Abstract
To explore whether super-resolution fluorescence microscopy is able to resolve topographic features of single cellular protein complexes, a two-photon 4Pi microscope was used to study the nuclear pore complex (NPC). The microscope had an axial resolution of 110–130 nm and a two-color localization accuracy of 5–10 nm. In immune-labeled HeLa cells, NPCs could be resolved much better by 4Pi than by confocal microscopy. When two epitopes of the NPC, one localized at the tip of the cytoplasmic filaments and the other at the ring of the nuclear basket, were immune-labeled, they could be clearly resolved in single NPCs, with the distance between them determined to be 152 ± 30 nm. In cells expressing a green fluorescent protein construct localized at the NPC center, the distances between the ring of the nuclear filaments and the NPC center was 76 ± 12 (Potorous tridactylus cells) or 91 ± 21 nm (normal rat kidney cells), whereas the distance between the NPC center and the tips of the cytoplasmic filaments was 84 ± 18 nm, all values in good agreement with previous electron or single-molecule fluorescence estimates. We conclude that super-resolution fluorescence microscopy is a powerful method for analyzing single protein complexes and the cellular nanomachinery in general.
INTRODUCTION
The nuclear pore complex (NPC) is a large cellular structure spanning the nuclear envelope. It mediates the transport of all kinds of molecules between cell nucleus and cytoplasm and therefore plays an important role in cell function. In particular, the NPC selectively exports fully processed messenger ribonucleoprotein particles from the nucleus while retaining biosynthetic precursors. The NPC also selectively imports into the nucleus proteins that are necessary for the maintenance, replication, and transcription of the genetic material. Thus, most basic functions of the cell, as well as many pathological conditions, are somehow related to the activity of the NPC. For these reasons, the NPC has become a major object of biological, biochemical, and biophysical studies (for review, see (1–5)).
A deeper understanding of the NPC requires detailed information on both its molecular structure and function. By proteomic techniques it has been shown that both yeast (6) and mammalian (7) NPCs consist of ∼30 different polypeptides referred to as nucleoporins, or nups. Nups occur in multiples of eight to yield a total number of ∼500 polypeptides and a total mass of ∼40 MDa (yeast) and 60 MDa (mammals) per NPC. Structural aspects of the NPC have been intensively studied by electron microscopic techniques culminating in three-dimensional (3D) models at a resolution of 6–10 nm (5,8,9). Although the NPC could not be analyzed as a whole by x-ray diffraction, several individual constituents could be crystallized and their structure solved at atomic resolution (10). This permitted hybrid methods to be applied which combine a variety of structural information from different sources to compute the most likely arrangement of subunits in the complex (11,12). According to such studies, the NPC is composed of a roughly cylindrical central scaffold having a diameter of ∼120 nm and a thickness of ∼80 nm. The central scaffold consists of three connected rings, the cytoplasmic ring, the spoke ring, and the nuclear ring. Eight filaments of ∼50 nm length are attached to the cytoplasmic face of the central scaffold. The nuclear face of the central scaffold also carries eight filaments; however, these are somewhat longer than the cytoplasmic filaments and fused at their distal endings to form the nuclear basket of the NPC. The molecular topography of the NPC, i.e., the precise arrangement of nups in relation to each other, has been intensively studied. It appears that the NPC, despite its large size and peptide number, has a relatively simple modular architecture (11,12). Eight types of protein fold seem to account for 85% of the amino acid residues. Nups seem to be arranged in three concentric layers around the central channel, with nups containing phenylalanine-glycine repeats and coiled-coiled folds forming the inner layer, nups containing α-propeller and α-solenoid folds the central layer, and nups containing transmembrane helices and cadherin folds the outer layer.
In the analysis of functional aspects of the NPC, fluorescence microscopy has played a major role. Thus, fluorescence microscopy has been used in combination with microinjection, cell fusion, and genetic manipulation to study nucleocytoplasmic transport in living cells. In combination with selectively permeabilized cells (13), fluorescence microscopy has revealed the participation of soluble transport factors referred to as karyopherins or importins/exportins/transportins and of the small GTPase Ran in nucleocytoplasmic transport. By attaching isolated nuclear envelopes to artificial nonporous substrates the transport of fluorescent substrates through single NPCs has been analyzed (14,15).
Collectively, these studies have demonstrated an apparent functional duality of the NPC. For inert molecules, the NPC functions as a molecular sieve with a pore diameter of 9–10 nm (14,16), and single-NPC experiments suggested that there is one such channel per NPC (14). However, for karyopherins and their complexes with transport substrates, the NPC appears to provide a patent channel of 30–50 nm diameter (17). Paradoxically, karyopherins and transport complexes are translocated through the 30- to 50-nm channel at high speed, although their translocation depends on tight binding to phenylalanine-glycine repeats of certain nups. Several models have been suggested to account for the transport functions of the NPC (6,18–21). Whether these models are mutually exclusive or can be incorporated into a unifying model remains to be seen.
Altogether, much has been learned about the NPC in recent years, but the mechanism of translocation through the NPC is still poorly understood. The ideal tool for the further analysis of the NPC would provide both structural and functional information at the nanoscopic level. Although this goal may appear ambitious or even utopian, it has been recently brought into reach. Progress in laser and light-detector technology provided the means for imaging single fluorescent molecules as diffraction-limited spots in solid matter (23), lipid bilayer membranes (24), free solution (25), and biological cells (26). By evaluating the intensity distribution of single-molecule images, the molecule center could be determined and tracked with an accuracy of up to 1 nm. Application of photobleaching methods and photoswitchable fluorescent proteins and synthetic dyes allowed researchers to extend single-molecule localization techniques to higher, physiological fluorophore concentrations (27–29) (for review, see (30)). More fundamentally, the classical resolution limits of fluorescence microscopy have been overcome by nonlinear structured-illumination techniques such as two-photon 4Pi microscopy (31,32) and stimulated emission depletion microscopy (33,34), and a resolution of 20 nm has been demonstrated (35). Fluorescence nanoscopy has also been combined with quantitative analytical techniques such as fluorescence photobleaching and fluorescence correlation spectroscopy (36,37) to assess dynamic processes at the nanometer level.
