Abstract
Natural variation of plant pathogen resistance is often quantitative. This type of resistance can be genetically dissected in quantitative resistance loci (QRL). To unravel the molecular basis of QRL in potato (Solanum tuberosum), we employed the model plant Arabidopsis thaliana for functional analysis of natural variants of potato allene oxide synthase 2 (StAOS2). StAOS2 is a candidate gene for QRL on potato chromosome XI against the oömycete Phytophthora infestans causing late blight, and the bacterium Erwinia carotovora ssp. atroseptica causing stem black leg and tuber soft rot, both devastating diseases in potato cultivation. StAOS2 encodes a cytochrome P450 enzyme that is essential for biosynthesis of the defense signaling molecule jasmonic acid. Allele non-specific dsRNAi-mediated silencing of StAOS2 in potato drastically reduced jasmonic acid production and compromised quantitative late blight resistance. Five natural StAOS2 alleles were expressed in the null Arabidopsis aos mutant under control of the Arabidopsis AOS promoter and tested for differential complementation phenotypes. The aos mutant phenotypes evaluated were lack of jasmonates, male sterility and susceptibility to Erwinia carotovora ssp. carotovora. StAOS2 alleles that were associated with increased disease resistance in potato complemented all aos mutant phenotypes better than StAOS2 alleles associated with increased susceptibility. First structure models of ‘quantitative resistant’ versus ‘quantitative susceptible’ StAOS2 alleles suggested potential mechanisms for their differential activity. Our results demonstrate how a candidate gene approach in combination with using the homologous Arabidopsis mutant as functional reporter can help to dissect the molecular basis of complex traits in non model crop plants.
Electronic supplementary material
The online version of this article (doi:10.1007/s00425-008-0737-x) contains supplementary material, which is available to authorized users.
Keywords: Black leg, Jasmonic acid, Late blight, Natural variation, Potato, Quantitative resistance, Soft rot
Introduction
Potato (Solanum tuberosum L.) cultivation suffers severe economic losses worldwide due to the destructive diseases late blight (caused by the Irish potato famine pathogen, the oömycete Phytophthora infestans) and stem black leg/tuber soft rot (resulting from bacterial infection by Erwinia carotovora ssp. atroseptica; Eca) (Kamoun 2001; Toth and Birch 2005). Genetic dissection of resistance to late blight has identified a number of R genes as well as quantitative resistance loci (QRL), whereas for resistance to Eca only QRL are known to date (reviewed in Gebhardt and Valkonen 2001). Quantitative disease resistance is polygenic, less pathogen race-specific and therefore considered more durable than R genes—mediated resistance in the field. To make the most powerful use of quantitative disease resistance for crop improvement via marker-assisted selection, identification of the genes and their natural alleles that cause resistance variation is necessary.
The potato is a difficult target for positional cloning of QTL. Tetrasomic inheritance and inbreeding depression in tetraploid potatoes, and self-incompatibility in diploid potatoes hamper generation and phenotypic analyses of homozygous plants required for ‘Mendelizing’ the QTL targeted for cloning (Mullins et al. 2006). An alternative strategy focuses on candidate genes that plausibly play a relevant role in the process under investigation, genetically co-localize with QTL for this trait and exist as natural allelic variants. Due to limited QTL mapping resolution and the presence of multiple physically linked genes in the region, functional analyses of the natural alleles are required to validate the candidate gene’s role (Weigel and Nordborg 2005). Functional comparison of natural alleles is facilitated when using a null mutant of the candidate gene’s functional homolog in a model organism for comparative complementation analysis (Fridman et al. 2004).
Candidates for underlying plant QRL are genes encoding proteins that function in various steps of plant–pathogen interactions, starting with pathogen recognition, followed by biosynthesis of defense signaling compounds and components of signal transduction pathways and ending with the synthesis of defense response molecules (Hammond-Kosack and Parker 2003). The wealth of knowledge gained from the model plant Arabidopsis thaliana (L.) Heynh provides a rich source of functionally characterized defense-related candidate genes. In a previous study we localized homologs of Arabidopsis defense genes on potato molecular maps (Pajerowska et al. 2005). Among others, the StAOS2 locus was identified as positional candidate. StAOS2 is located within a QRL region for P. infestans and Eca on chromosome XI detected in earlier mapping experiments (Oberhagemann et al. 1999; Zimnoch-Guzowska et al. 2000). The co-linear genomic region in the closely related tomato (S. lycopersicum) genome also harbors a QRL for P. infestans (Brouwer et al. 2004). StAOS2 encodes a potato homolog of the Arabidopsis allene oxide synthase (AOS), a member of the cytochrome P450 superfamily (CYP74A), which catalyzes the conversion of 13(S)-hydroperoxy-9(Z),11(E),15(Z)-octadecatrienoic acid [13(S)-HPOTE] to an unstable allene oxide ([12,13(S)-epoxy-9(Z),11,15(Z)-octadecatrienoic acid) (Liechti and Farmer 2002). The following reactions in the 13-lipoxygenase (13-LOX) pathway lead to the production of the signaling molecule jasmonic acid (JA) and its biologically active precursor 12-oxo-phytodienoic acid (OPDA) (Liechti and Farmer 2002).
Jasmonic acid is structurally related to mammalian inflammatory mediators, eicosanoids, and plays an important role in various plant stress responses. JA is required for insect resistance (McConn et al. 1997; Kessler et al. 2004) and induces expression of plant antimicrobial genes (Penninckx et al. 1996; Thomma et al. 1998). JA plays a major role in defense against E. carotovora ssp. carotovora in Arabidopsis (Norman-Setterblad et al. 2000). JA application inhibits sporangial germination and mycelial growth of P. infestans on potato (Cohen et al. 1993), while JA-deficient tomato plants are more susceptible to P. infestans than wild-type (Thaler et al. 2004). However, precise information on the genetic basis of potato resistance to P. infestans and Eca through JA signaling is lacking.
Besides StAOS2, the potato genome contains two additional members of the StAOS family, StAOS1 and StAOS3, which have been cloned and genetically mapped (Pajerowska et al. 2005). Unlike StAOS2, they did not localize within a map segment harboring known QRL. Based on EST data, StAOS1 and StAOS2 share high sequence homology and similar domain architecture (TIGR TGI database, ESTs no. NP451990 and TC128063 for StAOS1 and StAOS2, respectively). Studies on tomato AOS1 identified its affinity towards 13-hydroperoxylinolenic acid, similarly to StAOS2 and Arabidopsis AOS, and in contrast to SlAOS3 (a member of the CYP74C subgroup), which preferentially processes 9-hydroperoxylinolenic acid (Howe et al. 2000; Itoh et al. 2002). Moreover, StAOS3 has recently been found to be expressed exclusively in below-ground potato organs and involved in the 9-LOX pathway, not leading to JA production (Stumpe et al. 2006). Based on the available information on function and genomic position of AOS genes in potato and other plants, we selected StAOS2 on potato chromosome XI for further structural and functional analysis of natural variation. For comparing natural potato StAOS2 alleles at the functional level, we used an Arabidopsis aos knock-out mutant for quantitative complementation analysis. The general value of this approach is that it allows to study functional variation of candidate genes for complex traits from plant species, in which quantitative complementation analysis is difficult or impossible to perform. Such plants are polyploids with multiple alleles (e.g., potato, sugarcane), slow growing plants (shrubs and trees), self-incompatible plants (e.g., diploid potato) or plants difficult to transform fast and efficiently (e.g., sugar beet, legumes).
Materials and methods
See Supplemental text for details of the StAOS2 linkage analysis, dsRNAi-mediated silencing of StAOS2, Arabidopsis aos mutant complementation with StAOS2 alleles, wounding treatments, JA and OPDA extractions, RT-PCR and Q-RT-PCR, analysis of male fertility-related traits, and molecular modeling.
Cloning and sequencing of StAOS2 alleles
Two different forward primers were used to clone full-length StAOS2 alleles. Alleles StAOS2-1, StAOS2-6 and StAOS2-7 were cloned using StAOS2-F-A: 5′(GWF)taatggcattaacttcatctttttc3′ and StAOS2-R: 5′(GWR)cagcttttttcaaagaagttatag3′, while StAOS2-8 and StAOS2-12 were cloned with the primers: StAOS2-F-B:5′(GWF)taatggctttaacttcatttttttc3′ and StAOS2-R. Proofreading polymerase chain reactions and Gateway® recombination reactions were carried out as described (Pajerowska et al. 2005). PCR products were cloned into the pDONR201 Gateway® vector (Invitrogen) and sequenced on both strands. A consensus sequence for each allele was deduced from 10 independent clones sequenced per construct.
