Abstract
The ammonium permease Mep2 is required for the induction of pseudohyphal growth, a process in Saccharomyces cerevisiae that occurs in response to nutrient limitation. Mep2 has both a transport and a regulatory function, supporting models in which Mep2 acts as a sensor of ammonium availability. Potentially similar ammonium permease-dependent regulatory cascades operate in other fungi, and they may also function in animals via the homologous Rh proteins; however, little is known about the molecular mechanisms that mediate ammonium sensing. We show that Mep2 is localized to the cell surface during pseudohyphal growth, and it is required for both filamentous and invasive growth. Analysis of site-directed Mep2 mutants in residues lining the ammonia-conducting channel reveal separation of function alleles (transport and signaling defective; transport-proficient/signaling defective), indicating transport is necessary but not sufficient to sense ammonia. Furthermore, Mep2 overexpression enhances differentiation under normally repressive conditions and induces a transcriptional profile that is consistent with activation of the mitogen-activated protein (MAP) kinase pathway. This finding is supported by epistasis analysis establishing that the known role of the MAP kinase pathway in pseudohyphal growth is linked to Mep2 function. Together, these data strengthen the model that Mep2-like proteins are nutrient sensing transceptors that govern cellular differentiation.
INTRODUCTION
Ammonia is an important nutrient for many microorganisms and plants, and in animals the catabolism of amino acids produces ammonia as a by-product that must be excreted from the body to prevent its toxic accumulation. The Amt/Mep/Rh proteins form an evolutionary conserved family of permeases that mediate ammonium transport across cell membranes (reviewed in Andrade and Einsle, 2007). Structural and biochemical studies of bacterial members of this transporter family reveal that the Amt/Mep/Rh proteins form a trimeric complex that facilitates passive diffusion of ammonia gas (Khademi et al., 2004; Zheng et al., 2004; Andrade et al., 2005; Ishikita and Knapp, 2007). Conserved key residues required for ammonium translocation have been identified by site-directed mutagenesis and provide evidence that the mechanism of ammonium transport is evolutionarily conserved (Javelle, et al., 2006; Marini et al., 2006).
Previous studies support the model that certain members of the Amt/Mep/Rh family play a direct role in fungal development as ammonium receptors. The high-affinity ammonium permeases of Saccharomyces cerevisiae, Candida albicans, and Ustilago maydis are required for filamentous growth in response to low ammonium conditions (Lorenz and Heitman, 1998a; Smith et al., 2003; Biswas and Morschhäuser, 2005). The fission yeast Schizosaccharomyces pombe undergoes haploid invasive growth during ammonium limitation, and this is dependent on the ammonium permease Amt1, and, to a lesser extent, its paralogue, Amt2 (Mitsuzawa, 2006). Haploid invasive growth and mating by the basidiomycetous fungus Cryptococcus neoformans is induced by ammonium limitation and requires the high-affinity ammonium permease, Amt2 (Rutherford et al., 2008). However, the mechanisms that link ammonium transport to development are not understood. One hypothesis is that these permeases act as ammonium sensors, or transceptors, that regulate downstream effector molecules (Lorenz and Heitman, 1998a). This mechanism couples the physical process of nutrient transport to nutrient sensing and is distinct from a regulatory mechanism that responds to the changing metabolized levels of a transported nutrient (Holsbeeks et al., 2004).
Current evidence supports the hypothesis that fungal high-affinity permeases function as ammonium sensors. S. cerevisiae cells lacking the high-affinity permease Mep2 do not undergo pseudohyphal growth or exhibit any change in the activity of nitrogen metabolic enzymes (Lorenz and Heitman, 1998a). The C-terminal cytoplasmic domain of the C. albicans Mep2 protein is essential for the induction of filamentous growth but dispensable for transport activity (Biswas and Morschhäuser, 2005). Therefore, the roles of Mep2 in ammonium transport and induction of filamentous growth are separable. Independently of its role in pseudohyphal growth, Mep2 acts as a transceptor in the cAMP-independent activation of the protein kinase A (PKA) pathway after ammonium addition to starved cells (Van Nuland et al., 2006). This rapid response is not dependent on ammonium metabolism and can be induced by the transport of the nonmetabolizable ammonium analogue methylamine, which establishes the paradigm of Mep2 as a transceptor. It is not clear whether Mep2 has an equivalent transceptor function during pseudohyphal growth or whether it interacts with any of the established signal transduction pathways that are essential for this process. Epistasis analysis is consistent with the RAS-cAMP being a possible downstream target of Mep2 (Lorenz and Heitman, 1998a); however, a direct interaction between Mep2 and any signaling pathway has not been demonstrated in relation to pseudohyphal growth.
