Abstract
Recent work indicates that mitogen-activated protein kinase kinase (MEK)1 signaling at the G2/M cell cycle transition unlinks the contiguous mammalian Golgi apparatus and that this regulates cell cycle progression. Here, we sought to determine the role in this pathway of Golgi reassembly protein (GRASP)55, a Golgi-localized target of MEK/extracellular signal-regulated kinase (ERK) phosphorylation at mitosis. In support of the hypothesis that GRASP55 is inhibited in late G2 phase, causing unlinking of the Golgi ribbon, we found that HeLa cells depleted of GRASP55 show a fragmented Golgi similar to control cells arrested in G2 phase. In the absence of GRASP55, Golgi stack length is shortened but Golgi stacking, compartmentalization, and transport seem normal. Absence of GRASP55 was also sufficient to suppress the requirement for MEK1 in the G2/M transition, a requirement that we previously found depends on an intact Golgi ribbon. Furthermore, mimicking mitotic phosphorylation of GRASP55 by using aspartic acid substitutions is sufficient to unlink the Golgi apparatus in a gene replacement assay. Our results implicate MEK1/ERK regulation of GRASP55-mediated Golgi linking as a control point in cell cycle progression.
INTRODUCTION
The structural diversity of the Golgi apparatus among eukaryotes suggests two primary modes of organization. Cis-, medial-, and trans-Golgi cisternae, although unconnected in budding yeast, are typically present as membrane stacks. Whereas many cell types contain numerous scattered stacks positioned adjacent to endoplasmic reticulum (ER) exit sites, known as ministacks, mammalian cells and those from related metazoans exhibit a pericentrosomal Golgi in which ministacks are laterally linked into a ribbon. The in vivo requirements for Golgi stack formation remain unknown; however, recent work has begun to identify components necessary for linking cisternal stacks into a contiguous Golgi ribbon.
Depletion of the golgin Golgi matrix protein 130 kDa (GM130), or its binding partner Golgi reassembly protein (GRASP)65, by using RNA interference (RNAi) specifically disrupts formation of the Golgi ribbon (Puthenveedu et al., 2006). GM130 seems to recruit GRASP65 to Golgi membranes because Golgi localization of GRASP65 is lost upon GM130 knockdown, whereas GM130 remains Golgi localized in the absence of GRASP65 (Sutterlin et al., 2005; Puthenveedu et al., 2006). A mechanism for Golgi ribbon formation by GRASP65 is suggested by the finding that two postsynaptic density 95/disc-large/zona occludens-like domains at the N terminus of GRASP65 form oligomers that, in the presence of cytosol, can cross-link beads (Wang et al., 2003, 2005). Thus, the Golgi ribbon may be formed by GM130-dependent recruitment of GRASP65 to Golgi cisternal rims where GRASP65 transoligomer formation cross-bridges adjacent cis-cisternae and possibly primes them for soluble N-ethylmaleimide-sensitive factor attachment protein receptor-mediated membrane fusion (Puthenveedu et al., 2006).
The lateral contacts that form the Golgi ribbon seem dynamic. The C-terminal binding protein/brefeldin A-ADP-ribosylated substrate (CtBP/BARS) disrupts the Golgi ribbon, possibly catalyzing membrane fission at contact sites (Weigert et al., 1999; Bonazzi et al., 2005). Furthermore, the contacts that occur during ribbon formation are dependent on microtubule-based motility of Golgi membranes (Allan et al., 2002; Linstedt, 2004), and these contact zones are sites of extensive vesicle trafficking, which influences the integrity of the contacts (Marra et al., 2007).
Interestingly, GRASP65, and the closely related GRASP55, localize to cis- and medial-Golgi cisternae, respectively (Shorter et al., 1999), suggesting that GRASP65 may initiate lateral linkages at the cis-Golgi, whereas GRASP55 may then maintain ribbon formation in the medial-Golgi. Although GRASP55, similar to GRASP65, mediates assembly of Golgi stacks in an in vitro assay (Shorter et al., 1999), it is not yet known whether GRASP55 is required for Golgi ribbon formation in mammalian cells.
Testing the role of GRASP55 is particularly important because the protein has recently been implicated in Golgi breakdown in preparation for cell division and in cell division itself. Golgi ribbons become unlinked during late G2 phase (Colanzi et al., 2007; Feinstein and Linstedt, 2007) and a block in Golgi disassembly arrests or delays the G2/M transition (Sutterlin et al., 2002; Colanzi et al., 2007; Feinstein and Linstedt, 2007). Premitotic Golgi unlinking depends in part on the constitutive membrane fission activity of CtBP/BARS (Hidalgo Carcedo et al., 2004; Colanzi et al., 2007), and it is triggered by mitogen-activated protein kinase kinase (MEK)1 activation of extracellular signal-regulated kinase (ERK) (Shaul and Seger, 2006; Colanzi and Corda, 2007; Feinstein and Linstedt, 2007). GRASP55 is a likely target of this mitogen-activated protein kinase (MAPK) pathway because GRASP55 is a mitotic target of MEK-dependent phosphorylation by ERK at its T222 and T225 residues (Jesch et al., 2001) and because expression of an alanine-substituted version of GRASP55, GRASP55-T222,225A, blocks both G2 phase Golgi unlinking and mitotic entry (Feinstein and Linstedt, 2007).
To directly test the role of GRASP55 in Golgi ribbon formation, we inhibited GRASP55 expression using RNAi, and we analyzed Golgi organization, membrane trafficking, and cell cycle progression. The role of phosphoregulation of GRASP55 by ERK was tested using mutated versions of the protein designed to mimic either the phosphorylated or the nonphosphorylated state. The combined results indicate that GRASP55 plays a MEK/ERK-regulated role in Golgi ribbon formation and cell cycle progression.
MATERIALS AND METHODS
Reagents and Cell Culture
Primary antibodies used were as follows: anti-Myc 9e10 (Jesch et al., 2001), anti-giantin (Linstedt and Hauri, 1993), anti-p115 (Puthenveedu and Linstedt, 2001), anti-GM130 (Puthenveedu and Linstedt, 2001), anti-GP73 (Bachert et al., 2007), anti-GPP130 (Linstedt et al., 1997), and anti-GRASP55, which was generated by immunizing rabbits (Covance, Emeryville, CA) with purified glutathione transferase (GST)-GRASP55 (Jesch et al., 2001). Affinity-purified, conjugated secondary antibodies were purchased (Invitrogen, Carlsbad, CA). U0126 (used at 10 μM) was from Promega (Madison, WI), thymidine (used at 2 mM) was from EMD Biosciences (Darmstadt, Germany), and nocodazole (used at 500 nM) and olomoucine II (used at 10 μM) were from Sigma-Aldrich (St. Louis, MO). GRASP55-green fluorescent protein (GFP) was made by generating a ClaI–XbaI insert of enhanced green fluorescent protein (EGFP) by polymerase chain reaction (PCR) and inserting it into ClaI–Xba1 sites of pCB6-GRASP55 (Jesch et al., 2001) to generate a C-terminal tag. Monolayer HeLa cells were grown and synchronized using a double thymidine block (48 h in thymidine and then 12 h without then 12 h in thymidine) as described previously (Feinstein and Linstedt, 2007). When done in combination with RNAi, the first thymidine addition and the small interfering RNA (siRNA) transfection were performed concurrently, and the double thymidine protocol proceeded as normal.