In this study, 4Pi microscopy was used to examine the NPC topology at an axial resolution of ∼100 nm and a localization accuracy of <10 nm in fixed and immune-labeled specimens. We found that the distance between nups in single NPCs could be determined with an accuracy previously only available by electron microscopy. The procedures developed for the NPC can be applied to other cellular protein complexes.
MATERIALS AND METHODS
Reagents
Green fluorescent beads 106 nm in diameter (Fluoresbrite Plain YG 0.1 μm microspheres: excitation maximum, 441 nm; emission maximum, 486 nm) were obtained from Polysciences (Washington, DC). Red fluorescent beads 0.1 μm in diameter (TransFluoSpheres NeutrAvidin-labeled microspheres: excitation maximum, 488 nm; emission maximum, 605 nm) were purchased from Invitrogen (Carlsbad, CA).
Antibodies
The primary antibodies used were 1), polyclonal rabbit anti-Tpr; 2), polyclonal rabbit anti-NUP358 C-288 (directed against amino acid residues 2550–2840) (these antibodies were a gift from E. Coutavas, Rockefeller University of New York); and 3), polyclonal goat anti-Tpr C-terminus (Santa Cruz Biotechnology, Santa Cruz, CA).
Secondary antibodies (all from Invitrogen) were 1), Alexa488-labeled goat antirabbit; 2), Alexa594-labeled goat antirabbit; 3), Alexa488-labeled donkey antigoat antibody; and 4), Alexa594-labeled donkey antigoat antibody.
Cells
Potoroo kidney (PtK2, Potorous tridactylus) and rat kidney (NRK, Rattus norvegicus) cell lines stably expressing POM121-eGFP3 were obtained from J. Ellenberg (European Molecular Biology Laboratory, Heidelberg, Germany). Cells were grown in minimum Eagle's medium containing 10% fetal bovine serum (FBS), 2 mM glutamine, 1× nonessential amino acids, 100 U/ml streptomycin, 100 μg/ml penicillin, and 500 μg/ml G418 (Gibco, Carlsbad, CA) (PtK2), or in Dulbecco's modified Eagle's medium containing 10% FBS, 2 mM glutamine, 100 U/ml streptomycin, 100 μg/ml penicillin, and 500 μg/ml G418 (Gibco) (NRK). HeLa (human cervical carcinoma, epithelial-like) cells were grown in Dulbecco's modified Eagle's medium containing 10% FBS, 2 mM glutamine, 100 U/ml streptomycin, and 100 μg/ml penicillin. Cultures were maintained at 37°C in a humidified incubator with 5% CO2 atmosphere.
Immunostaining
Cells were removed from plastic dishes by trypsinization and replated on quartz coverslips of 220 μm thickness 24 h before use. Confluent cell monolayers were fixed in 4% paraformaldehyd for 20 min at room temperature, washed three times with phosphate-buffered saline (PBS, 138 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4, pH 7.4) and permeabilized in 0.25% Triton X-100 (Sigma-Aldrich, Steinheim, Germany) buffered with PBS for 10 min at room temperature. After washing three times with PBS, unspecific binding was blocked with 0.5% fish skin gelatin (Sigma-Aldrich, Steinheim, Germany) for 30 min at room temperature. Cells were then incubated with primary antibodies for 1 h at room temperature, washed for 20 min in PBS, and incubated with secondary antibodies for 1 h at room temperature. Finally, cells were washed again for 20 min in PBS and then covered with PBS-buffered glycerol (87% w/w).
Preparation of specimen for 4Pi microscopy
Great care was taken to avoid a deformation of the 4Pi point spread function (PSF) within the samples, which can be induced by refraction index differences. In essence, all components between the two opposing objective lenses had a refraction index of or close to 1.46. This was achieved by employing adjustable glycerol-immersion objectives with a numerical aperture of 1.35 and 100-fold magnification, using quartz coverslips, immersing cells in 87% PBS-buffered glycerol, and keeping the thickness of the glycerol layer <35 μm.
Confocal and 4Pi microscopy
Images were obtained with a commercial 4Pi microscope (TCS 4Pi microscope, type A, Leica Microsystems, Wetzlar, Germany). This is a confocal laser scanning microscope of type TCS SP2 incorporating one- and two-photon excitation, photon-counting by avalanche photodiodes, and a 4Pi attachment. Because of these features, the microscope could be used in the confocal mode with upright or inverted beam path and single- or two-photon excitation, or as a two-photon excitation 4Pi microscope. For two-photon excitation a modelocked Ti:Sapphire Laser (MaiTai, Spectra Physics, Mountain View, CA) with a pulse length stretched to 100 ps was used. The laser was tuned by a grating to a wavelength between 800 and 860 nm. The beam expander was set to 3. Fluorescence originating from the sample was passed through a filter cube (SP700, BS560, BP500–550, and BP607–683) and its intensity measured by photon-counting avalanche photodiodes (PerkinElmer, Foster City, CA). The detection pinhole was set to 0.74–0.88 Airy units.
Image improvement and processing
4Pi raw images were linearly brightened, rescaled, and linearly filtered by a subresolution mask employing either Leica TCS 4Pi software or the image processing program ImageJ (Wayne Rasband, National Institutes of Health, Bethesda, MD http://rsb.info.nih.gov/ij).
Ghost images, which arise in 4Pi microscopy because of side lobes of the PSF, can be eliminated by deconvolution. We usually used Leica software, which is based on a linear three-point or five-point deconvolution. In some cases we used the program Imspector, kindly provided by Andreas Schönle (cf. A. Schönle, 2006. Imspector Image Acquisition & Analysis Software, v0.1, http://www.imspector.de), which is able to perform nonlinear as well as three-point and five-point linear deconvolution.