In this report, we present evidence that Mep2 forms a multimeric complex localized to the cell membrane during pseudohyphal growth. Similar to S. pombe and C. neoformans, haploid S. cerevisiae cells undergo invasive growth in response to ammonium limitation and exhibit morphological changes analogous to those observed in pseudohyphal cells. We find that invasive growth requires Mep2, the Npr1 kinase, and elements of the PKA and mitogen-activated protein (MAP) kinase pathways. Furthermore, Mep2-dependent pseudohyphal and invasive growth requires two conserved histidine residues within the hydrophobic channel of Mep2 that have been predicted to be essential for ammonium translocation through the permease. Remarkably, constitutive Mep2 expression induces pseudohyphal growth on nitrogen-replete medium. Under these conditions, Mep2 induces a haploid- and diploid-specific transcriptional profile that includes genes known or predicted to be differentially regulated during pseudohyphal growth. Included within this group of genes are those controlled by the MAP kinase-regulated transcription factor Ste12. Consistent with a functional link between Mep2 and Ste12, constitutive MEP2 overexpression restores pseudohyphal growth in cells lacking PKA pathway elements but not in cells lacking Ste12. In accord with this finding, overexpression of Ste12, or of the MADS box transcription factor Mcm1, restores pseudohyphal growth in cells lacking Mep2 under ammonium limiting conditions. Therefore, the established role of the MAP kinase pathway in pseudohyphal growth may, in part, be due to its role as a downstream effector of the ammonium receptor function of Mep2. Collectively, these data further support a role for the Mep2 family of permeases as sensors of ammonium availability.
EXPERIMENTAL PROCEDURES
Strains and Growth Media
The S. cerevisiae strains used in this study are listed in Table 1. S. cerevisiae cells were grown in synthetic minimal medium with either 2% glucose, raffinose, or galactose as the carbon source. To induce pseudohyphal growth, cells were grown with 50 μM ammonium sulfate as the nitrogen source. Otherwise, cells were grown with the standard concentration of ammonium sulfate in yeast nitrogen base (5 g/l). Yeast cells were transformed as described previously (Schiestl and Gietz, 1989).
Table 1.
Yeast strains used in this study
| Strain | Reference |
|---|---|
| Congenic with ∑1278b | |
| HLY352 MATa/α ste12::LEU2/ste12::LEU2 | Liu et al. (1993) |
| MLY41 MATaura3-52 | Lorenz and Heitman (1997) |
| MLY54a MATaura3-52 npr1::LEU2 leu2::hisG | Lorenz and Heitman (1998a) |
| MLY54a/α MATa/α ura3-52/ura3-52 npr1::LEU2/npr1::LEU2 leu2::hisG/leu2::hisG | Lorenz and Heitman (1998a) |
| MLY61a/α MATa/α ura3-52/ura3-52 | Lorenz and Heitman (1997) |
| MLY104a MATaura3-52 mep1::LEU2 leu2::hisG | Lorenz and Heitman (1998a) |
| MLY108a MATaura3-52 mep2::G418 | Lorenz and Heitman (1998a) |
| MLY128a MATaura3-52 mep3::G418 | Lorenz and Heitman (1998a) |
| MLY132a MATaura3-52 gpa2::G418 | Lorenz and Heitman (1997) |
| MLY132a/α MATa/α ura3-52/ura3-52 gpa2::G418/gpa2::G418 | Lorenz and Heitman (1997) |
| MLY140a/α MATa/α ura3-52/ura3-52 ure2::G418/ure2::G418 | Lorenz and Heitman (1998a) |
| MLY183a MATaura3-52 tec1::G418 | Lorenz and Heitman (1998b) |
| MLY186a MATaura3-52 ras1::G418 | Lorenz unpublished |
| MLY187a MATaura3-52 ras2::G418 | Lorenz et al. (2000a) |
| MLY187a/α MATa/α ura3-52/ura3-52 ras2::G418/ras2::G418 | Lorenz et al. (2000a) |
| MLY216a MATaura3-52 ste12::G418 | Lorenz unpublished |
| MLY232a MATaura3-52 gpr1::G418 | Lorenz et al. (2000a) |
| MLY232a/α MATa/α ura3-52/ura3-52 gpr1::G418/gpr1::G418 | Lorenz et al. (2000a) |
| XPY5-1a MATaura3-52 tpk2::G418 | Pan and Heitman (1999) |
| XP5a/α MATa/α ura3-52/ura3-52 tpk2::G418/tpk2::G418 | Pan and Heitman (1999) |
| XPY14-1a MATaura3-52 tpk1::G418 tpk3::G418 | Pan and Heitman, (1999) |
| XPY207a MATaura3-52 she2::G418 | Pan and Heitman (2000) |
| Other strains | |
| SY1 MATaura3-52 leu2-3 his4-619 GAL sec6-4 | Novick et al. (1980) |
| Mep2-GFP MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 MEP2::GFP-HIS3 | Huh et al. (2003) |
Plasmids