Transfection
Transfection with siRNA was performed on 35-mm plates, containing coverslips as needed, according to manufacturer's recommendations (Invitrogen) by using 8 μl of Oligofectamine and 40 nM siRNA. After 24 h, the cells were refreshed with fresh growth medium. Assays were performed at 72 h after transfection. The siRNAs were obtained as purified duplexes from Ambion (Austin, TX). Human GRASP55 was targeted with aactgtcgagaagtgattatt (#514), aaaggcagacgctgcctcctc (#1206), and atctattcagccttatcgaaa (#443), with comparable results. Data are presented from experiments using #514. Myc epitope-tagged GRASP55 and GRASP55-T222,T225D plasmids were transfected using Transfectol (GeneChoice, Frederick, MD) according to the manufacturer's recommendations. When combined with RNAi, plasmid transfection was done 48 h after transfection with siRNA, and analysis was after an additional 24 h.
Microscopy and Image Analysis
Microscopy was performed as described previously (Puthenveedu et al., 2006) by using a spinning disk confocal system with a 100× oil immersion objective or a fluorescence bleaching-equipped epifluorescence microscope with a 100× oil immersion objective. Z-axis sectioning was at 0.3-μm intervals, and images were analyzed after maximum-value projection. Live imaging was performed at 37°C in Opti-MEM (Invitrogen) containing 10% serum. Images for a given experiment were captured using fixed parameters, and no modifications were performed between capture and analysis with ImageJ software (http://rsb.info.nih.gov/ij/). The images were thresholded using the autothreshold feature, and the number of objects per Golgi was determined using the Analyze Objects feature with a minimum size cutoff of 15 pixels. Fluorescence recovery after photobleaching was carried out at room temperature. Part of the Golgi was bleached using a single laser pulse, and images were acquired every 3 s as described previously (Puthenveedu et al., 2006). Fluorescence values in the bleached and an adjacent nonbleached area were measured using NIH Image. Fluorescence recovery is represented as the ratio of the bleached region to an adjacent unbleached region, normalized to the prebleach and immediate postbleach values. For live imaging after nocodazole treatment, cells were washed eight times in cold phosphate-buffered saline, transferred to a live cell chamber (Invitrogen), and then they were imaged using a confocal microscope. Stacks were captured every 30 s and collapsed into two-dimensional (2D) images for viewing. For colocalization analysis, images were collected at a fixed exposure, stacks were collapsed to 2D and colocalization was analyzed using the JACoP plugin for ImageJ. Lectin binding analysis was performed as described previously (Puthenveedu et al., 2006).
Vesicular Stomatitis Virus Glycoprotein (VSVG) Trafficking Assay
HeLa cells treated with siRNA were subsequently transfected with a plasmid encoding ts045 VSVG-GFP cDNA, and incubated at 40°C for 5 h. The cells were then shifted to 32°C for 0, 20, or 60 min. For determination of surface VSVG, live cells were stained with an antibody (8G5) directed against the luminal domain of VSVG (Sevier et al., 2000) for 10 min at 4°C, washed extensively, fixed, and stained by using Cy5-conjugated secondary antibodies. The ratio of surface-to-total fluorescence was used to calculate the extent of VSVG transport.
Electron Microscopy
Transmission electron microscopy was performed as described previously (Puthenveedu et al., 2006). To reconstruct three-dimensional (3D) representations of the Golgi, Photoshop (Adobe Systems, San Jose, CA) was used to draw freehand masks of Golgi membranes in each frame of contiguous serial sections of representative control and GRASP55-depleted cells. Each mask was aligned with adjacent masks based on the registration of cellular components in the sections from which they were made, and serial masks were reconstructed into 3D volumes by using Volocity software (PerkinElmer Life and Analytical Sciences, Boston, MA).
In Vitro GRASP55 Binding Assay and Immunoblotting
HeLa cells were transiently transfected with myc-tagged GRASP55 constructs (wild-type or GRASP55-T222,225D), and then they were lysed in HKT buffer (10 mM HEPES/KOH, pH 7.4, 100 mM KCl, 0.5% Triton X-100, 1 mM EDTA, 1 mM dithiothreitol, 0.2 mg/ml phenylmethylsulfonyl fluoride, 20 μg/ml leupeptin, and 20 μg/ml pepstatin A). Cell lysate (0.7 mg/ml) was mixed with 260 μg of GST-GRASP55 or GST protein purified from bacterial lysates and immobilized on 10 μl of glutathione agarose and rotated in HKT buffer for 4 h at 4°C. Lysate-incubated beads were then washed four times in 1 ml of cold HKT buffer, and recovery of myc-tagged proteins was determined by immunoblotting. Quantification was done by directly capturing enhanced chemiluminescence with a digital camera (LAS 3000; Fujifilm, Tokyo, Japan), and bands were compared after quantification and background subtraction by using the Profile function in ImageGuage (Fujifilm).
Statistical Analysis
The statistical significance of all comparisons was assessed by two-tailed Student's t tests, and, where indicated, nonoverlap of curves was estimated using root mean squared deviation.
RESULTS
GRASP55 Is Required for Golgi Ribbon Formation
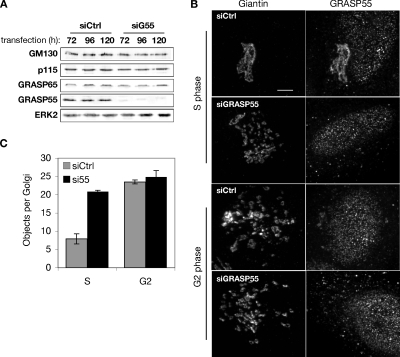
To determine whether GRASP55 contributes to maintaining the structure of interphase Golgi, we evaluated Golgi ribbon formation after depletion of GRASP55 by using each of three different siRNA oligonucleotides. Immunoblotting revealed that after 3, 4, or 5 d, knockdown effectiveness was ≥90% and that expression of other Golgi proteins, including p115 and GRASP65, was not affected (Figure 1A). Immunofluorescence was used to determine knockdown on a per-cell basis. Cells were first arrested and analyzed in S phase because the Golgi ribbon is intact during this stage of the cell cycle (Feinstein and Linstedt, 2007). Although the antibody specifically recognized GRASP55 by immunoblot, a moderate degree of nonspecific staining was observed by immunofluorescence, much of it nuclear. Nonetheless, a clear loss of Golgi-localized staining was evident in most cells after knockdown (Figure 1B). Significantly, the Golgi ribbon was disrupted in cells that lacked GRASP55 staining. The observed phenotype was similar to that previously noted after GM130 or GRASP65 knockdown (Puthenveedu et al., 2006), and quantification using the same image analysis protocol indicated a similar degree of Golgi fragmentation (Figure 1C).
Figure 1.
Depletion of GRASP55 mimics G2 phase Golgi fragmentation. (A) Extracts from HeLa cells lysed 72, 96, and 120 h after transfection with either control or GRASP55 siRNA were analyzed by immunoblotting using anti-GRASP55 or antibodies against the indicated proteins. (B) Control- or GRASP55 siRNA-transfected cells were synchronized in S phase using thymidine or at the G2/M transition point by using olomoucine II, and the Golgi was visualized by staining for giantin (left) or GRASP55 (right). Bar, 4 μm. (C) Distinct Golgi objects per cell were estimated using fixed acquisition parameters and image thresholding, followed by counting using the Analyze Objects function in ImageJ (n = 4, ±SEM).