Silencing StAOS2 in potato genotype G87
Three StAOS2 fragments were amplified by PCR using the specific primers (for details see Supplemental text). As a negative control, artificial sequence (Synthetic GeneAmplimer pAW109 RNA purchased from PerkinElmer, Wellesley, MA, USA) was used. The PCR fragments were cloned into pJawohl17, a binary Gateway®-compatible vector designed to produce double-stranded RNA in plants (B. Ülker, Durham University, School of Biological and Biomedical Sciences, Durham DH1 3LE, UK, personal communication). A. tumefaciens-mediated transformation of G87 was performed as described (Ballvora et al. 2002). Kanamycin-resistant transgenic plants and untransformed G87 were analyzed by semi-quantitative RT-PCR for StAOS2 transcript abundance. Transcripts were quantified by scanning ethidium bromide stained agarose gels on a PhosphoImager Typhoon 8600 and analyzing the images with ImageQuant 5.2 software (Molecular Dynamics). StAOS2 transcript levels were calibrated against the tubulin loading controls. Measurements were repeated three times on two independent biological replicates.
Phytophthora infestans infections
Three detached leaflets per genotype in each test were inoculated as described (Oberhagemann et al. 1999) with two 10 μl droplets of sporangial suspension (30–40 sporangia/μl) on both sides of the midrib. Disease symptoms were scored 7 dpi. DNA was extracted from infected plant material, pooling the leaflets of each genotype. Pathogen growth was monitored using P. infestans—ribosomal DNA specific primers (Judelson and Tooley 2000): Pinf-F 5′gaaaggcatagaaggtaga3′ and Pinf-R 5′taaccgaccaagtagtaaa3′). Intensities of P. infestans-specific amplicons were calibrated relative to potato tubulin DNA bands. Band intensities were quantified using GelDoc software (Bio-Rad, Munich, Germany) and converted in arbitrary units relative to the absolute values obtained from control G87 plants (=1 unit). The experiment was repeated three times with similar results.
Erwinia carotovora ssp. carotovora infections
Ecc strain WPP14 cultures (Yap et al. 2004) were grown in King’s B (KB) medium supplemented with 50 μg/ml rifampicin overnight and their concentration was determined spectrophotometrically. Bacterial cultures were washed and resuspended in 10 mM MgCl2 for in planta infiltrations. Culture containing 5 × 104 colony-forming units (OD600 nm = 0.000125) was infiltrated by syringe into three to four fully expanded leaves of soil-grown 3-week-old Arabidopsis plants (three independent transgenic lines per construct, 16 different plants per line/genotype). After bacterial infiltration, plants were maintained at 23°C and 100% RH for 3 days. Eight leaves were collected at 0, 24 and 72 h post-infiltration and the bacterial population was estimated by sampling two leaf discs (0.3 cm2 each), tissue grinding and colony plating onto KB medium supplemented with 50 μg/ml rifampicin. The experiment was repeated three times with similar results. For the graph shown in Fig. 5c, bacterial growth data from all three experiments were pooled.
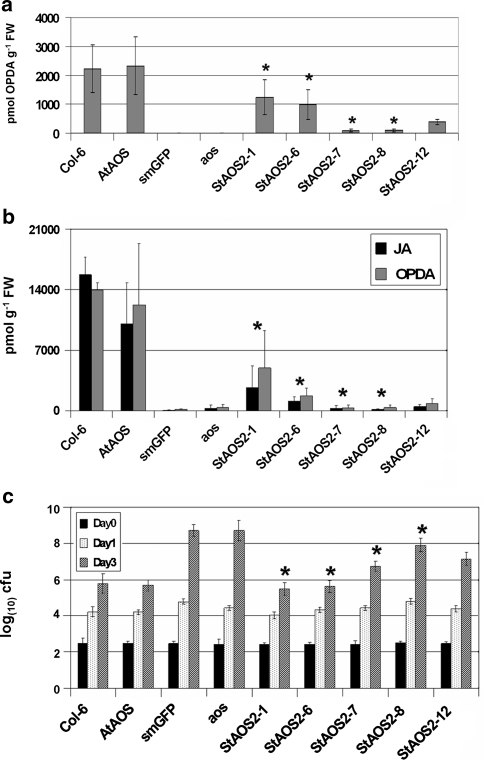
Fig. 5.
Basal and wound-induced OPDA and JA levels and resistance to Erwinia carotovora ssp. carotovora in Arabidopsis controls and StAOS2 allele complementation lines. a Basal levels of OPDA in controls (Col-6, AtAOS, aos, smGFP) and StAOS2 allele-specific complementation lines. b OPDA and JA levels 1.5 h after wounding in the same lines as in a. JA content differed ~20-fold between QR allele StAOS2-1 and QS allele StAOS2-8 of the ‘Erwinia’ population, and ~5 fold between QR allele StAOS2-6 and QS allele StAOS2-7 of the GC population (nested ANOVA, P < 0.005 for JA and P < 0.05 for OPDA content). FW fresh weight. *Significant in nested ANOVA (StAOS2-1 vs. StAOS2-8, StAOS2-6 vs. StAOS2-7). Error bars represent standard deviations of three independent measurements. c Growth (log scale of colony forming units = cfu) of Erwinia carotovora ssp. carotovora strain WPP14 at 0, 1 and 3 days post infection in the same lines as in a. Plants were inoculated with bacterial suspension at OD600 nm = 0.000125. Each data point represents the average cfu from 72 leaf discs representing three independent transgenic lines and three independent infection experiments, with error bars indicating 95% confidence intervals. *Significant in nested ANOVA (StAOS2-1 vs. StAOS2-8, StAOS2-6 vs. StAOS2-7) (P < 0.01). Bacterial growth was slower in plants expressing QR alleles StAOS2-1 or StAOS2-6 when compared to plants expressing QS alleles StAOS2-7 or StAOS2-8
Arabidopsis aos mutant complementation with StAOS2 alleles
Complementation constructs of StAOS2 alleles, AtAOS and smGFP were generated using MultiSite Gateway® Three-Fragment Vector Technology (Invitrogen). The AtAOS promoter and AtAOS gene were amplified from genomic DNA of Arabidopsis thaliana L. ecotype Col-6 (N8155, obtained from the Nottingham Arabidopsis Stock Centre) wild-type plants. Constructs were confirmed by sequencing, and recombined into the MultiSite Gateway® Three-Fragment vector pAM-PAT Multi (Invitrogen). Arabidopsis aos mutant plants (Park et al. 2002) were transformed by the inflorescence dipping method (Clough and Bent 1998) and transformants were selected on 0.01% Basta. Starting 3 days before transformation, flowers of aos plants were repeatedly sprayed with a fresh solution of 2 mM jasmonic acid (Sigma) with addition of 0.02% (v/v) Silwet L-77, to restore male fertility and allow seed set in male sterile plants. Full details of the complementation assay are given in the Supplemental Text.
Molecular modeling
The StAOS2-1 sequence was used as query to identify suitable modeling templates. A blast from Swiss-PdbViewer (Guex et al. 1999; Guex and Peitsch 1997) identified three possible templates (1z11, 1z10, 1cpt) covering at most the residues 339–379. Using the A. thaliana CYP74A (SwissProt entry Q96242), which shares 64% identity with StAOS2-1, additional templates (1e9x, 1x8v, 1u13, 1h5z, 1ea1, 1w0q, 1w0e, 1tqn, 1w0f, 1t2b) were identified. Taken together, they allowed to place StAOS2-1 residues 337–481. To define the position of remaining residues, segments of StAOS2-1 were used as a seed to Psi-Blast (Altschul et al. 1997; Schaffer et al. 2001). JPred (Cuff et al. 1998) secondary structure prediction was then used to further narrow down regions used as seed for Psi-Blast. In addition, StAOS2-1, A. thaliana CYP74A and S. lycopersicum CYP74C4 (SWISSPROT entries Q96242 and Q8S4C5) sharing 64 and 53% identity, respectively, with StAOS2-1 and only 48% between themselves were submitted to the fold recognition server nFOLD2 (L.J. McGuffin, University of Reading, Whiteknights, Reading RG6 6AS, UK, personal communication) (http://www.biocentre.rdg.ac.uk/bioinformatics/nFOLD/). Three to four of the best predictions for StAOS2-1, Q96242, and Q8S4C5 were superposed with Swiss-PdbViewer. The structural alignment was compared with the unambiguous sequence alignment of StAOS2-1, Q96242, and Q8S4C5 to identify regions with consensus predictions. These were then systematically mapped onto structurally equivalent residues from B. megaterium CYP102 (1jpza). This structure was chosen as main template because its active site contains a molecule of palmitoyl-glycine, which was used to facilitate the approximate placement of 13(S)-HPOTE (13-hydroperoxyoctadecatrienoic acid). A summary of the final mapping used to build a crude Carbon-Alpha model of StAOS2-1 is shown in Supplementary Table S2. Full details of the molecular modeling are given in the Supplementary Text.
