Abstract
Summary
The rth3 (roothairless 3) mutant is specifically affected in root hair elongation. We report here the cloning of the rth3 gene via a PCR-based strategy (amplification of insertion mutagenized sites) and demonstrate that it encodes a COBRA-like protein that displays all the structural features of a glycosylphosphatidylinositol anchor. Genes of the COBRA family are involved in various types of cell expansion and cell wall biosynthesis. The rth3 gene belongs to a monocot-specific clade of the COBRA gene family comprising two maize and two rice genes. While the rice (Oryza sativa) gene OsBC1L1 appears to be orthologous to rth3 based on sequence similarity (86% identity at the protein level) and maize/rice synteny, the maize (Zea mays L.) rth3-like gene does not appear to be a functional homolog of rth3 based on their distinct expression profiles. Massively parallel signature sequencing analysis detected rth3 expression in all analyzed tissues, but at relatively low levels, with the most abundant expression in primary roots where the root hair phenotype is manifested. In situ hybridization experiments confine rth3 expression to root hair-forming epidermal cells and lateral root primordia. Remarkably, in replicated field trials involving near-isogenic lines, the rth3 mutant conferred significant losses in grain yield.
Keywords: rth3, COBRA-like, root hairs, maize, mutant
Introduction
Root hairs enlarge the surface of the root to support the uptake of water and nutrients and the interaction with the abiotic and biotic rhizosphere (Gilroy and Jones, 2000). Unicellular root hairs are shaped by polarized growth of their tips (Kropf et al., 1998), a process mediated by vesicle fusion of membranes and exocytosis of cell wall material (Wasteneys and Galway, 2003). Root hair formation can be divided into two distinct phases: bulge formation or root hair initiation and the transformation of the bulge into the tip-growing apex of the emerging root hair (Ciamporova et al., 2003). Unlike alfalfa (Medicago truncatula), where all epidermal cells are trichoblasts which develop into root-hair-bearing cells (Sieberer and Emons, 2000), the root epidermis of most other higher plants is composed of trichoblasts and atrichoblasts which do not develop root hairs (Larkin et al., 2003). In Arabidopsis thaliana the position of epidermal cells in a cleft between two cortical cells determines the formation of root hairs (Dolan et al., 1994). In maize (Zea mays L.) the last division of surface cells produces two equally sized daughter cells, both of which can produce root hairs (Row and Reeder, 1957).
Despite the progress in characterizing the transcriptional networks which specify trichoblasts and atrichoblasts during epidermal patterning of the Arabidopsis root (Lee and Schiefelbein, 2002) only recently have a number of genes been identified that are involved in Arabidopsis root hair elongation. These genes have a variety of functions. The ROOT HAIR DEFECTIVE3 (RHD3) gene encodes for a GTP-binding protein (Wang et al., 1997) required for regulated cell enlargement, while the TINY ROOT HAIR1 (TRH1) gene encodes a potassium transporter, which may be involved in the spatial localization of potassium uptake during tip growth (Rigas et al., 2001). Recently, six Arabidopsis genes MORPHOGENESIS OF ROOT HAIR 1–6 (MRH1–6) have been identified (Jones et al., 2006) by comparing the transcriptomes of the root hair differentiation zones of the wild type and the root hair mutant root hair defective2 (rhd2), which carries a null mutation in a NADPH oxidase (Foreman et al., 2003). These genes encode a leucine-rich-repeat receptor-like kinase (MRH1), an armadillo-repeat containing kinesin-related protein (MRH2), an inositol-1,4,5-triphosphate 5 phosphatase-like protein (MRH3), a COBRA-like 9 gene (MRH4), a glycerophosphoryl diester phosphodiesterase-like protein (MRH5), and a gene with similarity to the Escherichia coli universal stress protein A (MRH6).
In maize, only three mutants, roothairless 1–3 (rth1, rth2 and rth3), that display root hair elongation phenotypes have been reported thus far (Wen and Schnable, 1994). The only cloned monocot gene that displays a root hair elongation phenotype, rth1, encodes a SEC3-like protein that is a member of the putative exocyst complex which tethers exocytotic vesicles prior to their fusion (Wen et al., 2005).
The COBRA-like gene family (Brady et al., 2007; Ching et al., 2006; Li et al., 2003; Roudier et al., 2002, 2005; Schindelman et al., 2001) is divided into two subgroups that are distinguished by an N-terminal stretch of 170 amino acids (Roudier et al., 2002) and contains 12 members in Arabidopsis, 11 members in rice (Oryza sativa) and currently nine members in the not yet completely sequenced maize genome (Brady et al., 2007). Within these subgroups there are clear differences between monocot and eudicot members, including the existence of a monocot-specific clade (Brady et al., 2007). Most COBRA-like proteins contain a predicted plant-specific glycosylphosphatidylinositol (GPI) anchoring site (Brady et al., 2007) which is connected through an amino acid designated ω to GPI anchors (Udenfriend and Kodukula, 1995). COBRA-like proteins follow a GPI secretion path and are found in Golgi vesicles and, finally, at the outer face of the cell wall (Roudier et al., 2002). Although the primary function of the COBRA-like gene superfamily needs to be fully determined, it appears that in general these genes are involved in various types of cell expansion and cell wall biosynthesis (Brady et al., 2007).
Two major types of cell walls can be distinguished in angiosperms according to their chemical composition (Carpita and McCann, 2000). In type I cell walls, which are characteristic of dicots, cellulose and hemicellulosic xyloglucans, which are present in approximately equimolar amounts, are embedded in a pectin matrix. The type II cell walls formed in maize cells are characterized by a low pectin content, mixed link glucans or glucuronoarabinoxylans as the major hemicellulosic component and a complex network of phenylpropanoids (Carpita and McCann, 2000).
Here, we describe the cloning and characterization of the rth3 gene encoding a COBRA-like protein that is unique to monocots and, based on the rth3 mutant phenotype (Wen and Schnable, 1994), is required for root hair elongation and normal grain yield.