Three-dimensional (3D) reconstructions of the nuclear envelope were derived from image stacks using Leica software.
Measurement of the distance between molecular sites in single protein complexes
To determine the distance between two different epitopes in single NPCs, we used a two-color localization method (38). Epitopes were labeled with different fluorophores that could be spectrally separated. Only those NPCs that were coaxially aligned with the optical axis of the microscope, i.e., the direction of highest resolution, were considered. The epitopes were localized and their distances determined using a new routine that provides for a true two-dimensional (2D) fit.
Concerning the new routine, it may be mentioned that the localization of subresolution particles by light microscopy requires fitting of the diffraction-limited image of the particle to Gaussian functions. Usually, a procedure is used in which a one-dimensional (1D) Gaussian is fitted to a 1D intensity profile of the particle and a second 1D Gaussian is fitted to another, orthogonal intensity profile. Although this procedure is sometimes referred to as a 2D fit, it basically is a 1D fit. The intensity profile is never taken into account as a whole, and difficulties arise when combining the two 1D Gaussians. Thus, this method usually leads to less accurate results than fitting a proper 2D Gaussian function to the whole intensity profile of the region of interest (ROI). This holds particularly for 4Pi microscopy, in which the PSF is intrinsically elliptical, having a full width at half maximum (FWHM) of ∼115–120 nm in the z direction of the optical axis and 220 nm in the focal plane (xy directions).
For these reasons, in cooperation with the company Ingo Lepper Software/Consulting (Münster, Germany), a true 2D fitting routine has been combined with a tool for distance measurements. The program is available from the aforementioned company as a commercial plugin for ImageJ. In this routine, the user selects a point of the image somewhere near the supposed center of the object in question. Then the routine determines the brightest pixel that has a given minimal number of bright neighboring pixels in a specified region around this point. This brightest pixel is the center of a rectangular ROI usually measuring 100 × 60 pixels. To the whole of this ROI, the routine fits an elliptical 2D Gaussian function. The fit process is based on a nonlinear least-squares routine according to the Levenberg-Marquardt method (39). A fit is abandoned if it does not converge, converges to a point outside the ROI, or is too flat. If the fit succeeds, the center of the fitted Gaussian function is taken to be the object center.
For distance measurements, this 2D fitting routine was applied independently to the two channels. If both fits succeeded, the coordinates of the two centers were further examined. The measurement was discarded if the centers did not have the same or very similar x coordinates. Otherwise, the z distance of the two coordinates was taken as the distance of the two colored proteins.
The new routine was tested by evaluating the distance between POM121 and Tpr in PtK2 cells (cf. Results). The 2D Gaussian routine yielded a distance of 76 ± 12 nm (n = 192 NPCs), whereas a value of 68 ± 34 nm was obtained when the same data were evaluated by the usual 1D Gaussian method.
RESULTS
Performance of the 4Pi microscope used in this study
In 4Pi microscopy (32), a confocal microscope is combined with an interferometer: a laser beam is split into two beams. The beams are focused by two opposing objectives into a common focal point. The phases of the two beams are adjusted to yield the characteristic and peculiar 4Pi PSF (for a graphic representation, see, e.g., (37)), consisting in direction of the optical axis of a sharp main peak and two side lobes. In the focal plane, the 4Pi PSF consists of a single peak that has virtually the same dimensions as the confocal PSF. At optimal conditions, the main peak of the PSF has a FWHM of ∼100 nm in the direction of the optical axis, and thus is ∼6–7-fold smaller than that of a confocal PSF. Employing normal one-photon excitation, the intensity of the side lobes is frequently larger than half that of the main peak. However, the relative size of the side lobes can be decreased to ∼30% by two-photon excitation. Scanning an object with the two-photon 4Pi PSF yields a stronger main image with an axial resolution of ∼100 nm and two weaker ghost images. If the intensity of the ghost images is <50% of the main image they can be removed by deconvolution. Another advantage of two-photon excitation on the side lobes is that it considerably increases their distance to the main peak.
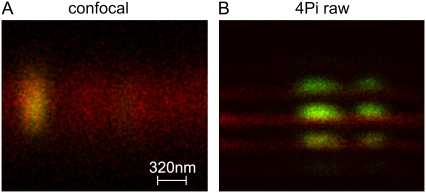
Since the performance of a given 4Pi microscope depends on a number of optical and mechanical parameters, we have checked the resolution as well as the localization accuracy of the instrument used in this study employing two-photon excitation. Using TransFluoSpheres of 0.1-μm diameter, the FWHM of the main peak of the PSF was found to be 115–120 nm in the axial direction and 200–250 nm in the focal plane. The peak intensity of the side lobes was ∼30% that of the main peak. The two-color localization accuracy was checked using a specimen in which a clean coverslip was coated with a thin layer of fluorescently labeled bovine serum albumin (BSA). Then, fluorescent beads of 0.1-μm diameter were deposited on the BSA layer. The BSA had been labeled either with Alexa488 (green fluorescence) or Alexa633 (red fluorescence). With green fluorescent BSA layers, red fluorescent spheres were used, and vice versa. An example is shown in Fig. 1. It can be recognized that bead and BSA layer can be clearly discriminated by 4Pi but not confocal microscopy. By fitting the intensity distribution of BSA layers and beads by Gaussians, the distance between the centers of the BSA layers and beads were determined. This yielded a localization accuracy (i.e., standard deviation of distance measurements) of consistently <10 nm. For repeated measurements of single beads, the localization accuracy was ∼3 nm. In other studies (40), a similar precision (1–4 nm) has been reported. In accordance with preceding studies (41), the localization accuracy did not depend on the fluorescence emission color of BSA layer or beads, indicating that any effect of chromatic aberration was ≤3 nm.
FIGURE 1.
