Abstract
Mouse models show that congenital neural tube defects (NTDs) can occur as a result of mutations in the platelet-derived growth factor receptor-α gene (PDGFRα). Mice heterozygous for the PDGFRα-mutation Patch, and at the same time homozygous for the undulated mutation in the Pax1 gene, exhibit a high incidence of lumbar spina bifida occulta, suggesting a functional relation between PDGFRα and Pax1. Using the human PDGFRα promoter linked to a luciferase reporter, we show in the present paper that Pax1 acts as a transcriptional activator of the PDGFRα gene in differentiated Tera-2 human embryonal carcinoma cells. Two mutant Pax1 proteins carrying either the undulated-mutation or the Gln → His mutation previously identified by us in the PAX1 gene of a patient with spina bifida, were not or less effective, respectively. Surprisingly, Pax1 mutant proteins appear to have opposing transcriptional activities in undifferentiated Tera-2 cells as well as in the U-2 OS osteosarcoma cell line. In these cells, the mutant Pax1 proteins enhance PDGFRα-promoter activity whereas the wild-type protein does not. The apparent up-regulation of PDGFRα expression in these cells clearly demonstrates a gain-of-function phenomenon associated with mutations in Pax genes. The altered transcriptional activation properties correlate with altered protein–DNA interaction in band-shift assays. Our data provide additional evidence that mutations in Pax1 can act as a risk factor for NTDs and suggest that the PDGFRα gene is a direct target of Pax1. In addition, the results support the hypothesis that deregulated PDGFRα expression may be causally related to NTDs.
Keywords: neural tube defects/embryonal carcinoma/osteosarcoma
Neural tube defects (NTDs) form a major group of congenital malformations with an average incidence of ≈1 per 1,000 pregnancies. NTDs are generally accepted to represent multifactorial traits with genetic and environmental factors contributing to their aetiology. To date, very little is known about the identity of the genetic factors involved, although several candidate genes have emerged from studies of mutant mouse strains which display NTD or related malformations (1).
Pax1 is a member of the Pax gene family of developmental control genes, which encode transcription factors that contain a DNA-binding “paired domain” (2). The gene is highly conserved between species and there is a 100% conservation between the paired domains of murine Pax1 and its human counterpart PAX1 at the amino acid level (3, 4). The mouse mutant undulated (un) represents a recessive missense mutation in the paired domain of the Pax1 gene, which results in a protein with altered DNA-binding affinity and transcriptional activity (5, 6). Analysis of the un phenotype has shown that Pax1 is essential for normal vertebral development (7, 8). Although these mice do not display spina bifida (sb), a high incidence of lumbar sb was observed in double mutants resulting from a cross between un and Patch (Ph) indicating that Pax1 can be involved in NTD aetiology (9). Moreover, we recently identified a missense mutation in the paired domain of the human PAX1 gene in a patient with sb, suggesting that mutations in this gene may also play a role in human NTD (10).
The PDGFRα gene is located within a 50–400-kb region that has been deleted in the mouse mutant Patch (11). Heterozygotes are characterized by patches of white fur and have an intact axial skeleton, whereas homozygous embryos display occult sb involving the entire spinal column and die during early embryogenesis (12). Targeted inactivation of the murine PDGFRα gene results in mice with sb at the thoracic level (13). Recently, we have cloned and characterized the human PDGFRα promoter and shown that the 5′-flanking region together with the noncoding exon-1 acts as a functional promoter for the 6.4-kb full length receptor transcript (14, 15).
The observed NTD-phenotype of the double-mutant mice with the (un/un, Ph/+) genotype, suggests an interaction between Pax1 and PDGFRα. This suggestion is supported by the observation that PDGFRα-expression overlaps with Pax1 expression during embryonic development, particularly in the sclerotome, which forms the vertebral column (9). PDGFRα-mutated mouse strains show no effect on Pax1-expression in the segmented paraxial mesoderm (13). Therefore, Pax1 may act as an upstream regulator of the PDGFRα. Consequently, both Pax1 and PDGFRα are considered to be candidate genes for sb.
Chalepakis et al. (6) have shown Pax1 to bind to a specific DNA sequence: recognition sequence 4 (RS4). In the present study, we show that the PAX1 mutation previously identified in a patient with sb, affects the RS4-binding properties of the protein. In addition, we demonstrate that wild-type (wt) and mutated Pax1 proteins have different effects on PDGFRα transcription in Tera-2 embryonal carcinoma cells in a differentiation-dependent manner and in the U-2 (OS) osteosarcoma cell line. The data obtained are discussed within the hypothesis that deregulated PDGFRα transcription may be causally related to NTD, including sb.
