The F primer was used with LBb1, 5′-GCGTGGACCGCTTGCTGCAACT-3′ to give a mutant band of 670 bp. xct-3 plants were genotyped in a multiplex reaction containing three primers: LBb1, F 5′-GGCAGTGATGAAGATGATGG-3′ and R 5′-GCCTCCCTCTCACTGAAACA -3′ to give a 674-bp band in Col and a mutant 520-bp band. toc1-1 (Millar et al., 1995; Somers et al., 1998b) and toc1-2 (Strayer et al., 2000) were introgressed into the Col CCR2pro:LUC background three and four times, respectively, before phenotypic testing. gi-200 and lhy-100 are previously reported EMS-generated mutant lines (Martin-Tryon et al., 2007).
CCR2pro:LUC Imaging
Imaging and analysis were performed as previously described (Martin-Tryon et al., 2007). Data are representative of at least three independent replicates.
Leaf Movement Rhythm Analysis
Plants were germinated on Murashige and Skoog (MS) medium containing 3% sucrose in plates held at a 45° angle in 12:12 LD cycles of 55 μmol m−2 s−1 for 2 d before being released to constant light conditions of 20 μmol m−2 s−1 provided by cool white fluorescent bulbs (Sylvania). Images were automatically acquired at 30-min intervals using LabView (National Instruments) and a PixelLink PL-A781 6.6 Mp camera equipped with a 16-mm Fujinon CF16HA-1 lens. The positions of cotyledon tips were analyzed using ImageJ (http://rsb.info.nih.gov/ij/) and normalized to the lowest position for each tip. Data represent the averages and se of 42 to 80 individual cotyledons and are indicative of two independent replicates.
qRT-PCR
Quantitative RNA profiles of 6-d-old seedlings were generated as previously described (Martin-Tryon et al., 2007) using two to three technical replicates for each sample normalized to PROTEIN PHOSPHATASE 2a subunit A3 (PP2a). Briefly, real-time qRT-PCR was performed using an iCycler (Bio-Rad) and SYBR Green I (Molecular Probes) on cDNA generated from each sample, a pool of 10 whole seedlings. Starting quantity was estimated from critical thresholds compared with the standard curve of amplification. All primer sets contain one primer that bridges an intron to reduce genomic amplification, melt curve analysis was performed following amplification to confirm specificity of products over primer dimers, and a no reverse transcriptase control was used to ensure products detected were from cDNA rather than genomic DNA. Data in Figure 7 are representative of two independent biological replicates, whereas data in Supplemental Figures 5 and 6 are internally replicated over the collection time course. Due to practical limitations, data presented in Supplemental Figure 1 online (LL) represent a single biological replicate (composed of a pool of 10 seedlings); however, similar findings are present for the two loss-of-function alleles xct-1 and xct-2, supporting the reproducibility of the expression profiles. XCT qRT-PCR primers are F 5′-TGGAGAAGAGGGTTAATATTCG-3′ and R 5′-GCAACTCCTCTTCCTCTTGC-3′, LHCB5 qRT-PCR primers are F 5′-CGGTGAAGTTGCTGGAGATTATGG-3′ and R 5′-TCGCATGGATCAATTCAAAA-3′, ELF4 qRT-PCR primers are F 5′-CAAAGCAACGTTCTTCGACA-3′ and R 5′-CGACAATCACCAATCGAGAA-3′, and LUX qRT-PCR primers are F 5′-CGGATTCGAAGAAGCAAAAG-3′ and R 5′-TCATCTCCATCACCGTTTGA-3′. Primers for LHY, CCA1, TOC1, GI, PRR3, PRR5, PRR7, PRR9, and CO are as described in Mockler et al. (2004). PP2a primers, F 5′-TAACGTGGCCAAAATGATGC-3′ and R 5′-GTTCTCCACAACCGATTGGT-3′, were described by Czechowski et al. (2005). XCT splice variation and expression normalized to UBQ-10 were monitored as previously described (Martin-Tryon et al., 2007) using XCT-specific primers flanking the mutation and insertion sites F 5′-TCAAGAAGGAAACAGTGGGTTT-3′ and R 5′-TGTTCTCACGTCTTCATGCAC-3′.