Characterization of resolution and localization accuracy of the 4Pi microscope used in this study. Fluorescent beads (100 nm in diameter, green) were deposited on a thin layer of BSA labeled with Alexa633 (red). (A) Confocal microscopy does not permit resolution of the position of beads on the layer. (B) Using 4Pi microscopy, the position of the bead relative to the layer is clearly resolved. An evaluation of the PSF yielded a resolution of 110–130 nm. A localization of beads and layers by a single-particle localization algorithm yielded an accuracy of 3–10 nm for 4Pi microscopy. The length scale in A holds for both the horizontal (xy) and the vertical (z) directions in A and B.
Although the removal of side lobes by deconvolution is mandatory in most 4Pi studies, it appears to be dispensable when studying topographic details of single submicroscopic protein complexes. In that case, the displacement between main image and ghost images is usually larger than the distance between subunits of the protein complex. Therefore we tested whether the two-color localization accuracy depends on deconvolution. We observed that in identical specimens, the standard deviation of distance measurements increased by ∼1.5 nm after deconvolution, i.e., deconvolution has a slightly negative effect on localization accuracy. Consequently, the distance measurements reported below were all based on raw, i.e., nondeconvolved images.
Visualization and quantification of NPCs by one-color 4Pi microscopy
HeLa cells were grown on coverslips to confluency. After fixation, cells were incubated consecutively with a primary antibody against Nup358, the major component of the cytoplasmic filaments of the NPC, and a secondary Alexa488-labeled antibody. xz scans were obtained by two-photon 4Pi microscopy. Image stacks were used to generate 3D reconstructions (Fig. 2).
FIGURE 2.
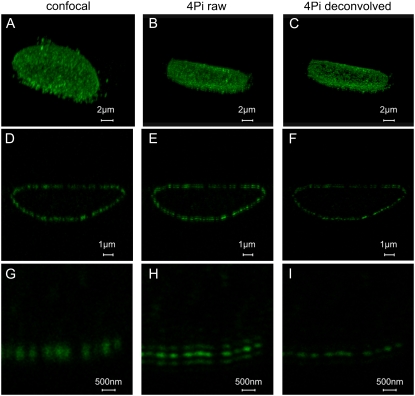
Imaging of NPCs in whole nuclei. HeLa cells were labeled by secondary immune fluorescence techniques using a primary antibody against the NPC protein Nup358 and an Alexa 488-labeled secondary antibody. (A–C) 3D reconstruction of the distribution of NPCs in the nuclear envelope, derived from a series of axial optical sections by confocal (A) and 4Pi (B) microscopy. The effect of deconvolution is shown in C. (D–F) Axial sections of a HeLa cell nucleus. The confocal image (D) is compared with the 4Pi raw image (E) and the deconvolved 4Pi image (F). (G–I) A detail from the same nucleus, comparing the confocal (G) with the 4Pi raw (H) and deconvolved (I) images. The length scales hold for both the horizontal (xy) and the vertical (z) directions.
By employing a small zoom factor (Fig. 2, A–F), whole nuclei were imaged. A comparison of confocal and 4Pi images clearly demonstrates that 4Pi microscopy discriminates NPCs better than confocal microscopy. Since the density of NPCs is relatively large in HeLa cells, 4Pi images were improved by deconvolution.
Series of xz sections permitted us to reconstruct the 3D distribution of NPCs in the nuclear envelope (Fig. 2, A–C). Under confocal conditions, resolution is lower, and therefore, single NPCs appear blurred in the axial direction.
A representative optical section through a HeLa cell nucleus is shown in Fig. 2, D–F. Here, a close inspection reveals that 4Pi microscopy depends in a very sensitive manner on optical path length in the specimen. In the lower nuclear membrane the 4Pi raw images of the NPC consist of strong main peaks and weak side lobes. In the upper nuclear membrane, however, one of the side lobes is almost as strong as the main peak, making the PSF asymmetric. Therefore, deconvolution is quite successful on the lower (Fig. 2, H and I), but less so on the upper, nuclear envelope. An improvement in such cases could probably be achieved by phase compensation. Despite these restrictions, it appears that deconvolved 4Pi images of whole-cell nuclei could be used to count the number of NPCs per nucleus. This is an important parameter concerning the biosynthesis and insertion of fresh NPC into the nuclear envelope during interphase.
Localization of epitopes in single NPCs by two-color 4Pi microscopy
In these experiments, HeLa cells were labeled with antibodies against both Nup358 and Tpr. Tpr is a major component of the nuclear basket. The anti-Tpr antibody we used was directed against an epitope in the C-terminus of Tpr, which, according to immunoelectron microscopic studies (42), is localized at the distal part of the nuclear filaments, close to the nuclear ring. The secondary antibodies against Nup358 and Tpr both were labeled with Alexa488 (green fluorescence) or Alexa593 (red fluorescence), thus permitting us to swap colors between Nup358 and Tpr epitopes. In addition, the orientation of NPCs could be swapped by imaging either the upper or lower part of the nuclear envelope.
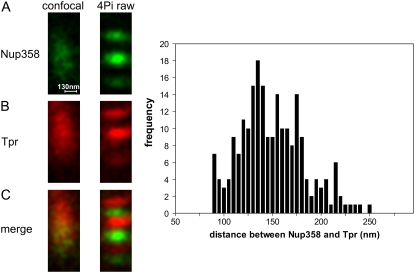
A representative example is shown in Fig. 3. In confocal sections (Fig. 3, A–C, left), the green (Nup358) and red (Tpr) fluorescent epitopes overlap to a large extent. However, in 4Pi sections (Fig. 3, A–C, right), the epitopes are clearly separated.
FIGURE 3.
Resolution of different epitopes in single NPCs and determination of their distance from each other. In HeLa cells, epitopes of the NPC proteins Nup358 and Tpr, which are localized at the tips of the cytoplasmic filaments and the ring of the nuclear basket (cf. Fig. 4 A), were labeled by secondary immune fluorescence methods and imaged by confocal and two-color two-photon 4Pi microscopy. (A–C) Representative images of a single NPC, derived from the green Nup358-associated fluorescence (A), the red Tpr-associated fluorescence (B), and their superposition (C), are shown, comparing confocal (left) with 4Pi (right) images. These 4Pi images were used to determine the distance between the two epitopes as 152 ± 30 nm (n = 218), as shown in the graph at the right. The length scale in A holds for both the horizontal (xy) and vertical (z) directions in A–C.