Results
The rth3 mutant is specifically affected in root hair elongation and grain yield
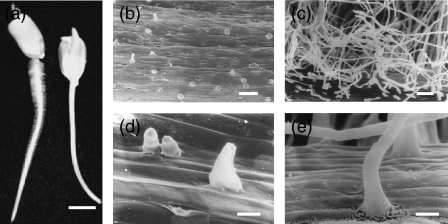
The rth3 mutant (Figure 1a) was previously isolated from Mutator transposon stocks (Wen and Schnable, 1994). In contrast to the wild type, the mutant rth3 is affected in root hair morphology in that it initiates root hair primordia but fails to elongate them properly (Figure 1b,d; wild type, Figure 1c,e). Samples of below-ground crown roots and above-ground brace roots that were excavated from mutant plants near the time of anthesis showed no evidence of root hair elongation, confirming that the rth3 mutation remained stable during field growth. Moreover, the rth3 mutant did not display any apparent aberrant phenotype in the aboveground portion of the plant (including trichome formation) when grown under field or greenhouse conditions (data not shown). To assess how impaired root hairs negatively affect grain yield, three independent yield trials over 2 years were conducted on per se homozygous mutants versus closely related homozygous wild-type plants (see Experimental procedures). The rth3 mutants indeed showed reductions in grain yield of 42, 37 and 19%, respectively, in the three trials (Table 1).
Figure 1.
The rth3 mutant is affected in root hair elongation. (a) Three-day-old wild-type seedling (left) and rth3 mutant (right). (b–e) Scanning electron microscopic images of 3-day-old wild type (c, e) and rth3 (b, d) demonstrate that root hairs are initiated in the mutant but fail to elongate. Size bars: (a) 10 mm; (b, c) 20 μm; (d, e) 200 μm.
Table 1.
Average yield differences between wild-type and rth3 mutant plants
| Yield | IA 97a | IA 94b | NE and KS 94c |
|---|---|---|---|
| Wild type (q ha−1) | 34.03 ± 8.51 | 35.01 ± 12.14 | 44.29 ± 17.69 |
| rth3 (q ha−1) | 19.63 ± 5.39 | 22.22 ± 12.53 | 35.87 ± 20.52 |
| Yield reduction in rth3d (%) | −42.3* | −36.5* | −19.0 |
The experiment was performed in Ames and Crawfordsville, IA in 1997 and included 30 lines that were homozygous rth3 and 30 wild-type (wt) lines.
The experiment was performed at Ames, Ankeny, and Fairfield, IA in 1994 and included 21 lines that were homozygous rth3 and 19 wt lines.
The experiment was performed at North Platte, NE and Garden City, KS in 1994 and included 20 lines that were homozygous rth3 and 20 wt lines.
[(yield rth3/yield wt) − 1] × 100.
t-test: P < 0.01.
Mapping, cloning and sequencing of the rth3 gene
To better understand the role of rth3 in root hair elongation we first mapped and then cloned the gene via a PCR-based approach. Mapping of the rth3 locus placed this gene on chromosome 1S (Wen and Schnable, 1994) between the B–A translocation breakpoints TB1-24464 (0.51) and TB1sb (0.05) in the vicinity of the pericarp color 1 (p1) gene (0–3.6 cm) and the RFLP marker ias8 (2.3 ± 1.3 cm). Cloning of the rth3 gene was performed with the amplification of insertion mutagenized sites (AIMS) system, which allows for the detection of Mutator (Mu) transposon flanking sequences that co-segregate with the mutant phenotype (Frey et al., 1998). This co-segregation analysis with genomic DNA of 126 F1 progeny of the cross Rth3-1/Rth3-1× rth3-1/Rth3-1 revealed a 170-bp band (Figure 2a) that was amplified from each of the 69 heterozygous rth3/Rth3 plants that contained the mutant allele but from none of their 57 homozygous wild-type (Rth3/Rth3) siblings. The 170-bp fragment of the mutant allele was obtained with the primer pair Mu Sel and Hpa II Sel/T from Hpa II-digested DNA (for details refer to Frey et al., 1998). Screening of a seedling cDNA library using the 170-bp fragment as a hybridization probe revealed a full-length rth3 transcript of 2450 bp (GenBank accession number AY265855), which contains an open reading frame that encodes for a 667 amino acid protein of 71.4 kDa (GenBank accession number AAQ81633). A B73 genomic clone with a size of about 10 kb, containing the complete cDNA sequence, was identified and sequenced via the Tn1000-based system (Morgan et al., 1996). Comparison of the genomic and cDNA sequences established that the rth3 gene lacks introns.
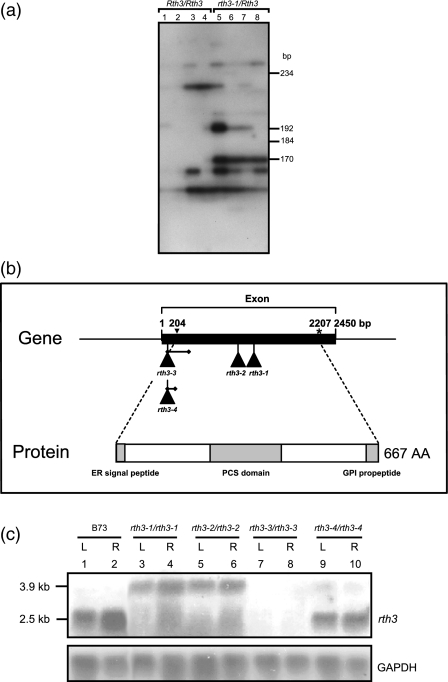
Figure 2.
Cloning and structural features of the rth3 gene. (a) Identification of a 170-bp DNA fragment that flanks a Mu transposon and that co-segregates with the mutant rth3 phenotype in amplification of insertion mutagenized sites experiments. Lanes 1–4: genomic DNA of plants homozygous for wild–type rth3 alleles. Lanes 5–8: genomic DNA isolated from plants heterozygous for the mutant rth3-1 allele. (b) Structures of the rth3 gene and RTH3 protein. Triangles indicate the positions of the Mu transposon insertions in the various rth3 alleles. Horizontal bars above the triangles indicate sequences deleted adjacent to the transposon insertion sites in the rth3-3 and rth3-4 alleles. The start and stop codons are at positions 204 and 2207 of the cDNA (GenBank accession number AY265855). (c) Ribonucleic acid gel blot analyses of the four rth3 mutant alleles with RNA isolated from leaves (L) and roots (R) demonstrate that rth3-3 represents a null allele. The 3.9-kb band in alleles rth3-1 and rth3-2 represents rth3 transcripts containing 1.4-kb Mu transposon insertions. The 2.8-kb band in allele rth3-4 is caused by a 294-bp deletion of rth3 sequences and the insertion of a truncated 658-bp Mu1 transposon. The wild-type allele (B73) displays the expected 2.5-kb band.