Alignment, Phylogenetic Analysis, and Similarity Matrix
Alignment of protein sequences was performed with AlignX (Vector NTI;, Informax) and ClustalW in Mega4 (Tamura et al., 2007) before Mega4 was used for phylogenetic analysis. Evolutionary relationships were deduced using the neighbor-joining method (Saitou and Nei, 1987) with bootstrap values (10,000 replicates) (Felsenstein, 1985) with the Schizosaccharomyces pombe gene designated as the outgroup since it has the lowest degree of homology to all the other sequences. Evolutionary distances were computed with the JTT matrix-based method (Jones et al., 1992) and represent amino acid substitutions per site. Pairwise deletion was used to eliminate gaps in pairwise sequence comparisons. The similarity matrix was generated in Mega4 using pairwise sequence comparison of 10 sequences to determine the p-distance, the number of amino acid substitutions per site. Identity is shown as 1 − (p-distance)*100.
Rescue and Overexpression Constructs
Two types of genomic rescue constructs were generated under the control of the native promoter, ∼2 kb of the upstream region of XCT. Using the primers F 5′-AACCAGCGGCCGCGATAGTAATTAAATA-3′ and R 5′-ATAACGCGGCCGCCATAGAAACCCTCGCAAAACG-3′, XCT was cloned into pMLBART (B. Janssen, unpublished data; a BASTA-resistant derivative of pART27; Gleave, 1992) using NotI sites. An additional rescue construct and an overexpression construct were generated using the pEarleyGate (pEG; Earley et al., 2006) vectors pEG 301 and pEG 101 and LR clonase (Invitrogen) after cloning the genomic XCT fragment in pENTR/D-TOPO (Invitrogen). Genomic fragments for the constructs were generated by high fidelity PCR with Pfu Ultra High Fidelity enzyme (Stratagene) according to the manufacturer's instructions for long fragments; the native promoter pEG 301 fragment was flanked by F 5′-CACCGGATGCGATAGTAATTAAATA-3′ and R 5′-ATCCCCATGGACTGTGTACC-3′, and the 35S promoter pEG 101 fragment was flanked by F 5′-CACCATGTCGGGTATGGGTGACGGC-3′ and R 5′-ATCCCCATGGACTGTGTACC-3′. The native promoter XCTpro:XCT-YFP-HA pEG301/101 construct was generated by subcloning the XCT promoter and 5′ region from the XCT pEG301 construct into the pEG101 backbone containing the remaining XCT sequence and tags. All clones were verified by sequence analysis.
YFP Imaging
xct-2 plants containing the XCTpro:XCT-YFP-HA pEG301/101 construct were imaged using an Eclipse 600 microscope (Nikon) and a Plan-Apo ×60 objective with the YFP filter Set (Chroma). Images were collected with MetaMorph software (Molecular Devices) and an Orca100 camera (Hamamatsu). Plants generated by five independent transformation events, all with rescued period, produced identical localization patterns.