Results
Natural potato AOS2 alleles
First we examined whether StAOS2 exhibited allelic variation in two unrelated diploid potato mapping populations that had been instrumental for identifying QRL on the short arm of potato chromosome XI. One population (called ‘GC’) had been assessed in the field for quantitative resistance of foliage to complex races of P. infestans (Oberhagemann et al. 1999), whereas another population (known as ‘Erwinia’) had been evaluated for quantitative resistance of foliage and tubers to Eca (Zimnoch-Guzowska et al. 2000). Sequence analysis of a gene specific amplicon from within the StAOS2 gene in both mapping populations showed that all four parents were heterozygous for single nucleotide polymorphisms (SNPs) and that the SNPs segregating in the two F1 families were different, indicating presence of various StAOS2 alleles in the parents. We scored SNPs diagnostic for each of the eight parental alleles (four heterozygous parents with two alternative alleles each) in the ‘GC’ and ‘Erwinia’ families and confirmed their linkage to the QRL. Two alleles, designated StAOS2-1 and StAOS2-8, were descended from the maternal parent DG83 of the ‘Erwinia’ family and distinguished by a substitution C versus T at position 692 (SNP692). The SNP692 locus was more closely linked to the QRL for Eca in the ‘Erwinia’ population than flanking markers (Supplementary Table S1). Allele StAOS2-1 showed linkage to increased resistance to Eca whereas allele StAOS2-8 was linked to increased susceptibility. A second pair of alleles, named StAOS2-6 and StAOS2-7, were descended from the maternal parent G87 of the ‘GC’ family and diagnosed by a polymorphism A versus G at position 679 (SNP679). Allele StAOS2-6 was linked (Supplementary Table S1) to increased late blight resistance whereas StAOS2-7 was linked to increased susceptibility. The paternal StAOS2 alleles in both populations did not discriminate between QRL effects. Based on linkage to quantitative resistance or susceptibility, we classified the alleles into three categories: the ‘quantitative resistant’ (QR) StAOS2-1 and StAOS2-6, the ‘quantitative susceptible’ (QS) StAOS2-7 and StAOS2-8, and the indiscriminate paternal alleles (further referred to as ‘neutral’).
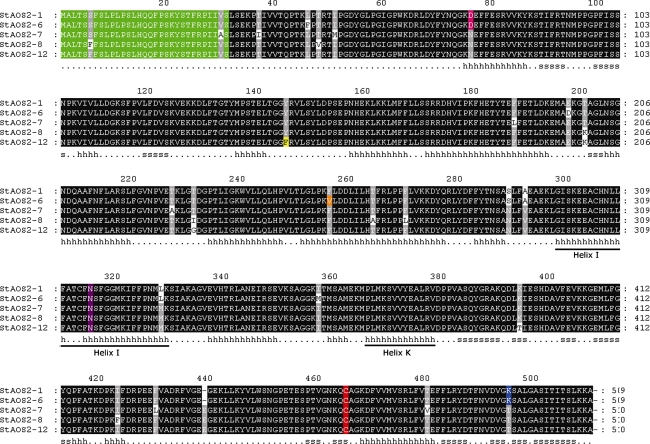
To gain insight into molecular variation of StAOS2, the full-length sequences of two QR alleles (StAOS2-1 and StAOS2-6), two QS alleles (StAOS2-7 and StAOS2-8) and one neutral allele StAOS2-12 (originating from the paternal parent of population ‘Erwinia’) were cloned by PCR from genomic DNA of carrier individuals of the ‘Erwinia’ or ‘GC’ populations and sequenced. All StAOS2 alleles were composed of a single exon coding for 509 or 510 amino acids and showed 97% sequence similarity to each other. The average SNP frequency in the coding sequence was one every 22 base pairs, which is in agreement with the polymorphism levels in potato reported previously (Rickert et al. 2002; Simko et al. 2006). Similarly, the observed frequency of synonymous versus non-synonymous mutations in the StAOS2 sequences (0.41) was comparable to the average for potato coding regions (0.42) (Simko et al. 2006). Deduced protein sequence alignments (Fig. 1) exposed variation at 25 amino acid positions and one insertion/deletion polymorphism (InDel) of a single amino acid. Five amino acid substitutions (D76N, S289N, A292V, L328M, K495T) and a deletion E439 (DelE439) were specific for the QR alleles, none was specific for the QS alleles and three substitutions (Y145F, T232I/G, K394T) occurred only in the neutral allele StAOS2-12.
Fig. 1.
Deduced amino acid sequences alignment of the StAOS2 alleles. Black-shaded boxes represent conserved amino acid residues. Chloroplast targeting signal peptide is indicated in green color. Conserved heme-binding C464 and catalytic residue N315 are highlighted in red and purple, respectively. Amino acid substitutions close to the enzyme catalytic site, D76N, K494T, Y145F and F256V, are shown in pink, blue, yellow, and orange, respectively. The secondary structure based on the predicted alignment to Bacillus megaterium CYP102 (1jpza) is indicated below the sequences (h helix, s strand). Helix I (the oxygen-binding pocket; amino acids 298–329) and Helix K (bearing the EXXR motif; amino acids 366–378) are underlined
Subcellular localization and function of StAOS2
A handful of known plant AOS enzymes are localized in chloroplasts (Vick and Zimmerman 1987; Laudert et al. 1996; Maucher et al. 2000; Froehlich et al. 2001). A putative chloroplast signal peptide of 31 amino acids was predicted in all StAOS2 alleles (Fig. 1). Heterologous and constitutive expression of full-length StAOS2 fused to the coding sequence of green fluorescent protein (GFP) in Nicotiana benthamiana leaves showed correct targeting to chloroplasts (Fig. 2). Recently, StAOS2 was also shown to be localized in potato chloroplasts using confocal microscopy of GFP-fused StAOS2 protein in A. thaliana suspension cells, as well as by chloroplast fractionation and Western blotting (Farmaki et al. 2007).
Fig. 2.