The analysis was then carried out for cells arrested at late G2 phase of the cell cycle when the ribbon undergoes MEK-dependent unlinking (Feinstein and Linstedt, 2007). As expected, control cells exhibited an unlinked Golgi, and the appearance of the Golgi in these G2 cells was indistinguishable from cells depleted of GRASP55 and fixed at either S phase or G2 phase (Figure 1, B and C). Thus, knockdown of GRASP55 is sufficient to unlink the Golgi, suggesting that inhibition of the protein could be the mechanism of late G2 phase Golgi fragmentation.
Fluorescence recovery after photobleaching was used to test for continuities among the fluorescent Golgi objects present before and after GRASP55 knockdown. As expected, continuity was readily demonstrated in control cells by the rapid recovery of fluorescence in bleached areas of the Golgi ribbon (Figure 2A). In contrast, fluorescent Golgi objects in GRASP55-depleted cells, even when they seemed optically contiguous, failed to recover significantly from photobleaching. These results were quantified for multiple experiments (Figure 2B), and representative movies are present in Supplemental Figure S1 and S2 movies.
Figure 2.
GRASP55 depletion severs the connections between Golgi stacks and inhibits normal protein glycosylation. (A) Representative cells stably expressing GalNAcT2-GFP were tested for Golgi integrity by photobleaching a Golgi segment with a brief laser pulse and measuring the return of Golgi fluorescence to the bleached spot. Bar, 10 μm. Corresponding movies are provided (see Supplemental Figures S1 and S2). (B) Fluorescence recovery was quantified by dividing GFP fluorescence within the bleached spot by the fluorescence within a nearby unbleached region in the same Golgi object, after subtracting background fluorescence from both. Each curve was further normalized by bracketing values between a minimum of 0 and a maximum of 1 (n = 3, ≥12 cells each, ±SEM). (C) To test the glycolytic processing of Golgi cargo, live, nonpermeabilized, control- or GRASP55-depleted cells were stained with fluorescently tagged GSII lectin and then rinsed, fixed and processed for immunofluorescence by using GP73 as a Golgi marker. Bar, 4 μm.
An earlier study found that disruption of the Golgi ribbon correlated with discontinuities in Golgi enzyme distribution and perturbed sialylation, resulting in an increase in nonsialylated proteins detectable on the cell surface (Puthenveedu et al., 2006). This was also the case for cells exhibiting an unlinked Golgi due to GRASP55 depletion. Compared with control transfected cells, cells transfected with GRASP55 siRNA exhibited a striking increase in surface binding of Cy5-labeled GS-II lectin purified from Griffonia simplicifolia, which selectively binds terminal N-acetyl-d-glucosamine (Figure 2C). This provides additional evidence that Golgi linking is important for proper glycolytic processing, at least in cultured mammalian cells.
Live Imaging of Golgi Reassembly in the Absence of GRASP55
To evaluate the importance of GRASP55 in the assembly of Golgi membranes, two live imaging assays were performed using a GFP-tagged copy of the Golgi enzyme N-acetylgalactosaminyl transferase-2 (GalNAcT2-GFP) as a Golgi marker. First, de novo biogenesis out of the ER was examined using washout of brefeldin A and H89 (Puri and Linstedt, 2003). Brefeldin A redistributes Golgi components into the ER and IC-like compartments, and H89 causes collapse of the IC-like compartments into the ER. The kinetics of Golgi assembly was indistinguishable for control and GRASP55 knockdown cells (data not shown). In terms of the appearance of the Golgi during reassembly, the only noted difference was that control cells formed a juxtanuclear ribbon, whereas GRASP55 knockdown cells accumulated Golgi fragments that failed to link into a ribbon. Thus, within the limits of this assay, GRASP55 was required for ribbon formation, but not for earlier steps in Golgi assembly.
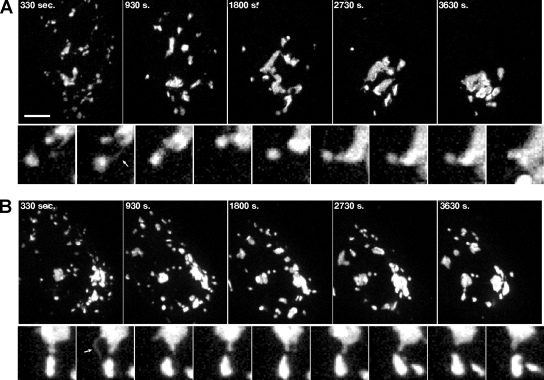
In the second assay, Golgi assembly was visualized after nocodazole washout. Nocodazole treatment disrupts microtubules, causing reorganization of the Golgi into multiple dispersed ministacks positioned adjacent to ER exit sites. On nocodazole washout of control cells, the ministacks rapidly moved to a juxtanuclear position and reestablished the ribbon (Supplemental Figure S3 movie). Inward movement was also apparent in GRASP55 knockdown cells, although the final degree of Golgi membrane coalescence was impaired and the Golgi membranes remained fragmented (Supplemental Figure S4 movie). Interestingly, numerous membrane tubules were observed during reassembly of control as well as GRASP55-depleted Golgi. In control cells, these tubules often created an apparent tubular “bridge” between two Golgi objects. The bridge was relatively stable over time, and it frequently resulted in one of the connected Golgi objects moving toward the other and seeming to fuse (Figure 3A, see inset showing tubule marked by arrow and followed by Golgi coalescence). In contrast, in GRASP55 depleted cells, the Golgi membrane tubules rarely formed an apparent stable contact and movement of one object toward another followed by apparent fusion was never observed (Figure 3B, see inset showing tubular extension but absence of Golgi coalescence).
Figure 3.
GRASP55 is required for the reformation of Golgi ribbons. (A and B) HeLa cells treated with nocodazole for 3 h to redistribute the Golgi to peripheral ministacks were rinsed repeatedly, and then they were transferred to chambers for live imaging. Confocal image stacks were collected every 30 s and collapsed into 2D images for analysis (larger frames). Smaller frames represent enlargements shown at 60-s intervals to illustrate tubulation and membrane capture that was frequently observed during Golgi reassembly. Movies are representative of at least three independent experiments per treatment. Corresponding movies are provided (Supplemental Figures S3 and S4).
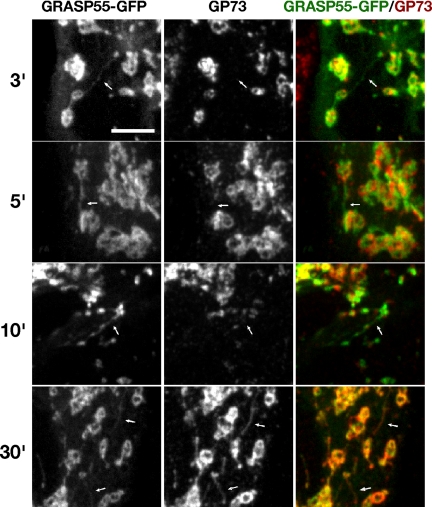
This observation suggested that Golgi ribbon formation may be mediated by tubular contacts, a potentially efficient mechanism akin to microtubule search and capture for cisternal membranes to probe for cognate fusion partners. Furthermore, GRASP55 may mediate the formation of a stable contact between a tubular cisternal extension and a cognate cisterna. To test whether GRASP55 is present in the observed tubules, we first imaged GRASP55-GFP during nocodazole washout. Although the labeling was relatively weak in cells with properly targeted GRASP55-GFP, abundant GRASP55-GFP tubules were evident forming connections between discrete Golgi objects during the earliest stages of postnocodazole Golgi assembly (Supplemental Figure S5 movie). Next, we examined fixed time points to test whether GRASP55-GFP might be enriched in the tubules relative to the cisternal Golgi marker GP73. Indeed, during the early phases of Golgi assembly, tubules were labeled primarily with GRASP55 (Figure 4, 3–10 min). In contrast, tubules observed at later time points, particularly in slowly reassembling Golgi, often contained detectable amounts of both proteins (Figure 4, 30 min). Thus, GRASP55 is present in tubular extensions that form just before Golgi membrane coalescence, suggesting that assembly of dispersed mammalian Golgi stacks is initiated by contact via tubules labeled with GRASP55 and that this early contact may form a basis for the rapid and cognate fusion of Golgi membranes into a contiguous ribbon.