Immunoblot
xct-2 or xct-2 plants containing XCTpro:XCT-YFP-HA pEG301/101 were germinated on filter paper (Whatman) on MS media containing 3% sucrose in white light and then held in 12:12 LD cycles for 5 d before collection. Samples of five seedlings each were collected directly to liquid nitrogen and stored at −80°C until analysis. XCT-YFP-HA immunoblots of whole cell extracts were performed according to standard procedures using the conjugated anti-HA-peroxidase antibody (Roche) at a 1:1000 dilution. Rubisco was monitored on stripped blots previously probed for HA using 1:2000 diluted primary rabbit anti-cotton Rubisco IgG (a gift from W.J. Lucas) and secondary goat anti-rabbit IgG-HRP (SC-2054; Santa Cruz Biotechnology) at 1:10,000 dilution. SuperSignal West Femto Maximum Sensitivity Substrate (Pierce) was used to react with the peroxidase, and chemiluminescence was detected with BioMax Light Film (Kodak). Signal quantification was performed from exposures that were not saturated and where detection was within the linear range. Each lane was loaded with 1% of the total soluble protein extracted from each sample, and Ponceau stain was used to monitor loading and transfer of proteins. Similar results were observed with at least three independent replicates. Quantification of XCT protein was determined using ImageQuant software (GE Healthcare) to analyze scanned immunoblots. Anti-HA signal was normalized to Ponceau-stained proteins 62 to 83 kD in size, which were quantified before immunodetection was performed. Three to four replicate blots were sequentially quantified for Ponceau and XCT protein before normalization and averaging to determine XCT protein levels.
Period and Hypocotyl Fluence Rate Response Curves
The response of circadian period and hypocotyl length after 6 d of growth to increased fluence rate was determined as previously described (Martin-Tryon et al., 2007). Difference in slope of period fluence rate response was determined by the interaction between genotype and fluence in ANOVA for general linear fixed-effects models. Response curves are representative of three independent replicates, with 5 to 20 individuals in each condition, in each replicate. Rescued hypocotyl phenotypes are representative of three rescued lines generated by independent transformation events.
Phase Response Curve
After germination on MS media containing 3% sucrose and entrainment in 12:12 LD cycles of 55 μmol m−2 s−1 for 6 d, plants containing the CCR2pro:LUC (Strayer et al., 2000) reporter were moved to constant darkness at subjective dawn (CT 0) for imaging. A subset of individuals (n = 16) received a single 3-h pulse of 40 μmol m−2 s−1 red light at various time points over the course of the next 48 h. After the pulse, plants were returned to constant darkness for imaging. Phase estimates were generated from the time of the first peak of LUC activity after the pulse, corrected to circadian time, and plotted against the circadian time of the light pulse for each genotype as previously described (Covington et al., 2001) and are representative of two independent replicates each with 12 to 16 individuals.
Etiolated Pulse Response
Seeds were plated on MS media containing 3% sucrose and stratified for 3 d at 4°C before a 12-h white light pulse of 50 μmol m−2 s−1 to induce germination. Plates were then wrapped in foil and held at 22°C for 108 h before release at subjective dawn (CT 0) to constant red light of 40 μmol m−2 s−1 or constant white light of 50 μmol m−2 s−1, and controls were held in darkness at 22°C until collection. Samples were collected in liquid nitrogen, and RNA was prepared with TRIzol reagent (Invitrogen) for analysis by qRT-PCR. Data are representative of two independent biological replicates and three technical replicates for each sample.
Flowering
Stratified seeds were germinated and grown individually on soil at 22°C in short-day cycles of 8 h light at 200 μmol m−2 s−1 and 16 h of darkness or long-day cycles of 16 h light at 100 μmol m−2 s−1 and 8 h of darkness; light was provided by cool white fluorescent bulbs (Sylvania and Phillips). Rosette leaf number was counted when a 1-cm bolt was present. Data are representative of two independent replicates each with 18 individuals per genotype in short days and 9 to 25 individuals per genotype in long days.
Accession Numbers
Sequence data for XCT and other XAP5 homologs can be found in the GenBank/EMBL data libraries under accession numbers At2g21150 (Arabidopsis XCT), Os09g0535300 (Oryza sativa), CAB46282 (Mus musculus XAP5), mCG48748 (M. musculus XAP5 like), XP_001183761 (Strongylocentrotus purpuratus), NP_004690 (Homo sapiens XAP5), NP_036267 (H. sapiens XAP5 like), NP_001080084 (Xenopus laevis), NP_001017636 (Danio rerio), NP_651595 (Drosophila melanogaster), NP_506625 (Caenorhabditis elegans), and NP_587947 (S. pombe). Sequence data for PP2a can be found under accession number At1g13320.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. qRT-PCR Analysis of Expression Profiles of Clock Components.