Confirmation of rth3 cloning via independently generated alleles
We identified additional mutant alleles to confirm that the rth3 phenotype was generated by a mutation in the gene with the GenBank accession number AY265855. Phenotypic screens for root hair defective mutants yielded three additional rth3 alleles. Allele rth3-2 was isolated from a screen of selfed Mu transposon stocks. Two additional alleles, rth3-3 and 3-4, were identified from a direct Mu transposon tagging experiment using rth3-1 and the subsequent screening of 62 100 F1 progenies (see Experimental procedures). Each of these newly induced alleles displayed the same roothairless phenotype as the reference allele rth3-1. All four alleles contained a Mu transposon insertion in the transcribed region of the single exon of the rth3 gene (Figure 2b). With reference to the rth3 sequence deposited in GenBank (AY265855), the Mu8 transposon inserted in allele rth3-1 at position 1287 and is flanked by the typical direct target site duplication (TSD) of 9 bp. In the rth3-2 allele a 9-bp TSD flanks a 1.4-kb Mu1 transposon inserted at position 1070. Allele rth3-3 is characterized by a 294-bp deletion from base pair 84 to 377 in which a truncated 658-bp fragment of the 5′-end of a Mu1 transposon was inserted. A direct TSD flanking the transposon is not present in this allele. Finally, allele rth3-4 contains a 107-bp deletion of the region defined by positions 84–190 of AY265855 in which a 1.4-kb Mu1 transposon was inserted that was not flanked by a direct TSD.
Based on RNA samples isolated from 2-week-old leaves and roots of the wild type (B73) and the four rth3 mutant alleles, the rth3 gene was expressed in wild-type (B73) roots and leaves (Figure 2c). Seedlings homozygous for the rth3-3 mutant allele did not accumulate rth3 transcripts in either their roots or leaves. Hence, the rth3-3 allele represents an apparently null allele. Seedlings homozygous for the rth3-1 and rth3-2 mutant alleles displayed a transcript of 3.9 kb when hybridized with the 170-bp rth3 probe identified in the AIMS experiments. The 3.9-kb band is consistent with the presence of a 1.4-kb Mu transposon in these rth3 alleles. The origins of the smaller hybridizing smears from these mutants are not known, but they could potentially represent multiple transcripts derived from alternative transcript initiation sites as has been observed in some other Mu insertion alleles (e.g. Cui et al., 2003). Seedlings homozygous for the rth3-4 mutant allele displayed a band of ∼2.8 kb, which is consistent with the deletion of 294 bp together with the insertion of a truncated 658-bp Mu1 transposon fragment. As expected, only the 2.5-kb band was detected in wild-type (B73) seedlings.
The rth3 gene encodes a putative GPI-anchored COBRA-like protein
Sequence similarity searches using the BlastX algorithm (Altschul et al., 1997) revealed that the RTH3 protein displays significant similarity to members of the COBRA gene family. Consistent with the predicted COBRA relationship, the RTH3 protein contains all features of a putative GPI-anchor protein (Udenfriend and Kodukula, 1995). First, the RTH3 protein contains a hydrophilic central portion flanked by cleavable hydrophobic sequences including a N-terminal signal peptide (amino acids 1–23) for targeting the protein to the endoplasmic reticulum and a C-terminal propeptide (amino acids 649–667) required for GPI linkage. Moreover, the sequence motifs of a ω amino acid serine to which the GPI anchor is linked and the ω + 2 amino acid glycine followed by a spacer of five amino acids containing a proline and a subsequent stretch of 12 hydrophobic amino acids meet all sequence requirements of a GPI-anchored protein. Furthermore, the RTH3 protein contains a central cysteine-rich domain between amino acids 419 and 462, which includes eight cysteine residues, although the CCVS domain that appears in Arabidopsis GPI-anchored COBRA-like proteins is only conserved as CCVT. The cysteine-rich region also contained a number of predicted N-glycosylation sites which are frequently associated with GPI-anchored proteins (Roudier et al., 2002). Moreover, only a weak similarity to a cellulose-binding domain II has been identified between amino acid residues 224 to 274 (E-value 0.009). Finally, like all members of the COBRA gene family, the RTH3 protein also contains a central phytochelatine synthase (PCS) domain. Phytochelatine synthetases are known to play a role in plant detoxification from heavy metals (Cobbett, 2000). However, wild type and rth3 mutants did not display any phenotypic differences when grown in 50 and 100 μm CdCl2 (data not shown). In addition, expression of the RTH3 protein in the yeast yap1 and ycf1 mutants (Li et al., 1997) grown on medium containing 50–100 μm CdCl2 did not complement the cadmium-sensitive phenotype of these mutants (data not shown). These results imply that there is only a structural but not a functional relationship of the RTH3 protein with known PCSs.
The rth3 gene belongs to a monocot-specific clade of the COBRA gene family
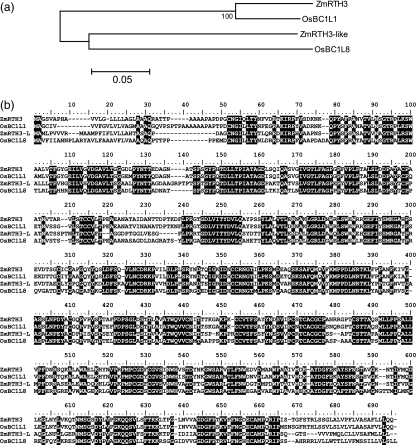
The COBRA gene family can be divided into two subgroups, which are characterized by the presence or absence of an N-terminal stretch of 170 amino acids (Brady et al., 2007). The rth3 gene belongs to a monocot-specific clade of the subgroup that contains the 170 N-terminal amino acid residues. This clade is composed of the maize genes rth3 and rth3-like and the rice genes OsBC1L1 and OsBC1L8 (Figure 3a). All four genes contain only a single exon. Identity among the members of this clade at the protein level is between 62% and 64% (Figure 3b). Remarkably, RTH3 and OsBC1L1 share a sequence identity of 86%. Most probably the rth3 and OsBC1L1 genes are orthologs, not only because of their exceptional degree of sequence similarity but also because rth3 (chromosome 1S) and OsBC1L1 (chromosome 3) map on syntenic regions of the maize and rice genomes (Gale and Devos, 1998). The closest maize relative of rth3, the rth3-like gene, displays an overall sequence identity to RTH3 of 64% at the protein level.
Figure 3.