Figure 4.
GRASP55 selectively labels tubules that connect reassembling Golgi elements. To determine the relative abundance of golgins and resident proteins on cisternal bridging tubules, HeLa cells expressing GRASP55-GFP were fixed at designated time points after nocodazole washout, and then they were processed for immunofluorescence by using and antibody against the cis-Golgi resident protein GP73. Arrows, tubules labeled with either GRASP55 or GP73. Results are representative of three separate experiments. A movie of GRASP55-GFP labeled Golgi assembly after nocodazole washout is provided as a supplemental movie (see Supplemental Figure S5).
Golgi Stacking, Compartmentalization, and Transport Do Not Require GRASP55
Transmission electron microscopy was used to characterize the GRASP55 knockdown phenotype in further detail. Both control and GRASP55-depleted cells exhibited Golgi membranes as flattened stacks with vesicles gathered near the stack rims (Figure 5, A and B). Comparable numbers of flattened cisternae per stack were observed (our unpublished observation). However, stack profiles were significantly shorter in GRASP55-depleted cells. This was clearly evident in the computed average stack length, which decreased from 1.5 μm in controls to 0.5 μm in cells after GRASP55 knockdown (Figure 5C), and also in a histogram reporting the distribution of measured Golgi stack lengths for one representative experiment (Figure 5D). Note that because two-dimensional transmission electron micrograph (TEM) thin sections often do not contain the full length of control Golgi ribbons, the difference is likely underestimated. To give a more thorough comparison of the difference between control and knockdown Golgi ribbons, serial sections were acquired, and they are presented after reconstruction into 3D volumes (Figure 5, E and F; also see Supplemental Figures S6 and S7 movies). There was a clear fragmentation evident of the Golgi ribbon into short stacks that rarely coaligned with one another, in striking contrast to the collinear arrays of Golgi stacks forming the ribbon in control cells.
Figure 5.
Transmission electron microscopy of GRASP55-depleted Golgi. (A and B) Representative TEM thin sections showing Golgi from control and GRASP55-depleted cells. Bar, 500 nm. (C) Quantitative representation of Golgi ribbon length in thin sections from control and GRASP55-depleted cells. The length of membrane profiles was estimated using the freehand line tool in ImageJ (n = 3, ≥20 profiles each, ±SEM). (D) Golgi stack lengths from a representative experiment were divided into size class bins and presented in histogram format to illustrate the distinct distributions of stack lengths in the presence (black diamonds) and absence (gray squares) of GRASP55. Note that despite the variation evident in the histogram, the error in panel C is small because the average stack length was consistent across multiple experiments. (E) Masks were drawn over Golgi membranes taken from a contiguous stack of TEM serial sections (representative sections are shown in insets) and reassembled into a 3D volume. The full 3D representations are provided as supplemental movies (see Supplemental Figures S6 and S7).
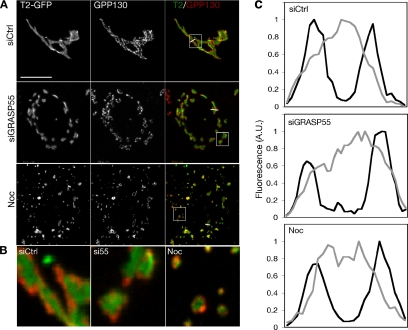
Colocalization analysis was used to estimate marker compartmentalization based on comparison of cis-localized GPP130 with GalNAcT2-GFP. As expected if the two labels indeed mark distinct regions of each physical object, every GPP130-positive object was colabeled with adjacent but not fully overlapping GalNAcT2-GFP (Figure 6A). Adjacent rather than overlapping staining was also observed in the unlinked Golgi objects present in cells after either GRASP55 knockdown or nocodazole treatment (Figure 6A). The distinct localization of the markers is visible in enlargements (Figure 6B), and it is quantitatively demonstrated in transect lines drawn across Golgi objects (Figure 6C). A separate quantitative analysis using Pearson's coefficient, which measures the pixel-by-pixel correlation in gray intensities for two labels, also did not show a significant difference between the treatments (unpublished data). These results, together with the morphological evidence of stacked cisternae present (Figure 5), indicate that stacking and apparent compartmentalization of resident proteins do not require GRASP55.
Figure 6.
Evidence of marker compartmentalization after GRASP55 depletion. (A) HeLa cells expressing the medial resident protein GalNAcT2-GFP and either control (top) or GRASP55 depleted (middle) or treated with nocodazole for 3 h (bottom) were fixed and processed for immunofluorescence by using antibodies against the cis-Golgi marker GPP130. Bar, 4 μm. (B) An enlarged selection from each of the RGB images above (white squares) is presented to emphasize the physically distinct cis and medial staining in all three treatments. (C) Representative linear transects through Golgi (white lines in A) showing the distinct fluorescence intensity profiles for GPP130 (black traces) and GalNAcT2-GFP (gray traces).
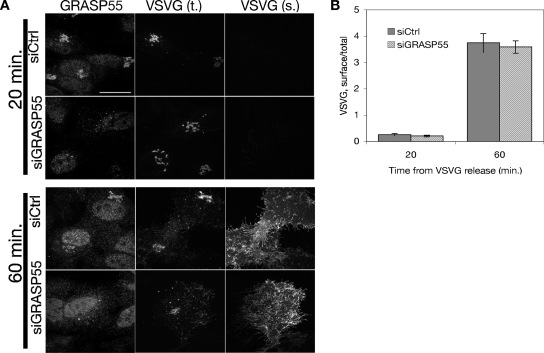
Anterograde cargo transport was assayed using the cargo marker ts045 VSVG-GFP, which unfolds at its restrictive temperature (40°C) and arrests in the ER. Typically, VSVG concentrates in the Golgi roughly 20 min after transfer to a permissive temperature (32°C), and it reaches the plasma membrane by 60–90 min. Indeed, in both control and GRASP55-depleted cells, VSVG accumulated in the Golgi at 20 min after release, and it was present at the surface within 60 min (Figure 7A). The data, quantified as surface/total VSVG fluorescence, support the conclusion that VSVG trafficking is normal in the absence of GRASP55-mediated Golgi linking (Figure 7B). Although it remains possible that a subtler defect exists outside the scope of our analysis, we observed no gross defect in anterograde traffic.
Figure 7.
Anterograde traffic through the Golgi in GRASP55-depleted cells. (A) Golgi transport was measured after release of ts045 VSVG-GFP from a 40°C ER block by shift to 32°C. Total VSVG was measured using GFP fluorescence, whereas surface VSVG was measured by staining live cells with a monoclonal antibody against a lumenal fragment of VSVG. (B) Surface and total fluorescence values were quantified and represented as ratios to indicate the relative amount of VSVG on the plasma membrane. Data shown are representative values from one of three experiments.