MATERIALS AND METHODS
Pax1 Constructs.
Expression constructs containing the full-length murine Pax1 cDNA as well as the full-length Pax1-undulated construct (6) were kindly provided by R. Balling (Munich, Germany). To generate the sb-Pax1 mutant, the Gln at position 42 of the paired domain was replaced by a His corresponding to the mutation previously found in a patient with sb (9). Mutagenesis experiments were carried out by using the Altered Sites System kit (Promega) according to the manufacturer’s protocol by using oligonucleotide primer 5′-GAGACCCGCAGGTGCCTACTGAT-3′. Presence of the mutation (bold) was confirmed by dyedeoxy termination cycle sequencing (ABI) of the constructs on an ABI370A automated sequencer.
In Vitro Transcription/Translation.
The synthesis of wt Pax1, sb-Pax1, and un-Pax1 proteins was performed by using the TnT–T7-Coupled Promega Rabbit Reticulocyte Lysate System (Promega) according to the manufacturer’s protocol. For this purpose, a T7-promoter was introduced by PCR on Pax1 expression constructs, which encode Pax1 (wt-Pax1), un-Pax1, or sb-derived Pax1 (sb-Pax1), respectively. Forward primer (including T7 promoter and start codon) 5′-CGCTAATACGACTCACTATAGGAACAGACCACCATGGAGCAGACGTACCGAAGTGAAC-3′ and reverse primer 5′-GGCTGTGGCTCTGTGAGAG-3′ (located in 3′ untranslated region; ref. 6) were used to generate the wt-Pax1-, un-Pax1-, and sb-Pax1-encoding templates. These templates were used subsequently for in vitro transcription/translation.
Generation of Pax1-Specific Antibodies.
Pax1-specific polyclonal antibodies were obtained by immunizing two rabbits with a Pax1-specific peptide coupled to maleimide-activated keyhole-limpet-hemocyanin (Pierce) according to the manufacturer’s instructions and established protocols (16). The Pax1-specific peptide (NH2)-FKHREGTDRKPPSPG-(COOH) corresponds to amino acid number 327–341 of the Pax1-protein (6). A total of 1 mg of coupled protein was mixed with incomplete Freund’s adjuvans and used for initial immunization. Booster injections with 100 μg of coupled protein were given in 4-week intervals. Total serum was collected 11 days after the fourth boost, designated as 289-IV, and used in these studies.
Western Blotting.
Protein extracts were separated on a 10% SDS-polyacrylamide gel and subsequently transferred to a nitro-cellulose membrane, which then was pretreated for 20 min with blocking buffer (0.4% gelatine/3% BSA/10 mM Tris, pH 7.5/350 mM NaCl). The membrane is incubated subsequently with the Pax1 antibody (diluted 1:200 in RIA buffer: 10 mM Tris, pH 7.5/160 mM NaCl/1% Triton/0.1% SDS/0.5% sodiumdesoxycholate) for 1 hr at room temperature and treated with goat-anti-rabbit alkaline phosphatase conjugate for 1 hr at room temperature (Bio-Rad; dilution 1:5,000 in RIA buffer). The membrane was then treated with APB buffer (100 mM Tris pH 9.5; 100 mM NaCl; 5 mM MgCl2) and subsequently stained by using bromochloroindolylphosphate and nitrobluetetrazolin as substrates.
Electrophoretic Mobility Shift Assay.
In these assays, a 32P-3′ end-labeled PRS4 oligonucleotide was used as described (6) (5′-TGGGCTCACCGTTCCGCTCTAGATATCTCGA-3′). The assays were performed essentially as described (17, 18).
Cell Culture.
Tera-2 embryonal carcinoma (EC) cells (Clone 13) were grown in α modification of minimal essential medium lacking nucleosides and deoxynucleosides, supplemented with 10% (vol/vol) fetal calf serum and 44 mM NaHCO3 in a 7.5% CO2 atmosphere at 37°C. Undifferentiated Tera-2 EC cells were seeded at high density (5.0 × 104/cm2), and differentiation of cells was induced by the addition of retinoic acid (RA) (5 μM) 12 h after the cells were seeded at low density (5.0 × 103/cm2), and maintained in this medium for 7 days before further use. U-2 OS cells were seeded at high density (5.0 × 104/cm2) and grown in DMEM/nutrient mix F12 (1:1) with supplements and conditions as mentioned above.
