Abstract
Malondialdehyde (MDA) and its reactive equivalent, base propenal, are products of oxidative damage to lipids and DNA, respectively; they are mutagenic in bacterial and mammalian systems and MDA is carcinogenic in rats. MDA adducts of deoxyguanosine (M1dG), deoxyadenosine (OPdA) and deoxycytidine (OPdC) have been characterized. We have developed site-specific syntheses of M1dG and OPdA adducted oligonucleotides that rely on a post-synthetic modification strategy. This work provides an alternative route to the M1dG adducted oligonucleotide and to date, the only viable strategy for the site-specific synthesis of OPdA modified oligonucleotides. The stability of the modified oligonucleotides was examined by UV thermal melting studies (Tm). In contrast to the M1dG adduct, OPdA caused very little change in the Tm.
Introduction
Malondialdehyde (MDA, Figure 1) or reactive equivalents of MDA such as base propenals are produced endogenously through free radical degradation of polyunsaturated fatty acids and DNA or as a by-product of prostaglandin biosynthesis (1–3). MDA and base propenal react with DNA to form adducts of dGuo (M1dG), dAdo (OPdA) and dCyd (OPdC). Recent studies in E. coli indicate that base propenal is likely to be the major endogenous source of M1dG adducts (4–6). MDA and base propenal are bis-electrophiles and can undergo two reactions with nucleophilic groups of DNA or proteins. Such reactivity can lead to cyclic DNA adducts, DNA-DNA cross-links or DNA-protein cross-links. Recent results have shown that the dGuo adduct of acrolein, a lower oxidation state homologue of MDA, can mediate interstrand DNA-DNA and DNA-peptide cross-links (7–10).
Figure 1.
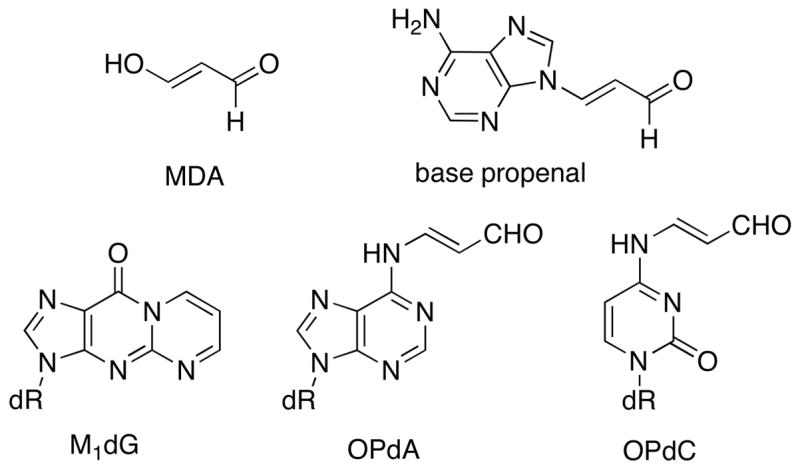
Structures of NMD, base propenal and their nucleoside adducts
MDA and base propenal are mutagenic in bacteria and mammalian cells and MDA is carcinogenic in rats (11–16). The miscoding properties of M1dG have been evaluated with site-specifically adducted oligonucleotides (17). The major base-pair substitution mutation by M1dG after replication in COS-7 cells and E. coli were G → T transversions (17, 18). Of particular interest, M1dG was shown to induce -1 and -2 frameshift mutations when incorporated in a reiterated (CG)4 sequence (18).
The mutagenic potential of the other MDA adducts, namely OPdA and OPdC have not been evaluated by site-specific mutagenesis. Several lines of evidence indicate that these adducts contribute substantially to the overall mutagenic spectrum of MDA. For instance, in random mutagenesis experiments in SOS-induced E. coli, a significant proportion of A→G and C→T transitions were observed in addition to G→T transversions (19). Additionally, replication of an MDA-treated double-strand vector in human kidney cells gave 63% insertion or deletion mutations; the remaining base-pair substitution mutations were at CG base pairs (14). Although M1dG has been shown to be a substrate for nucleotide excision repair (NER), the mutational frequency did not increase when NER deficient cells were used, suggesting that the deletions and insertions were caused by DNA lesions other than M1dG.
MDA adducts of dAdo and dCyd are base-labile. In order for these adducts to be site-specifically incorporated into oligonucleotides, a strategy must be devised that avoids the strong alkaline conditions generally used for the deprotection step in solid-phase oligonucleotide synthesis. Building on previous synthetic efforts in our laboratory (20), we report here an alternative approach to the synthesis of M1dG and OPdA-adducted oligonucleotides in which the modification is site-specifically incorporated after oligonucleotide assembly and deprotection, a synthetic approach that has been termed as a post-oligomerization or post-synthetic modification strategy (21–31).
Experimental Procedures
All commercially obtained chemicals were used as received. 4-Amino-3-phenylselanyl-1,2-butanediol (1) was synthesized as previous described (20). The reactions of oligonucleotides were carried out in plastic vials. MALDI-TOF mass spectra (negative ion) of modified oligonucleotides were obtained on a Voyager Elite DE instrument (Perseptive Biosystems) at the Vanderbilt Mass Spectrometry Resource Center using a 3-hydroxypicolinic acid (HPA) matrix containing ammonium hydrogen citrate (7 mg/mL) to suppress sodium and potassium adducts.
Synthesis of 5′-d(GCTAGC-(4)-AGTCC)-3′ (7)
Oligonucleotide 6 (10 A260 units) was mixed in a plastic test tube with diisopropylethylamine (50 μL), DMSO (100 μL), and 1 (1 mg, 3.8 mmol). The reaction mixture was stirred at 55 °C for 1 day. HPLC analysis showed complete conversion of the starting material (6). The solvents were removed in vacuo with a centrifugal evaporator. The residue was then dissolved in 5% acetic acid (500 μL) and the mixture stirred for 2 h at room temperature. The mixture was neutralized with 1 M NaOH and purified by reversed-phase HPLC (gradient A) to give modified oligonucleotide 7 (6.5 A260 units, ~65%). MS (MALDI-TOF) m/z calcd for 3888.5, found 3888.7. The presence of the modified base was confirmed by enzymatic hydrolysis of 7 and HPLC analysis of the resulting nucleosides using 4 as an authentic sample (20).
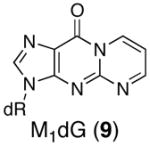
Synthesis of 5′-d(GCTAGC-(M1dG)-AGTCC)-3′ (9)
A solution of NaIO4 (20 mM) was added to a solution of oligonucleotide 7 (5.0 A260 units) in 0.5 % AcOH solution (300 μL), and the reaction mixture was stirred at room temperature for 7 h. The reaction resulted in a single product as observed by reversed phase HPLC (gradient A). Purification gave by reversed-phase HPLC (gradient A) gave M1dG-modified oligonucleotide 9 (2.75 A260 units, ~55%). MS (MALDI-TOF) m/z calcd for 3680.6, found 3681.4. The presence of the modified base was confirmed by enzymatic hydrolysis of 9 and HPLC analysis of the resulting nucleosides using M1dG as an authentic standard (20).
Synthesis of 5′-d(TCGTT-(5)-TTGCT)-3′ (11)
The oligonucleotide 10 (35 A260 units) was mixed in a plastic test tube with diisopropylethylamine (50 μL), DMSO (100 μL), and 1 (1 mg, 3.8 mmol). The reaction mixture was stirred at 65 °C for 1 day. HPLC analysis (gradient A) showed complete disappearance of 10. The solution was directly purified by reversed-phase HPLC (gradient A) to give modified oligonucleotide 11 (29 A260 units, ~83%). MS (MALDI) m/z calcd for 3554.5, found 3553.6. The presence of the modified base was confirmed by enzymatic hydrolysis of 11 and HPLC analysis of the resulting nucleosides using 5 as an authentic standard.
Synthesis of 5′-d(TCGTT-(OPdA)-TTGCT)-3′ (12)
A solution of NaIO4 (40 mM) was added to a solution of oligonucleotide 11 (29 A260 units) in 0.2 % acetic acid solution (400 μL), and the reaction mixture was stirred at room temperature over 10 h. The reaction mixture was purified directly by reversed-phase HPLC (Gradient A) to give M1A-modified oligonucleotide 12 (9.4 A260 units, ~32%). MS (MALDI-TOF) m/z calcd for 3365.6, found 3366.8. The presence of the OPdA modification was confirmed by enzymatic digestion of 12 and HPLC analysis of the nucleosides with detection at both 254 and 320 nm and comparison to an authentic standard of OPdA (20).
Synthesis of 5′-d(CAGTC-OPdA-CTAGA)-3′ (13)
Oligonucleotide 13 (5.5 A260 units, ~24 %) was prepared from the corresponding oligonucleotide containing the 6-chloropurine base (23 A260 units) following the procedure described for the synthesis of oligonucleotides 11 and 12 with the following modification: the initial adduction reaction with 1 was conducted at 55 °C for 10 h. MS (MALDI-TOF) m/z calcd for 3377.6, found 3377.5.