These results unequivocally demonstrate that it is possible to resolve the nanoscopic substructure of single-protein complexes by light microscopy. It should be recalled, however, that the NPC has an octagonal symmetry with regard to its central axis and that eight copies of both Nup358 and Tpr are present in each NPC. The eight copies of Nup358 and Tpr, respectively, are arranged around the central axis of the NPC on a radius of 80–120 nm and thus far have not been resolved.
Images such as the one shown in Fig. 3 were used to determine the distance between the Nup358 and Tpr, employing the 2D Gaussian fitting routine described in Materials and Methods. The outcome is shown in the panel at the right of Fig. 3. Measurements for a total of n = 218 NPCs yielded a broad distribution with a mean value of 152 ± 30 nm. Since the localization accuracy of the technique itself, as measured with beads on thin layers, was much better than 30 nm, we suppose that the broad distribution might represent a certain variability between individual nups. It is interesting that deviations from perfect rotational symmetry in individual NPCs have been observed very recently by cryoelectron microscopic tomography (9).
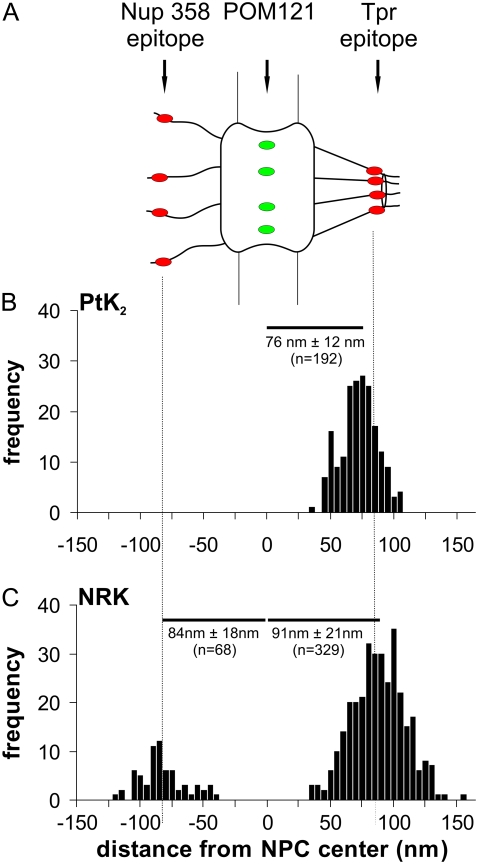
In a further series of experiments, distance measurements were extended to cells constitutively expressing a green fluorescent protein (GFP) fusion protein. We used cells in which the NPC protein POM121, which anchors the NPC in the nuclear envelope and is localized in or very close to the center plane of the NPC, was fused to three molecules of enhanced GFP (eGFP). Thus, PtK2 cells expressing POM121-GFP were fixed and labeled with the anti-Tpr antibody. After reaction with an Alexa594-labeled, secondary anti-Tpr antibody, NPCs were imaged by 4Pi microscopy and the distance between green (POM121-GFP) and red (Tpr) spots in single NPCs was determined as described above for the Nup358/Tpr pair. Results are shown in Fig. 4 B. The mean distance between POM121 and Tpr was found to be 76 ± 12 nm (n = 192). This may be compared with post-embedding immune gold electron microscopic studies of HeLa cells, which used a comparable anti-Tpr and determined the distance between Tpr and the midplane of the NPC to be 76 nm (42).
FIGURE 4.
Distance between molecular sites in single NPCs as determined by two-color localization 4Pi microscopy. (A) Sketch of the NPC indicating the positions of the sites that were fluorescently labeled: an epitope on Nup358 at the tips of the cytoplasmic filaments, a site at the center of the NPC (GFP construct of POM121), and an epitope on Tpr at the distal part of the nuclear filaments. (B) Distances measured in single NPCs of PtK2 cells between the Tpr epitope and POM121-GFP, represented as the mean ± SD of n measurements. (C) Distances measured in single NPCs of NRK cells between the Tpr epitope and POM121-GFP, as well as between POM121-GFP and the Nup358 epitope, represented as the mean ± SD of n measurements.
We also measured the distance between POM121 and Tpr in NRK cells using the same Tpr antibody as with PtK2 cells. A value of 91 ± 21 nm (n = 329) was obtained (Fig. 4 C). Furthermore, the mean distance between POM121 and Nup358 in NRK cells was measured to be 84 ± 18 nm (n = 68). In single-molecule measurements of HeLa cells expressing POM121-GFP, Nup358 was located 72 nm from the midplane (43), whereas immune electron microscopy on isolated rat liver nuclear envelopes using an antibody against Nup358 residues 2550–2837 suggests a distance of 59 nm (44).
DISCUSSION
This study was inspired by observations in recent genomic and proteomic studies (45–47) that proteins occur predominantly in complexes. Protein complexes cooperate in functional modules (48) and form, on the level of the whole cell, one coherent giant network (49,50). The giant protein complex network can be assumed to “run” the cell, directing all processes, including the organization, maintenance, and use of the genetic material. It therefore appears justified to consider as a working hypothesis that the protein complex network is the true fundament of cell function and, by implication, of human health and disease (51,52). Based on the number of genes in the human genome and the average number of subunits per protein complex, it can be roughly estimated that the human cell contains 4000–5000 different protein complexes. Currently, only a small percentage of the assumed large number of protein complexes has been characterized on a molecular level by techniques such as electron microscopy, x-ray diffraction, nuclear magnetic resonance techniques, and computational methods. Most of these techniques require chemical or cryogenic fixation of cells or rely on biochemical isolation or recombinant reconstitution of protein complexes. However, the protein complex network is a highly dynamic entity that exists only in the living cell. Therefore, new techniques are urgently required to facilitate the study of structural and functional properties of protein complexes and their network in living cells.