The monocot-specific clade of the N-terminal 170 amino acid residue containing subgroup of COBRA-like proteins. (a) Phylogenetic relationship of the two maize and rice genes comprising the monocot-specific clade of COBRA-like genes. (b) The four monocot-specific COBRA-like proteins display extensive stretches of similarity.
Massively parallel signature sequencing expression profiles of the rth3 gene display a variable spatial and temporal expression pattern which is distinct from that of rth3-like
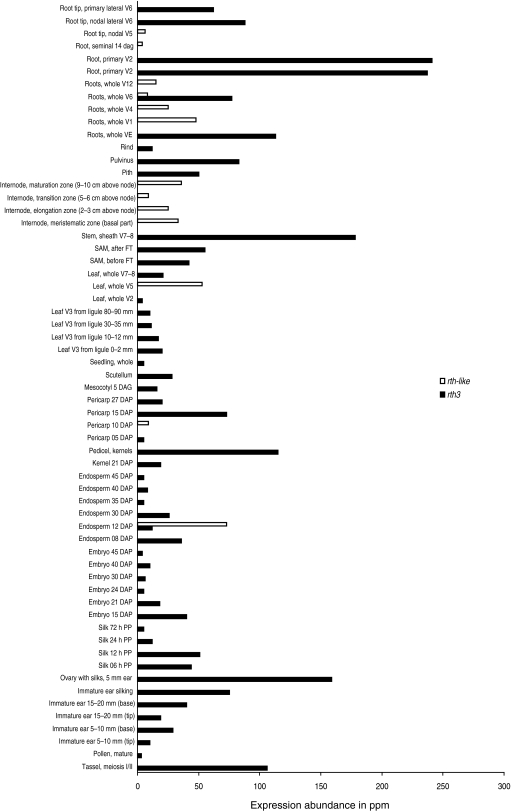
To obtain a detailed picture of the expression patterns of the rth3 and rth3-like genes during development, DuPont's (http://www2.dupont.com/) massively parallel signature sequencing (MPSS; Brenner et al., 2000) database was surveyed (Figure 4). Transcripts of rth3 were detected in 48 different samples of the inbred line B73 representing a wide variety of tissues and developmental stages of the maize plant. The young primary root (V2 stage), which was represented by two independent libraries, displayed the highest rth3 transcript abundance (about 240 p.p.m.) consistent with the observed root hair phenotype of the rth3 mutant. Within reproductive tissues, rth3 exhibited its highest expression in ‘ovaries with silk’ of young 5-mm ears (160 p.p.m.). Lack of expression in this tissue in the mutant might partially explain the significantly reduced grain yield of the rth3 mutant. In contrast to rth3, the rth3-like gene is expressed in only a limited number of tissues including different stages of root development and all zones of the internode. Moreover, rth3 transcript accumulates in 12-days after pollination (DAP) endosperm, 10-DAP pericarp and V5 leaves. No expression of the rth3-like gene was detected in reproductive tissue and embryos. Hence the closely related genes rth3 and rth3-like display complementary expression patterns. While expression in the internode was only detected in rth3-like, both genes exhibit a mutually exclusive expression pattern in distinct phases of root, pericarp and leaf development except for 12-DAP endosperm where both rth3 and rth3-like are expressed.
Figure 4.
Relative expression levels (in p.p.m.) of the rth3 and rth3-like genes in 50 different tissues and developmental stages from wild-type B73 inbred using the Solexa massively parallel signature sequencing system.
In situ hybridization experiments demonstrate rth3 expression in trichoblasts and lateral root primordia in the differentiation zone of the primary root
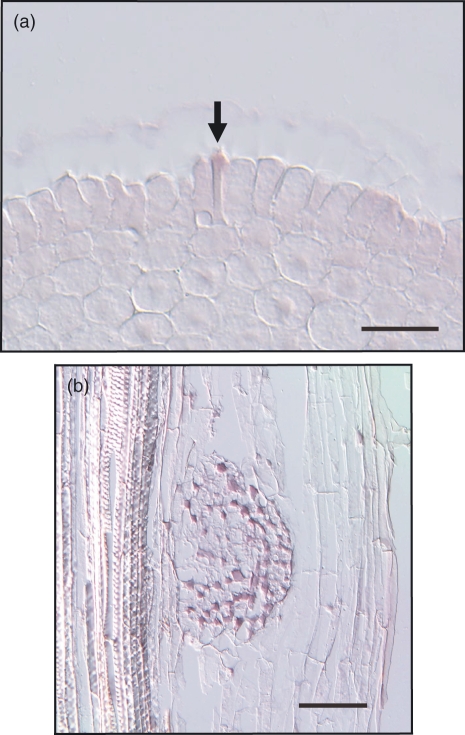
The MPSS analyses demonstrated that rth3 displays its strongest expression in young primary roots where the rth3 root hair phenotype is manifested. Hence, we performed in situ hybridization experiments to obtain a detailed understanding of the spatial expression patterns of rth3 in the root hair zone of the primary root. Consistent with the mutant phenotype, rth3 transcripts were detected in the apical tips of elongating, root-hair-forming, epidermal cells (Figure 5a). Additionally, rth3 was also expressed in young lateral root primordia, which are not associated with a mutant phenotype in rth3 (Figure 5b).
Figure 5.
In situ localization of rth3 transcripts in the differentiation zone of maize primary roots reveal expression in distinct epidermal cells (a) and in emerging lateral root primordia (b). Size bars: (a) 50 μm; (b) 100 μm.
Discussion
In recent years a number of maize mutants affected in various aspects of root development have been identified (summarized in Hochholdinger et al., 2004a, Hochholdinger et al., 2004b). However, only two of the genes impaired in these mutants have been cloned (Taramino et al., 2007; Wen et al., 2005). Among these, the rth1 gene, which like rth3 displays a root hair phenotype, encodes a sec3 homolog involved in exocytotic vesicle fusion (Wen et al., 2005). While the rth3 mutant is specifically affected in root hair elongation, the rth1 mutant displays pleiotropic effects during plant development (Wen and Schnable, 1994).