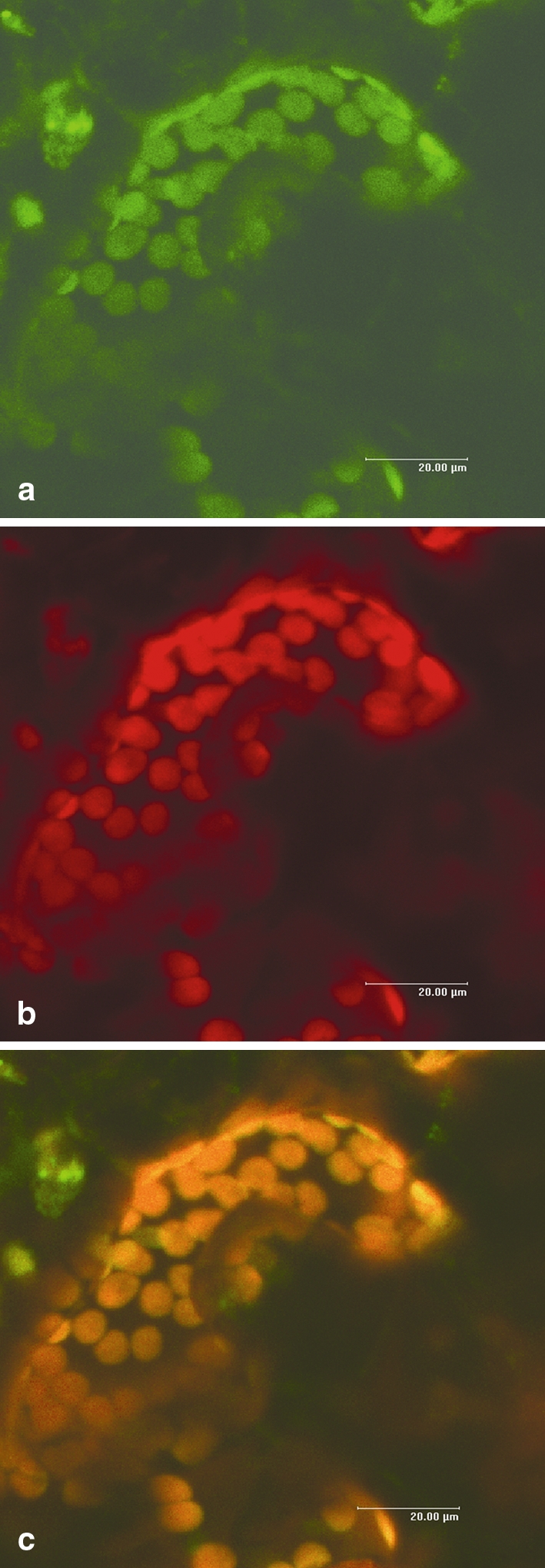
Subcellular localization of StAOS2. Confocal laser scanning microscope photographs of a N. benthamiana leaf epidermal guard cell transformed with the fusion construct 2x35S::StAOS2-GFP. a GFP fluorescence (emission spectrum 490–560 nm). b Chlorophyll auto-fluorescence (emission spectrum 650–700 nm). c Overlay of a and b, demonstrating perfect co-localization of StAOS2-GFP with chloroplasts
To demonstrate that potato StAOS2 encodes a protein with a similar function in JA biosynthesis as the Arabidopsis AOS (Park et al. 2002), we generated stable StAOS2-dsRNAi silencing lines in potato genotype G87. Since ESTs for both StAOS1 and StAOS2 genes were found in tissues derived from aerial parts of potato plants (TIGR TGI database), we designed the dsRNAi construct A (lines AOS-A1 and AOS-A2 in Fig. 3) that shares 79% sequence homology with StAOS1, and construct C (line AOS-C1 in Fig. 3) sharing only low sequence homology with StAOS1 and therefore being StAOS2-specific. Ten of 44 dsRNAi lines expressing AOS-A or AOS-C constructs accumulated no more than 10% StAOS2 transcript when compared to controls (untransformed G87 and G87 transformed with an artificial sequence-dsRNAi construct that does not share homology with any known plant gene). In contrast to the EST data, StAOS1 transcripts were not detectable in leaf tissue of wild-type G87 plants as well as in StAOS2-RNAi transgenic lines expressing silencing constructs AOS-A or AOS-C, possibly due to extremely low levels of StAOS1 mRNA in the plant material grown under the conditions tested (data not shown). Nevertheless, we selected for further analysis two independent StAOS2–RNAi lines carrying construct AOS-A and one line expressing construct AOS-C that contained the lowest StAOS2 transcript levels.
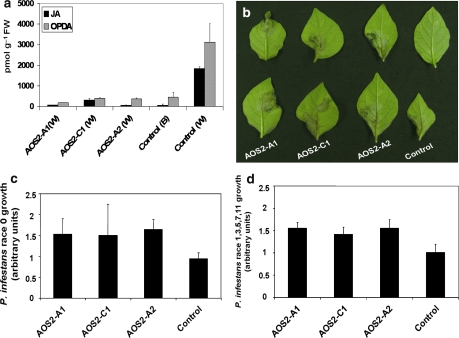
Fig. 3.
Phenotypes of StAOS2 silenced potato G87 plants. a JA and OPDA levels in Control (G87 expressing a synthetic gene RNAi construct) before [Control (B)] and 30 min after wounding [Control (W)], and in StAOS2-dsRNAi silenced lines AOS2-A1 (W), AOS2-C1 (W) and AOS2-A2 (W) 30 min after wounding. JA and OPDA are reduced to basal level in StAOS2 silenced plants (nested ANOVA, P < 0.002). b Leaflets of Control and StAOS2-dsRNAi silenced lines AOS2-A1, AOS2-C1 and AOS2-A2 seven days post infection (dpi) with P. infestans race 0. StAOS2 silenced plants are more susceptible showing lesions. Increased growth of P. infestans race 0 (c) and race 1,3,5,7,11 (d) at 7 dpi on StAOS2-dsRNAi silenced lines AOS2-A1, AOS2-C1 and AOS2-A2 compared to the Control (nested ANOVA, P < 0.004). Error bars represent standard deviations of three independent experiments
Since StAOS2 transcript levels and JA/OPDA do not increase during a compatible potato—P. infestans interaction (Weber et al. 1999; Gobel et al. 2002; TIGR SGEdb, study ID 62, probe STMCR05), we took advantage of wounding treatment that mimicks herbivory and is known to cause rapid up-regulation of AtAOS expression and induction of JA biosynthesis (Creelman et al. 1992). Plants of the three dsRNAi lines and control plants were wounded, and JA and OPDA were extracted and quantified. As shown in Fig. 3a, both compounds were barely detectable in unwounded plants, but rapidly and substantially increased in the controls after wounding. In contrast, wounded dsRNAi plants accumulated only ~3% JA and ~6% OPDA when compared to the wound-induced controls (nested ANOVA, P < 0.002). This provided direct evidence that StAOS2 is the functional potato equivalent of the AtAOS required for JA biosynthesis during the early wounding response.
Genotype G87 carries none of the 11 race specific R (resistance) genes that were introgressed into cultivated potato from its wild relative S. demissum (Wastie 1991), but does display quantitative disease resistance to complex races of P. infestans in the field (Oberhagemann et al. 1999). If jasmonate signaling via StAOS2 plays a role in the potato resistance response to late blight, then the reduction or elimination of StAOS2 transcripts in G87 is expected to result in increased disease susceptibility. To test this, we assessed detached leaves of the three dsRNAi lines with the lowest wound-induced JA and OPDA levels for quantitative resistance to P. infestans race 0 (virulent only on plants lacking all 11 S. demissum R genes) and race 1, 3, 5, 7, 11 (virulent on plants having the R genes R1, R3, R5, R7 and R11). After 1 week, a clear difference in disease progression was observed between StAOS2-silenced plants and the controls (Fig. 3b). Growth of P. infestans race 0 (Fig. 3c) and race 1, 3, 5, 7, 11 (Fig. 3d) was higher in all three StAOS2-dsRNAi lines when compared to controls (nested ANOVA, P < 0.004). This not only confirmed a functional role of StAOS2 in the potato resistance response to P. infestans infection, but also provided genetic evidence for the importance of jasmonate-mediated defenses against late blight. Moreover, we established that StAOS2, but not StAOS1, is required for jasmonates accumulation and restricting P. infestans growth in the G87 background.