Synthesis of 5′-d(CAGTG-OPdA-GTACA)-3′ (14)
Oligonucleotide 14 (6.3 A260 units, ~18 %) was prepared from the corresponding oligonucleotide containing the 6-chloropurine base (35 A260 units) following the procedure described for the synthesis of oligonucleotides 11 and 12 with the following modification: the initial adduction reaction with 1 was conducted at 55 °C for 10 h. MS (MALDI-TOF) m/z calcd for 3417.7, found 3418.6.
Synthesis of 5′-d(GCAAAAA-OPdA-AAAACATGG)-3′ (15)
The corresponding oligonucleotide containing the 6-chloropurine base (21.4 A 260 units) was mixed in a plastic test tube with diisopropylethylamine (80 μL), DMSO (150 μL), and 1 (2 mg, 7.6 mmol). The reaction mixture was stirred at 55 °C for 24 h. An additional portion of 1 (2 mg, in 50 μL DMSO) was added and the reaction stirred at 55 °C for an additional 24 h. The solvents were removed in vacuo with a centrifugal evaporator and the residue dissolved in 500 μL of water. Purification by reversed-phase HPLC (gradient B) gave the corresponding N6-(2-phenylselanyl-3,4-butanediol)-dAdo adducted oligonucleotide (11.5 A260 units, ~53.7%). MS (MALDI-TOF) m/z calcd for [M - H]− 5494.08, found 5493.5
An aqueous solution of NaIO4 (20 mM) was added to a solution of modified oligonucleotide (11.5 A260 units) in 0.05 M, pH 7.0 phosphate buffer, (500 μL) and the reaction mixture was stirred at room temperature for 10 min. The mixture was purified directly by HPLC (gradient C) to give oligonucleotide 15 (2.7 A260 units, 23.5%). MS (MALDI-TOF) m/z calcd for [M - H]− 5304.9, found 5306.4. The presence of the modified base was confirmed by enzymatic hydrolysis of 15 and HPLC analysis of the resulting nucleosides using OPdA as an authentic standard (20).
Oligonucleotide Purification
The purification of oligonucleotides were performed on a Beckman HPLC system (32 Karat software version 3.1, pump module 125) with a diode array UV detector (module 168) monitoring at 260 nm using Phenomenex Luna 5μ C8 column (250 mm × 10 mm i.d., 3 mL/min for purification) with 0.1 M aqueous ammonium formate and acetonitrile. HPLC gradients:
initially 1% acetonitrile, 15 min linear gradient to 10 % acetonitrile, 5 min linear gradient to 20 % acetonitrile, 5 min isocratic at 20% acetontrile, 3 min linear gradient to 100 % acetonitrile, 4 min isocratic in 10 0% acetonitrile followed by 3 min linear gradient to initial conditions.
initially 1% acetonitrile, 5 min linear gradient to 5 % acetonitrile, 25 min linear gradient to % acetonitrile, 3 min linear gradient to 99% acetonitrile, isocratic at 99 % acetonitrile for 6 min, and 3 min linear gradient to the initial conditions, isocratic at 1 % acetonitrile for 3 min.
initially 1% acetonitrile, 5 min linear gradient to 5 % acetonitrile, 20 min linear gradient to 5.7 % acetonitrile, 3 min linear gradient to 99% acetonitrile, isocratic at 99% acetonitrile for 5 min, and then a 3 min linear gradient to the initial conditions, isocratic at 1% acetonitrile for 2 min.
Melting temperature determinations (Tm)
Melting temperatures of modified and unmodified duplexes were determined as previously described (32).
Enzyme digestion
Enzyme digestions were preformed as previously described (33). Reversed phase HPLC analysis of the enzyme digestion reactions were performed using gradient A. Modified oligonucleotides were identified by comparison of the HPLC retention time and diode array UV spectrum with those of authentic standards, which were synthesized as previously described (20).
Sequencing of oligonucleotide 15
The oligonucleotide (0.03 OD) in 20 μL of ammonium citrate buffer (40 mM, pH 9.4) containing 20 mM MgSO4 was incubated at 37 °C with 2 milliunits of phosphodiesterase I. Aliquots (4 μL) were removed after 0, 10, 20, 30 and 40 min and frozen at −20 °C. The aliquots were combined and desalted using a Millipore C18 ZipTip. The solution is then used for MALDI-TOF analysis. An identical analysis was performed using phosphodiesterase II.
Crosslinking reaction of oligonucleotide 9
The ability of M1dG-modified oligonucleotide 9 to form an interstrand crosslink was evaluated using a previously established protocol (8, 9, 33).
Results and Discussion
Synthesis of M1dG and OPdA adducted oligonucleotides
We previously described syntheses of MDA adducts M1dG, OPdA, and OPdC (Figure 1) at the nucleoside level (20). The synthetic approach contained the elements required for the synthesis of site-specifically adducted oligonucleotides via a post-oligomerization strategy. Although less common than the adducted-phosphoramidite strategy, the post-oligomerization strategy is ideally suited for alkaline-labile adducts that would not survive the strongly basic conditions used for oligonucleotide deprotection (9). The “activated” nucleosides 2 and 3 required for the synthesis of the dGuo and dAdo adducts (Scheme 1), respectively, were reacted with an MDA synthon (1) via a nucleophilic aromatic substitution reaction to give the modified nucleosides 4 and 5. Since 1 was synthesized as a mixture of stereoisomers, the products of this reaction were a mixture of four diastereomers. Oxidation with sodium periodate cleaved the diol to give the corresponding aldehyde. Periodate also oxidized the phenyl selenide to the corresponding selenoxide, which spontaneously eliminated to give the MDA-adducted nucleosides M1dG and OPdA. In the case of the dGuo adduct, the N2-3-oxopropenyl group undergoes dehydrative cyclization to give the M1dG nucleoside. NMR and UV analysis showed that the dAdo adduct of MDA existed in the open-chain N6-3-oxopropenyl form.
Scheme 1.
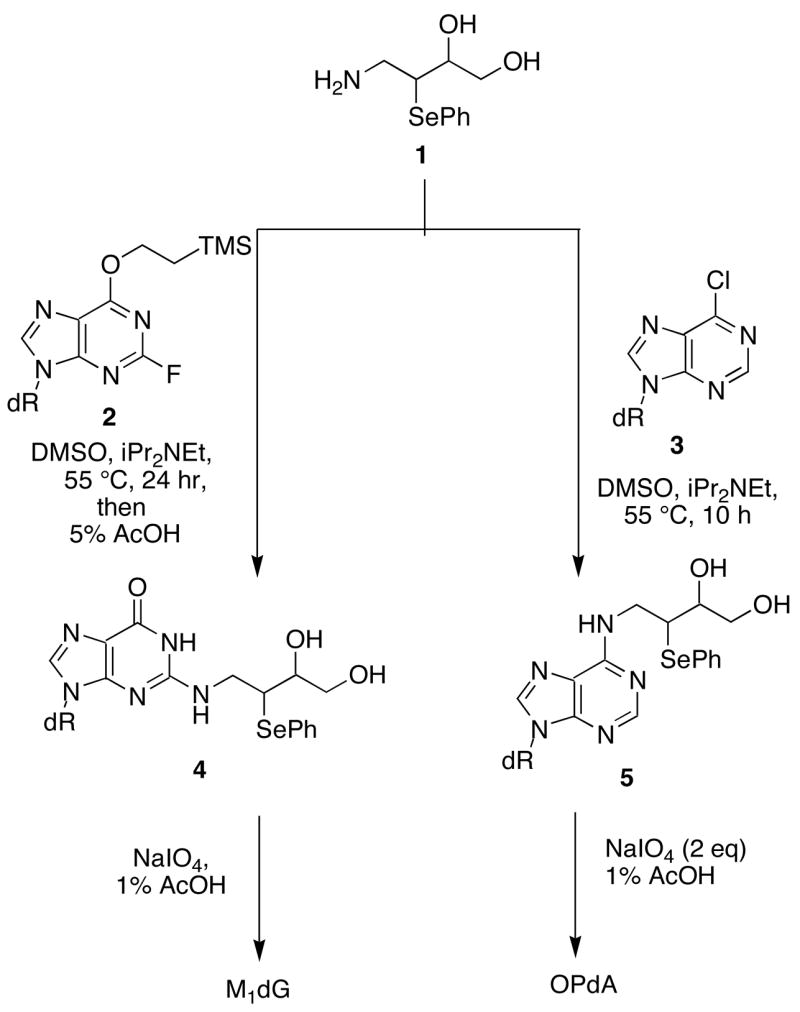
Non-biomimetic synthesis of M1dG and OPdA.