The rth3 gene encodes a 71.4 kDa protein that displays significant similarity with proteins of the COBRA-like gene superfamily. A predicted GPI anchor is present in all maize COBRA-like genes (Brady et al., 2007), but was predicted for only 9 of 12 COBRA-like genes from Arabidopsis and for only 9 of 11 rice COBRA-like genes. Another characteristic feature of COBRA-like proteins is the presence of a central PCS domain (Figure 2b). Phytochelatines play a critical role in cadmium detoxification of plants by sequestering phytochelatine–cadmium complexes into vacuoles (Cobbett, 2000). Functional tests for PCS activity of the rth3 gene by complementation of the yeast cadmium-sensitive mutants ycf1 and yap1 (Li et al., 1997) were negative, and the growth behavior of wild-type versus mutant seedlings in cadmium solution also did not display any differences (data not shown) that could be related to a function of RTH3 in cadmium detoxification. This demonstrates that rth3 does not contain a functional PCS domain. In addition, phylogenetically, functional PCS proteins are only distantly related to COBRA-like proteins. The complementation of a yeast mutant deficient in a PCS by Arabidospsis COBRA (Leuchter et al., 1998) was attributed to the cysteine-rich region of the protein that could bind metal ions (Roudier et al., 2002), not to the PCS domain of the COBRA protein.
COBRA-like proteins can be divided into two subgroups based on the presence of an N-terminal stretch of 170 amino acid residues (Roudier et al., 2002). The RTH3 protein contains this additional N-terminal domain. Within these subgroups there are significant differences between monocot and dicot COBRA-like proteins. These differences complicate efforts to identify orthologous genes between Arabidopsis and rice or maize (Brady et al., 2007). On the other hand, for most maize genes a likely rice orthologous counterpart could be assigned based on phylogenetic reconstructions (Brady et al., 2007). The rth3 gene belongs to a monocot-specific clade composed of two maize and two rice genes. The rth3 gene is the first member of this monospecific clade that has been characterized based on a specific mutant phenotype. Most likely OsBC1L1 is the rice homolog of rth3, not only because of the high degree of identity at the protein level (86%) but also because the two genes map to syntenic regions of the maize and rice genomes. On the other hand, the closest maize relative, rth3-like appears not to be a functional homolog of rth3 despite the structural similarity on the protein sequence level. The MPSS expression data revealed expression of rth3 in 48 different tissue types. The rth3-like gene, however, was only expressed in one of these tissues (14-DAP endosperm) but in addition was also transcribed at different stages of root and internode development.
Thus far, only a few of the Arabidopsis, rice and maize COBRA-like genes have been functionally characterized based on their mutant phenotypes. Within the subgroup that lacks the N-terminal stretch of 170 amino acids the AtCOBRA gene is required for the oriented deposition of cellulose microfibrils during anisotropic expansion of most organs during post-embryonic development (Roudier et al., 2005). Moreover, the AtCOBL4 gene that is one of the few COBRA-like proteins without a GPI anchor that is required for cellulose biosynthesis in the secondary cell wall (Brown et al., 2005; Persson et al., 2005). In monocots, the rice brittle culm 1 like (Li et al., 2003) and the maize gene brittle stalk 2 (Ching et al., 2006) genes affect the mechanical strength of the plants. These genes belong to the same clade within the subgroup that lacks the 170-amino-acid N-terminus and are most likely orthologs. Moreover, they are currently the only example for which an Arabidopsis ortholog (AtCOBL4) has been postulated (Brady et al., 2007).
In the second subgroup whose members are defined by the presence of the N-terminal stretch of 170 amino acids, only AtCOBL9 is currently associated with a mutant phenotype. Arabidopsis cobl9 mutant root hairs are shorter and wider than wild-type root hairs and burst soon after the establishment of tip growth at an unpredictable point of the root hair surface (Jones et al., 2006). Although the rth3 mutant also displays a root hair phenotype, its phenotype differs from that of Arabidopsis cobl9 mutants. Unlike Arabidopsis cobl9 mutants, the root hairs of rth3 seedlings do not burst, but instead cease elongation soon after initiation. The rth3 gene belongs to a different, and monocot-specific, clade of subgroup II than the Arabidopsis cobl9 gene. Moreover, only a relatively low degree of sequence similarity (namely 49%) exists between AtCOBL9 and RTH3 on the protein level. In contrast, the confirmed orthologs AtCOBL4 versus ZmBK2 and OsBC1 exhibited sequence identities between the monocot and dicot members of between 72% and 77%, respectively. Finally, according to MPSS data, the rth3 gene is expressed in almost all tissues, with highest expression in young primary roots. In contrast, a RT-PCR analysis of AtCOBL9 did not detect expression in 2- to 7-week-old roots and leaves but only in 7-week-old flowers (Roudier et al., 2002), and this only after 30–40 cycles of PCR. This difference in gene expression is also confirmed by more detailed microarray data of AtCOBL9 (Brady et al., 2007) which again indicate very low expression of this gene in the radicle and roots as compared to other organs. This discrepancy between the sequence and gene expression level might imply that rth3 and AtCOBL9 are not orthologous although both genes display root hair phenotypes. The functional differences between these genes might be related to the different composition of Arabidopsis (class I) and maize (class II) cell walls (Carpita and McCann, 2000) and hence alternative functions of these cell wall proteins in the distinct cell wall context.
Despite the specific root hair phenotype of the rth3 mutant, the gene is expressed in all analyzed wild-type tissues and all studied developmental stages between embryogenesis and post-embryonic vegetative and generative development. Two peaks of expression might be associated with the observed phenotype. First, the highest expression of rth3 was observed in young (V2) primary roots. Such young roots bear a high concentration of root hairs. Lack of rth3 transcripts in these roots might therefore be directly associated with the observed root hair phenotype. During generative development the highest expression was observed in ovaries with silks from young 5-mm ears. Again this expression pattern might be directly associated with the reduced yield that is observed in rth3 mutant plants. However, reduced yield might also alternatively be indirectly associated with the reduced nutrient transport in the plant due to the significant reduction of the absorbing surface of the root hair, which provides up to 77% of the root surface in crops (Parker et al., 2000). Normal rth3 gene function may require expression beyond a certain threshold level that is only surpassed in young primary roots and perhaps in young ovaries, which might explain the reduced yield of rth3 mutants.
In situ hybridization experiments demonstrated that rth3 is expressed in epidermal cells that develop into root hair. This is in line with the observed phenotype of the rth3 mutant which does not allow proper elongation of root hairs in the absence of the rth3 gene product. Remarkably, rth3 expression was also detected in lateral root primordia, although no lateral root defects were observed in rth3 mutants. This implies that rth3 might also play a role in the complex molecular networks that are involved in lateral root formation but that the rth3 function might be redundant in lateral root formation. Although the rth3 gene is expressed in all analyzed tissues and developmental stages, based on the very specific phenotype of the rth3 mutant and the lack of a significant cellulose-binding domain it is unlikely that RTH3 is a general regulator of cell expansion and cell wall biosynthesis. More likely, the rth3 gene plays a specific role in the tightly coordinated network of genes responsible for root hair elongation in the epidermis while the spatial and temporal expression of other COBRA-like genes might be required for the regulation of cell expansion in other plant organs. Future identification of the interaction partners of RTH3 will enhance our understanding of the molecular networks involved in root hair elongation in monocots.