The plasmids used in this study are listed in Table 2. All manipulations of DNA fragments and plasmids were carried out using standard procedures and the Escherichia coli strain DH5α (Sambrook et al., 1989). All DNA sequences encoding Mep2, epitope-tagged Mep2, and mutant derivatives of these were generated and inserted into the vector pRS316 (URA3) by homologous recombination (Ma et al., 1987). In each case, the MEP2 gene included its own promoter (1 kb), and, except in the Mep2-green fluorescent protein (GFP) fusion encoding plasmid and its mutant derivatives, the MEP2 terminator. The Mep2-GFP encoding sequences were amplified from the relevant strain of the yeast GFP clone collection to include the ADH1-terminating sequences (Huh et al., 2003). All Mep2-FLAG fusions contain the N4Q mutation to prevent glycosylation of Mep2 (Marini and André, 2000). MCM1, including its own promoter and terminator, was inserted into the vector pRS426 (URA3) by homologous recombination. All newly generated plasmid inserts were verified by DNA sequencing. The plasmid pYes2.1-Mep2, a kind gift from Mike Perlin (University of Louisville, Louisville, KY), was used to express MEP2 to high levels from the galactose-inducible promoter (Smith et al., 2003).
Table 2.
Plasmids used in this study
| Plasmid | Function | Reference |
|---|---|---|
| pJRH1 | Mep2-GFP CEN | This study |
| pJRH2 | Mep2-D186A-GFP CEN | This study |
| pJRH3 | Mep2-H194A-GFP CEN | This study |
| pJRH4 | Mep2-H348A-GFP CEN | This study |
| pJRH7 | Mep2-N4Q-FLAG CEN | This study |
| pJRH8 | Mep2-N4Q-H194A-FLAG CEN | This study |
| pJRH9 | Mep2-N4Q-H348A-FLAG CEN | This study |
| pJRH10 | Mep2-N4Q-D186A-FLAG CEN | This study |
| pJRH11 | Mep2-N4Q-T288A-FLAG CEN | This study |
| pJRH17 | MCM1 2μ | This study |
| pML151 | MEP2 2μ | Lorenz and Heitman (1998a) |
| pYES2.1-MEP2 | GAL-MEP2 2μ | Smith et al. (2003) |
| pNC252 | GAL-STE12 CEN | Liu et al. (1993) |
| pSC4 | STE12 CEN | Fields and Herskowitz (1987) |
| pSL1509 | STE11-4 CEN | Stevenson et al. (1992) |
| pMW2 | RAS2Val19 CEN | Ward et al. (1995) |
All novel plasmids were generated using pRS316 (URA3) except pJRH17 (pRS426-URA3).
Membrane Preparation, Immunoprecipitation, and Western Immunoblotting
The preparation of cell membranes was carried out as described previously (Galan et al., 1996). Briefly, yeast cells in the exponential growth phase were harvested by centrifugation, washed in distilled water, and suspended in lysis buffer (0.1 M Tris-HCl, pH 7.5, 0.15 M NaCl, 5 mM EDTA) and a mixture of proteinase inhibitors (cocktail IV; Calbiochem, San Diego, CA) and 0.5 mM phenylmethylsulfonyl fluoride. Glass beads (0.45 μm in diameter) were added, and the cells were lysed by vigorous vortex mixing for 3 min. The resulting homogenate was diluted threefold with lysis buffer and centrifuged at 3000 rpm for 3 min at 4°C. The plasma membrane-enriched fraction was collected by centrifugation for 45 min at 12,000 rpm at 4°C and then suspended in lysis buffer and trichloroacetic acid (10%). For Western analysis, the precipitates were neutralized and dissolved in 20 μl of 1 M Tris base plus 80 μl of sample buffer (100 mM Tris-HCl, pH 6.8, 4 mM EDTA, 4% SDS, 20% glycerol, and 0.02% bromphenol blue) containing 2% 2-mercaptoethanol and heated at 37°C for 15 min. For immunoprecipitation, 2-mercaptoethanol was omitted from the samples, which were diluted with 0.6 ml of TNET buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, and 1% Triton X-100) plus the proteinase inhibitors described. Western analysis was carried out using antibodies, either monoclonal ANTI-FLAG M2 (Sigma-Aldrich, St. Louis, MO), monoclonal anti-GFP (Roche Diagnostics, Indianapolis, IN), or anti-Pma1 (Cardenas et al., 1990), and the relevant horseradish peroxidase-conjugated anti-IgG second antibody, followed by ECL chemiluminescence (GE Healthcare, Chalfont St. Giles, United Kingdom). In the experiments using the npr1Δ strain, relative abundance of Mep2-FLAG was quantified using Quantity One version 4.6.2. (Bio-Rad, Hemel Hempstead, United Kingdom). Immunoprecipitation was carried out as described previously (Harashima and Heitman, 2002) by using EZview Red ANTI-FLAG M2 affinity gel (Sigma-Aldrich). Native Mep2 was resolved by suspending membrane samples in 1× Tris-glycine native sample buffer and separating on 6% NOVEX Tris-glycine gels with Tris-glycine native running buffer (Invitrogen).