Transfection, Luciferase, and β-Galactosidase Assays.
The −944/+118 human PDGFRα-promoter construct (14, 15) and expression vectors containing no insert, wt-Pax1-, sb-Pax1-, and un-Pax1-encoding inserts, respectively, were transiently transfected into Tera-2 cells or U-2 OS cells by using the calcium phosphate coprecipitation method (19). Luciferase activity was detected 48 h after transfection (Luciferase assay kit, Promega). The luciferase activity was corrected for transfection efficiency by measuring the β-galactosidase activity due to a cotransfected cytomegalovirus promoter-driven lacZ gene construct. Transfections were done in duplicate with different batches of DNA.
RESULTS
Production of wt and Mutant Pax1.
To test the functionality of the different Pax1 proteins, full-length murine Pax1 proteins were generated in an in vitro transcription/translation system. In addition to the wt-Pax1 protein (wt-Pax1), two NTD-associated mutant proteins were synthesized also. The first one (sb-Pax1) contained the Gln → His substitution that was previously identified by us at position 42 of the paired domain in a patient with sb (10). The second mutant protein (un-Pax1) contains the Gly → Ser replacement at position 15 of the paired domain that is known to be responsible for the un phenotype in mice (5).
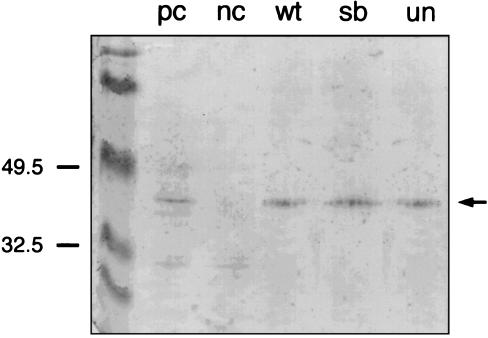
To show that full-length proteins are synthesized, we performed a Western blot analysis. For this purpose polyclonal antibodies were generated recognizing the C-terminal part of Pax1, which is not affected in the mutated proteins. These antibodies detect a protein of the predicted size of 40 kDa in protein extracts of the developing vertebral column of 13.5-day-old mouse embryos (Fig. 1). In contrast, no Pax1 protein was detectable in the extract of the Uns mouse embryo in which the Pax1 gene is deleted (7), showing that the antibodies are Pax1-specific. By using these antibodies, the three in vitro-generated Pax1 proteins are detectable at the same level as the wt-Pax1 protein (Fig. 1), thereby demonstrating that full-length proteins have been synthesized.
Figure 1.
In vitro transcription and translation of Pax1 and derived mutants. The in vitro transcription/translation products of wt-Pax1 (wt), sb-derived-Pax1 (sb), and un-Pax1 (un) were detected on Western blot by a specific anti-Pax1 antibody. As a positive (pc) and negative control (nc) total protein extract from the developing vertebral column of 13.5-day-old wt and a homozygous undulated shorttail (UnS) embryos were used, respectively. In UnS mutant mice, the Pax1 gene has been deleted completely. The antibody detects a predicted band of ≈40 kDa, which is not present in the negative control. No Pax1 was detected in the transcription/translation lysate (not shown).
Binding Properties of wt and Mutant Pax1.
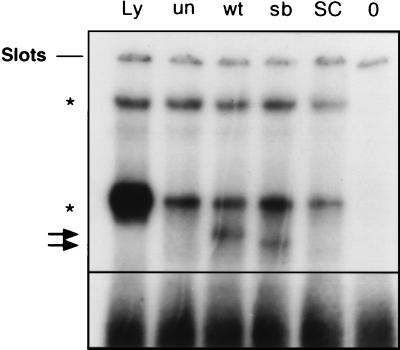
To study the DNA-binding ability of the different Pax proteins, electrophoretic mobility shift assays were carried out by using the PRS4 consensus-binding sequence for Pax1 (6). Fig. 2 shows that with wt-Pax1 the formation of a specific DNA–protein complex causes a clearly detectable band-shift. Using the antibody, Pax1 was identified within this complex (not shown). In contrast, no bandshift was detected with un-Pax1, which is in accordance with the earlier work of Chalepakis et al. (6). In this context one should realize, however, that un-Pax1 was shown to be able to bind to other sequences. Remarkably, sb-Pax1 induces a shift but with a different mobility from that induced by wt-Pax1. Because both proteins have an identical molecular size (Fig. 1), sb-Pax1 apparently binds to the DNA-sequence in a slightly different conformation compared with wt-Pax1. These altered binding characteristics may have consequences for the proper functioning of the Pax1 transcription factor.