Experimental procedures
Isolation of new rth3 alleles and maintenance of the mutant stocks
The rth3 reference allele rth3-1 described in Wen and Schnable (1994) was maintained by backcrossing heterozygous plants to the inbred line B73 over numerous generations as described previously (Wen and Schnable, 1994). An additional allele, rth3-2, was identified by a forward genetic screening of Mutator stocks at Pioneer Hi-Bred. The alleles rth3-3 and rth3-4 were obtained from the 62 100 progeny of a directly tagged population using rth3-1/rth3-1 as males crossed to females from highly active Mutator stocks (Rth3/Rth3 Mu X rth3-1/rth3-1).
Genetic mapping of the rth3 gene
After the initial B–A translocation mapping of rth3 to the short arm of chromosome 1 (Wen and Schnable, 1994), a higher-resolution mapping experiment was conducted using phenotypic markers. A stock carrying the phenotypic markers, sr1 (striate 1), which displays a pale green leaf phenotype, and P1-rr (pericarp color1) which regulates the synthesis of a red phlobaphene pigment in maize floral organs, with suffix rr indicating red pericarp and cob color, was crossed to rth3-1/rth3-1 plants with green leaves and a white pericarp and cob (P1-ww) (sr1 Rth3 P1-rr/sr1 Rth3 P1-ww× Sr1 rth3-1 P1-ww/Sr1 rth3-1 P1-ww). Progenies of this cross were backcrossed to both parent lines (sr1 Rth3 P1-rr (or P1-ww)/Sr1 rth3-1 P1-ww × Sr1 rth3-1 P1-ww/Sr1 rth3-1 P1-ww and sr1 Rth3 P1-rr (or P1-ww)/Sr1 rth3-1 P1-ww× sr1 Rth3 P1-rr/sr1 Rth3 P1-ww). The pale-green-leaf phenotype of sr1 was scored for each progeny of these crosses 1 month after germination in the field and the rth3 and p1 genotypes were identified from scoring the root hair phenotypes and the pigmentation of the pericarp and cob of their selfed-pollinated ears, respectively (Coe, 1994). In addition, the molecular markers npi286 and ias8 were used as to genotype Ben Burr's recombinant inbred lines (Burr and Burr, 1991; Burr et al., 1988).
The AIMS system to clone rth3
The rth3 gene was cloned using the PCR-based AIMS system as described in Frey et al. (1998). Homo- or heterozygosity of the analyzed plants was confirmed by selfing and subsequent segregation analysis of the progeny.
RNA gel blot analyses
Ribonucleic acid was isolated from 2-week-old leaves and roots by a phenol/SDS method (Ausubel et al., 1994). An oligo (dT)-cellulose column (Molecular Research Center, http://www.mrcgene.com/) was used to purify mRNA. Ten micrograms of mRNA per sample was subjected to electrophoresis and transferred to nylon membranes. Transfer, crosslinking, probe labeling, pre-hybridization, hybridization and post-hybridization washes of RNA gel blots were conducted as previously described (Stinard et al., 1993).
Survey of the massively parallel signature sequencing database
The Solexa MPSS technology (Brenner et al., 2000) allows for the quantification of 17-bp signature sequences beginning with the nucleotides GATC in populations of 2 × 105 to 2 × 106 cDNAs of a defined developmental stage of an organ. These 17-bp signature sequences almost always correspond to unique cDNAs by direct sequence matching, thus allowing for the quantification of the abundance of a particular cDNA in a sample representing a particular organ and developmental stage. The MPSS data were normalized and filtered according to the Solexa protocol.
In situ hybridization experiments
Five-day-old primary root samples were fixed at 4°C overnight in 4% formaldehyde in phosphate-buffered saline, dehydrated in an ethanol series and embedded in paraffin wax (Paraplast Plus; McCormick, http://www.mccormickscientific.com/). Embedded roots were sectioned using a Leica RM 2035 rotary microtome (http://www.leica.com/) and mounted on Super-Frost Plus slides (Menzel GmbH; http://www.mendel.de/). The template for the antisense rth3 in situ probe was amplified with primers RTH3F_as (5′-ACATGCGCGGGCCCCACTTTACAAGCG-3′) and RTH3RT7 (5′-GGGGGGTAATACGACTCACTATAGGGGTCGTCGCCCTCT-GCGC-3′) and corresponds to positions 19 to 668 of GenBank accession AY265855 followed by a T7 promoter site. The template for the sense probe was amplified accordingly using primers RTH3FT7 (5′-GGGGGGTAATACGACTCACTATAGGG-CTCTCCTCGGCACATGC-3′) and RTH3R_se (5′-CAGCTGGATGCCGTTGCAGCC-3′) with the T7 promoter preceding the rth3 sequence. Digoxigenin-labeled RNA probes were transcribed using T7 RNA polymerase (Roche, http://www.roche.com/) according to the manufacturer's instructions. The RNA in situ hybridizations were performed according to Jackson (1992). Nomarski images were taken using a Zeiss Axioplan 2 microscope (http://www.zeiss.com/) in combination with a Photometrics Cool Snap camera (Roper Scientific GmbH, http://www.roperscientific.com/).
Sequence analysis and phylogeny
Glycosylphosphatidylinositol-anchor prediction was performed with big-PI Predictor software (http://mendel.imp.ac.at/gpi/gpi_server.html; Eisenhaber et al., 1999). The signal peptide was predicted with WoLF PSORT software (http://wolfpsort.org/; Horton et al., 2006). The hydropathy plot according to Kyte and Doolittle (1982) was generated with ProtScale software (http://expasy.org/tools/protscale.html; Gasteiger et al., 2005). Cellulose-binding domain analysis was performed with Superfamily 1.69 software (http://supfam.mrc-lmb.cam.ac.uk/SUPERFAMILY/hmm.html; Gough et al., 2001). Prediction of N-glycosylation sites was performed via the NetNGlyc 1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc/). Phylogenetic analysis of the monocot-specific clade of COBRA-like proteins was performed as described in Zimmermann and Werr (2005). Pairwise alignment of the monocot-specific COBRA-like proteins for evaluation of sequence identity was performed with the MegAlign algorithm of the DNAstar software package (DNAstar Inc., http://www.dnastar.com/) via the Lipman–Pearson method.