Differential complementation of Arabidopsis aos mutant with StAOS2 alleles
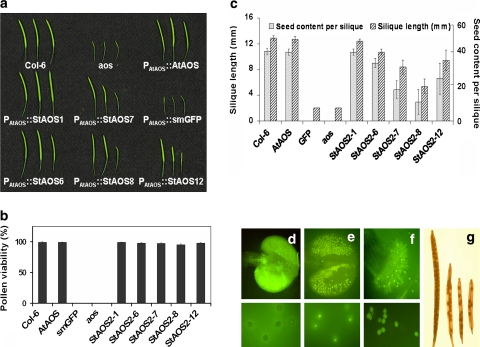
To examine the functional effects of natural variation in StAOS2 alleles, a transgenic complementation approach is required. In potato, however, the differential phenotypic effects of the transgenic alleles will need to be controlled for and separated from phenotypic effects of the endogenous alleles and genetic background, transgene position, dosage and consequently expression level, which might be of similar or even larger size than the transgene effect itself. To circumvent these limitations, we employed Arabidopsis as a plant host organism to quantitatively test the functionality of the StAOS2 alleles. We transformed the five StAOS2 alleles (StAOS2-1, StAOS2-6, StAOS2-7, StAOS2-8 and StAOS2-12) into an Arabidopsis aos null mutant, which contains a T-DNA insertion that knocks out the function of the single-copy AtAOS gene (At5g42650) and causes complete male sterility due to the block of JA biosynthesis (Park et al. 2002). The expression of all coding sequences, five potato StAOS2 alleles, the endogenous AtAOS gene (as a positive control) and the smGFP (soluble modified GFP) gene (as a negative control) was driven by the entire AtAOS 5′ regulatory region, 2.7 kb in length (P AtAOS). To eliminate position effects of T-DNA insertions on transgene expression levels that could influence the phenotype of the transgenic plants, StAOS2/AtAOS transcript levels were assayed by q-RT-PCR (Supplementary Fig. S1 and S2), and three independent T3 complementation lines per construct were selected on the basis of similar transcript accumulation detected in different StAOS2 and AtAOS transgenic plants. The shared regulatory element and the selection applied to transgenic plants ensured that differences observed could be attributed solely to the allelic variation in the StAOS2 coding sequence. No disease resistance-related phenotypes, but male sterility has been reported for the aos loss-of-function mutant plants (Park et al. 2002). The transgenic lines (three independent lines per construct, eight to ten plants per line) were therefore quantitatively scored for male fertility-related phenotypes and the values were subjected to statistical analyses. We observed differences in the restoration of silique length and seed set depending on the construct used for complementation (Fig. 4a). These differences were consistent among all individual plants representing each complementation construct. Construct P AtAOS ::AtAOS fully restored wild-type silique length, seed content per silique and pollen viability, whereas aos mutant and plants carrying P AtAOS ::smGFP remained sterile (Fig. 4b, c). All five P AtAOS ::StAOS2 constructs at least partially alleviated the aos mutant fertility-related phenotypes, demonstrating functionality of the StAOS2 alleles in Arabidopsis. However, the QR alleles StAOS2-1 and StAOS2-6 complemented the mutant phenotypes better than did the QS alleles StAOS2-7 and StAOS2-8. Silique length and seed content per silique of P AtAOS ::StAOS2-1 and P AtAOS ::StAOS2-6 were almost identical to those of P AtAOS ::AtAOS and Arabidopsis wild-type plants (Fig. 4a, c, Supplementary Fig. S3 and S4). Plants expressing the QS alleles (P AtAOS ::StAOS2-7 or P AtAOS ::StAOS2-8) developed siliques showing a size range from wild-type to the aos mutant on the same plant, with some siliques containing fewer than normal amounts of seed (Fig. 4g). This resulted in decreased seed yield when compared to plants expressing QR alleles (P < 0.001; Fig. 4c). No defect in pollen grain viability was found in any of the StAOS2 complementation lines, whereas the aos mutant and P AtAOS ::smGFP plants completely failed to develop vital pollen (Fig. 4b, d, Supplementary Fig. S5). We observed that P AtAOS ::StAOS2-1 and P AtAOS ::StAOS2-6 anthers, similarly to P AtAOS ::AtAOS and wild-type, dehisced just before or at the time of flower opening (Fig. 4f). However, breakage of stomium (future site of anther wall rupture and pollen release) was delayed in plants expressing QS alleles StAOS2-7 and StAOS2-8 (Fig. 4e), leaving the viable pollen grains trapped inside the indehiscent anther. Thus, aberrant anther dehiscence explained the large variation in silique length and seed content observed in the StAOS2 QS alleles complementation lines. Consistently in different phenotypic assessments, although not always significant, QR allele StAOS2-1 performed better than QR allele StAOS2-6. Plants carrying the neutral allele StAOS2-12 exhibited phenotypes intermediate between plants expressing QR and QS alleles (Fig. 4a, c).
Fig. 4.
Male fertility phenotypes of Arabidopsis aos complemented with StAOS2 alleles. a Siliques of P AtAOS ::StAOS2 allele complementation lines, P AtAOS ::AtAOS, P AtAOS ::smGFP, wild-type Col-6 and aos mutant plants. b Average pollen viability of wildtype (Col-6) and aos mutant complemented with AtAOS, GFP, potato QR alleles StAOS2-1, StAOS2-6, QS alleles StAOS2-6, StAOS2-8 or the neutral allele StAOS-12. c Average silique length and seed content per silique. d Indehiscent anther of aos mutant containing no viable pollen. e Indehiscent anther with mature pollen trapped inside, typical for lines complemented with QS alleles. f Col-6 stigma and fully viable pollen. g Differentially developed siliques of P AtAOS ::StAOS2-8, impaired in fertility
The role of JA in coordinating flower opening, stamen elongation, pollen maturation and anther dehiscence (Ishiguro et al. 2001) suggested that the differential complementation of the aos mutant by the QR and QS alleles was correlated with JA and OPDA content. Given that StAOS2 encodes an enzyme directly involved in JA biosynthesis, and since the Arabidopsis aos mutant completely lacks jasmonic acid (Park et al. 2002), quantification of both JA and OPDA provided the most sensitive assay for comparing the performance of the five StAOS2 alleles. JA and OPDA were measured in leaf tissue of Arabidopsis complementation lines, wild-type and aos mutant plants. Since the basal JA levels were barely detectable and could not be quantified reliably, we measured its precursor OPDA instead. As shown in Fig. 5a, endogenous OPDA levels in P AtAOS ::AtAOS and Arabidopsis wild-type plants reached ~2,200 pmol g−1 fresh weight, while OPDA was undetectable in the aos mutant and P AtAOS ::smGFP lines. Plants expressing QR alleles contained only half the amount of OPDA when compared to wild-type Arabidopsis (~1,100 pmol g−1 fresh weight; Fig. 5a). Evidently, this level was sufficient to rescue male sterile phenotype of aos, suggesting that a lower than Arabidopsis wild-type concentration of OPDA, and probably also JA, is adequate for complementation of reproductive traits. OPDA quantities were ~tenfold lower in plants expressing QS alleles than in plants expressing QR alleles (nested ANOVA, P < 0.05). After wounding, both JA and OPDA increased dramatically in wild-type and the P AtAOS ::AtAOS complementation lines, whereas the aos mutant and P AtAOS ::smGFP lines failed to accumulate both compounds (Fig. 5b). Under the same conditions, the QR alleles StAOS2-1 and StAOS2-6 mediated less OPDA and JA production than wounded wild-type plants, but contents were still ~tenfold higher than in plants expressing QS alleles (nested ANOVA, P < 0.005 for JA and P < 0.05 for OPDA content; Fig. 5b and Supplementary Fig. S6). As before, one of the QR alleles (StAOS2-1) performed better than the other (StAOS2-6) and the neutral allele StAOS2-12 produced intermediate levels of JA and OPDA. These results demonstrate the differential effect on JA and OPDA levels and therefore complementation of reproductive traits, when expressing either QR or QS StAOS2 alleles in the aos mutant background.
To correlate the differential JA and OPDA contents to levels of pathogen resistance, we infected wild-type plants, aos mutant and StAOS2 complementation lines with the necrotrophic bacterium Erwinia carotovora ssp. carotovora strain WPP14 (Ecc), closely related to Eca and virulent on both potato and Arabidopsis (Yap et al. 2004). Three days post inoculation, differential growth of Ecc was observed (Fig. 5c, Supplementary Fig. S7). The aos mutant and P AtAOS ::smGFP lines were highly susceptible to Ecc compared to wild-type and P AtAOS ::AtAOS plants. Bacterial growth in lines complemented with QR alleles P AtAOS ::StAOS2-1 and P AtAOS ::StAOS2-6 was similar to wild-type and significantly reduced when compared to lines complemented with QS alleles P AtAOS ::StAOS2-8 and P AtAOS ::StAOS2-7 (nested ANOVA, P < 0.01). This is consistent with the higher levels of OPDA and JA in plants complemented with QR StAOS2 alleles. Bacterial growth in the P AtAOS ::StAOS2-12 lines was intermediate between QR and QS alleles complementation lines. These experiments demonstrate that potato QR and QS StAOS2 alleles have a differential effect on resistance to Ecc in Arabidopsis.