Supplemental Figure 2. Analysis of XCT Expression in xct Mutants.
Supplemental Figure 3. Alignment of XAP5 Proteins from Plants and Animals.
Supplemental Figure 4. N- and C-Terminal Fusions between XCT and YFP Are Found in the Nuclei of Juvenile and Adult Plants.
Supplemental Figure 5. Short-Day (8:16 LD) Expression Profiles of Clock Genes in xct Mutants.
Supplemental Figure 6. The Phase of CONSTANS Expression is Unaltered in xct Mutants.
Supplemental Figure 7. Leaf Number at Bolting for Various Clock Mutants Grown in Long or Short Days.
Supplemental Figure 8. Etiolated xct Seedlings Do Not Have a Hypocotyl Phenotype.
Supplementary Material
Acknowledgments
We thank K. Nozue for assistance and use of the camera and software for monitoring leaf movement rhythms; J.N. Maloof for use of red and blue LED chambers; B. Liu for assistance with fluorescence microscopy; R. Rawat for technical assistance with immunoblots; W.J. Lucas for the anti-Rubisco antibody; S.A. Kay in whose lab the xct-1 mutation was first isolated; J.C. Lagarias, M.F. Covington, and J.N. Maloof for helpful discussions; and M.F. Covington, J. Callis, J.C. Lagarias, and J.N. Maloof for critical review of this manuscript. We also thank the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants and the ABRC for distribution of seed stocks. Support from the National Institutes of Health (Grant GM069418 to S.L.H.) and the National Science Foundation (Graduate Research Fellowship to E.L.M.-T.) is gratefully acknowledged.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Stacey L. Harmer (slharmer@ucdavis.edu).
Online version contains Web-only data.
References
- Alabadí, D., Oyama, T., Yanovsky, M.J., Harmon, F.G., Más, P., and Kay, S.A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293 880–883. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Aschoff, J. (1979). Circadian rhythms: Influences of internal and external factors on the period measured in constant conditions. Z. Tierpsychol. 49 225–249. [DOI] [PubMed] [Google Scholar]
- Beator, J., and Kloppstech, K. (1993). The circadian oscillator coordinates the synthesis of apoproteins and their pigments during chloroplast development. Plant Physiol. 103 191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry, J.O., Breiding, D.E., and Klessig, D.F. (1990). Light-mediated control of translational initiation of ribulose 1,5-bisphosphate carboxylase in amaranth cotyledons. Plant Cell 2 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry, J.O., Carr, J.P., and Klessig, D.F. (1988). mRNAs encoding ribulose 1,5-bisphosphate carboxylase remain bound to polysomes but are not translated in amaranth seedling transferred to darkness. Proc. Natl. Acad. Sci. USA 95 4190–4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashmore, A.R. (2003). Cryptochromes: Enabling plants and animals to determine circadian time. Cell 114 537–543. [PubMed] [Google Scholar]
- Chiurazzi, P., Hamel, B.C.J., and Neri, G. (2000). XLMR genes: Update 2000. Eur. J. Hum. Genet. 9 71–81. [DOI] [PubMed] [Google Scholar]
- Covington, M.F., Panda, S., Liu, X.L., Strayer, C.A., Wagner, D.R., and Kay, S.A. (2001). ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 13 1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski, T., Stitt, M., Altmann, T., Udvardi, M.K., and Scheible, W.-R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mairan, J. (1729). Observation botanique. Hist. Acad. Roy. Sci. 35–36.