Fluorescence microscopy has been shown previously to have the specificity and the analytical power for analyzing molecular processes in living cells (53). More recently, it has been shown that the spatial resolution of fluorescence microscopy, in contrast to previous contentions, is, in principle, unlimited (54,55). In this article, we asked whether structural features of single-protein complexes can be resolved and quantitatively assessed by fluorescence microscopy.
As an example, for a cellular protein complex, we chose the NPC because it plays an exceptionally important role in cell function and is sufficiently large and sophisticated to defy many current techniques, such as crystallization and x-ray diffraction analysis. We tested whether a commercial 4Pi microscope is able to resolve structural features in single NPCs. The study showed that 4Pi microscopy yields an axial resolution of ∼100 nm and, at the specific signal/noise ratio of present experiments, a localization accuracy <10 nm. In comparison with earlier determinations of localization accuracy in single-molecule studies and superresolution microscopy of biological samples, the value obtained in this study is quite good, although substantially higher values have been reported in special cases (56,57). In single NPCs, different epitopes, labeled by secondary immune fluorescence, were localized with a standard deviation of 12–30 nm, part of which can be attributed to a certain intrinsic structural variability of the NPC. A similar resolution and localization accuracy has previously been achieved only by immunoelectron microscopy. Recent cryoelectron microscopic studies of the NPC (9) have reached a substantially larger resolution (up to 5.6 nm), lacking, however, molecular specificity and relying on averaging and computational resolution improvement.
Thus, our question as to whether structural features of single NPCs can be resolved by fluorescence microscopy received an unequivocally positive answer. Nevertheless, the study is only a starting point. Important questions remain about, in particular, the extension to living cells, the further improvement of resolution and localization accuracy, and the application to dynamic processes.
The extension of recent studies to living cells is straightforward. The NPC seems to tolerate the fusion of nups with fluorescent proteins, and permanent cell lines have been created expressing fluorescent protein constructs of virtually any nup. In this study, glycerol-immersion objectives were used, and the refractive index of the specimen was adjusted to that of glycerol. However, meanwhile, water-immersion objectives have become available that are suitable for 4Pi microscopy and can be applied to cells in physiological media. Our first live-cell experiments using these objectives yielded just slightly worse resolution in the z direction (130 nm), whereas the localization accuracy was not affected within measuring accuracy. Meanwhile also, we have adapted continuous fluorescence microphotolysis and fluorescence correlation spectroscopy to 4Pi microscopy (37), enabling kinetic studies in the microsecond to minute range. The application of 4Pi microscopy to other topics in cell biology, including live cell studies, has been reviewed elsewhere (58).
Super-resolution light microscopy has made substantial progress. In particular, stimulated emission depletion (STED) microscopy has been largely improved, so that it now achieves a resolution of 20 nm (35). A commercial STED microscope has even become available very recently. However, STED microscopy is most easily installed in a manner improving resolution in the xy plane, not in the z direction. Thus, the STED PSF assumes a cylindrical shape with a small diameter (e.g., 20 nm) but a considerable length (≥600 nm). In contrast, the 4Pi PSF measures ∼250 nm by 100 nm. Furthermore, STED is usually restricted to a narrow range of excitation wavelengths, severely restricting the choice of fluorophores. The installation of two-color fluorescence in STED microscopy is possible (59), although laborious and costly. Thus, 4Pi microscopy remains a valid tool also in the presence of STED microscopy. Most desirable is the ability to combine multicolor 4Pi microscopy with one-color STED microscopy in a single instrument.
In essence, this article shows that structural aspects of single cellular protein complexes can be studied by fluorescence microscopy. Certainly, this possibility will be rapidly improved, expanded, and used to have a strong impact on biology and medicine.
Acknowledgments
We thank Dr. E. Coutavas for providing anti-Nup 358 and anti-Tpr antibodies.
This work was supported by the National Institutes of Health, Bethesda, MD, grant No. 1 R01 GM071329-01, and the Deutsche Forschungsgemeinschaft, grant PE-138/19-1.
Editor: Denis Wirtz.
References
- 1.Cook, A., F. Bono, M. Jinek, and E. Conti. 2007. Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 76:647–671. [DOI] [PubMed] [Google Scholar]
- 2.Tran, E. J., and S. R. Wente. 2006. Dynamic nuclear pore complexes: life on the edge. Cell. 125:1041–1053. [DOI] [PubMed] [Google Scholar]
- 3.Hetzer, M., T. C. Walther, and I. W. Mattaj. 2005. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu. Rev. Cell Dev. Biol. 21:347–380. [DOI] [PubMed] [Google Scholar]
- 4.Peters, R. 2006. Introduction to nucleocytoplasmic transport: molecules and mechanisms. Methods Mol. Biol. 322:235–258. [DOI] [PubMed] [Google Scholar]
- 5.Stoffler, D., K. Schwarz-Herion, U. Aebi, and B. Fahrenkrog. 2006. Getting across the nuclear pore complex: new insights into nucleocytoplasmic transport. Can. J. Physiol. Pharmacol. 84:499–507. [DOI] [PubMed] [Google Scholar]
- 6.Rout, M. P., J. D. Aitchison, A. Suprapto, K. Hjertaas, Y. Zhao, and B. T. Chait. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148:635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cronshaw, J. M., A. N. Krutchinsky, W. Zhang, B. T. Chait, and M. J. Matunis. 2002. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158:915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beck, M., F. Forster, M. Ecke, J. M. Plitzko, F. Melchior, G. Gerisch, W. Baumeister, and O. Medalia. 2004. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 306:1387–1390. [DOI] [PubMed] [Google Scholar]