Protein Localization
Cells containing plasmid expressed Mep2-GFP were grown in synthetic minimal media and examined for protein localization under a fluorescent microscope (Axioskop2 Plus; Carl Zeiss, Thornwood, NY).
Pseudohyphal and Invasive Growth
Cells undergoing pseudohyphal and invasive growth assays were grown for 6 d on low-ammonium SLAD medium (50 μM ammonium sulfate) and investigated as described previously (Harashima et al., 2006).
Microarray Analysis
S288C haploid and ∑1278b diploid wild-type strains containing a 2μ plasmid consisting of a GAL1/10-driven MEP2 gene were grown concurrently with isogenic strains containing an empty vector to mid-log phase in selective liquid medium plus 2% raffinose. MEP2 gene expression was induced for 3 h in 2% galactose. Comparison of the S288C and ∑1278b diploid wild-type strains was carried out in synthetic complete medium with 2% glucose. RNA preparation, hybridization, image acquisition, and data processing for microarrays were performed as described previously (Grigull et al., 2004). Each microarray experiment was replicated in dye reverse.
RESULTS
Mep2 Forms a Multimeric Complex In Vivo
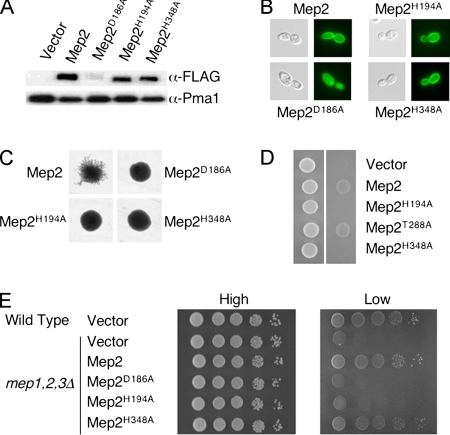
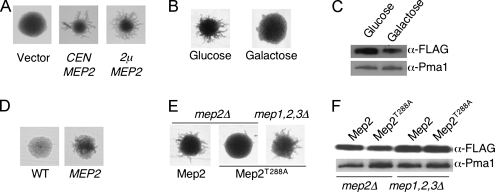
To facilitate studies of the role of Mep2 in the initiation of pseudohyphal growth, a plasmid was generated expressing Mep2 fused to a C-terminal FLAG epitope (Mep2-N4Q-FLAG) from the MEP2 endogenous promoter. In this allele, the fourth asparagine residue of Mep2 was substituted with glutamine, which prevents glycosylation but does not significantly influence the ability of Mep2 to transport ammonium or induce pseudohyphal growth (Marini and André, 2000). The Mep2-N4Q-FLAG protein is functional; transformation of cells lacking endogenous Mep2 with the Mep2-N4Q-FLAG plasmid restored pseudohyphal growth and growth under ammonium limiting conditions (Figure 5).
Figure 5.
Mep2 transport function is linked to pseudohyphal differentiation. (A) The levels of Mep2-N4Q-FLAG and derived mutants within the membrane fraction of cells grown in low- (50 μM) ammonium sulfate medium were quantified by Western analysis. Levels of the plasma membrane H+-ATPase Pma1 served as a loading control. (B) Localization of Mep2 was visualized using diploid mep2Δ/mep2Δ cells that contained plasmids expressing wild-type and mutant GFP-tagged Mep2 that had been grown in low-ammonium sulfate (50 μM) medium for 2 h. (C) Pseudohyphal growth was assayed using diploid mep2Δ/mep2Δ cells that contained plasmids expressing wild-type and mutant Mep2 that were grown on low ammonium sulfate (50 μM) medium for 6 d. (D) Haploid mep2Δ mutant cells containing relevant plasmids were spotted onto low-ammonium sulfate (50 μM) medium lacking uracil and grown for 6 d. Panels show cells before (left) and after (right) cells have been washed off the surface of the medium to reveal the extent of invasive growth. (E) Wild-type and mutant cells containing the relevant plasmids were grown overnight in complete medium (CMD-ura), washed, and serially diluted onto medium containing high or low (50 μM) levels of ammonium sulfate.