Figure 2.
Electrophoretic mobility shift assays with Pax1 and derived mutants. In vitro transcription/translation products of wt-Pax1, sb-derived Pax1, and un-Pax1 were assayed for binding activity on the PRS4 probe derived from the e5 site in the Drosophila even-skipped promoter. The lanes Ly, un, wt, sb, SC, and 0, correspond to incubation of the probe with reticulocyte lysate, un-Pax1, wt-Pax1, sb-Pax1, and wt-Pax1 + unlabeled PRS4 probe in a 500-fold excess, and H2O, respectively. The Pax1 bands are indicated with arrows. Nonspecific bands are indicated with asterisks.
Effect of wt and Mutant Pax1 on PDGFRα Transcription.
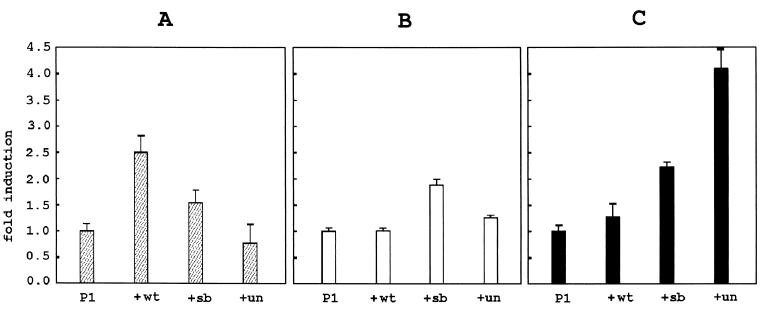
As already mentioned, the phenotype of (un/un, Ph/+) double mutants suggests that Pax1 and PDGFRα participate in the same functional cascade. One possibility is that Pax1 acts as an upstream regulator of PDGFRα. To assess the effect of the various Pax1 proteins on the transcriptional activity of the PDGFRα promoter, plasmids encoding these proteins were cotransfected with the −944/+118 human PDGFRα promoter linked to the luciferase gene. Transfection studies were carried out in Tera-2 cells. In their undifferentiated state, these EC cells resemble cells in early stages of human development. Only after in vitro differentiation by RA these cells express a functional PDGFRα (20). No endogenous expression of Pax1 was observed in both Tera-2 EC and Tera-2 RA cells by reverse transcription–PCR analysis (not shown). Because Pax1 is one of the earliest markers for sclerotome differentiation, additional transfection experiments were carried out in the U-2 OS osteosarcoma cell line. These cells have endogenous PDGFRα-expression (21). Fig. 3A shows that wt-Pax1 induces up to threefold stimulation of promoter activity in Tera-2 RA cells. Un-Pax1 is ineffective in increasing promoter activity in these cells, whereas sb-Pax1 has intermediate activity. These results show that an intact Pax1 protein is essential to efficiently promote PDGFRα transcription in Tera-2 RA cells. Interestingly, in Tera-2 EC cells a significant increase in activity of the PDGFRα promoter was observed with sb-Pax1 whereas only little effect was seen with un-Pax1 (Fig. 3B). Fig. 3C shows that in U-2 OS sb-Pax1 and un-Pax both induce promoter activity, the latter being the most potent activator under these conditions, whereas wt-Pax is again ineffective. These results show that wt and mutated Pax1 proteins can have opposing transcriptional activities, depending on the specific transcriptional background in different cell types. Moreover, it shows that mutations in Pax1 can result in a gain-of-function with respect to transcriptional activity of the protein.
Figure 3.
Regulation of transcriptional activity of the PDGFRα promoter by Pax-1 and derived mutants. The activity of the −944/+118 promoter luciferase construct (P1) is regulated differently by wt-Pax1, sb-Pax1, and un-Pax1 in RA-differentiated Tera-2 RA cells (A, hatched bars), undifferentiated Tera-2 EC cells (B, open bars), and U-2 OS (C, black bars). Values are presented as mean promoter activity relative to the activity of the −944/+118 clone cotransfected with the expression construct without a Pax1 insert, which was set to 1. Error bars indicate the sample SD after four luciferase/β-galactosidase measurements.