- 9.Beck, M., V. Lucic, F. Forster, W. Baumeister, and O. Medalia. 2007. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature. 449:611–615. [DOI] [PubMed] [Google Scholar]
- 10.Schwartz, T. U. 2005. Modularity within the architecture of the nuclear pore complex. Curr. Opin. Struct. Biol. 15:221–226. [DOI] [PubMed] [Google Scholar]
- 11.Devos, D., S. Dokudovskaya, R. Williams, F. Alber, N. Eswar, B. T. Chait, M. P. Rout, and A. Sali. 2006. Simple fold composition and modular architecture of the nuclear pore complex. Proc. Natl. Acad. Sci. USA. 103:2172–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alber, F., S. Dokudovskaya, L. M. Veenhoff, W. Zhang, J. Kipper, D. Devos, A. Suprapto, O. Karni-Schmidt, R. Williams, B. T. Chait, A. Sali, and M. P. Rout. 2007. The molecular architecture of the nuclear pore complex. Nature. 450:695–701. [DOI] [PubMed] [Google Scholar]
- 13.Adam, S. A., R. S. Marr, and L. Gerace. 1990. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 111:807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keminer, O., and R. Peters. 1999. Permeability of single nuclear pores. Biophys. J. 77:217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keminer, O., J. P. Siebrasse, K. Zerf, and R. Peters. 1999. Optical recording of signal-mediated protein transport through single nuclear pore complexes. Proc. Natl. Acad. Sci. USA. 96:11842–11847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paine, P. L., L. C. Moore, and S. B. Horowitz. 1975. Nuclear envelope permeability. Nature. 254:109–114. [DOI] [PubMed] [Google Scholar]
- 17.Pante, N., and M. Kann. 2002. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol. Biol. Cell. 13:425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ribbeck, K., and D. Gorlich. 2001. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 20:1320–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frey, S., R. P. Richter, and D. Gorlich. 2006. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science. 314:815–817. [DOI] [PubMed] [Google Scholar]
- 20.Lim, R. Y., N. P. Huang, J. Koser, J. Deng, K. H. Lau, K. Schwarz-Herion, B. Fahrenkrog, and U. Aebi. 2006. Flexible phenylalanine-glycine nucleoporins as entropic barriers to nucleocytoplasmic transport. Proc. Natl. Acad. Sci. USA. 103:9512–9517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peters, R. 2005. Translocation through the nuclear pore complex: selectivity and speed by reduction-of-dimensionality. Traffic. 6:421–427. [DOI] [PubMed] [Google Scholar]
- 22.Reference deleted in proof.
- 23.Moerner, W. E., and L. Kador. 1989. Optical detection and spectroscopy of single molecules in a solid. Phys. Rev. Lett. 62:2535–2538. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt, T., G. J. Schutz, W. Baumgartner, H. J. Gruber, and H. Schindler. 1996. Imaging of single molecule diffusion. Proc. Natl. Acad. Sci. USA. 93:2926–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kubitscheck, U., O. Kuckmann, T. Kues, and R. Peters. 2000. Imaging and tracking of single GFP molecules in solution. Biophys. J. 78:2170–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kues, T., A. Dickmanns, R. Luhrmann, R. Peters, and U. Kubitscheck. 2001. High intranuclear mobility and dynamic clustering of the splicing factor U1 snRNP observed by single particle tracking. Proc. Natl. Acad. Sci. USA. 98:12021–12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gordon, M. P., T. Ha, and P. R. Selvin. 2004. Single-molecule high-resolution imaging with photobleaching. Proc. Natl. Acad. Sci. USA. 101:6462–6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Betzig, E., G. H. Patterson, R. Sougrat, O. W. Lindwasser, S. Olenych, J. S. Bonifacino, M. W. Davidson, J. Lippincott-Schwartz, and H. F. Hess. 2006. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 313:1642–1645. [DOI] [PubMed] [Google Scholar]
- 29.Qu, X., D. Wu, L. Mets, and N. F. Scherer. 2004. Nanometer-localized multiple single-molecule fluorescence microscopy. Proc. Natl. Acad. Sci. USA. 101:11298–11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moerner, W. E. 2007. New directions in single-molecule imaging and analysis. Proc. Natl. Acad. Sci. USA. 104:12596–12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanninen, P. E., S. W. Hell, J. Salo, E. Soini, and C. Cremer. 1995. 2-Photon excitation 4Pi confocal microscope: enhanced axial resolution microscope for biological research. Appl. Phys. Lett. 66:1698–1700. [Google Scholar]
- 32.Hell, S. W., and E. H. K. Stelzer. 1992. Fundamental improvement of resolution with a 4Pi-confocal fluorescence microscope using 2-photon excitation. Opt. Commun. 93:277–282. [Google Scholar]
- 33.Hell, S. W., and J. Wichmann. 1994. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 19:780–782. [DOI] [PubMed] [Google Scholar]
- 34.Hell, S. W., P. E. Hanninen, M. Schrader, T. Wilson, and E. Soini. 1995. Resolution beyond the diffraction limit: 4Pi-confocal, STED, and GSD. Zool. Stud. 34:70. [Google Scholar]
- 35.Donnert, G., C. Eggeling, and S. W. Hell. 2007. Major signal increase in fluorescence microscopy through dark state relaxation. Nat. Methods. 4:81–86. [DOI] [PubMed] [Google Scholar]
- 36.Eggeling, C., L. Kastrup, H. Blom, and S. W. Hell. 2005. Enhancing the applicability of fluorescence fluctuation spectroscopy through reduced focal volumes generated by stimulated emission depletion. Biophys. J. 88:662a. (Abstr.) [Google Scholar]
- 37.Arkhipov, A., J. Hüve, M. Kahms, R. Peters, and K. Schulten. 2007. Continuous fluorescence microphotolysis and correlation spectroscopy using 4Pi microscopy. Biophys. J. 93:4006–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Esa, A., P. Edelmann, G. Kreth, L. Trakhtenbrot, N. Amariglio, G. Rechavi, M. Hausmann, and C. Cremer. 2000. Three-dimensional spectral precision distance microscopy of chromatin nanostructures after triple-colour DNA labelling: a study of the BCR region on chromosome 22 and the Philadelphia chromosome. J. Microsc. 199:96–105. [DOI] [PubMed] [Google Scholar]
- 39.Press, W. H., W. T. Vetterling, B. P. Flannery, and S. A. Teukolsky. 1992. Numerical Recipes in C: The Art of Scientific Computing. Cambridge University Press, Cambridge, UK.