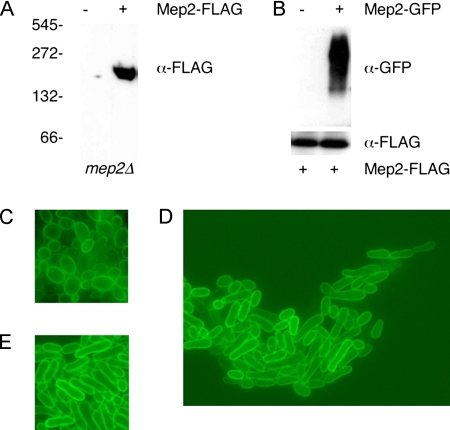
The bacterial Amt ammonium permeases form trimeric membrane protein complexes (Conroy et al., 2004; Zheng et al., 2004). Potential multimeric forms of S. cerevisiae Mep2 have also been detected following SDS electrophoresis (Marini and André, 2000). We observed that Mep2-N4Q-FLAG migrated at a size consistent with that of a trimeric complex under native electrophoresis conditions (Figure 1A). To confirm that Mep2 monomers interact in vivo, Mep2-N4Q-FLAG was coexpressed with Mep2 fused to green fluorescent protein. Immunoprecipitation of Mep2-N4Q-FLAG resulted in coimmunoprecipitation of Mep2-GFP, confirming the two epitope-tagged Mep2 derivatives interact (Figure 1B).
Figure 1.
Mep2 forms a multimeric complex and is membrane localized in cells undergoing pseudohyphal growth. (A) Membrane fractions from haploid mep2Δ cells containing a plasmid expressing Mep-N4Q-FLAG or a control vector that had been grown in low-ammonium medium (SLAD) were resolved under native conditions, and the FLAG epitope was detected by Western analysis. (B) Mep2-FLAG was immunoprecipitated from control cells and cells expressing an Mep2-GFP fusion protein that had been grown in SLAD medium. Diploid MLY108a/α cells (mep2Δ/mep2Δ) expressing Mep2-GFP were grown on low-ammonium medium (SLAD) plates for 6 d. The Mep2 fusion protein was localized by direct epifluorescence microscopy in cells at the center of the colony (C), within cells that had formed pseudohyphae (D), and at the edge of the colony (E).
Mep2 Localization on the Cell Membrane Requires the Exocyst Complex
Diploid S. cerevisiae cells undergo polarized growth during pseudohyphal growth that is, in some respects, similar to polarized growth of haploid cells during mating. In response to mating pheromone, cells develop a mating projection, or shmoo, that grows toward the mating partner to enable cell–cell contact (Jackson and Hartwell, 1990). Proteins important for polarized growth during mating are localized to the shmoo (Bagnat and Simons, 2002). Because pseudohyphal growth involves polarized growth, the extent to which Mep2 is localized to the growing tip of a filament during pseudohyphal growth was examined.
Cells expressing the Mep2-GFP fusion protein were grown on solid agar under pseudohyphal conditions. Within the center of the colony, cells were oval in shape (Figure 1C), whereas cells at the colony edge exhibited the elongated morphology characteristic of pseudohyphal growth (Figure 1E). In both cases, Mep2-GFP was expressed and localized throughout the cell membrane. In addition, Mep2-GFP was localized throughout the cell membrane of cells that had grown away from the center of the colony (Figure 1D). Unlike the signaling components important for mating, Mep2-GFP does not exhibit any preferential localization to any discrete area in the cell membrane or at the growing tip of the cell during pseudohyphal growth.