- Devlin, P.F., and Kay, S.A. (2000). Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell 12 2499–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Z., Millar, A.J., Davis, A.M., and Davis, S.J. (2007). TIME FOR COFFEE encodes a nuclear regulator in the Arabidopsis thaliana circadian clock. Plant Cell 19 1522–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd, A.N., Salathia, N., Hall, A., Kévei, E., Tóth, R., Nagy, F., Hibberd, J.M., Millar, A.J., and Webb, A.A.R. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309 630–633. [DOI] [PubMed] [Google Scholar]
- Earley, K.W., Haag, J.R., Pontes, O., Opper, K., Juehne, T., Song, K., and Pikaard, C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45 616–629. [DOI] [PubMed] [Google Scholar]
- Emery, P., and Reppert, S.M. (2004). A rhythmic Ror. Neuron 43 443–446. [DOI] [PubMed] [Google Scholar]
- Farré, E.M., Harmer, S.L., Harmon, F.G., Yanovsky, M.J., and Kay, S.A. (2005). Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol. 15 47–54. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution Int. J. Org. Evolution 39 783–791. [DOI] [PubMed] [Google Scholar]
- Gleave, A.P. (1992). A versatile binary vector system with a T-DNA organizational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20 1203–1207. [DOI] [PubMed] [Google Scholar]
- Gould, P.D., Locke, J.C.W., Larue, C., Southern, M.M., Davis, S.J., Hanano, S., Moyle, R., Milich, R., Putterill, J., Millar, A.J., and Hall, A. (2006). The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18 1177–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, R.M., Tingay, S., Wang, Z.-Y., and Tobin, E.M. (2002). Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 129 576–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, A., Bastow, R.M., Davis, S.J., Hanano, S., McWatters, H.G., Hibberd, V., Doyle, M.R., Sung, S., Halliday, K.J., Amasino, R.M., and Millar, A.J. (2003). The TIME FOR COFFEE gene maintains the amplitude and timing of Arabidopsis circadian clocks. Plant Cell 15 2719–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, E.R., Petracek, M.E., Dickey, L.F., and Thompson, W.F. (2001). The 5′ end of the pea ferredoxin-1 mRNA mediates rapid and reversible light-directed changes in translation in tobacco. Plant Physiol. 125 770–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer, S.L., and Kay, S.A. (2005). Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. Plant Cell 17 1926–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen, S.P., Schultz, T.F., Pruneda-Paz, J.L., Borevitz, J.O., Ecker, J.R., and Kay, S.A. (2005). LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. USA 102 10387–10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton, P., Park, K.-J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C.J., and Nakai, K. (2007). WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed]
- Johnson, C.H. (2005). Testing the adaptive value of circadian systems. Methods Enzymol. 393 818–837. [DOI] [PubMed] [Google Scholar]
- Jones, D.T., Taylor, W.R., and Thornton, J.M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8 275–282. [DOI] [PubMed] [Google Scholar]
- Kang, X., Chong, J., and Ni, M. (2005). HYPERSENSITIVE TO RED AND BLUE 1, a ZZ-type zinc finger protein, regulates Phytochrome B–mediated red and Cryptochrome-mediated blue light responses. Plant Cell 17 822–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, T., Murakami, M., Nakamura, Y., Ito, S., Nakamichi, N., Yamashino, T., and Mizuno, T. (2007). Mutants of circadian associated PRR genes display a novel and visible phenotype as to light responses during de-etiolation of Arabidopsis thaliana seedlings. Biosci. Biotechnol. Biochem. 71 834–839. [DOI] [PubMed] [Google Scholar]
- Kawaguchi, R., and Bailey-Serres, J. (2002). Regulation of translational initiation in plants. Curr. Opin. Plant Biol. 5 460–465. [DOI] [PubMed] [Google Scholar]