Mutation Mimicking ERK Phosphorylation Disrupts GRASP55 Activity
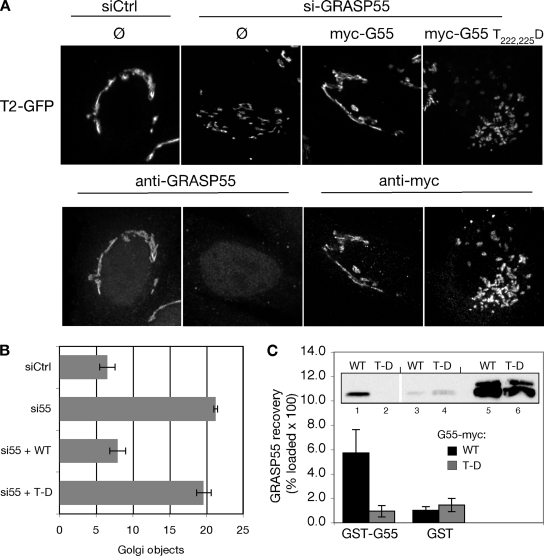
If GRASP55 actively promotes linkage of the mammalian Golgi ribbon, then it is possible that premitotic phosphorylation of the protein by ERK is the first necessary step in preparing the Golgi for mitosis. To test the influence of GRASP55 phosphorylation on Golgi linking, we substituted aspartic acids in place of two threonines known to be mitotically phosphorylated by ERK (Jesch et al., 2001). Gene replacement after knockdown was carried out as described previously (Puthenveedu and Linstedt, 2004), with myc-tagged GRASP55 wild-type or myc-tagged GRASP55-T222,225D constructs. As expected, the Golgi ribbon was fragmented after knockdown, and expression of the GRASP55 wild-type construct restored an intact Golgi ribbon (Figure 8A). Significantly, although GRASP55-T222,225D was equally expressed and also targeted to Golgi membranes, expression of this construct failed to rescue the ribbon structure. This finding was supported by quantitative analysis of multiple experiments (Figure 8B).
Figure 8.
Aspartic acid substitution at threonines 222 and 225 inhibits Golgi linking and oligomerization activities of GRASP55. (A) Golgi ribbon integrity was assessed in control (siCtrl) or GRASP55 (siGRASP55) knockdown cells without gene replacement (Ø) or after replacement with myc-tagged GRASP55 or GRASP55 T222,225D. Images show GalNAcT2-GFP fluorescence (top) and in the same cells either anti-GRASP55 staining or anti-myc staining (bottom). Bar, 4 μm. (B) The number of Golgi objects per cell was counted using the automated protocol described in the Figure 1 legend (n = 4, ±SEM). (C) Extracts from HeLa cells transfected with either wild-type GRASP55 (WT) or GRASP55 T222,225D (T-D) were incubated with GST-GRASP55 (lanes 1 and 2) or GST alone (lanes 3 and 4) immobilized on glutathione-agarose beads. Recovery was determined by immunoblotting with anti-myc antibodies. Values shown represent the percentage of loaded protein (lanes 5–6) bound to beads × 100 (n = 5, ±SEM).
Based on its similarity to GRASP65, it could be proposed that homo-oligomerization of GRASP55 cross-bridges membranes to form the Golgi ribbon, and this activity might be inhibited by phosphorylation (Wang et al., 2003, 2005; Puthenveedu et al., 2006; Feinstein and Linstedt, 2007). To test whether GRASP55 can interact with itself, purified GST-tagged GRASP55 immobilized on agarose beads was incubated with extracts prepared from HeLa cells transiently expressing myc-tagged GRASP55. Background binding was estimated using an equivalent volume of beads coated with GST alone. Consistent with GRASP55 self-interaction, myc-tagged GRASP55 was specifically recovered by GST-GRASP55 (Figure 8C). In contrast, no binding above background was detected when the extract contained myc-tagged GRASP55-T222,225D (Figure 8C). The failure of GRASP55-T222,225D to either rescue Golgi linking in vivo or to form oligomers in vitro suggests that the cross-bridging activity of GRASP55 is regulated by phosphorylation.
MEK/ERK Mediates Mitotic Progression via GRASP55 Phosphorylation
As described in the Introduction, MEK1 promotes the G2/M transition by mediating unlinking of the Golgi ribbon in late G2 phase of the cell cycle (Feinstein and Linstedt, 2007). We hypothesize that the mechanism involves MEK pathway phospho-inhibition of GRASP55-mediated Golgi linking. GRASP55 is a mitotic target of the MEK/ERK pathway (Jesch et al., 2001), and as shown above, a phospho-mimic version of GRASP55 fails to link the Golgi. Furthermore, a nonphosphorylatable version of GRASP55 blocks both G2 Golgi unlinking and normal G2/M kinetics (Feinstein and Linstedt, 2007).
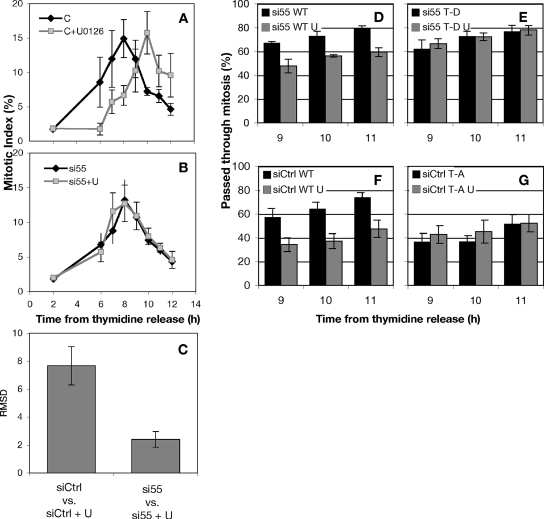
If the MEK inhibitor U0126 delays cyclin-dependent protein kinase (CDK)1 activation by preventing MEK/ERK inhibition of GRASP55, then GRASP55 knockdown would be expected to suppress this inhibition. As a test, control and GRASP55-depleted cells were synchronized, and then they were allowed to progress into M phase in the presence or absence of U0126. U0126 induced a 2-h delay in mitotic entry in control cells (Figure 9A), but it failed to induce a delay in cells depleted of GRASP55 (Figure 9B). Analysis by root mean squared deviation (RMSD) measurement, which estimates the net difference between two data series, indicated that the difference between normal and MEK-inhibited mitotic progression was reduced dramatically when GRASP55 was depleted (Figure 9C). Furthermore, we did not observe any defect in cell division as measured by the prevalence of mitotic cells in the unsynchronized population (our unpublished observation). These data suggest that the primary role of MEK/ERK signaling in facilitating CDK1 activation is via the Golgi and that the MEK/ERK pathway may directly unlink the Golgi by phospho-inhibition GRASP55.
Figure 9.
MEK1 promotes mitosis at G2/M by phosphorylation of GRASP55 at T222 and T225. (A) The percentage of cells in mitosis (mitotic index) was measured after thymidine release. The MEK1/2 inhibitor U0126 or 0.05% dimethyl sulfoxide carrier were added at 6 h after release. Tandem experiments were carried out in cells transfected with control siRNA (A and B) or siRNA against GRASP55 (B). (C) The degree to which MEK1 inhibition delays mitosis was quantified using an RMSD comparison of control and U0126-treated cells. (D–G) A postmitotic couplet assay was used to estimate the abundance of postmitotic HeLa cells expressing wild-type (D) or nonphosphorylatable GRASP55 (G55 T222,225A; E). Conversely, a functional gene replacement strategy was used (Puthenveedu et al., 2006) to compare wild-type expressors (F) with cells expressing a mutant GRASP55 designed to mimic mitotic ERK phosphorylation (G55 T222,225D; G).