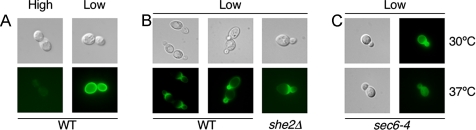
We next examined whether Mep2 might be transiently localized during its initial expression after the transition from high- to low-ammonium media (Marini et al., 1997) (Figure 2A). Under these conditions, Mep2-GFP localizes to the entire cell membrane within 2 h (Figure 2A). In budding cells, however, Mep2-GFP is initially (within 30 min) localized to small daughter cells, and in larger daughter cells, to the bud neck (Figure 2B). Longer incubation periods resulted in Mep2-GFP becoming gradually localized throughout more of the cell membrane, until Mep2-GFP covered the entire cell membrane by 2 h. The polarized transport of vesicles to the mother-bud neck or the bud tip is dependent on the actin cytoskeleton and the Myo2 myosin motor protein (Pruyne et al., 1998; Karpova et al., 2000). An alternative mechanism to ensure daughter-cell–specific protein expression is myosin-dependent mRNA transport from mother to daughter cell (Takizawa et al., 1997). To distinguish between these two mechanisms, the localization of Mep2-GFP was analyzed in cells lacking the She2 protein and cells that contain the temperature-sensitive sec6-4 allele. She2 is an RNA binding protein essential for daughter-cell–specific mRNA transport (Long et al., 2000), whereas Sec6 is an integral component of the exocyst complex that mediates polarized targeting of secretory vesicles to sites of active exocytosis (TerBush et al., 1996; TerBush and Novick, 1995). Mep2-GFP exhibited a wild-type pattern of localization in a she2Δ mutant strain; however, this pattern of localization was absent in sec6-4 cells grown at the restrictive temperature (Figure 2C). Thus, Mep2 distribution on the cell membrane involves the exocyst complex.
Figure 2.
Mep2 is localized to the cell membrane via the exocyst complex. Direct epifluorescence microscopy of cells expressing Mep2-GFP was performed with wild-type cells that were grown in high-ammonium medium, pelleted, washed and then grown in high- or low (50 μM)-ammonium sulfate medium for 2 h (A), wild-type and she2Δ cells that were transferred from growth in high-ammonium to low-ammonium medium and grown for 20 min (B), and sec6-4 cells that were grown at the permissive and nonpermissive temperature in low-ammonium medium (C).
Mep2 Is Correctly Localized in Mutants Lacking Npr1 and Ure2 and Is Required for Pseudohyphal Growth on Nitrogen Sources Other Than Ammonium
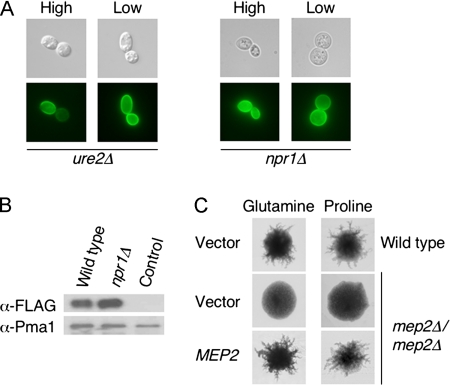
To further analyze the link between Mep2 localization and pseudohyphal signaling, the localization of Mep2 was analyzed in mutants lacking Ure2 and Npr1 that do not form pseudohyphae (Lorenz and Heitman, 1998a). Ure2 is a regulatory protein that binds to the Gln3 transcription factor and thereby represses transcription of nitrogen catabolite-regulated genes (Blinder et al., 1996). The Npr1 kinase regulates sorting and localization of the general amino acid permease Gap1 (De Craene et al., 2001). We observed that Mep2-GFP is normally localized to the plasma membrane in both ure2 and npr1 mutant strains when grown under high and low levels of ammonium (Figure 3A). Therefore, the loss of either Ure2 or Npr1 results in the derepression of MEP2 gene expression, and the lack of pseudohyphal growth of these mutants cannot be attributed to the lack of Mep2 localization to the cell membrane. In addition, the GFP fluorescence observed in npr1Δ cells expressing Mep2-GFP was greater than in wild-type cells, and Mep2-N4Q-FLAG expression was also ∼30% higher in these cells as detected by Western blotting (Figure 3B).
Figure 3.
Mep2 is correctly localized in mutants lacking Npr1 and Ure2, and it is required for pseudohyphal growth on nitrogen sources other than ammonium. (A) Epifluorescence microscopy of ure2Δ and npr1Δ cells containing Mep2-GFP that had been grown in high-ammonium medium, pelleted, washed, and then grown in high- or low-ammonium sulfate medium for 2 h. (B) The levels of Mep2-N4Q-FLAG within the membrane fraction of wild-type and npr1Δ cells grown in low (50 μM) ammonium sulfate medium were quantified by Western analysis. Levels of the plasma membrane H+-ATPase Pma1 served as a loading control. Cells not expressing Mep2-N4Q-FLAG served as a negative control. (C) Diploid wild-type and mep2Δ cells containing either a control plasmid or low copy plasmid expressing MEP2 from its own promoter were grown with either glutamine (100 μM) or proline (100 μM) as the sole nitrogen source for 6 d.