- 40.Schmidt, M., M. Nagorni, and S. W. Hell. 2000. Subresolution axial distance measurements in far-field fluorescence microscopy with precision of 1 nanometer. Rev. Sci. Instrum. 71:2742–2745. [Google Scholar]
- 41.Kano, H., S. Jakobs, M. Nagorni, and S. W. Hell. 2001. Dual-color 4Pi-confocal microscopy with 3D-resolution in the 100 nm range. Ultramicroscopy. 90:207–213. [DOI] [PubMed] [Google Scholar]
- 42.Krull, S., J. Thyberg, B. Bjorkroth, H. R. Rackwitz, and V. C. Cordes. 2004. Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Mol. Biol. Cell. 15:4261–4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kubitscheck, U., D. Grunwald, A. Hoekstra, D. Rohleder, T. Kues, J. P. Siebrasse, and R. Peters. 2005. Nuclear transport of single molecules: dwell times at the nuclear pore complex. J. Cell Biol. 168:233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu, J., M. J. Matunis, D. Kraemer, G. Blobel, and E. Coutavas. 1995. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J. Biol. Chem. 270:14209–14213. [DOI] [PubMed] [Google Scholar]
- 45.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 415:141–147. [DOI] [PubMed] [Google Scholar]
- 46.Gavin, A. C., P. Aloy, P. Grandi, R. Krause, M. Boesche, M. Marzioch, C. Rau, L. J. Jensen, S. Bastuck, B. Dumpelfeld, A. Edelmann, M. A. Heurtier, V. Hoffman, C. Hoefert, K. Klein, M. Hudak, A. M. Michon, M. Schelder, M. Schirle, M. Remor, T. Rudi, S. Hooper, A. Bauer, T. Bouwmeester, G. Casari, G. Drewes, G. Neubauer, J. M. Rick, B. Kuster, P. Bork, R. B. Russell, and G. Superti-Furga. 2006. Proteome survey reveals modularity of the yeast cell machinery. Nature. 440:631–636. [DOI] [PubMed] [Google Scholar]
- 47.Krogan, N. J., G. Cagney, H. Yu, G. Zhong, X. Guo, A. Ignatchenko, J. Li, S. Pu, N. Datta, A. P. Tikuisis, T. Punna, J. M. Peregrin-Alvarez, M. Shales, X. Zhang, M. Davey, M. D. Robinson, A. Paccanaro, J. E. Bray, A. Sheung, B. Beattie, D. P. Richards, V. Canadien, A. Lalev, F. Mena, P. Wong, A. Starostine, M. M. Canete, J. Vlasblom, S. Wu, C. Orsi, S. R. Collins, S. Chandran, R. Haw, J. J. Rilstone, K. Gandi, N. J. Thompson, G. Musso, P. St. Onge, S. Ghanny, M. H. Lam, G. Butland, A. M. Altaf-Ul, S. Kanaya, A. Shilatifard, E. O'Shea, J. S. Weissman, C. J. Ingles, T. R. Hughes, J. Parkinson, M. Gerstein, S. J. Wodak, A. Emili, and J. F. Greenblatt. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 440:637–643. [DOI] [PubMed] [Google Scholar]
- 48.Hartwell, L. H., J. J. Hopfield, S. Leibler, and A. W. Murray. 1999. From molecular to modular cell biology. Nature. 402:C47–C52. [DOI] [PubMed] [Google Scholar]
- 49.Barabasi, A. L., and Z. N. Oltvai. 2004. Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 5:101–113. [DOI] [PubMed] [Google Scholar]
- 50.Ma'ayan, A., R. D. Blitzer, and R. Iyengar. 2005. Toward predictive models of mammalian cells. Annu. Rev. Biophys. Biomol. Struct. 34:319–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peters, R. 2006. Checking and fixing the cellular nanomachinery: towards medical nanoscopy. Trends Mol. Med. 12:83–89. [DOI] [PubMed] [Google Scholar]
- 52.Peters, R. 2006. Nanoscopic medicine: the next frontier. Small. 2:452–456. [DOI] [PubMed] [Google Scholar]
- 53.Day, R. N., and F. Schaufele. 2005. Imaging molecular interactions in living cells. Mol. Endocrinol. 19:1675–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gustafsson, M. G. L. 2005. Nonlinear structured-illumination microscopy: Wide-field fluorescence imaging with theoretically unlimited resolution. Proc. Natl. Acad. Sci. USA. 102:13081–13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hell, S. W. 2004. Strategy for far-field optical imaging and writing without diffraction limit. Phys. Lett. A. 326:140–145. [Google Scholar]
- 56.Martin, S., A. V. Failla, U. Spori, C. Cremer, and A. Pombo. 2004. Measuring the size of biological nanostructures with spatially modulated illumination microscopy. Mol. Biol. Cell. 15:2449–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yildiz, A., and P. R. Selvin. 2005. Fluorescence imaging with one nanometer accuracy: application to molecular motors. Acc. Chem. Res. 38:574–582. [DOI] [PubMed] [Google Scholar]
- 58.Egner, A., and S. W. Hell. 2005. Fluorescence microscopy with super-resolved optical sections. Trends Cell Biol. 15:207–215. [DOI] [PubMed] [Google Scholar]
- 59.Donnert, G., J. Keller, C. Wurm, S. Rizzoli, V. Westphal, A. Schoenle, R. Jahn, S. Jakobs, C. Eggeling, and S. W. Hell. 2007. Two-color far-field fluorescence nanoscopy. Biophys. J. 92:L67–L69. [DOI] [PMC free article] [PubMed] [Google Scholar]