Phosphoramidite reagents containing the required “activated” O6-(2-(trimethylsilylethyl)-2-fluorohypoxanthine (2) and 6-chloropurine (3) bases have been previously synthesized and incorporated in oligonucleotides via standard solid-phase methods (25, 34–36). The site-specific synthesis of an M1dG-adducted oligonucleotide is shown in Scheme 2. Reaction of amine 1 with oligonucleotide 6 containing activated base 2 gave the modified oligonucleotide 7. Upon treatment of 7 with excess sodium periodate, we initially observed two products (8 and 9) with one being converted to the other over time (Panel B, Figure 2). The UV spectrum of the transient intermediate, as observed via a diode array detector during HPLC analysis, possessed a long wavelength absorbance suggesting its identity to be the open chain N2-(3-oxopropenyl) derivative of deoxyguanosine (Panel B, Figure 2). It has been demonstrated that the open chain form undergoes isomerization of the trans-double bond, ring closure and dehydration to M1dG; the dehydration step occurs under general acid catalysis (37, 38). When the periodate oxidation of 7 was carried out in 5 % acetic acid, conversion of 8 to 9 was faster. Oligonucleotides 7 and 9 were characterized by MALDI-TOF mass spectrometry (Table 1) and enzymatic digestion (Figure 3). The modified nucleosides were observed upon HPLC analysis of the enzymatic digestion reaction.
Scheme 2.
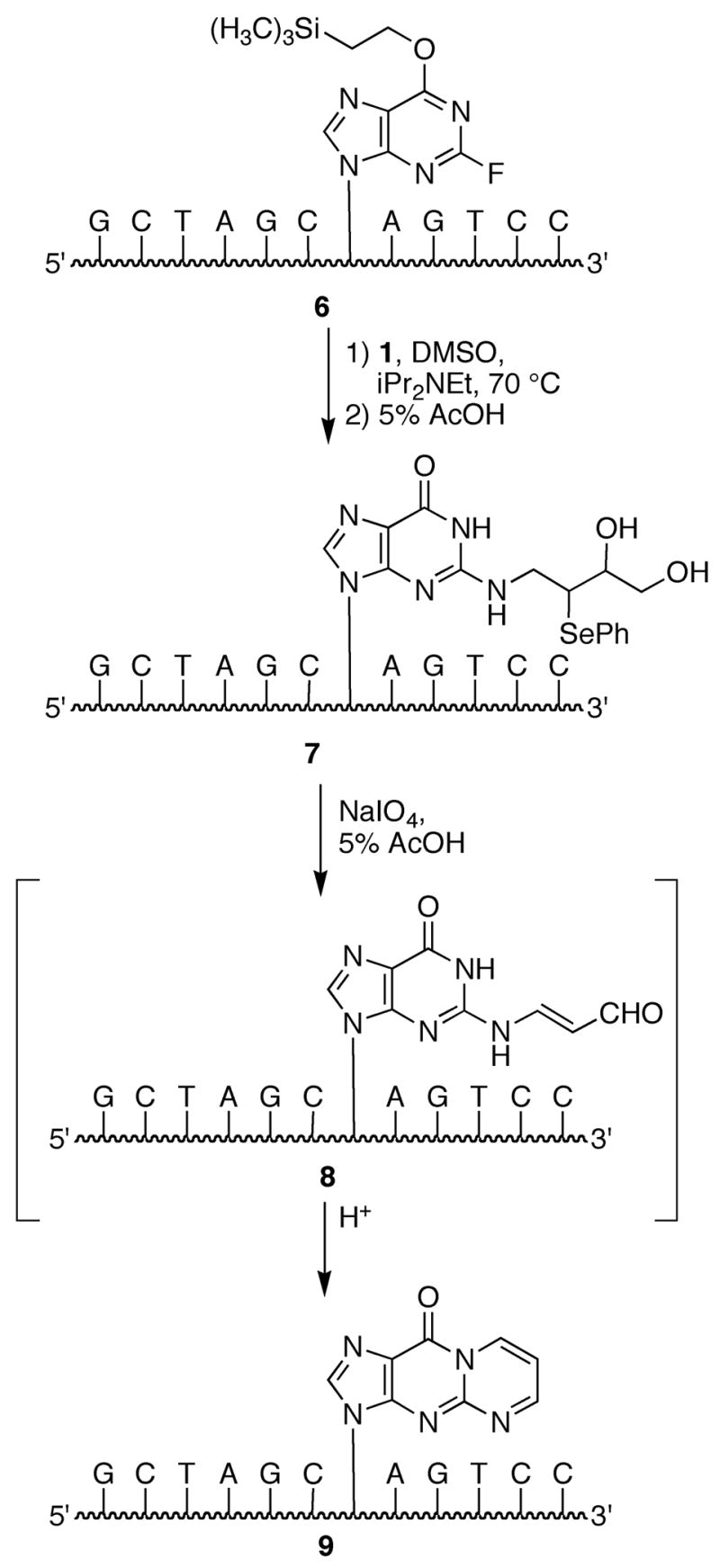
Synthesis of an M1dG-containing oligonucleotide using a post-synthetic modification strategy.
Figure 2.
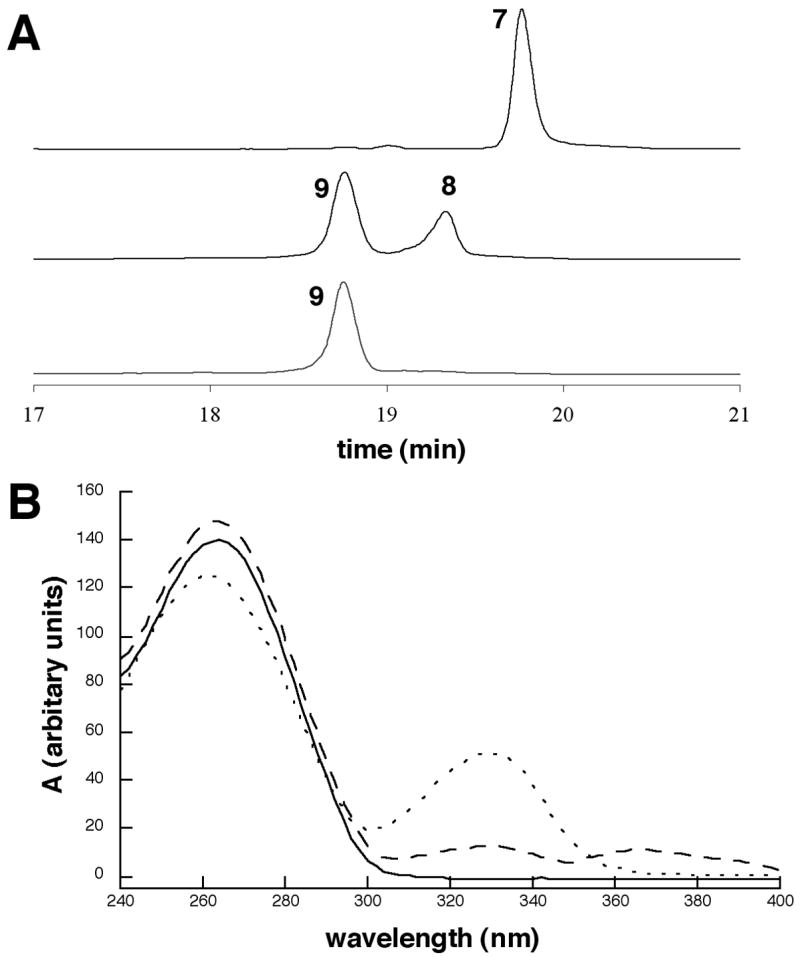
A. HPLC trace of the conversion of oligonucleotide 7 to 9 upon periodate treatment. The middle trace is the periodate cleavage reaction of 7 after 8 h. The bottom trace is the periodate cleavage of 7 in 5% acetic acid solution after 7 h. B. UV spectra of oligonucleotides 7 (——), 8, (- - - -), and 9 (— — —) as obtained by a diode array detector during HPLC analysis.
Table 1.
M1dG- and OPdA-modified oligonucleotides synthesized in this studied and their mass spectral characterization.
| Oligonucleotide | m/z (da) | |
|---|---|---|
| calcd | observed | |
| 5′-GCTAGC-M1dG-AGTCC-3′ (9) | 3680.6 | 3681.4 |
| 5′-TCGTT-OPdA-TTGCT-3′ (12) | 3365.6 | 3366.8 |
| 5′-CAGTC-OPdA-CTAGA-3′ (13) | 3377.6 | 3377.5 |
| 5′-CAGTG-OPdA-GTAGA-3′ (14) | 3417.7 | 3418.6 |
| 5′-GCAAAAA-OPdA-AAAACATGG-3′ (15) | 5304.9 | 5306.4 |
Figure 3.