Our previous studies reported that Mep2 induced pseudohyphal growth in response to growth on ammonium alone and not when cells were grown on alternative sources of nitrogen (Lorenz and Heitman, 1998a). This suggested that ammonium transport through Mep2 is required for the initiation of pseudohyphal growth. Conversely, studies in C. albicans have found that Mep2 is required for the induction of filamentous growth in response to nitrogen sources other than ammonium (Biswas and Morschhäuser, 2005). We reanalyzed the growth of diploid mep2Δ mutants on agar media containing either glutamine or proline as a nitrogen source. Under these conditions, wild-type cells produced pseudohyphae, whereas the diploid mep2Δ mutant strain did not (see Discussion). Expression of the MEP2 gene in the diploid mep2Δ strain from a centromeric plasmid restored the ability of the mutant strain to undergo pseudohyphal growth (Figure 3C). Therefore, consistent with what has been described in C. albicans, Mep2 is required for the initiation of pseudohyphal growth on nitrogen sources other than ammonium.
Mep2 Induces Haploid Invasive Growth in Response to Ammonium Limiting Conditions
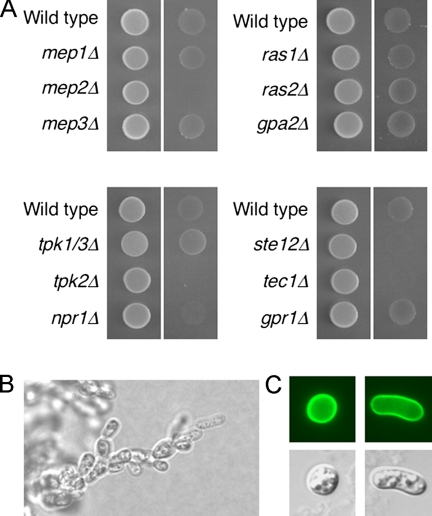
Dimorphic growth by S. cerevisiae in response to ammonium limitation has traditionally been investigated using diploid cells. Haploid S. cerevisiae cells undergo invasive growth when grown in rich medium or in response to fusel alcohols (Roberts and Fink, 1994; Lorenz et al., 2000b), but their response to ammonium levels has not been extensively studied. The finding that the basidiomycetous fungus C. neoformans and the fission yeast S. pombe undergo haploid invasive growth in response to ammonium limitation suggests that this pattern of growth may be conserved within fungi (Mitsuzawa, 2006; Rutherford et al., 2008). Growth of wild-type cells on low-ammonium media resulted in invasive growth and was dependent on the presence of Mep2 but not on the presence of the Mep1 or Mep3 ammonium permeases (Figure 4A). Complementation of the mep2Δ mutation with MEP2 regulated by its native promoter from a low copy number centromeric plasmid restored invasive growth (Figure 5D). Microscopic examination of the areas of invasive growth revealed formation of chains of elongated cells (Figure 4B). Consistent with the morphology of cells undergoing pseudohyphal growth, some haploid cells exhibited an elongated cell shape when grown under ammonium Limiting conditions (Figure 4C).
Figure 4.
Mep2 is required for haploid invasive growth under nitrogen limiting conditions. (A) Haploid wild-type and mutant cells containing a control plasmid were spotted onto low ammonium sulfate (50 μM) solid medium lacking uracil and grown for 6 d. Panels show cells before (left) and after (right) cells have been washed off the surface of the plate to reveal the extent of invasive growth. (B) The morphology of invading cells was visualized with a Zeiss Axiophot 2 Plus fluorescence microscope revealing, in some areas, filaments of pseudohyphal-like chains of cells. (C) Wild-type cells expressing Mep2-GFP that had undergone haploid invasive growth exhibit either morphology consistent with cell cycle arrest or pseudohyphal-like elongation.
The extent of invasive growth under low ammonium conditions was then analyzed in mutant strains lacking genes involved in diploid pseudohyphal growth. Tpk1, Tpk2, and Tpk3 are the PKA catalytic subunits, and Ste12 and Tec1 are transcription factors regulated by the MAP kinase pathway. Mutants lacking Npr1, Tpk2, Ste12, or Tec1 exhibited reduced invasive growth under ammonium limiting conditions. Interestingly, a tpk1 tpk3 double mutant exhibited increased invasive growth under the same conditions. The link between ammonium-responsive invasive growth is consistent with the finding that cAMP can restore invasive growth in a S. pombe mutant that lacks amt1Δ (Mitsuzawa, 2006). The Gpr1 G protein-coupled receptor and the Gα subunit Gpa2 activate the cAMP–PKA pathway and are required for pseudohyphal growth. Ras2 is a guanosine triphosphate-binding protein that activates both the cAMP–PKA and the MAP kinase pathways (reviewed in Pan et al., 2000). Mutants lacking Gpr1, Gpa2, Ras1, or Ras2 exhibited invasive growth equivalent to wild-type haploid cells under ammonium limiting conditions (Figure 4A).