- Kevei, É., Gyula, P., Fehér, B., Tóth, R., Viczián, A., Kircher, S., Rea, D., Dorjgotov, D., Schäfer, E., Millar, A.J., Kozma-Bognár, L., and Nagy, F. (2007). Arabidopsis thaliana circadian clock is regulated by the small GTPase LIP1. Curr. Biol. 17 1456–1464. [DOI] [PubMed] [Google Scholar]
- Kikis, E.A., Khanna, R., and Quail, P.H. (2005). ELF4 is a phytochrome-regulated component of a negative feedback loop involving the central oscillator components CCA1 and LHY. Plant J. 44 300–313. [DOI] [PubMed] [Google Scholar]
- Kim, J.-Y., Song, H.-R., Taylor, B.L., and Carré, I.A. (2003). Light-regulated translation mediates gated induction of the Arabidopsis clock protein LHY. EMBO J. 22 935–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, W.Y., Fujiwara, S., Suh, S.S., Kim, J., Kim, Y., Han, L., David, K., Putterill, J., Nam, H.G., and Somers, D.E. (2007). ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449 356–360. [DOI] [PubMed] [Google Scholar]
- Lechner, E., Achard, P., Vansiri, A., Potuschak, T., and Genschik, P. (2006). F-box proteins everywhere. Curr. Opin. Plant Biol. 9 631–638. [DOI] [PubMed] [Google Scholar]
- Liu, X.L., Covington, M.F., Frankhauser, C., Chory, J., and Wagner, D.R. (2001). ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke, J.C.W., Kozma-Bognar, L., Gould, P.D., Fehér, B., Kevei, É., Nagy, F., Turner, M.S., Hall, A., and Millar, A.J. (2006). Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol. Syst Biol. 2 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke, J.C.W., Southern, M.M., Kozma-Bognár, L., Hibberd, V., Brown, P.E., Turner, M.S., and Millar, A.J. (2005). Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol. Syst Biol. 1 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz, W., Gillmor, C.S., and Scheible, W.-R. (2000). Positional cloning in Arabidopsis: Why it feels good to have a genome initiative working for you. Plant Physiol. 123 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Tryon, E.L., Kreps, J.A., and Harmer, S.L. (2007). GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol. 143 473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más, P., Alabadí, D., Yanovsky, M.J., Oyama, T., and Kay, S.A. (2003. a). Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más, P., Kim, W.Y., Somers, D.E., and Kay, S.A. (2003. b). Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 464 567–570. [DOI] [PubMed] [Google Scholar]
- Matsuyama, A., Arai, R., Yashiroda, Y., Shirai, A., Kamata, A., Sekido, S., Kobayashi, Y., Hashimoto, A., Hamamoto, M., Hiraoka, Y., Horinouchi, S., and Yoshida, M. (2006). ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 24 841–847. [DOI] [PubMed] [Google Scholar]
- Mazzarella, R., Pengue, G., Yoon, J., Jones, J., and Schlessinger, D. (1997). Differential expression of XAP5, a candidate disease gene. Genomics 45 216–219. [DOI] [PubMed] [Google Scholar]
- McWatters, H.G., Bastow, R.M., Hall, A., and Millar, A.J. (2000). The ELF3 zeitnehmer regulates light signaling to the circadian clock. Nature 408 716–720. [DOI] [PubMed] [Google Scholar]
- Michelmore, R.W., Paran, I., and Kesseli, R.V. (1991). Identification of markers linked to disease resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions using segregating populations. Proc. Natl. Acad. Sci. USA 88 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.J. (2004). Input signals to the plant circadian clock. J. Exp. Bot. 55 277–283. [DOI] [PubMed] [Google Scholar]
- Millar, A.J., Carré, I.A., Strayer, C.A., Chua, N.-H., and Kay, S.A. (1995). Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267 1161–1163. [DOI] [PubMed] [Google Scholar]