Molecular modeling of StAOS2
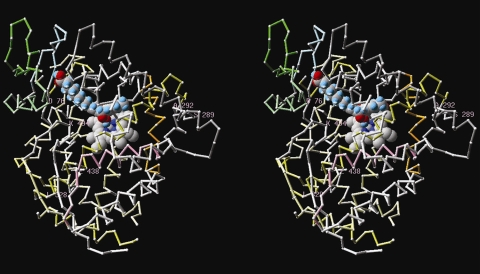
In an attempt to predict which amino acids of the StAOS2 protein are located near the crystallized cytochrome P450 active site and thus may impinge on the enzyme’s performance, we constructed the first molecular model for StAOS2 using information from various sequence-related, crystallized cytochrome P450 proteins (Fig. 6 and Supplementary Table S2). The model indicates that elements common to most CYP450 proteins are in place in StAOS2. We have identified the C464 (amino acid numbering according to the StAOS2-1 isoform) as the conserved cysteine that binds the iron atom of the heme group, and the common ExxR motif in helix K represented by E374–R377 that binds to the conserved R434. Finally, residues G298-K329 are predicted to correspond to Helix I bearing a [AG]-[AG]-X-X-T motif, in which the threonine has been postulated to play a role in substrate recognition and enzyme efficiency (Clark et al. 2006). In StAOS2-1, we propose that N315 plays a similar role based on the recently published structure of a coral AOS (Oldham et al. 2005) and full conservation of Asn in various plant cytochrome P450 sequences similar to StAOS2-1 (not shown). We focused our attention on the residues predicted to be located near the CYP450 active site and we noticed that the most likely candidate residues to explain the functional difference between the QR and QS alleles are K494T and D76N (Fig. 7a, b). In the QR alleles, K494 likely resides on the tip of the last beta-turn, which sits in the active site just above the heme, close to N315, although it is located outside the region that can be aligned with absolute confidence because of the large sequence variation among cytochromes. Interestingly, D76 is also predicted to be located in the same general region and could possibly contact K494. Hence K494 and D76 would be ideally situated to act in concert and directly affect substrate recognition or even catalysis (Fig. 7a, b), suggesting that both residues might be functionally relevant for the QR phenotype. Naturally occurring non-conservative changes of D76 by N and K494 by T would, in turn, affect the structure of the active site in the QS alleles and may reduce their substrate affinity (Fig. 7b). Although an influence of the other polymorphic residues cannot be excluded, they are all predicted to be located far from the active site (Fig. 6), usually in surface loops, and are unlikely to have impact on the enzymatic activity as much as the two candidate residues D76N and K494T. The model also showed that substitution F256V is the best candidate to explain the slightly inferior performance of QR allele StAOS2-6 versus StAOS2-1 based on its location relative to the substrate (data not shown). Moreover, the model indicated that the Y145F residue, one of three substitutions specific for the neutral allele StAOS2-12, is also located in a region adjacent to the active site (Fig. 7c) and might be responsible for the intermediate phenotypic performance of this allele.
Fig. 6.
3-D-structure of the StAOS2 protein in complex with heme and the substrate molecule. Steroscopic view of the predicted spatial localization of residues conserved within QR alleles StAOS2-1 and StAOS2-6, which differ in QS alleles StAOS2-7 and StAOS2-8. heme and the 13-(S)HPOTE (13-hydroperoxyoctadecatrienoic acid) substrate molecule placed manually based on the position occupied by the palmitoyl-glycine molecule present in Bacillus megaterium CYP102 structure (1jpza) appear in spacefilling. All positions are indicative only. Numbering is according to StAOS2-1; color scheme according to Table S2
Fig. 7.
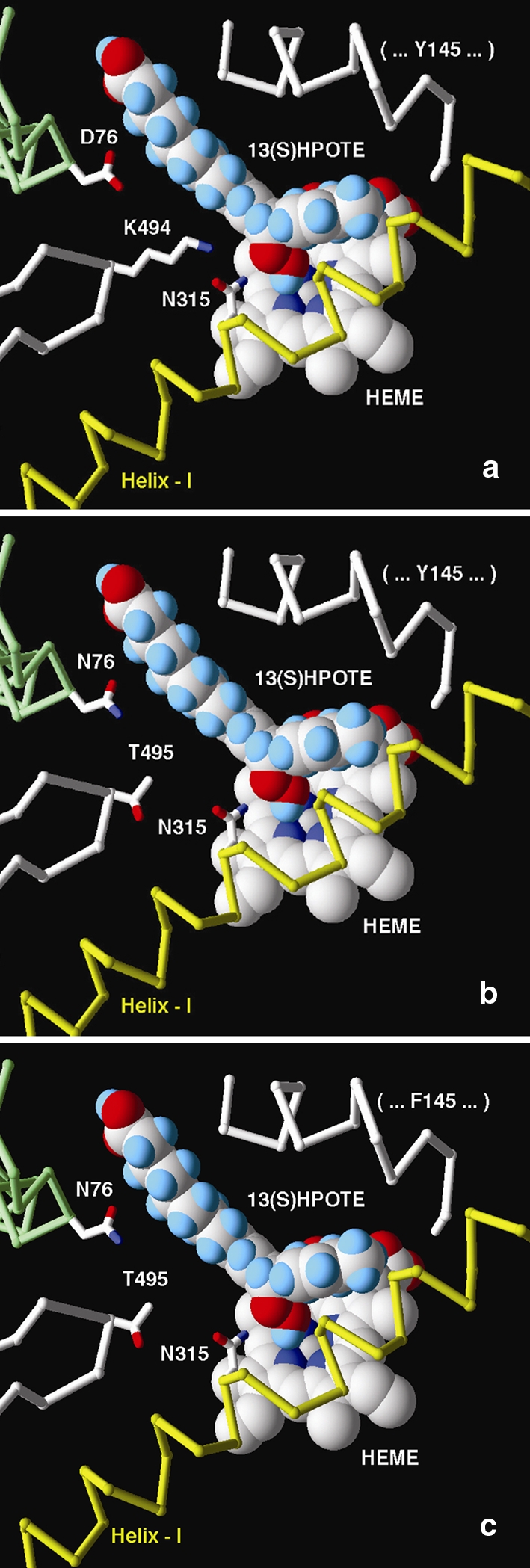
Rough allele models of the StAOS2 active site. a QR alleles StAOS2-1 and StAOS2-6; b QS alleles StAOS2-7 and StAOS2-8; c Neutral allele StAOS2-12. For clarity, only parts of the model and amino acid positions relevant for discussion are shown. The 13-(S)HPOTE (13-hydroperoxyoctadecatrienoic acid) substrate molecule was placed manually based on the position occupied by the palmitoyl-glycine molecule present in Bacillus megaterium CYP102 structure (1jpza). All positions are indicative only
Discussion
Our experiments corroborate the model of StAOS2 being one of the genes that contribute by natural variation to quantitative disease resistance in potato. Allele non-specific dsRNAi-mediated silencing of StAOS2 compromised JA biosynthesis and quantitative late blight resistance in transgenic potato. This confirmed that StAOS2 is a functional homolog of Arabidopsis AOS and participates in the resistance response to P. infestans, similar to tomato (Thaler et al. 2004). From a larger number of natural StAOS2 alleles found in potato germ plasm (unpublished results from this laboratory) we selected four specific alleles that could be associated with either increased resistance or susceptibility to P. infestans or Eca based on QTL mapping experiments. These alleles were cloned and subjected to functional complementation analysis in the Arabidopsis aos knock-out mutant. We quantified their differential ability to restore male sterile mutant phenotypes. We provide evidence that both basal and induced levels of JA vary in the aos mutant depending on which StAOS2 allele is expressed under the control of the endogenous Arabidopsis AOS promoter. We further demonstrate that the multiplication rate of E. carotovora is reduced in StAOS2 allele complementation lines containing higher levels of JA compared with those producing lower amounts of JA. All complementation phenotypes evaluated in the Arabidopsis aos mutant were in agreement with the potato ‘quantitative resistant’ (QR) alleles StAOS2-1 and StAOS2-6 being more effective in JA production than the ‘quantitative susceptible (QS) alleles StAOS2-7 and StAOS2-8. Structural models of StAOS2 QR and QS alleles identified amino acid residues that might play a role in the observed functional differences. Two non-synonymous amino acid substitutions close to the StAOS2 substrate binding site could possibly change the enzyme’s substrate affinity and other kinetic properties. This could lead to variable basal levels of the signaling molecules OPDA and JA, which might influence quantitative defense responses in a concentration dependent manner. Site-directed mutagenesis of specific amino acid positions and complementation analysis with such mutant alleles will provide direct evidence for the molecular mechanism of the observed functional differences.