DISCUSSION
In the present study, we have shown that the Gln → His mutation in PAX1, previously identified by us in a patient with sb, is associated with altered in vitro DNA-binding properties of the full length protein. Our results suggest that this mutation affects the functionality of the PAX-1 protein and, therefore, may have contributed to the phenotype observed in the patient.
In addition, we investigated the influence of Pax1 on PDGFRα transcription. We showed that wt-Pax1 protein enhances PDGFRα transcription in differentiated Tera-2 human embryonal carcinoma cells (Fig. 3A), whereas two mutant Pax1 proteins carrying either the undulated mutation (un-Pax1) or the Gln → His mutation (sb-Pax1), were not or less effective, respectively. Surprisingly, Pax1 mutant proteins appear to have opposing transcriptional activities, depending on the cell differentiation status in the Tera-2 cells, as well as in the U-2 OS osteosarcoma cell line. (Fig. 3 B and C). These activities of the mutated Pax-1 proteins are the first examples of mutations in a Pax gene that can be associated with a gain-of-function phenomenon. This could have yet unknown consequences during embryonic development and may play a role in the pathogenic mechanism underlying specific types of NTD.
The above observations support the hypothesis that Pax1 is an upstream regulator of PDGFRα and that deregulated PDGFRα gene transcription may be causally related to sb. In this way, it can be explained that sb is not only observed in the targeted PDGFRα null mutation (13) and in homozygous Patch (Ph/Ph) mice (12) but also in Patch (Ph/+) heterozygotes with an additional Pax1 mutation (un/un) (9). Accordingly, the sb phenotype in the patient carrying the sb-Pax1 mutation may be the result of impaired regulation of PDGFRα transcription. In this context, the gain-of-function phenomenon could be very important for effectively deregulating PDGFRα-expression and thus could contribute as well to the sb phenotype. It is well realized, however, that NTDs including sb are a multifactorial trait and that other genes and/or environmental influences are likely to be involved.
We have previously shown that the human PDGFRα gene is able to give rise to multiple mRNA products as a consequence of alternative promoter use and alternative splicing (18, 22). In Tera-2 EC cells, a 1.5-kb and a 5.0-kb transcript have been identified, which are transcribed from a promoter in intron 12 of the gene (18). In contrast, in Tera-2 RA cells a 6.4-kb and a 3.0-kb transcript are present, for which transcription is driven by the promoter upstream of exon 1. Because the 6.4-kb transcript generates the functional, full length α-receptor, this promoter has been used for the current activation studies with Pax1. The −994/+118 region of the human PDGFRα promoter contains the larger part of the information required for tissue-specific regulation in transgenic mice, as previously shown by us (15). This promoter fragment contains multiple putative Pax1-binding site core elements (5′-GTTCC-3′; ref. 6).
Additional studies will have to indicate whether Pax1 actually binds to the PDGFRα promoter directly, or rather mediates its effect via interaction with other components of the transcription machinery. In the case of direct binding of the proteins to the PDGFRα promoter, the different activities could be explained by an altered cofactor dependency of the mutated proteins. In the case of an indirect influence of Pax1 proteins on PDGFRα-promoter activity, at least for undulated, it also might be possible that other genes are activated because of its altered DNA-binding specificity (6).
In summary, the present results emphasize the relevance of PDGFRα expression regulation during vertebral column development and support the model that impaired expression of this gene may be causally related to sb. Evidence from the transfection studies suggests that Pax1 is indeed an upstream regulator of PDGFRα, and mutations in Pax1 may result in a gain-of-function of the protein. The different activities of the wt and mutant Pax1 proteins correlate with an altered conformation of the corresponding protein–DNA complex in the bandshift assays. The present data corroborate our earlier hypothesis that mutations in PAX1 can act as risk factors in NTD but also present the PDGFRα gene as the central gene involved. Additional studies of this gene have to be performed to investigate whether mutations in PDGFRα contribute to human NTD, particularly to sb.
Acknowledgments
We thank Neeltje Arts and José Hendriks for technical assistance. F.H. and E.M. are members of INTEGER, the International Neural Tube Embryology, Genetics and Epidemiology Research consortium to identify genes that predispose to NTDs. This study was supported by the Dutch Prinses Beatrix Fonds Grants 96-0212 and 97-0107.