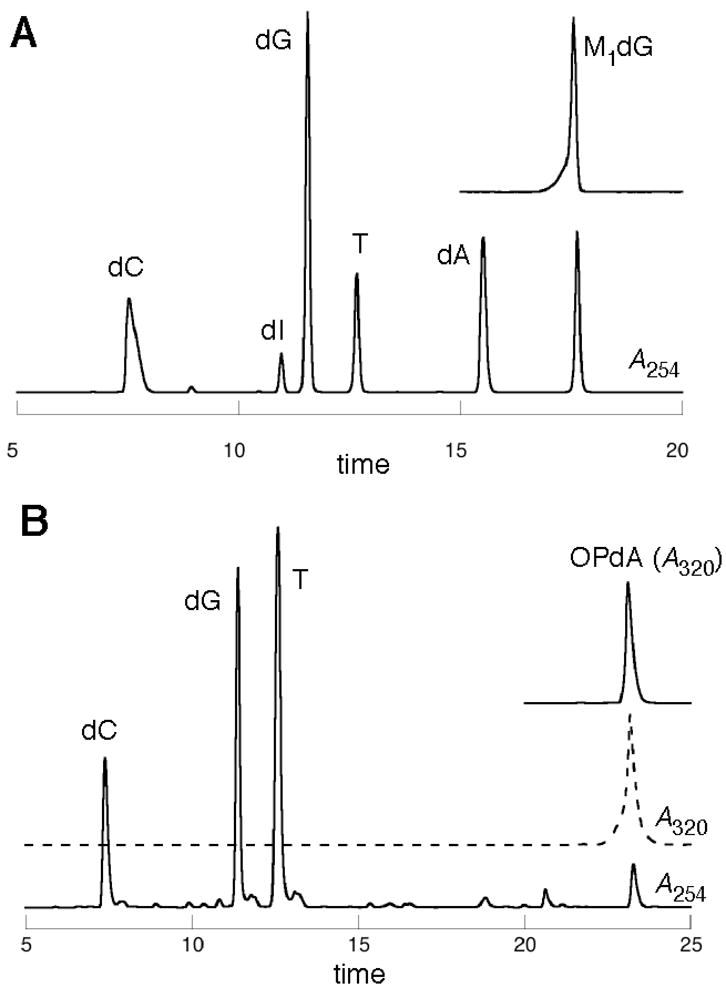
HPLC analysis of the enzymatic digestion of modified oligonucleotides. A. oligonucleotide 9 with detection at 254 (——) and 320 (- - - -) nM and a with a M1dG standard; B. oligonucleotide 12 OPdA standard (320 nM).
The synthesis of an oligonucleotide site-specifically modified with a dAdo adduct of MDA (OPdA) was achieved according to Scheme 3 starting from oligonucleotide 10 containing activated base 3. Nucleophilic aromatic substitution of 10 with amine 1 gave oligonucleotide 11. For the synthesis of the OPdA nucleoside we found that excess periodate gave unidentified by-products and the best yields were obtained with stoichiometric oxidant in the presence of trace acetic acid, although the role of the acetic acid was unclear. However, the oxidation of oligonucleotide 11 was very inefficient under these conditions. We therefore used excess NaIO4 for the conversion of 11 to 12; although by-products were observed for this reaction, they appeared to be less problematic than at the nucleoside level. MDA-adducted oligonucleotide 12 possessed a strong UV absorbance at ~320 nm, which is characteristic of the open-chain OPdA moiety (39). This oligonucleotide was further characterized by MALDI-TOF mass spectrometry (see supporting information) and enzymatic digestion (Figure 3, panel B), which showed the presence of the desired modified nucleoside.
Scheme 3.
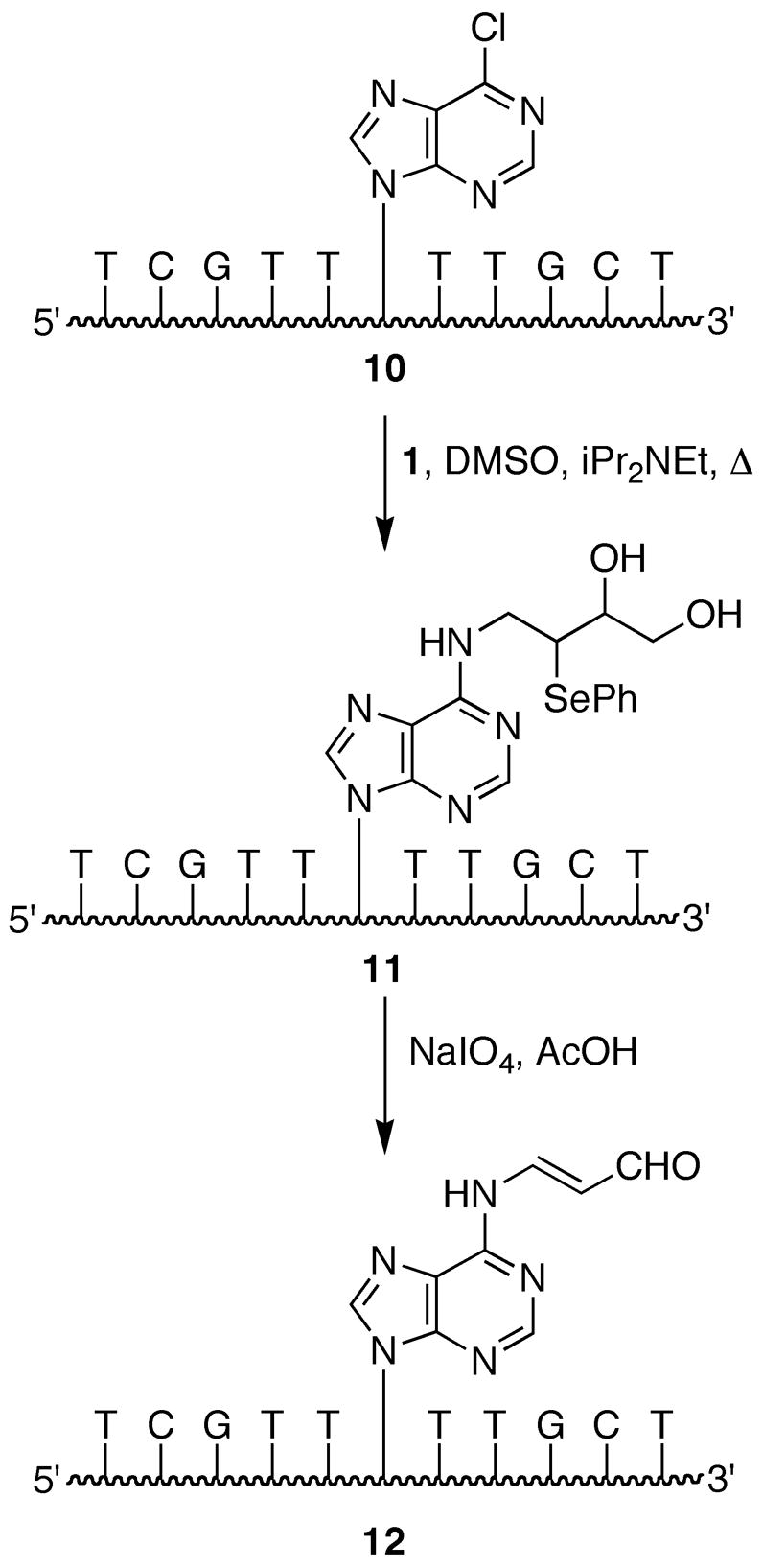
Synthesis of an OPdA-containing oligonucleotide using a post-synthetic modification strategy.
Three other oligonucleotides that were site-specifically modified with OPdA (13–15) were synthesized by an identical strategy and are listed in Table 3 along with their thermal melting temperatures (Tm). Of note is oligonucleotide 15, which possesses an OPdA adduct in a run of ten dAdo’s. Reiterated sequences are prone to frameshift mutations, which are important contributors to human cancer, particularly among individuals with inherited defects in mismatch repair genes (40). There are numerous genes with long runs of dAdo’s (≥ dAdo8) in their coding regions that are mutated in individuals with compromised mismatch repair. For instance, the coding regions for TGFβRII, AIM2, Caspase-5, and SEC63 all contain runs of ten dAdo’s. It has also been demonstrated that oxidative stress can stimulate frameshift mutations in reiterated sequences (41, 42). This opens the possibility that DNA modification by MDA and related bis-electrophiles could lead to frameshift mutations.
Table 3.
Thermal melting temperatures (Tm) of OPdA-modified oligonucleotidesa
| Oligonucleotide | Tm (unmodified) | ΔTm |
|---|---|---|
| 5′-TCGTT-OPdA-TTGCT-3′ (12) 5′-AGCAA——T——AACGA-5′ | 46° C (43° C) | 3° C |
| 5′-CAGTC-OpdA-CTAGA-3′ (13) 3′-GTCAG T——GATCT-5′ | 44° C (44° C) | 0° C |
| 5′-CAGTG-OPdA-GTAGA-3′ (14) 3′-GTCAC T——CATCT-5′ | 46° C (45° C) | 1° C |
| 5′-GCAAAAA-OPdA-AAAACATGG-3′ (15) 3′-CGTTTTT——T——TTTTGTACC-5′ | 55° C (56° C) | −1° C |
| 5′-GCAAAAA-OPdA-AAAACATGG-3′ (15) 3′-CGTTTTT—————TTTTGTACC-5′ | 51° C (50° C) | 1° C |
| 5′-GCAAAAA-OPdA-AAAACATGG-3′ (15) 3′-CGTTTTT—————TTTGTACC-5′ | 48° C (47° C) | 1° C |
Conditions: 100 mM NaCl, 10 mM, pH 7 sodium phosphate, 50 μM EDTA, 0.5 A260/mL of each oligonucleotide. The temperature was raised 1° C/min.