- Mizoguchi, T., Wright, L., Fujiwara, S., Cremer, F., Lee, K., Onouchi, H., Mouradov, A., Fowler, S., Kamada, H., Putterill, J., and Coupland, G. (2005). Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17 2255–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler, T.C., Yu, X., Shalitin, D., Parikh, D., Michael, T.P., Liou, J., Huang, J., Smith, Z., Alonso, J.M., Ecker, J.R., Chory, J., and Lin, C. (2004). Regulation of flowering time in Arabidopsis by K homology domain proteins. Proc. Natl. Acad. Sci. USA 101 12759–12764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue, K., and Maloof, J.N. (2006). Diurnal regulation of plant growth. Plant Cell Environ. 29 396–408. [DOI] [PubMed] [Google Scholar]
- Onai, K., and Ishiura, M. (2005). PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells 10 963–972. [DOI] [PubMed] [Google Scholar]
- Osterlund, M.T., Wei, N., and Deng, X.W. (2000). The roles of photoreceptor systems and the COP1-targeted destabilization of HY5 in light control of Arabidopsis seedling development. Plant Physiol. 124 1520–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper, A.E., Seong-Kim, M., Hebst, S.M., Ivey, K.N., Kwak, S.J., and Broyles, D.E. (2001). shl, a new set of Arabidopsis mutants with exaggerated developmental responses to available red, far-red, and blue light. Plant Physiol. 127 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piano, F., Schetter, A.J., Morton, D.G., Gunsalus, K.C., Reinke, V., Kim, S.K., and Kemphues, K.J. (2002). Gene clustering based on RNAi phenotypes of ovary-enriched genes in C. elegans. Curr. Biol. 12 1959–1964. [DOI] [PubMed] [Google Scholar]
- Ruckle, M.E., Demarco, S.M., and Larkin, R.M. (2007). Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 19 3944–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Salomé, P.A., and McClung, C.R. (2004). The Arabidopsis thaliana clock. J. Biol. Rhythms 19 425–435. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Devlin, P.F., and Kay, S.A. (1998. a). Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282 1488–1490. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Kim, Y.W., and Geng, R. (2004). The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell 16 769–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers, D.E., Webb, A.A.R., Pearson, M., and Kay, S.A. (1998. b). The short period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 125 485–494. [DOI] [PubMed] [Google Scholar]
- Staiger, D., Allenbach, L., Salathia, N., Fiechter, V., Davis, S.J., Millar, A.J., Chory, J., and Fankhauser, C. (2003). The Arabidopsis SRR1 gene mediates phyB signaling and is required for normal circadian clock function. Genes Dev. 17 256–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer, C.A., Oyama, T., Schultz, T.F., Raman, R., Somers, D.E., Más, P., Panda, S., Kreps, J.A., and Kay, S.A. (2000). Cloning of the Arabidopsis clock gene TOC1, an auto-regulatory response regulator homolog. Science 289 768–771. [DOI] [PubMed] [Google Scholar]
- Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24 1596–1599. [DOI] [PubMed] [Google Scholar]
- Tang, L., Bhat, S., and Petracek, M.E. (2003). Light control of nuclear gene mRNA abundance and translation in tobacco. Plant Physiol. 133 1979–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman, J.M., Zhu, T., Chang, H.-S., Wang, X., and Quail, P.H. (2001). Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc. Natl. Acad. Sci. USA 98 9437–9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, J.M., Cufr, C.A., Denzel, M.A., and Neff, M.M. (2005). The Dof transcription factor OBP3 modulates phytochrome and cryptochrome signaling in Arabidopsis. Plant Cell 17 475–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, M.W., and Kay, S.A. (2001). Time zones: A comparative genetics of circadian clocks. Nat. Rev. Genet. 2 702–715. [DOI] [PubMed] [Google Scholar]
- Zeilinger, M.N., Farré, É.M., Taylor, S.R., Kay, S.A., and Doyle, F.J. (2006). A novel computational model of the circadian clock in Arabidopsis that incorporates PRR7 and PRR9. Mol. Syst Biol. 2 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.