Grain yield tests
Grain yield (quintal/hectare) of wild type was compared to that of the rth3 mutant in three different experiments in Iowa, Kansas and Nebraska over 2 years (1994 and 1997). Yield plots were planted at a density of approximately 69 000 plants ha−1 in two rows per plot in a split-plot complete random design and replicated at least three times at all locations. The experiments were conducted with no visible water stress, and pests and weeds were adequately controlled throughout the season. Selfed progeny from rth3-1/Rth3 heterozygotes were again self-pollinated. The root hair phenotypes of approximately 15 kernels from each twice selfed family were examined as described (Wen and Schnable, 1994). Families that did not segregate for root hair mutants (Rth3/Rth3) and those that did not segregate for non-mutants (rth3-1/rth3-1) were separately pooled to comprise the mutant and wild-type controls used in the yield tests. To ensure stability of the mutant phenotype in field-grown plants, the root clumps and brace root samples from several randomly chosen mutant and wild-type plants near flowering time were excavated from several observation plots and gently washed with tap water. Small sections of above-ground brace roots, below-ground primary and lateral roots were placed in sterile water in Petri dishes and examined stereoscopically for evidence of root hairs.
Acknowledgments
Supported by a grant from Pioneer Hi-Bred International, Inc. to PSS, State of Iowa and Hatch Act funds, and grants to FH from the Deutsche Forschungsgemeinschaft (DFG) (HO2249/4, HO2249/6 and the SFB446 project B16).
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley and Sons; 1994. [Google Scholar]
- Brady SM, Song S, Dhugga KS, Rafalski JA, Benfey PN. Combining expression and comparative evolutionary analysis. The COBRA gene family. Plant Physiol. 2007;143:172–187. doi: 10.1104/pp.106.087262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S, Johnson M, Bridgham J, et al. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- Brown DM, Zeef LAH, Ellis J, Goodacre R, Turner SR. Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell. 2005;17:2281–2295. doi: 10.1105/tpc.105.031542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr B, Burr FA. Recombinant inbreds for molecular mapping in maize: theoretical and practical considerations. Trends Genet. 1991;7:55–60. doi: 10.1016/0168-9525(91)90232-F. [DOI] [PubMed] [Google Scholar]
- Burr B, Burr FA, Thompson KH, Albertson MC, Stuber CW. Gene mapping with recombinant inbreds in maize. Genetics. 1988;118:519–526. doi: 10.1093/genetics/118.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita N, McCann M. The cell wall. In: Buchanan B, Gruissem W, Jones R, editors. Biochemistry and Molecular Biology of Plants. Rockville, MD: American Society of Plant Physiologists; 2000. pp. 52–109. [Google Scholar]
- Ching A, Dhugga KS, Appenzeller L, Meeley R, Bourett TM, Howard RJ, Rafalski A. Brittle stalk 2 encodes a putative glycosylphosphatidylinositol-anchored protein that affects mechanical strength of maize tissues by altering the composition and structure of secondary cell walls. Planta. 2006;224:1174–1184. doi: 10.1007/s00425-006-0299-8. [DOI] [PubMed] [Google Scholar]
- Ciamporova M, Dekankova K, Hanackova Z, Peters P, Ovecka M, Baluska F. Structural aspects of bulge formation during root hair initiation. Plant and Soil. 2003;255:1–7. [Google Scholar]
- Cobbett CS. Phytochelatines and their roles in heavy metal detoxification. Plant Physiol. 2000;123:825–832. doi: 10.1104/pp.123.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe E. Genetic experiments and mapping. In: Freeling M, Walbot V, editors. The Maize Handbook. New York: Springer; 1994. pp. 189–197. [Google Scholar]
- Cui X, Hsia A-P, Liu F, Ashlock DA, Wise RP, Schnable PS. Alternative transcription initiation sites and polyadenylation sites are recruited during Mu suppression at the rf2a locus of maize. Genetics. 2003;163:685–698. doi: 10.1093/genetics/163.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L, Duckett CM, Grierson C, Linstead P, Schneider K, Lawson E, Dean C, Poethig S, Roberts K. Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development. 1994;120:2465–2474. [Google Scholar]
- Eisenhaber B, Bork P, Eisenhaber F. Prediction of potential GPI-modification sites in proprotein sequences. J. Mol. Biol. 1999;292:741–758. doi: 10.1006/jmbi.1999.3069. [DOI] [PubMed] [Google Scholar]
- Foreman J, Demidchik V, Bothwell JH, Mylona P, Miedema H, Torres MA, Linstead P, Costa S, Brownlee C, Jones JD, Davies JM, Dolan L. Nature. Vol. 422. 2003. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth; pp. 442–446. [DOI] [PubMed] [Google Scholar]
- Frey M, Stettner C, Gierl A. A general method for gene isolation in tagging approaches: amplification of insertion mutagenised sites (AIMS) Plant J. 1998;13:717–722. [Google Scholar]
- Gale MD, Devos KM. Plant comparative genetics after 10 years. Science. 1998;282:656–659. doi: 10.1126/science.282.5389.656. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The Proteomics Protocols Handbook. Totowa: Humana Press; 2005. pp. 571–607. [Google Scholar]
- Gilroy S, Jones DL. Through form to function: root hair development and nutrient uptake. Trends Plant Sci. 2000;5:56–60. doi: 10.1016/s1360-1385(99)01551-4. [DOI] [PubMed] [Google Scholar]
- Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Park WJ, Sauer M, Woll K. From weeds to crops: genetic analysis of root development in cereals. Trends Plant Sci. 2004a;9:42–48. doi: 10.1016/j.tplants.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Woll K, Sauer M, Dembinsky D. Genetic dissection of root formation in maize (Zea mays) reveals root-type specific developmental programs. Annals Bot. 2004b;93:359–368. doi: 10.1093/aob/mch056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Park K-J, Obayashi T, Nakai K. Protein Subcellular Localization Prediction with WoLFPSORT. Taipei: World Scientific Publishing Co. Pte. Ltd; 2006. pp. 39–48. [Google Scholar]