Oligonucleotide 15 was initially characterized by MALDI-TOF mass spectrometry and enzymatic digestion (see supporting information). To confirm that OPdA modification was in the desired location, 15 was sequenced via controlled enzymatic digestion and MALDI-TOF mass spectrometry (43). The oligonucleotide was separately digested with phosphodiesterase I (snake venom phosphodiesterase) and II (bovine spleen phosphodiesterase). Aliquots were taken at various time points and initially frozen; they were then combined for MALDI-TOF analysis, which provided a mass ladder corresponding to the sequential loss of nucleotides from either the 3′- or 5′-end. Since each nucleotide has a unique mass, the sequence of the adducted oligonucleotide could be readily determined. Figure 5 shows the MALDI-TOF analysis from controlled digestion of 15 (m/z 5305.97 Da) with phosphodiesterase I, which possesses 3′→5′ exonuclease activity. A complementary analysis with phosphodiesterase II allowed for sequencing of the oligonucleotide from the 5′-direction and thus provided the sequence of the entire oligonucleotide (see supporting information).
Figure 5.
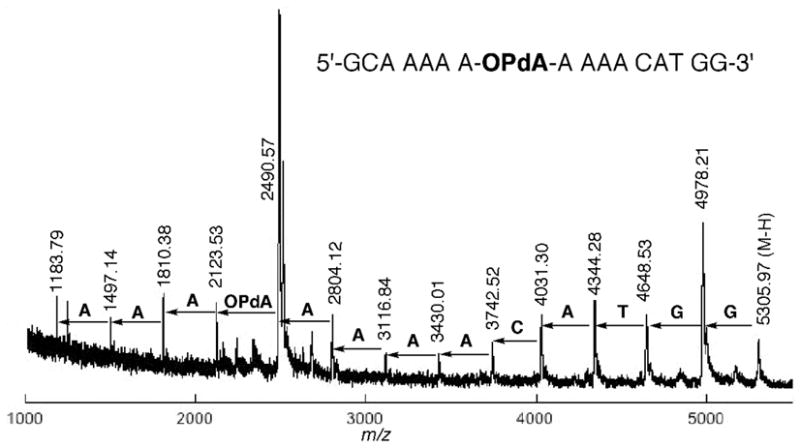
Sequencing of oligonucleotide 15 by partial digestion with phosphodiesterase I (snake venom phosphodiesterase) and MALDI-TOF mass spectrometry.
Melting (Tm) studies of M1dG and OPdA adducted oligonucleotides
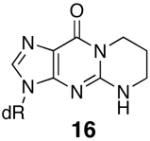
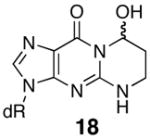
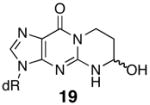
The thermal melting temperatures of the MDA-modified oligonucleotides are given in Tables 2 and 3. The M1dG-containing oligonucleotide 9 is significantly destabilized and its Tm is nineteen degrees lower than that of the unadducted oligonucleotide (Table 2). We have previously examined a number of related dGuo adducts in this sequence, including both regioisomers of the acrolein adduct (18 and 19) and the 1,N2-etheno adduct (20) (32). When this sequence contained the major acrolein adduct of dGuo, 8-hydroxy-3,5,6,7-tetrahydro-9H-imidazo[1,2-a]purine-9-one (18), the Tm was twelve degrees lower than unadducted. NMR studies have demonstrated that both M1dG and the major acrolein adduct exist in the ring-opened N2-3-oxopropenyl (21, OPdG) and N2-3-oxopropyl (22) forms (Scheme 4), respectively when paired with a complementary dCyd (44–46). The difference of seven degrees in Tm’s between these adducted oligonucleotides is somewhat surprising given the structural similarities of the open chain forms. One explanation is that ring closure of the OPdG (21) is more facile than for 22 and ring closure to M1dG promotes denaturation of the duplex (38). The 1,N2-propano-dGuo (PdG, 16) adduct is a saturated analog of M1dG but cannot undergo the ring-opening chemistry. NMR studies have shown the PdG base to be in a syn comformation and involved in Hoogsteen pairing with a complementary Cyt (47–49). The Tm for the PdG adduct was identical to M1dG (Table 2). An alternative explanation for the large destabilization caused by M1dG is that it actually exists in the ring-closed form with a syn geometry, similar to the PdG-adduct. However, the nineteen degree destabilization for the duplex of 9 is in the same range as previously reported for an M1dG-modified oligonucleotide containing the hisD3052 gene sequence, which melted fourteen degree lower than the unmodified oligonucleotide (50, 51). In this case, NMR analysis showed that the adduct was in the open N2-3-oxopropenyl form. The hisD3052 oligonucleotide duplex containing the PdG adduct melted eleven degrees lower than the corresponding M1dG-containing oligonucleotide (50, 51).
Table 2. T.
m of M1dG and structurally related dGuo adducts in duplex 5′-GCT AGC XAG TCC-3′ • 5′-GGACTCGCTACG-3′a
| X= | Tm | ΔTm |
|---|---|---|
| dG | 59 °C | |
|
40 °Cb | −19 °C |
|
40 °Cb | −19 °C |
|
43° Cb | −17 °C |
|
47 °Cc | −12 °C |
|
40 °Cc | −19 °C |
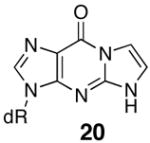
|
41 °Cc | −18 °C |
Conditions for Tm: 100 mM NaCl, 10 mM pH 7 sodium phosphate, 50 μM EDTA, 0.5 A260/mL of each oligonucleotide. The temperature was raised 1° C per minute
This work
reference (32)
Scheme 4.
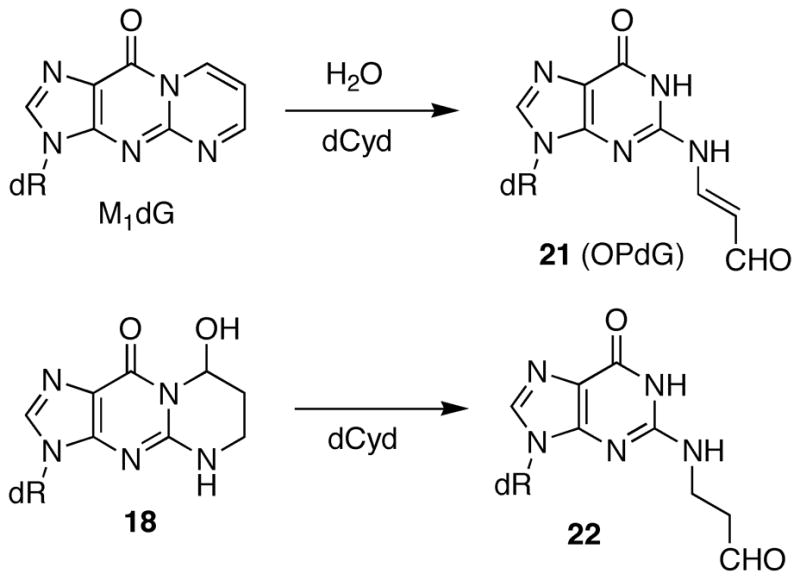
Structural similarities between the MDA and acrolein adducts of deoxyguanosine.
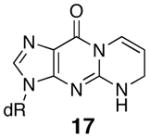
The regioisomeric acrolein adduct, 6-hydroxy-3,5,6,7-tetrahydro-9H-imidazo[1,2-a]purine-9-one (19), also caused the Tm to be lower by 19° compared to unadducted and is the same as for the M1G adduct. There is little driving force for ring opening of this regioisomer since this would yield an N1-3-oxopropyl-Gua base, which would also severely interfere with Watson-Crick base pairing. NMR studies have shown this regioisomer exists in the ring-closed form in duplex DNA with a syn conformation about the glycosidic bond like that of the PdG adduct (52). The 1,N2-etheno-dGuo adduct (20) and partially reduced M1dG adduct 17 show similar Tm values as the M1dG adduct and also are unable to undergo ring opening. Structural studies of oligonucleotides containing 20 and 17 have not been reported to date.
We observed that the OPdA adduct had little effect on the thermal stability of the oligonucleotide and actually caused a modest stabilization in most cases, a significant contrast to the M1dG adduct (Table 3). The OPdA-containing oligonucleotide 15 was also hybridized to complements in which one or two dThd’s across from the adduct were deleted to mimic potential one- and two-base deletion products (Table 3). These duplexes were less stable than those with a full-length complement by 4° and 7° C, respectively, but were slightly more stable than when the corresponding unadducted oligonucleotide was hybridized to a complement containing a one- or two-base deletion. The modest destabilization due to the N6-(3-oxopropenyl) group can be attributed to its relatively small size and that it can adopt a conformation that would not interfere with Watson-Crick hydrogen bonding to a complimentary dThd (Figure 6). The effect of the N6-(2,3,4-trihydroxybutyl)-dAdo adduct derived from butadiene diepoxide, on the thermal stability of an oligonucleotide duplex has also been reported to be modest; the Tm’s were 5–8° C lower than unmodified depending on the stereochemistry of the adduct (53, 54). The modest destabilization as measured by Tm might suggest that the OPdA adduct causes minimal local distortion of the double helix and therefore may be repaired less efficiently than M1dG.