Performing functional complementation analysis in a model organism, which is more accessible to experimentation than the target organism, is the method of choice when searching for causal genes in humans (Nebert et al. 2000). In this sense we made use of the Arabidopsis model in two ways. First, functional dissection of plant defense signaling networks in Arabidopsis provided a list of candidate genes, which were ranked by the map positions of potato sequence homologs relative to QRL that had been mapped in potato (Pajerowska et al. 2005). Second, an Arabidopsis knock-out mutant for the chosen candidate gene was used to assess differential functionality of natural candidate gene alleles. Quantitative complementation analysis in the heterologous system Arabidopsis enabled us to separate the StAOS2 alleles’ effects from the rest of the genetic network for quantitative resistance, and to quantify and compare allele effects in a homogenous genetic background. The equivalent analysis in transgenic potato plants would be impractical, due to a number of reasons. The phenotypic effects of the StAOS2 alleles are defined relative to each other, not in absolute terms like mutant and wild type. When transformed into potato, the quantitative effect of a transgenic StAOS2 allele needs to be separated from effects of the endogenous StAOS2 alleles. The dominance or additive effect of resident alleles relative to the transgenic allele is unknown, genotype dependent and might be obscured by natural variation at other loci affecting the resistance phenotype. Near isogenic introgression lines for StAOS2 alleles, needed as genetic background of complementation with StAOS2 alleles, are not available due to self-incompatibility of diploid and severe inbreeding depression of tetraploid potatoes.
StAOS2 in one of a currently unknown number of candidate loci for the QRL on the short arm of potato chromosome XI. There is no assembled and annotated genomic sequence available at present for this region, neither in potato nor in the co-linear tomato, which would allow predictions of other candidate genes. Unless high resolution QTL mapping reaches the single gene level (Fridman et al. 2000), it cannot be excluded that genes physically linked to StAOS2 are causal for quantitative disease resistance. High-resolution QTL mapping in potato has the same constraints as outlined above and does not obliterate the necessity for functional complementation analysis. Conversely, an efficient reporter organism for functional complementation analysis of candidate alleles as shown here can, at least in part, replace the investment of time and labor required for high resolution QTL mapping and complementation analysis in crop plants. Currently, we perform association mapping for late blight resistance including the StAOS2 locus in populations of individuals related by descent, which may increase the genetic resolution in the genomic region of interest.
The StAOS2-linked QRL on chromosome XI is one of at least twenty QRL distributed in the potato genome (Gebhardt and Valkonen 2001). Moreover, each QRL may be composed of more than one gene and different QRL can act in concert to enhance the resistance phenotype (Bormann et al. 2004; Caromel et al. 2005). To disentangle the complex genetic network of plant quantitative resistance, further efforts in QRL cloning and allele mining are required, before ‘precision breeding’ for quantitative disease resistance becomes a reality.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors are grateful to M. Koornneef (MPI for Plant Breeding Research, Cologne, Germany), J. Dangl (University of North Carolina, Chapel Hill, NC, USA), X. Dong and N. Spivey (Duke University, NC, USA) for their comments on the manuscript, E. Schmelzer (MPI for Plant Breeding Research, Cologne, Germany) for suggestions on confocal microscopy and P. infestans infections, J. Dangl for providing the Ecc strain WDD14, O. Miersch (Leibniz Institute for Plant Biochemistry, Halle, Germany) for GC-MS analysis of JA and OPDA content, B. Ülker (Durham University, UK) for providing the pJawohl17 vector, D. Caldelari-Guex (Duke University, NC, USA) for discussions on molecular modeling, H. Henselewski and A. Weinel for technical assistance. This work was supported by the International Max Planck Research School fellowships to KMPM and MSM and the Deutsche Forschungsgemeinschaft (DFG) grants SPP1067 and SFB648 to SR.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- dsRNAi
Double stranded RNA interference
- Eca
Erwinia carotovora ssp. atroseptica
- Ecc
Erwinia carotovora ssp. carotovora
- JA
Jasmonic acid
- OPDA
12-oxo-phytodienoic acid
- QTL
Quantitative trait loci
- QRL
Quantitative resistance loci
- SNP
Single nucleotide polymorphism
- StAOS2
Solanum tuberosum allene oxide synthase 2
Footnotes
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballvora A, Ercolano MR, Weiss J, Meksem K, Bormann CA, Oberhagemann P, Salamini F, Gebhardt C. The R1 gene for potato resistance to late blight (Phytophthora infestans) belongs to the leucine zipper/NBS/LRR class of plant resistance genes. Plant J. 2002;30:361–371. doi: 10.1046/j.1365-313X.2001.01292.x. [DOI] [PubMed] [Google Scholar]
- Bormann CA, Rickert AM, Ruiz RA, Paal J, Lubeck J, Strahwald J, Buhr K, Gebhardt C. Tagging quantitative trait loci for maturity-corrected late blight resistance in tetraploid potato with PCR-based candidate gene markers. Mol Plant Microbe Interact. 2004;17:1126–1138. doi: 10.1094/MPMI.2004.17.10.1126. [DOI] [PubMed] [Google Scholar]
- Brouwer DJ, Jones ES, St Clair DA. QTL analysis of quantitative resistance to Phytophthora infestans (late blight) in tomato and comparisons with potato. Genome. 2004;47:475–492. doi: 10.1139/g04-001. [DOI] [PubMed] [Google Scholar]
- Caromel B, Mugniery D, Kerlan MC, Andrzejewski S, Palloix A, Ellisseche D, Rousselle-Bourgeois F, Lefebvre V. Resistance quantitative trait loci originating from Solanum sparsipilum act independently on the sex ratio of Globodera pallida and together for developing a necrotic reaction. Mol Plant Microbe Interact. 2005;18:1186–1194. doi: 10.1094/MPMI-18-1186. [DOI] [PubMed] [Google Scholar]
- Clark JP, Miles CS, Mowat CG, Walkinshaw MD, Reid GA, Daff SN, Chapman SK. The role of Thr268 and Phe393 in cytochrome P450 BM3. J Inorg Biochem. 2006;100:1075–1090. doi: 10.1016/j.jinorgbio.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cohen Y, Gisi U, Niderman T. Local and systemic protection against Phytophthora infestans induced in potato and tomato plants by jasmonic acid and jasmonic acid methyl ester. Phytopathology. 1993;83:1054–1062. doi: 10.1094/Phyto-83-1054. [DOI] [Google Scholar]
- Creelman RA, Tierney ML, Mullet JE. Jasmonic acid/methyl jasmonate accumulate in wounded soybean hypocotyls and modulate wound gene expression. Proc Natl Acad Sci USA. 1992;89:4938–4941. doi: 10.1073/pnas.89.11.4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- Farmaki T, Sanmartin M, Jimenez P, Paneque M, Sanz C, Vancanneyt G, Leon J, Sanchez-Serrano JJ. Differential distribution of the lipoxygenase pathway enzymes within potato chloroplasts. J Exp Bot. 2007;58:555–568. doi: 10.1093/jxb/erl230. [DOI] [PubMed] [Google Scholar]
- Fridman E, Pleban T, Zamir D. A recombination hotspot delimits a wild-species quantitative trait locus for tomato sugar content to 484 bp within an invertase gene. Proc Natl Acad Sci USA. 2000;97:4718–4723. doi: 10.1073/pnas.97.9.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman E, Carrari F, Liu Y-S, Fernie AR, Zamir D. Zooming in on a quantitative trait for tomato yield using interspecific introgressions. Science. 2004;305:1786–1789. doi: 10.1126/science.1101666. [DOI] [PubMed] [Google Scholar]
- Froehlich JE, Itoh A, Howe GA. Tomato allene oxide synthase and fatty acid hydroperoxide lyase, two cytochrome P450 s involved in oxylipin metabolism, are targeted to different membranes of chloroplast envelope. Plant Physiol. 2001;125:306–317. doi: 10.1104/pp.125.1.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt C, Valkonen JP. Organization of genes controlling disease resistance in the potato genome. Annu Rev Phytopathol. 2001;39:79–102. doi: 10.1146/annurev.phyto.39.1.79. [DOI] [PubMed] [Google Scholar]