Because absence of GRASP55 suppressed the MEK requirement at G2/M, we next tested whether GRASP55 phosphorylation itself would be sufficient to suppress the MEK requirement. Endogenous GRASP55 was replaced with myc-tagged wild-type or the phospho-mimic version (GRASP55-T222,225D), and cell cycle progression kinetics was determined in the presence or absence of U0126. In transiently transfected cells, gene replacement after knockdown is only evident in a fraction of the total population. Therefore, a previously described morphological assay was used to assess cell cycle progression restricted to transfected cells (Feinstein and Linstedt, 2007). The assay determines the fraction of myc-positive cells that have passed through mitosis at various times after release from an S phase block. Cultures were grown at low density and sparsely transfected so that postmitotic cells were clearly identified as a pair of closely adjacent cells with a similar level of myc staining. Postmitotic pairs were scored as a single cell that had progressed through mitosis. As expected U0126 addition caused a significant decrease in the number of postmitotic cells expressing wild-type GRASP55 as a replacement construct at 9, 10, and 11 h after thymidine release (Figure 9D). In striking contrast, knockdown cells replaced with myc-tagged GRASP55-T222,225D seemed unaffected by U0126 addition (Figure 9E). As a control for this experiment, a nonphosphorylatable version of GRASP55, also myc-tagged (GRASP55-T222,225A), was transiently expressed in otherwise normal cells. Consistent with published observations (Feinstein and Linstedt, 2007), and compared with matched controls (Figure 9F), GRASP55-T222,225A expression decreased the prevalence of postmitotic cells at the 9-, 10-, and 11-h time points regardless of the presence or absence of U0126 (Figure 9G). Thus, constitutive GRASP55 phosphorylation bypassed the MEK requirement at G2/M. The converse also proved true, in that preventing GRASP55 phosphorylation recapitulated the effect of MEK inhibition. Together, these observations indicate that MEK/ERK phosphorylation of GRASP55 is the means by which MEK promotes G2/M.
DISCUSSION
Recent results begin to elucidate a novel step in mitotic progression that requires Golgi-specific signaling via the mitogen-activated protein kinase kinase MEK1 to alleviate a block of G2/M progression (Colanzi et al., 2000; Sutterlin et al., 2002; Colanzi et al., 2007; Feinstein and Linstedt, 2007). Specifically, in late G2 phase MEK1 activity triggers unlinking of the Golgi ribbon, and this facilitates activation of CDK1 (Feinstein and Linstedt, 2007). An important putative player is GRASP55, which is the primary known Golgi target for mitotic ERK signaling (Jesch et al., 2001) and the phosphorylation of which seems important for G2 phase Golgi fragmentation (Feinstein and Linstedt, 2007). The present study shows that GRASP55 depletion causes unlinking of the Golgi and that under this condition MEK signaling is no longer required for timely CDK1 activation. Golgi functions distinct from lateral linking such as compartmentalization, stacking and anterograde cargo transport did not seem to require GRASP55. Thus, GRASP55 is specifically required for Golgi ribbon formation. Furthermore, a version of GRASP55 containing aspartic acid substitutions at the known ERK phosphorylation sites failed to link the Golgi. GRASP55-T222,225D was also defective in oligomer formation, supporting a model in which the homotypic fusion underlying Golgi ribbon formation depends on membrane cross-bridging by GRASP55 transoligomers.
This model for the function of mammalian GRASP proteins largely agrees with previous work on Golgi assembly (Barr et al., 1997; Shorter et al., 1999; Wang et al., 2003, 2005), but its relevance to other described functions for GRASP55 and GRASP65 remains to be determined. GRASP55 binds and may facilitate transport of the transmembrane growth factor α (Kuo et al., 2000), and both GRASP proteins bind members of the p24 protein family, thereby inhibiting p24 recycling to the ER (Barr et al., 2001). The GRASP homologues in Dictyostelium and Drosophila melanogaster, both of which organisms have one rather than two GRASP proteins, do not organize the nonlinked Golgi but rather contribute to unconventional secretion (Kinseth et al., 2007; Schotman et al., 2008), suggesting that linking of Golgi cisternae may be a later step in the evolutionary development of these proteins. Whether these selective transport functions of the GRASP protein constitute an inseparable part of its structural function, e.g., in membrane tethering, or whether selective transport represents a second and distinct function remains a compelling and unanswered question.
MEK1/ERK/GRASP55 regulation of Golgi linking in mammalian cultured cells may constitute a Golgi structure-dependent cell cycle “checkpoint” at G2/M. Injecting peptides corresponding to the CDK1-phosphorylated fragment of GRASP65 caused a mitotic arrest that was proposed to result from a block in mitotic Golgi fragmentation (Sutterlin et al., 2002). The separate findings that MEK-ERK signaling is required for fragmentation of the Golgi during G2/M (Colanzi et al., 2000; Shaul and Seger, 2006) and that MEK-ERK signaling promotes mammalian G2/M progression (Wright et al., 1999) were integrated into a single model by the demonstration that the G2/M transition requires prefragmentation of the Golgi apparatus mediated by MEK1 (Feinstein and Linstedt, 2007). It is worth noting that none of the studies thus far have shown that inhibiting MEK/ERK fragmentation of the Golgi causes an absolute block in mitosis, rather the timing of mitotic entry is delayed by several hours. Indeed, mitotic Golgi fragmentation is observed even in the absence of MEK signaling (Feinstein and Linstedt, 2007), supporting the hypothesis that a second pathway operating through CDK1 ultimately vesiculates the Golgi (Lowe et al., 1998). Consistent with this, in a permeabilized cell assay Golgi disassembly involves sequential activation of ribbon fragmentation by MEK and vesiculation by CDK1 (Kano et al., 2000). Given that CtBP/BARS promotes Golgi fission (Corda et al., 2006) and CtBP/BARS activity is required for the G2/M transition (Colanzi et al., 2007), the evidence presented supports a model in which MEK1/ERK-mediated phospho-inhibition of GRASP55 tips a dynamic structural balance in favor of CtBP/BARS-driven Golgi fissioning, thereby allowing timely activation of CDK1.
Knockdown of either GRASP55 (Figure 1) or GRASP65 (Puthenveedu et al., 2006) unlinks the Golgi, indicating that each protein is required for ribbon formation. The localization of GRASP65 and GRASP55 to cis- and medial-Golgi cisternae (Shorter et al., 1999), respectively, suggests that GRASP65 helps to establish lateral cisternal contacts, which are then maintained by GRASP55. There may also be a sequential relationship in the regulation of the two proteins at the onset of mitosis. Evidence suggests that GRASP55 is phosphorylated in late G2 before CDK1 activation, and, therefore, that its regulation occurs upstream of GRASP65 phosphorylation by CDK1 and CDK1-activated PLK1 (Elia et al., 2003a,b; Wang et al., 2003; Preisinger et al., 2005; Feinstein and Linstedt, 2007). If so, GRASP65 phosphorylation might be considered redundant with respect to unlinking the Golgi. Nevertheless, because these events occur in rapid succession GRASP65 phosphorylation likely contributes to rapid Golgi disassembly. Furthermore, as mentioned above, reagents including GRASP65 C-terminal peptides block or delay progression into mitosis, arguing that, at least under certain conditions, GRASP65 regulation is not redundant. It could be that GRASP65 phosphorylation feeds back somehow to sustain CDK1 activity or that the GRASP65 reagents blocked PLK1, and hence CDK1 activity, or that there is a role of GRASP65 in an early mitotic event having to do with spindle assembly. Alternatively, interfering with GRASP65 function by using these reagents may strengthen Golgi linkages such that they depend less on GRASP55. Clearly, further work is needed to clarify the exact relationship between the two GRASP proteins and their regulation at mitosis.