Figure 6.
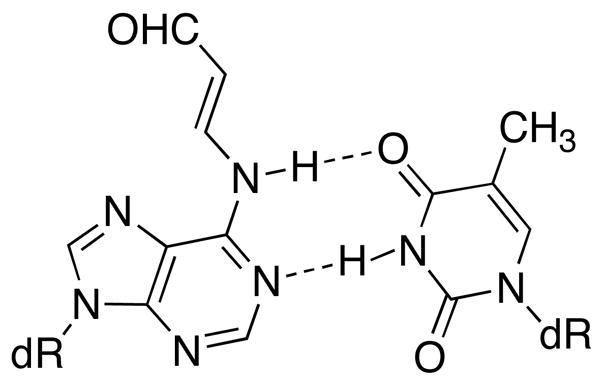
Conformation of OPdA that does not interfere with Watson-Crick hydrogen bonding.
Interstrand cross-linking of the M1dG adducted oligonucleotides
M1dG has been shown to undergo a ring-opening reaction to OPdG in duplex DNA and the mechanism of this process has been studied in detail (16, 37, 44, 51). It is likely that the ring-opening chemistry is important in defining the biological processing of this lesion. As such, M1dG can be viewed as a reactive intermediate within DNA. It had been hypothesized that OPdG could form DNA cross-links and such cross-links contribute to the genotoxicity of MDA (14, 55). We have previously examined the ability of the 1,N2-dGuo adduct of acrolein (18) as well as the higher congeners crotonaldehyde and 4-hydroxynonenal, to form interstrand DNA cross-links in the identical sequence context as oligonucleotide 9 (8, 9, 33). Like M1dG, the acrolein adduct undergoes a ring-opening reaction in duplex DNA when paired opposite a dCyd (45, 46). The acrolein adduct is a lower oxidation state homologue of the MDA adduct and the structural similarities between the two can be most readily seen in the ring-opened form (Scheme 4). The interstrand DNA cross-linking reaction for the acrolein, crotonaldhyde and HNE adducts were followed and quantified by capillary gel electrophoresis (CGE) with UV detection, and the acrolein and crotonaldehyde DNA cross-links were recently been characterized by NMR (56, 57). When the OPdG-modified oligonucleotide 9 was subjected to our cross-linking reaction conditions, we observed no cross-link formation by our previously established assay. It should be noted that the acrolein, crotonaldehyde and HNE cross-linked oligonucleotides had high melting temperatures (> 90° C), which certainly aided in their detection by CGE. Therefore it is possible that 9 formed a transient interstrand cross-links that was labile under the denaturing conditions of the CGE analysis. Alternatively, these results may indicate that the MDA adduct of dGuo is not involved in DNA-DNA crosslink formation and suggests that the OPdA and OPdC adducts are the relevant crosslinking lesions.
Summary
We have developed site-specific syntheses of oligonucleotides that contain the MDA adducts M1dG and OPdA using a post-synthetic modification strategy. This involved the reaction of MDA synthon 1 with oligonucleotides containing either the O6-(2-trimethylsilylethyl)-2- fluorohypoxanthine or 6-chloropurine activated base followed by periodate oxidation (Schemes 2 and 3). Thermal melting (Tm) studies indicated that M1dG caused significant destabilization of the DNA duplex, whereas the OPdA had little effect on the thermal stability. M1dG-modified oligonucleotides were previously synthesized using the adducted phosphoramidite approach; however, the synthesis of the M1dG nucleoside suffers from low overall yield and difficult purification. The synthesis outlined in this work is likely to improve the availability of M1dG-adducted oligonucleotides and is to date the only viable route to OPdA-adducted oligonucleotides. This will allow the assessment of the relative contribution of the OPdA adduct to the genotoxicity of MDA.
Supplementary Material
Supporting Information Available: HPLC analyses of the enzymatic digestion of oligonucleotides 7, 11, 13 and 15; MALDI-TOF mass spectra of oligonucleotides 7, 9, 11, 12, 13, 14, and 15; MALDI-TOF sequencing ladder of 15 from phosphodiester PII digestion; and Tm meltng curves for 9, 12, 13, 14, 15, 16, and 17.
Figure 4.
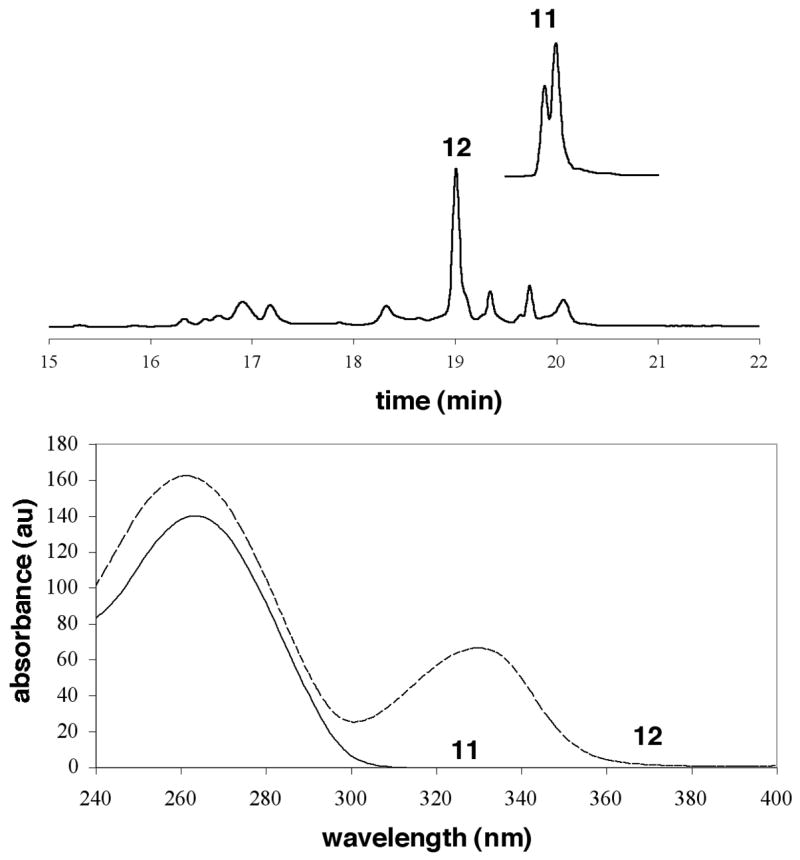
Panel A: HPLC trace for the conversion of oligonucleotide 11 to 12 upon periodate treatment. Panel B: Comparison of the UV spectra of oligonucleotides 11 and 12.
Acknowledgments
This work was supported by the National Institutes of Health through research grants CA87819 (LJM), ES05355 (TMH), and ES11331 (CJR) and center grant ES00267.
Footnotes
Abbreviations: MDA, malondialdehyde; M1dG, 3-(2-deoxy-β-D-erythro-pentofuranosyl)pyrimido[1,2-a]purin-10(3H)-one; OPdG, N2-(3-oxo-1-propenyl)-2′-deoxyguanosine; OPdA, N6-(3-oxo-1-propenyl)-2′-deoxyadenosine; OPdC, N4-(3-oxo-1-propenyl)-2′-deoxycytidine; PdG, 3-(2-deoxy-β-D-erythro-pentofuranosyl)-4,6,7,8-tetrahydro-pyrimido[1,2-a]purin-10(3H)-one, THF, tetrahydrofuran; AcOH, acetic acid; DMSO, dimethylsulfoxide; iPr2NEt, diisopropylethylamine CGE, capillary gel electrophoresis.