- Gobel C, Feussner I, Hamberg M, Rosahl S. Oxylipin profiling in pathogen-infected potato leaves. Biochim Biophys Acta. 2002;1584:55–64. doi: 10.1016/s1388-1981(02)00268-8. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Guex N, Diemand A, Peitsch MC. Protein modelling for all. Trends Biochem Sci. 1999;24:364–367. doi: 10.1016/S0968-0004(99)01427-9. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Parker JE. Deciphering plant–pathogen communication: fresh perspectives for molecular resistance breeding. Curr Opin Biotechnol. 2003;14:177–193. doi: 10.1016/S0958-1669(03)00035-1. [DOI] [PubMed] [Google Scholar]
- Howe GA, Lee GI, Itoh A, Li L, DeRocher AE. Cytochrome P450-dependent metabolism of oxylipins in tomato. Cloning and expression of allene oxide synthase and fatty acid hydroperoxide lyase. Plant Physiol. 2000;123:711–724. doi: 10.1104/pp.123.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro S, Kawai-Oda A, Ueda J, Nishida I, Okada K. The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell. 2001;13:2191–2209. doi: 10.1105/tpc.13.10.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh A, Schilmiller AL, McCaig BC, Howe GA. Identification of a jasmonate-regulated allene oxide synthase that metabolizes 9-hydroperoxides of linoleic and linolenic acids. J Biol Chem. 2002;277:46051–46058. doi: 10.1074/jbc.M207234200. [DOI] [PubMed] [Google Scholar]
- Judelson HS, Tooley PW. Enchanced polymerase chain reaction methods for detecting and quantifying Phythophthora infestans in plants. Phytopathology. 2000;90:1112–1119. doi: 10.1094/PHYTO.2000.90.10.1112. [DOI] [PubMed] [Google Scholar]
- Kamoun S. Nonhost resistance to Phytophthora: novel prospects for a classical problem. Curr Opin Plant Biol. 2001;4:295–300. doi: 10.1016/S1369-5266(00)00176-X. [DOI] [PubMed] [Google Scholar]
- Kessler A, Halitschke R, Baldwin IT. Silencing the jasmonate cascade: induced plant defenses and insect populations. Science. 2004;305:665–668. doi: 10.1126/science.1096931. [DOI] [PubMed] [Google Scholar]
- Laudert D, Pfannschmidt U, Lottspeich F, Hollander-Czytko H, Weiler EW. Cloning, molecular and functional characterization of Arabidopsis thaliana allene oxide synthase (CYP 74), the first enzyme of the octadecanoid pathway to jasmonates. Plant Mol Biol. 1996;31:323–335. doi: 10.1007/BF00021793. [DOI] [PubMed] [Google Scholar]
- Liechti R, Farmer EE. The jasmonate pathway. Science. 2002;296:1649–1650. doi: 10.1126/science.1071547. [DOI] [PubMed] [Google Scholar]
- Maucher H, Hause B, Feussner I, Ziegler J, Wasternack C. Allene oxide synthases of barley (Hordeum vulgare cv. Salome): tissue specific regulation in seedling development. Plant J. 2000;21:199–213. doi: 10.1046/j.1365-313x.2000.00669.x. [DOI] [PubMed] [Google Scholar]
- McConn M, Creelman RA, Bell E, Mullet JE, Browse J. Jasmonate is essential for insect defense in Arabidopsis. Proc Natl Acad Sci USA. 1997;94:5473–5477. doi: 10.1073/pnas.94.10.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins E, Milbourne D, Petti C, Doyle-Prestwich BM, Meade C. Potato in the age of biotechnology. Trends Plant Sci. 2006;11:254–260. doi: 10.1016/j.tplants.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Dalton TP, Stuart GW, Carvan MJ., 3rd “Gene-swap knock-in” cassette in mice to study allelic differences in human genes. Ann N Y Acad Sci. 2000;919:148–170. doi: 10.1111/j.1749-6632.2000.tb06876.x. [DOI] [PubMed] [Google Scholar]
- Norman-Setterblad C, Vidal S, Palva ET. Interacting signal pathways control defense gene expression in Arabidopsis in response to cell wall-degrading enzymes from Erwinia carotovora. Mol Plant Microbe Interact. 2000;13:430–438. doi: 10.1094/MPMI.2000.13.4.430. [DOI] [PubMed] [Google Scholar]
- Oberhagemann P, Chatot-Balandras C, Bonnel E, Schäfer-Pregl R, Wegener D, Palomino C, Salamini F, Gebhardt C. A genetic analysis of quantitative resistance to late blight in potato: towards marker assisted selection. Mol Breed. 1999;5:399–415. doi: 10.1023/A:1009623212180. [DOI] [Google Scholar]
- Oldham ML, Brash AR, Newcomer ME. The structure of coral allene oxide synthase reveals a catalase adapted for metabolism of a fatty acid hydroperoxide. Proc Natl Acad Sci USA. 2005;102:297–302. doi: 10.1073/pnas.0406352102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowska KM, Parker JE, Gebhardt C. Potato homologs of Arabidopsis thaliana genes functional in defense signaling—identification, genetic mapping, and molecular cloning. Mol Plant Microbe Interact. 2005;18:1107–1119. doi: 10.1094/MPMI-18-1107. [DOI] [PubMed] [Google Scholar]
- Park JH, Halitschke R, Kim HB, Baldwin IT, Feldmann KA, Feyereisen R. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 2002;31:1–12. doi: 10.1046/j.1365-313X.2002.01328.x. [DOI] [PubMed] [Google Scholar]
- Penninckx IA, Eggermont K, Terras FR, Thomma BP, De Samblanx GW, Buchala A, Metraux JP, Manners JM, Broekaert WF. Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell. 1996;8:2309–2323. doi: 10.1105/tpc.8.12.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickert AM, Premstaller A, Gebhardt C, Oefner PJ (2002) Genotyping of SNP’s in a polyploid genome by pyrosequencing. Biotechniques 32:592–593, 596–598, 600 passim [DOI] [PubMed]
- Schaffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simko I, Haynes KG, Jones RW. Assessment of linkage disequilibrium in potato genome with single nucleotide polymorphism markers. Genetics. 2006;173:2237–2245. doi: 10.1534/genetics.106.060905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpe M, Gobel C, Demchenko K, Hoffmann M, Klosgen RB, Pawlowski K, Feussner I. Identification of an allene oxide synthase (CYP74C) that leads to formation of alpha-ketols from 9-hydroperoxides of linoleic and linolenic acid in below-ground organs of potato. Plant J. 2006;47:883–896. doi: 10.1111/j.1365-313X.2006.02843.x. [DOI] [PubMed] [Google Scholar]
- Thaler JS, Owen B, Higgins VJ. The role of the jasmonate response in plant susceptibility to diverse pathogens with a range of lifestyles. Plant Physiol. 2004;135:530–538. doi: 10.1104/pp.104.041566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma B, Eggermont K, Penninckx I, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA. 1998;95:15107–15111. doi: 10.1073/pnas.95.25.15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth IK, Birch PR. Rotting softly and stealthily. Curr Opin Plant Biol. 2005;8:424–429. doi: 10.1016/j.pbi.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Vick BA, Zimmerman DC. Pathways of fatty acid hydroproxide metabolism in spinach leaf chloroplasts. Plant Physiol. 1987;85:1073–1078. doi: 10.1104/pp.85.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wastie RL. Breeding for resistance. Adv Plant Pathol. 1991;7:193–223. [Google Scholar]
- Weber H, Chetelat A, Caldelari D, Farmer EE. Divinyl ether fatty acid synthesis in late blight-diseased potato leaves. Plant Cell. 1999;11:485–494. doi: 10.1105/tpc.11.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Nordborg M. Natural variation in Arabidopsis. How do we find the causal genes? Plant Physiol. 2005;138:567–568. doi: 10.1104/pp.104.900157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap MN, Barak JD, Charkowski AO. Genomic diversity of Erwinia carotovora subsp carotovora and its correlation with virulence. Appl Environ Microbiol. 2004;70:3013–3023. doi: 10.1128/AEM.70.5.3013-3023.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimnoch-Guzowska E, Marczewski W, Lebecka R, Flis B, Schäfer-Pregl R, Salamini F, Gebhardt C. QTL Analysis of new sources of resistance to Erwinia carotovora ssp. atroseptica in potato done by AFLP, RFLP, and Resistance-Gene-Like markers. Crop Sc. 2000;40:1156–1167. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.