Another outstanding question concerns the step directly upstream of MEK1-ERK in G2 phase. Raf1 likely activates MEK1 (Colanzi et al., 2003), but the well-understood pathways to MEK1 activation seem to be unlikely in this case. M phase MEK1 activity is uncoupled from growth factor receptors (Dangi and Shapiro, 2005) and Golgi structure does not seem to be controlled by growth factor/MAPK signaling at other cell cycle stages. Intriguingly, distinct scaffolding proteins mediate MEK-ERK signaling on the Golgi (Torii et al., 2004), and it is an ERK1 splice variant, ERK1c, that seems to drive Golgi fragmentation (Shaul and Seger, 2006). Thus, Golgi fragmentation likely depends on a specialized MAPK pathway responsive to a nontraditional upstream signal.
Intriguingly, recent work has suggested specific advantages conferred by coalescence of distributed ministacks into a pericentriolar ribbon. Defective lateral linking of the Golgi apparatus in HeLa cells is associated with nonuniform distribution of Golgi resident enzymes and, importantly, Golgi-specific defects in protein glycosylation (Puthenveedu et al., 2006). Similar glycosylation defects were observed in the present study when GRASP55-mediated Golgi linking was inhibited. Thus, Golgi linking may be required to accommodate the complex protein glycosylation requirements that arose during metazoan evolution (Drickamer and Taylor, 1998). Diverse processes seem to depend on precise control of protein glycan “codes.” Glycosyltransferase knockouts frequently cause strong developmental defects in mammals (Lowe and Marth, 2003; Angata et al., 2006), and altered protein glycosylation is frequently associated with malignant cancer (Sato and Furukawa, 2007).
An important consequence of forming a contiguous Golgi ribbon is that the ribbon must be disassembled to allow Golgi partitioning into daughter cells during mitosis. The coupling of Golgi unlinking to CDK1 activation could thus help ensure that the ribbon is broken before mitotic entry. In this sense the GRASP proteins may have diversified from a more primordial function in tethering membranes during homotypic fusion into factors participating in the control of cell cycle progression.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Fred Lanni for generous assistance with fluorescence recovery after photobleaching microscopy and members of the Linstedt laboratory for critical reading and advice. Funding was provided by grants RSG-03-148-01-CSM and GM-56779 (to A.D.L.).
Abbreviations used:
- CDK
cyclin-dependent protein kinase
- GalNAcT2
N-acetylgalactosaminyl transferase-2
- GFP
green fluorescent protein
- GM130
Golgi matrix protein 130 kDa
- GRASP
Golgi reassembly protein
- GST
glutathione transferase
- MAPK
mitogen-activated protein kinase
- RNAi
RNA interference
- siRNA
small interfering RNA.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-11-1200) on April 23, 2008.
REFERENCES
- Allan V. J., Thompson H. M., McNiven M. A. Motoring around the Golgi. Nat. Cell Biol. 2002;4:E236–E242. doi: 10.1038/ncb1002-e236. [DOI] [PubMed] [Google Scholar]
- Angata K., Lee W., Mitoma J., Marth J. D., Fukuda M. Cellular and molecular analysis of neural development of glycosyltransferase gene knockout mice. Methods Enzymol. 2006;417:25–37. doi: 10.1016/S0076-6879(06)17003-2. [DOI] [PubMed] [Google Scholar]
- Bachert C., Fimmel C., Linstedt A. D. Endosomal trafficking and proprotein convertase cleavage of cis Golgi protein GP73 produces marker for hepatocellular carcinoma. Traffic. 2007;8:1415–1423. doi: 10.1111/j.1600-0854.2007.00621.x. [DOI] [PubMed] [Google Scholar]
- Barr F. A., Preisinger C., Kopajtich R., Korner R. Golgi matrix proteins interact with p24 cargo receptors and aid their efficient retention in the Golgi apparatus. J. Cell Biol. 2001;155:885–891. doi: 10.1083/jcb.200108102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr F. A., Puype M., Vandekerckhove J., Warren G. GRASP65, a protein involved in the stacking of Golgi cisternae. Cell. 1997;91:253–262. doi: 10.1016/s0092-8674(00)80407-9. [DOI] [PubMed] [Google Scholar]
- Bonazzi M., et al. CtBP3/BARS drives membrane fission in dynamin-independent transport pathways. Nat. Cell Biol. 2005;7:570–580. doi: 10.1038/ncb1260. [DOI] [PubMed] [Google Scholar]
- Colanzi A., Carcedo C. H., Persico A., Cericola C., Turacchio G., Bonazzi M., Luini A., Corda D. The Golgi mitotic checkpoint is controlled by BARS-dependent fission of the Golgi ribbon into separate stacks in G2. EMBO J. 2007;26:2465–2476. doi: 10.1038/sj.emboj.7601686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colanzi A., Corda D. Mitosis controls the Golgi and the Golgi controls mitosis. Curr. Opin. Cell Biol. 2007;19:386–393. doi: 10.1016/j.ceb.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Colanzi A., Deerinck T. J., Ellisman M. H., Malhotra V. A specific activation of the mitogen-activated protein kinase kinase 1 (MEK1) is required for Golgi fragmentation during mitosis. J. Cell Biol. 2000;149:331–339. doi: 10.1083/jcb.149.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colanzi A., Sutterlin C., Malhotra V. RAF1-activated MEK1 is found on the Golgi apparatus in late prophase and is required for Golgi complex fragmentation in mitosis. J. Cell Biol. 2003;161:27–32. doi: 10.1083/jcb.200208099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corda D., Colanzi A., Luini A. The multiple activities of CtBP/BARS proteins: the Golgi view. Trends Cell Biol. 2006;16:167–173. doi: 10.1016/j.tcb.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Dangi S., Shapiro P. Cdc2-mediated inhibition of epidermal growth factor activation of the extracellular signal-regulated kinase pathway during mitosis. J. Biol. Chem. 2005;280:24524–24531. doi: 10.1074/jbc.M414079200. [DOI] [PubMed] [Google Scholar]
- Drickamer K., Taylor M. E. Evolving views of protein glycosylation. Trends Biochem. Sci. 1998;23:321–324. doi: 10.1016/s0968-0004(98)01246-8. [DOI] [PubMed] [Google Scholar]
- Elia A. E., Cantley L. C., Yaffe M. B. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 2003a;299:1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- Elia A. E., Rellos P., Haire L. F., Chao J. W., Ivins F. J., Hoepker K., Mohammad D., Cantley L. C., Smerdon S. J., Yaffe M. B. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 2003b;115:83–95. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- Feinstein T. N., Linstedt A. D. Mitogen-activated protein kinase kinase 1-dependent Golgi unlinking occurs in G2 phase and promotes the G2/M cell cycle transition. Mol. Biol. Cell. 2007;18:594–604. doi: 10.1091/mbc.E06-06-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo Carcedo C., Bonazzi M., Spano S., Turacchio G., Colanzi A., Luini A., Corda D. Mitotic Golgi partitioning is driven by the membrane-fissioning protein CtBP3/BARS. Science. 2004;305:93–96. doi: 10.1126/science.1097775. [DOI] [PubMed] [Google Scholar]