References
- 1.Marnett LJ. Oxy radicals, lipid peroxidation and DNA damage. Toxicology. 2002;181–182:219–222. doi: 10.1016/s0300-483x(02)00448-1. [DOI] [PubMed] [Google Scholar]
- 2.Marnett LJ. Chemistry and biology of DNA damage by malondialdehyde. IARC Sci Publ. 1999;150:17–27. [PubMed] [Google Scholar]
- 3.Marnett LJ. Lipid peroxidaion– DNA damage by malondialdehyde. Mutat Res. 1999;424:83–95. doi: 10.1016/s0027-5107(99)00010-x. [DOI] [PubMed] [Google Scholar]
- 4.Zhou X, Taghizadeh K, Dedon PC. Chemical and biological evidence for base propenals as the major source of the endogenous M1dG adduct in cellular DNA. J Biol Chem. 2005;280:25377–25382. doi: 10.1074/jbc.M503079200. [DOI] [PubMed] [Google Scholar]
- 5.Dedon PC, Plastaras JP, Rouzer CA, Marnett LJ. Indirect mutagenesis by oxidative DNA damage: Formation of the pyrimidopurinone adduct of deoxyguanosine by base propenal. Proc Natl Acad Sci USA. 1998;95:11113–11116. doi: 10.1073/pnas.95.19.11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grollman AP, Takeshita M, Pillai KM, Johnson F. Origin and cytotoxic properties of base propenals derived from DNA. Cancer Res. 1985;45:1127–1131. [PubMed] [Google Scholar]
- 7.Sanchez AM, Kozekov ID, Harris TM, Lloyd RS. Formation of inter- and intrastrand imine type DNA-DNA cross-links through secondary reactions of aldehydic adducts. Chem Res Toxicol. 2005;18:1683–1690. doi: 10.1021/tx0500528. [DOI] [PubMed] [Google Scholar]
- 8.Kozekov ID, Nechev LV, Moseley MS, Harris CM, Rizzo CJ, Stone MP, Harris TM. DNA interchain crosslinks formed by acrolein and crotonaldehyde. J Am Chem Soc. 2003;125:50–61. doi: 10.1021/ja020778f. [DOI] [PubMed] [Google Scholar]
- 9.Kozekov ID, Nechev LV, Sanchez A, Harris CM, Lloyd RS, Harris TM. Interchain crosslinking of DNA mediated by the principal adduct of acrolein. Chem Res Toxicol. 2001;14:1482–1485. doi: 10.1021/tx010127h. [DOI] [PubMed] [Google Scholar]
- 10.Kurtz AJ, Lloyd RS. 1,N2-Deoxyguanosine adducts of acrolein, crotonaldehyde, and trans-4-hydroxynonenal cross-link to peptides via Schiff base linkages. J Biol Chem. 2003;278:5970–5976. doi: 10.1074/jbc.M212012200. [DOI] [PubMed] [Google Scholar]
- 11.Yau TM. Mutagenicity and cytotoxicity of malondialdehye in mammalian cells. Mech Aging Dev. 1979;11:137–144. doi: 10.1016/0047-6374(79)90031-9. [DOI] [PubMed] [Google Scholar]
- 12.Basu AK, Marnett LJ. Unequivocal demonstration that malondialdehyde is a mutagen. Carcinogenesis. 1983;4:331–333. doi: 10.1093/carcin/4.3.331. [DOI] [PubMed] [Google Scholar]
- 13.Plastaras JP, Riggins JN, Otteneder M, Marnett LJ. Reactivity and mutagenicity of endogenous DNA oxopropenylating agents: Base propenals, malondialdehyde, and Nε-oxopropenyllysine. Chem Res Toxicol. 2000;13:1235–1242. doi: 10.1021/tx0001631. [DOI] [PubMed] [Google Scholar]
- 14.Niedernhofer LJ, Daniels JS, Rouzer CA, Greene RE, Marnett LJ. Malondialdehyde, a product of lipid peroxidation, is mutagenic in human cells. J Biol Chem. 2003;278:31426–31433. doi: 10.1074/jbc.M212549200. [DOI] [PubMed] [Google Scholar]
- 15.Spalding JW. Toxicology and carcinogenesis studies of malondialdehyde sodium salt (3-hydrox-2-propenal, sodium salt) in F344/N rats and B6C3F1 mice. NTP Technical Report. 1988;331:5–13. [PubMed] [Google Scholar]
- 16.Riggins JN, Marnett LJ. Mutagenicity of the malondialdehyde oligomerization products 2-(3′-oxo-1′-propenyl)-malondialdehyde and 2,4-dihydroxymethylene-3-(2,2-dimethoxyethyl)glutaraldehyde in Salmonella. Mutat Res. 2001;497:153–157. doi: 10.1016/s1383-5718(01)00253-4. [DOI] [PubMed] [Google Scholar]
- 17.Fink SP, Reddy GR, Marnett LJ. Mutagenicity in Escherichia coli of the major DNA adduct derived from the endogenous mutagen malondialdehyde. Proc Natl Acad Sci USA. 1997;94:8652–8657. doi: 10.1073/pnas.94.16.8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.VanderVeen LA, Hashim MF, Shyr Y, Marnett LJ. Induction of frameshift and base pair substitution mutations by the major DNA adduct of the endogenous carcinogen malondialdehyde. Proc Nat Acad Sci USA. 2003;100:14247–14252. doi: 10.1073/pnas.2332176100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benamira M, Johnson K, Chaudhary A, Bruner K, Tibbetts C, Marnett LJ. Induction of mutations by replication of malondialdehyde-modified M13 DNA in Escherichia coli: determination of the extent of DNA modifications, genetic requirements for mutagenesis, and types of mutations induced. Carcinogenesis. 1995;16:93–99. doi: 10.1093/carcin/16.1.93. [DOI] [PubMed] [Google Scholar]
- 20.Wang H, Marnett LJ, Harris TM, Rizzo CJ. A novel synthesis of malondialdehyde adducts of deoxyguanosine, deoxyadenosine and deoxycytidine. Chem Res Toxicol. 2004;17:144–149. doi: 10.1021/tx034174g. [DOI] [PubMed] [Google Scholar]
- 21.Narukulla R, Shuker DEG, Xu YZ. Post-synthetic and site-specific modification of endocyclic nitrogen atoms of purines in DNA and its potential for biological and structural studies. Nucl Acids Res. 2005;33:1767–1778. doi: 10.1093/nar/gki315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kierzek E, Kierzek R. The synthesis of oligoribonucleotides containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines via post-synthetic modification of precursor oligomers. Nucl Acids Res. 2003;31:4461–71. doi: 10.1093/nar/gkg632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shishkina IG, Johnson F. A new method for the postsynthetic generation of abasic sites in oligomeric DNA. Chem Res Toxicol. 2000;13:907–912. doi: 10.1021/tx000108s. [DOI] [PubMed] [Google Scholar]
- 24.Khullar S, Varaprasad CV, Johnson F. Postsynthetic generation of a major acrolein adduct of 2′-deoxyguanosine in oligomeric DNA. J Med Chem. 1999;42:947–950. doi: 10.1021/jm980605u. [DOI] [PubMed] [Google Scholar]
- 25.DeCorte BL, Tsarouhtsis D, Kuchimanchi S, Cooper MD, Horton P, Harris CM, Harris TM. Improved strategies for postoligomerization synthesis of oligodeoxynucleotides bearing structurally defined adducts at the N2-position of deoxygunaosine. Chem Res Toxicol. 1996;9:630–637. doi: 10.1021/tx9501795. [DOI] [PubMed] [Google Scholar]
- 26.Coleman RS, Kesicki EA. Synthesis and postsynthetic modification of oligodeoxynucleotides containing 4-thio-2′-deoxyuridine (D(S4)U) J Am Chem Soc. 1994;116:11636–11642. [Google Scholar]
- 27.Xu YZ, Zheng Q, Swann PF. Synthesis by post-synthetic substitution of oligomers comntaing guansine modified at the 6-position with S-, N-, O-derivatives. Tetrahedron. 1992;48:1729–1740. [Google Scholar]
- 28.Xu YZ, Zhang Q, Swann PF. Synthesis of DNA containing modified bases by postsynthetic substitution. Synthesis of oliomers containg 4-substituted thymine, 5-methylcytosine, N4-(dimethylamino)-5-methylcytosine and 4-thiothymine. J Org Chem. 1992;57:3839–3845. [Google Scholar]
- 29.Kim SJ, Stone MP, Harris CM, Harris TM. A postoligomerization synthesis of oligodeoxynucleotides containing polycyclic aromatic hydrocarbon adducts at the N6-position of deoxyadenosine. J Am Chem Soc. 1992;114:5480–5481. [Google Scholar]
- 30.Gao H, Fathi R, Gaffney BL, Goswami B, Kung PP, Rhee Y, Jin R, Jones RA. 6-O-(Pentafluororphenyl)-2′-deoxyguanosine: a versitile synthon for nucleoside and oligonucleotide synthesis. J Org Chem. 1992;57:6954–6959. [Google Scholar]
- 31.Macmillan AM, Verdine GL. Engineering tethered DNA molecules by the convertible nucleoside approach. Tetrahedron. 1991;47:2603–2616. [Google Scholar]