- Jackson D. In situ hybridization in plants. In: Gurr SR, McPherson MJ, Bowles DJ, editors. Molecular Plant Pathology: A Practical Approach. Oxford: Oxford University Press; 1992. pp. 163–174. [Google Scholar]
- Jones MA, Raymond MJ, Smirnoff N. Analysis of the root-hair morphogenesis transcriptome reveals the molecular identity of six genes with roles in root-hair development in Arabidopsis. Plant J. 2006;45:83–100. doi: 10.1111/j.1365-313X.2005.02609.x. [DOI] [PubMed] [Google Scholar]
- Kropf DL, Bisgrove SR, Hable WE. Differing roles of the cytoskeleton in intercalary growth and tip growth of plant cells. Curr. Opin. Cell Biol. 1998;10:117–122. doi: 10.1016/s0955-0674(98)80094-x. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle R. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Larkin JC, Brown ML, Schiefelbein J. How do cells know what they want to be when they grow up? Lessons from epidermal patterning in Arabidopsis. Annu. Rev. Plant. Biol. 2003;54:403–430. doi: 10.1146/annurev.arplant.54.031902.134823. [DOI] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J. Cell pattern in the Arabidopsis root epidermis determined by lateral inhibition with feedback. Plant Cell. 2002;14:611–618. doi: 10.1105/tpc.010434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuchter R, Wolf K, Zimmermann M. Isolation of an Arabidopsis cDNA complementing a Schizosaccharomyces pombe mutant deficient in phytochelatin synthesis. Plant Physiol. 1998;117:331–333. [Google Scholar]
- Li ZS, Lu YP, Zhen RG, Szczypka M, Thiele DJ, Rea PA. A new pathway for vacuolar cadmium sequestration in Saccharomyces cerevisiae: YCF1-catalyzed transport of bis(glutathionato)cadmium. Proc. Natl Acad. Sci. USA. 1997;94:42–47. doi: 10.1073/pnas.94.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Qian Q, Zhou Y, et al. BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell. 2003;15:2020–2031. doi: 10.1105/tpc.011775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan BA, Conlon FL, Manzanares M, Millar JBA, Kanuga N, Sharpe J, Krumlauf R, Smith JC, Sedgwick SG. Transposon tools for recombination DNA manipulation: characterization of transcriptional regulators from yeast, Xenopus, and mouse. Proc. Natl Acad. Sci. USA. 1996;93:2801–2806. doi: 10.1073/pnas.93.7.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Cavell AC, Dolan L, Robert K, Grierson CS. Genetic interactions during root hair morphogenesis in Arabidopsis. Plant Cell. 2000;12:1961–1974. doi: 10.1105/tpc.12.10.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Wei H, Milne J, Page GP, Somerville CR. Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc. Natl Acad. Sci. USA. 2005;102:8633–8638. doi: 10.1073/pnas.0503392102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigas S, Debrosses G, Haralampidis K, Vicente-Aqullo F, Feldman KA. TRP1 encodes a potassium transporter required for tip growth in Arabidopsis root hairs. Plant Cell. 2001;13:139–151. doi: 10.1105/tpc.13.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier F, Schindelman G, DeSalle R, Benfey PN. The COBRA family of putative GPI-anchored proteins in Arabidopsis. A new fellowship in expansion. Plant Physiol. 2002;130:538–548. doi: 10.1104/pp.007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier F, Fernandez AG, Fujita M, et al. COBRA, an Arabidopsis extracellular glycosyl-phosphatidyl inositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell. 2005;17:1749–1763. doi: 10.1105/tpc.105.031732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Row HC, Reeder JR. Root-hair development as evidence of relationships among genera of gramineae. Am. J. Bot. 1957;44:596–601. [Google Scholar]
- Schindelman G, Morikami A, Jungm J, Baskin TI, Carpita NC, Derbyshire P, McCann MC, Benfey PN. COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 2001;15:1115–1127. doi: 10.1101/gad.879101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieberer B, Emons AMC. Cytoarchitecture and pattern of cytoplasmic streaming in root hairs of Medicago truncatula during development and deformation by nodulation factors. Protoplasma. 2000;214:118–127. [Google Scholar]
- Stinard PS, Robertson DS, Schnable PS. Genetic isolation, cloning, and analysis of a Mutator-induced, dominant antimorph of the maize amylose extender1 locus. Plant Cell. 1993;5:1555–1566. doi: 10.1105/tpc.5.11.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taramino G, Sauer M, Stauffer J, Multani D, Niu X, Sakai H, Hochholdinger F. The rtcs gene in maize (Zea mays L.) encodes a lob domain protein that is required for postembryonic shoot-borne and embryonic seminal root initiation. Plant J. 2007;50:649–659. doi: 10.1111/j.1365-313X.2007.03075.x. [DOI] [PubMed] [Google Scholar]
- Udenfriend S, Kodukula K. How glycosyl-phosphatidylinositol-anchored membrane-proteins are made. Annu. Rev. Biochem. 1995;64:563–591. doi: 10.1146/annurev.bi.64.070195.003023. [DOI] [PubMed] [Google Scholar]
- Wang H, Lockwood SK, Hoeltzel MF, Schiefelbein JW. The ROOT HAIR DEFECTIVE3 gene encodes an evolutionarily conserved protein with GTP-binding motifs and is required fro regulated cell enlargement in Arabidopsis. Genes Dev. 1997;11:799–811. doi: 10.1101/gad.11.6.799. [DOI] [PubMed] [Google Scholar]
- Wasteneys GO, Galway ME. Remodeling the cytoskeleton for growth and form: an overview with some new views. Annu. Rev. Plant. Biol. 2003;54:691–722. doi: 10.1146/annurev.arplant.54.031902.134818. [DOI] [PubMed] [Google Scholar]
- Wen TJ, Schnable PS. Analyses of mutants of three genes that influence root hair development in Zea mays (Gramineae) suggest that root hairs are dispensable. Am. J. Bot. 1994;81:833–842. [Google Scholar]
- Wen TJ, Hochholdinger F, Sauer M, Bruce W, Schnable PS. The roothairless1 gene of maize encodes a homolog of sec3, which is involved in polar exocytosis. Plant Physiol. 2005;138:1637–1643. doi: 10.1104/pp.105.062174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann R, Werr W. Pattern formation in the monocot embryo as revealed by NAM and CUC3 orthologues from Zea mays L. Plant Mol. Biol. 2005;58:669–685. doi: 10.1007/s11103-005-7702-x. [DOI] [PubMed] [Google Scholar]