ABBREVIATIONS
- NTDs
neural tube defects
- PDGFRα
platelet-derived growth factor receptor α
- EC
embryonal carcinoma
- RA
retinoic acid
- wt
wild type
- un
undulated
- sb
spina bifida
- Patch
Ph
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Harris M J, Juriloff J M. Teratology. 1997;56:177–187. doi: 10.1002/(SICI)1096-9926(199709)56:3<177::AID-TERA1>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 2.Strachan T, Read A P. Curr Opin Genet Dev. 1994;4:427–438. doi: 10.1016/0959-437x(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 3.Burri M, Tromvoukis Y, Bopp D, Frigerio G, Noll M. EMBO J. 1989;8:1183–1190. doi: 10.1002/j.1460-2075.1989.tb03490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schnittger S, Rao V V, Deutsch U, Gruss P, Balling R, Hansmann I. Genomics. 1992;14:740–744. doi: 10.1016/s0888-7543(05)80177-6. [DOI] [PubMed] [Google Scholar]
- 5.Balling R, Deutsch U, Gruss P. Cell. 1988;55:531–535. doi: 10.1016/0092-8674(88)90039-6. [DOI] [PubMed] [Google Scholar]
- 6.Chalepakis G, Fritsch R, Fickenscher H, Deutsch U, Goulding M, Gruss P. Cell. 1991;66:873–884. doi: 10.1016/0092-8674(91)90434-z. [DOI] [PubMed] [Google Scholar]
- 7.Wallin J, Wilting J, Koseki H, Fritsch R, Christ B, Balling R. Development (Cambridge, UK) 1994;120:1109–1121. doi: 10.1242/dev.120.5.1109. [DOI] [PubMed] [Google Scholar]
- 8.Balling R, Neubuser A, Christ B. Semin Cell Dev Biol. 1996;7:129–136. [Google Scholar]
- 9.Helwig U, Imai K, Schmahl W, Thomas B E, Varnum D S, Nadeau J H, Balling R. Nat Genet. 1995;11:60–63. doi: 10.1038/ng0995-60. [DOI] [PubMed] [Google Scholar]
- 10.Hol F A, Geurds M P A, Chatkupt S, Shugart Y Y, Balling R, Schrander-Stumpel C T R M, Johnson W G, Hamel B C J, Mariman E C M. J Med Genet. 1996;8:655–660. doi: 10.1136/jmg.33.8.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stephenson D A, Mercola M, Anderson E, Wang C Y, Stiles C D, Bowen-Pope D F, Chapman V M. Proc Natl Acad Sci USA. 1991;88:6–10. doi: 10.1073/pnas.88.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Payne J, Shibasaki F, Mercola M. Dev Dyn. 1997;209:105–116. doi: 10.1002/(SICI)1097-0177(199705)209:1<105::AID-AJA10>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 13.Soriano P. Development (Cambridge, UK) 1997;124:2691–2700. doi: 10.1242/dev.124.14.2691. [DOI] [PubMed] [Google Scholar]
- 14.Afink G B, Nister M, Stassen B H G J, Joosten P H L J, Rademakers P J H, Bongcam-Rudloff E, Van Zoelen E J J. Oncogene. 1995;10:1667–1672. [PubMed] [Google Scholar]
- 15.Zhang X Q, Afink G B, Svensson K, Jacobs J J L, Guenther T, Forsberg-Nilsson K, Van Zoelen E J J, Westermark B, Nistér M. Mech Dev. 1998;70:167–180. doi: 10.1016/s0925-4773(97)00190-1. [DOI] [PubMed] [Google Scholar]
- 16.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 17.Schreiber E, Matthias P, Mueller M M, Schaffner W. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kraft H J, Mosselman S, Smits H A, Hohenstein P, Piek E, Chen Q, Artzt K, van Zoelen E J J. J Biol Chem. 1996;22:12873–12878. doi: 10.1074/jbc.271.22.12873. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsh E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 20.Mosselman S, Claesson-Welsh L, Kamphuis J S, van Zoelen E J J. Cancer Res. 1994;54:220–225. [PubMed] [Google Scholar]
- 21.Betsholtz C, Westermark B, Ek B, Heldin C-H. Cell. 1984;39:447–457. doi: 10.1016/0092-8674(84)90452-5. [DOI] [PubMed] [Google Scholar]
- 22.Mosselman S, Looijenga L H J, Gillis A L M, van Rooijen M A, Kraft H J, van Zoelen E J J, Oosterhuis J W. Proc Natl Acad Sci USA. 1996;93:2884–2888. doi: 10.1073/pnas.93.7.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]