- Jesch S. A., Lewis T. S., Ahn N. G., Linstedt A. D. Mitotic phosphorylation of Golgi reassembly stacking protein 55 by mitogen-activated protein kinase ERK2. Mol. Biol. Cell. 2001;12:1811–1817. doi: 10.1091/mbc.12.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano F., Takenaka K., Yamamoto A., Nagayama K., Nishida E., Murata M. MEK and Cdc2 kinase are sequentially required for Golgi disassembly in MDCK cells by the mitotic Xenopus extracts. J. Cell Biol. 2000;149:357–368. doi: 10.1083/jcb.149.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinseth M. A., Anjard C., Fuller D., Guizzunti G., Loomis W. F., Malhotra V. The Golgi-associated protein GRASP is required for unconventional protein secretion during development. Cell. 2007;130:524–534. doi: 10.1016/j.cell.2007.06.029. [DOI] [PubMed] [Google Scholar]
- Kuo A., Zhong C., Lane W. S., Derynck R. Transmembrane transforming growth factor-alpha tethers to the PDZ domain-containing, Golgi membrane-associated protein p59/GRASP55. EMBO J. 2000;19:6427–6439. doi: 10.1093/emboj/19.23.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linstedt A. D. Positioning the Golgi apparatus. Cell. 2004;118:271–272. doi: 10.1016/j.cell.2004.07.015. [DOI] [PubMed] [Google Scholar]
- Linstedt A. D., Hauri H. P. Giantin, a novel conserved Golgi membrane protein containing a cytoplasmic domain of at least 350 kDa. Mol. Biol. Cell. 1993;4:679–693. doi: 10.1091/mbc.4.7.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linstedt A. D., Mehta A., Suhan J., Reggio H., Hauri H. P. Sequence and overexpression of GPP130/GIMPc: evidence for saturable pH-sensitive targeting of a type II early Golgi membrane protein. Mol. Biol. Cell. 1997;8:1073–1087. doi: 10.1091/mbc.8.6.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J. B., Marth J. D. A genetic approach to Mammalian glycan function. Annu. Rev. Biochem. 2003;72:643–691. doi: 10.1146/annurev.biochem.72.121801.161809. [DOI] [PubMed] [Google Scholar]
- Lowe M., Rabouille C., Nakamura N., Watson R., Jackman M., Jamsa E., Rahman D., Pappin D. J., Warren G. Cdc2 kinase directly phosphorylates the cis-Golgi matrix protein GM130 and is required for Golgi fragmentation in mitosis. Cell. 1998;94:783–793. doi: 10.1016/s0092-8674(00)81737-7. [DOI] [PubMed] [Google Scholar]
- Marra P., Salvatore L., Mironov A., Jr, Di Campli A., Di Tullio G., Trucco A., Beznoussenko G., Mironov A., De Matteis M. A. The biogenesis of the Golgi ribbon: the roles of membrane input from the ER and of GM130. Mol. Biol. Cell. 2007;18:1595–1608. doi: 10.1091/mbc.E06-10-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preisinger C., Korner R., Wind M., Lehmann W. D., Kopajtich R., Barr F. A. Plk1 docking to GRASP65 phosphorylated by Cdk1 suggests a mechanism for Golgi checkpoint signalling. EMBO J. 2005;24:753–765. doi: 10.1038/sj.emboj.7600569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri S., Linstedt A. D. Capacity of the Golgi apparatus for biogenesis from the endoplasmic reticulum. Mol. Biol. Cell. 2003;14:5011–5018. doi: 10.1091/mbc.E03-06-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu M. A., Bachert C., Puri S., Lanni F., Linstedt A. D. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat. Cell Biol. 2006;8:238–248. doi: 10.1038/ncb1366. [DOI] [PubMed] [Google Scholar]
- Puthenveedu M. A., Linstedt A. D. Evidence that Golgi structure depends on a p115 activity that is independent of the vesicle tether components giantin and GM130. J. Cell Biol. 2001;155:227–238. doi: 10.1083/jcb.200105005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu M. A., Linstedt A. D. Gene replacement reveals that p115/SNARE interactions are essential for Golgi biogenesis. Proc. Natl. Acad. Sci. USA. 2004;101:1253–1256. doi: 10.1073/pnas.0306373101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Furukawa K. Sequential action of Ets-1 and Sp1 in the activation of the human β-1,4-galactosyltransferase V gene involved in abnormal glycosylation characteristic of cancer cells. J. Biol. Chem. 2007;282:27702–27712. doi: 10.1074/jbc.M611862200. [DOI] [PubMed] [Google Scholar]
- Schotman H., Karhinen L., Rabouille C. dGRASP-mediated noncanonical integrin secretion is required for Drosophila epithelial remodeling. Dev. Cell. 2008;14:171–182. doi: 10.1016/j.devcel.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Sevier C. S., Weisz O. A., Davis M., Machamer C. E. Efficient export of the vesicular stomatitis virus G protein from the endoplasmic reticulum requires a signal in the cytoplasmic tail that includes both tyrosine-based and di-acidic motifs. Mol. Biol. Cell. 2000;11:13–22. doi: 10.1091/mbc.11.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul Y. D., Seger R. ERK1c regulates Golgi fragmentation during mitosis. J. Cell Biol. 2006;172:885–897. doi: 10.1083/jcb.200509063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J., Watson R., Giannakou M. E., Clarke M., Warren G., Barr F. A. GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. EMBO J. 1999;18:4949–4960. doi: 10.1093/emboj/18.18.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterlin C., Hsu P., Mallabiabarrena A., Malhotra V. Fragmentation and dispersal of the pericentriolar Golgi complex is required for entry into mitosis in mammalian cells. Cell. 2002;109:359–369. doi: 10.1016/s0092-8674(02)00720-1. [DOI] [PubMed] [Google Scholar]
- Sutterlin C., Polishchuk R., Pecot M., Malhotra V. The Golgi-associated protein GRASP65 regulates spindle dynamics and is essential for cell division. Mol. Biol. Cell. 2005;16:3211–3222. doi: 10.1091/mbc.E04-12-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii S., Kusakabe M., Yamamoto T., Maekawa M., Nishida E. Sef is a spatial regulator for Ras/MAP kinase signaling. Dev. Cell. 2004;7:33–44. doi: 10.1016/j.devcel.2004.05.019. [DOI] [PubMed] [Google Scholar]
- Wang Y., Satoh A., Warren G. Mapping the functional domains of the Golgi stacking factor GRASP65. J. Biol. Chem. 2005;280:4921–4928. doi: 10.1074/jbc.M412407200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Seemann J., Pypaert M., Shorter J., Warren G. A direct role for GRASP65 as a mitotically regulated Golgi stacking factor. EMBO J. 2003;22:3279–3290. doi: 10.1093/emboj/cdg317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigert R., et al. CtBP/BARS induces fission of Golgi membranes by acylating lysophosphatidic acid. Nature. 1999;402:429–433. doi: 10.1038/46587. [DOI] [PubMed] [Google Scholar]
- Wright J. H., Munar E., Jameson D. R., Andreassen P. R., Margolis R. L., Seger R., Krebs E. G. Mitogen-activated protein kinase kinase activity is required for the G(2)/M transition of the cell cycle in mammalian fibroblasts. Proc. Natl. Acad. Sci. USA. 1999;96:11335–11340. doi: 10.1073/pnas.96.20.11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.