- 32.Goodenough AK, Kozekov ID, Zang H, Choi J-Y, Guengerich FP, Harris TM, Rizzo CJ. Site-specific synthesis and polymerase bypass of oligonucleotides containing a 6-hydroxy-3,5,6,7-tetrahydro-9H-imidazo[1,2-a]purin-9-one base, an intermediate in the formation of 1, N2-etheno-2′-deoxyguanosine. Chem Res Toxicol. 2005;18:1701–1714. doi: 10.1021/tx050141k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H, Kozekov ID, Harris TM, Rizzo CJ. Site-specific synthesis and reactivity of oligonucleotides containing stereochemically defined 1,N2-deoxyguanosine adducts of trans-4-hyrdoxynonenal. J Am Chem Soc. 2003;123:5687–5700. doi: 10.1021/ja0288800. [DOI] [PubMed] [Google Scholar]
- 34.Harris CM, Zhou L, Strand EA, Harris TM. New strategy for the synthesis of oligodeoxynucleotides bearing adducts at exocyclic amino sites of purine nucleosides. J Am Chem Soc. 1991;113:4328–4329. [Google Scholar]
- 35.Kim HY, Nechev L, Zhou LA, Tamura P, Harris CM, Harris TM. Synthesis and adduction of fully deprotected oligodeoxynucleotides containing 6-chloropurine. Tetrahedron Lett. 1998;39:6803–6806. [Google Scholar]
- 36.Nechev LV, Harris CM, Harris TM. Synthesis of nucleosides and oligonucleotides containing adducts of acrolein and vinyl chloride. Chem Res Toxicol. 2000;13:421–429. doi: 10.1021/tx990167+. [DOI] [PubMed] [Google Scholar]
- 37.Riggins JN, Daniels JD, Rouzer CA, Marnett LJ. Kinetic and thermodynamic analysis of the hydrolytic ring-opening of the malondialdehyde-deoxyguanosine adduct, 3-(2′-deoxy-β-D-erythro-pentofuranosyl)- [1,2-α]purin- 10(3H)-one. J Am Chem Soc. 2004;126:8237–8243. doi: 10.1021/ja040009r. [DOI] [PubMed] [Google Scholar]
- 38.Riggins JN, Pratt DA, Voehler M, Daniels JS, Marnett LJ. Kinetics and mechanism of the general-acid-catalyzed ring-closure of the malondialdehdye-DNA adduct, N2-(3-oxo-1-propenyl)deoxyguanosine (N2OPdG–), to 3-(2′-deoxy-β-D-erythro-pentofuranosyl)pyrimido[1,2-α]purin- 10(3H)-one (M1dG) J Am Chem Soc. 2004;126:10571–10581. doi: 10.1021/ja040010q. [DOI] [PubMed] [Google Scholar]
- 39.Stone K, Ksebati MB, Marnett LJ. Investigation of the adducts formed by reaction of malondialdehyde with adenosine. Chem Res Toxicol. 1990;3:33–38. doi: 10.1021/tx00013a006. [DOI] [PubMed] [Google Scholar]
- 40.Duval A, Hamelin R. Mutations at Coding Repeat Sequences in Mismatch Repair-deficient Human Cancers. Toward a New Concept of Target Genes for Instability. Cancer Res. 2002;62:2447–2454. [PubMed] [Google Scholar]
- 41.Jackson AL, Chen R, Loeb LA. Induction of microsatellite instability by oxidative DNA damage. Proc Nat Acad Sci USA. 1998;95:12468–12473. doi: 10.1073/pnas.95.21.12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gasche C, Chang CL, Rhees J, Goel A, Boland CR. Oxidative stress increases frameshift mutations in human colorectal cancer cells. Cancer Res. 2001;61:7444–7448. [PubMed] [Google Scholar]
- 43.Tretyakova N, Matter B, Ogdie A, Wishnok JS, Tannenbaum SR. Locating nucleobase lesions within DNA sequences by MALDI-TOF mass spectral analysis of exonuclease ladders. Chem Res Toxicol. 2001;14:1058–70. doi: 10.1021/tx010062i. [DOI] [PubMed] [Google Scholar]
- 44.Mao H, Schnetz-Boutaund N, Weisenseel JP, Marnett LJ, Stone MP. Duplex DNA catalyzes the chemical rearrangement of a malondialdehyde deoxyguanosine adduct. Proc Natl Acad Sci USA. 1999;96:6615–6620. doi: 10.1073/pnas.96.12.6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de los Santos C, Zaliznyak T, Johnson F. NMR characterization of a DNA duplex containing the major acrolein-derived deoxyguanosine adduct of γ-OH-1,-N2-propano-2′-deoxyguanosine. J Biol Chem. 2001;276:9077–9082. doi: 10.1074/jbc.M009028200. [DOI] [PubMed] [Google Scholar]
- 46.Kim H-Y, Voehler M, Harris TM, Stone MP. Detection of an interchain carbinolamine cross-link formed in a CpG sequence by the acrolein DNA adduct γ-OH-1, N2-propano-2′-deoxyguanosine. J Am Chem Soc. 2002;124:9324–9325. doi: 10.1021/ja020333r. [DOI] [PubMed] [Google Scholar]
- 47.Singh US, Moe JG, Reddy GR, Weisenseel JP, Marnett LJ, Stone MP. 1H NMR of an oligodeoxynucleotide containing a propanodeoxyguanosine adduct positioned in a (CG)3 frameshift hotspot of salmonella typhimurium hisD3052: Hoogsteen base-pairing at pH 5.8. Chem Res Toxicol. 1993;6:825–836. doi: 10.1021/tx00036a012. [DOI] [PubMed] [Google Scholar]
- 48.Weisenseel JP, Moe JG, Reddy GR, Marnett LJ, Stone MP. Structure of a duplex oligodeoxynucleotide containing propanodeoxyguanosine opposite a two-base deletion in the (CpG)3 frame shift hotspot of Salmonella typhimurium hisD3052 determined by 1H NMR and restrained molecular dynamics. Biochemistry. 1995;34:50–64. doi: 10.1021/bi00001a007. [DOI] [PubMed] [Google Scholar]
- 49.Weisenseel JP, Reddy GR, Marnett LJ, Stone MP. Structure of the 1,N2-propanodeoxyguanosine adduct in a three-base DNA hairpin loop derived from a palindrome in the Salmonella typhimurium hisD3052 gene. Chem Res Toxicol. 2002;15:140–159. doi: 10.1021/tx010107f. [DOI] [PubMed] [Google Scholar]
- 50.Schnetz-Boutaud NC, Saleh S, Marnett LJ, Stone MP. The exocyclic 1,N2-deoxyguanosine pyrimidopurinone M1G is a chemically stable DNA adduct when placed opposite a two base deletion in the (CpG)3 frameshift hotspot of the Salmonella typhimurium hisD3052 gene. Biochemistry. 2001;40:15638–15649. doi: 10.1021/bi011242u. [DOI] [PubMed] [Google Scholar]
- 51.Mao H, Reddy GR, Marnett LJ, Stone MP. Solution structure of an oligodeoxynucleotide containing the malondialdehyde-deoxyguanosine adduct N2-[3-oxo-1-propenyl]-dG (ring-opened M1G) positioned in a (CpG)3 frameshift hotspot of the Salmonella typhimurium hisD3052 gene. Biochemistry. 1999;38:13491–13501. doi: 10.1021/bi9910124. [DOI] [PubMed] [Google Scholar]
- 52.Lukin M, de Los Santos C. NMR structures of damaged DNA. Chem Rev. 2006;106:607–686. doi: 10.1021/cr0404646. [DOI] [PubMed] [Google Scholar]
- 53.Nechev LV, Zhang M, Tsarouhtsis D, Tamura PJ, Wilkinson AS, Harris CM, Harris TM. Synthesis and characterization of nucleosides and oligonucleotides bearing adducts of butadiene epoxides on adenine N6 and guanine N2. Chem Res Toxicol. 2001;14:379–388. doi: 10.1021/tx000241k. [DOI] [PubMed] [Google Scholar]
- 54.Merritt WK, Scholdberg TA, Nechev LV, Harris TM, Harris CM, Lloyd RS, Stone MP. Stereospecific structural perturbations arising from adenine N6 butadiene triol adducts in duplex DNA. Chem Res Toxicol. 2004;17:1007–1017. doi: 10.1021/tx049908j. [DOI] [PubMed] [Google Scholar]
- 55.Basu AK, Marnett LJ, Romano LJ. Dissociation of malondialdehyde mutagenicity in salmonella typhimurium from its ability to induce interstrand dna cross-links. Mutat Res. 1984;129:39–46. doi: 10.1016/0027-5107(84)90121-0. [DOI] [PubMed] [Google Scholar]
- 56.Cho Y-J, Wang H, Kozekov ID, Kurtz AJ, Jacob J, Voehler M, Smith J, Harris TM, Lloyd RS, Rizzo CJ, Stone MP. Stereospecific formation of interstrand carbinolamine DNA cross-links by crotonaldehyde- and acetaldehyde-derived α-CH3-γ-OH-1,N2-propano-2′-deoxyguanosine adducts in the 5′-CpG-3′ sequence. Chem Res Toxicol. 2005;19:195–208. doi: 10.1021/tx050239z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cho YJ, Kim HY, Huang H, Slutsky A, Minko IG, Wang H, Nechev LV, Kozekov ID, Kozekova A, Tamura P, Jacob J, Voehler M, Harris TM, Lloyd RS, Rizzo CJ, Stone MP. Spectroscopic characterization of interstrand carbinolamine cross-links formed in the 5′-CpG-3′ sequence by the acrolein-derived γ-OH-1,N2-propano-2′-deoxyguanosine DNA Adduct. J Am Chem Soc. 2005;127:17686–17696. doi: 10.1021/ja053897e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Available: HPLC analyses of the enzymatic digestion of oligonucleotides 7, 11, 13 and 15; MALDI-TOF mass spectra of oligonucleotides 7, 9, 11, 12, 13, 14, and 15; MALDI-TOF sequencing ladder of 15 from phosphodiester PII digestion; and Tm meltng curves for 9, 12, 13, 14, 15, 16, and 17.