Ammonium Transport Is Linked to Pseudohyphal and Invasive Growth
Transceptors bind and/or transport nutrients as part of their sensing mechanism. If Mep2 functions as a transceptor, then mutations that affect transport would be predicted to play a role in pseudohyphal differentiation. In contrast, if Mep2 signals in the absence of its ligand, then mutations that prevent Mep2 from binding or transporting ammonia/ammonium might mimic the ligand free receptor state and activate pseudohyphal growth. These opposing models were tested by identifying and mutating residues involved in Mep2 transport function. An aspartate residue in AmtB that is conserved throughout the Mep/Amt/Rh family is required for ammonium transport (Javelle et al., 2004). In addition, two conserved histidine residues that line the hydrophobic channel of the ammonium permease family are essential for ammonium transport by AmtB (Javelle et al., 2006). These residues are equivalent to D186, H194, and H348 in Mep2 of S. cerevisiae.
Individual alanine substitutions of residues D186, H194, and H348 were generated in the Mep2-N4Q-FLAG and Mep2-GFP fusion proteins. The H194A and H348A Mep2 proteins were stably expressed based on Western blot analysis, and they were also localized to the surface of the plasma membrane in the context of a GFP fusion protein (Figure 5, A and B, and Table 3). In contrast, the Mep2D186A mutant protein was unstable, and instead of plasma membrane localization, it seemed to be localized to endogenous membranes, consistent with a recent report (Marini et al., 2006). Cells expressing the Mep2H194A and Mep2H348A proteins did not undergo pseudohyphal growth in response to low ammonium, even though both proteins were stably expressed and correctly localized (Figure 5C) (Table 3). Under ammonium limiting conditions, the Mep2H194A protein did not support growth of a strain lacking the three Mep permeases of S. cerevisiae, whereas cells expressing Mep2H348A grew to the same extent as wild-type cells. Thus, neither the transport-defective H194A mutant nor the transport-proficient H348A mutant support pseudohyphal differentiation, indicating that transport is not sufficient for signaling, and that signaling requires functions in addition to transport (Figure 5E and Table 3).
Table 3.
Summary of phenotypes of diploid (pseudohyphal growth, growth on low ammonium) and haploid (invasive growth) mep1,2,3Δ cells expressing wild-type or mutant Mep2 permeases (see Fig. 5)
| Mep2 localized to membrane | Pseudohyphal growth | Growth on low ammonium | Invasive growth | |
|---|---|---|---|---|
| Mep2 | + | + | + | + |
| Mep2D186A | − | − | − | NT |
| Mep2H194A | + | − | − | − |
| Mep2H348A | + | − | + | − |
In addition, the ability of these Mep2 mutants to support haploid invasive growth under ammonium limiting conditions was assessed. Consistent with the analysis of diploid pseudohyphal growth, haploid cells expressing the Mep2H194A and Mep2H348A proteins failed to undergo invasive growth in response to low ammonium. Conversely, expression of a Mep2T288A mutant protein that exhibits defects in pseudohyphal growth supported haploid invasive growth under the same conditions (Figure 5D). The T288 residue of Mep2 is a potential PKA phosphorylation site that may link the PKA signaling cascade to Mep2 signaling activities under certain conditions (Smith et al., 2003).
Mep2 Levels Correlate with Extent of Pseudohyphal Growth
Analysis in C. albicans has established that the ability of Mep2 to induce pseudohyphal growth is dependent on its expression levels (Biswas and Morschhäuser, 2005). In accord with these studies, we have found that in S. cerevisiae, Mep2 expression levels are important for its regulatory function. MEP2 expression from a high copy plasmid resulted in more robust pseudohyphal growth compared with expression from a low copy number centromeric plasmid (Figure 6A). Similarly, growth of cells on galactose as opposed to glucose resulted in a significant reduction in the level of pseudohyphal growth, and this was correlated with reduced Mep2-N4Q-FLAG expression (Figure 6, B and C).
Figure 6.
Mep2 function correlates with its expression levels. Pseudohyphal growth was assayed after growth for 6 d by using diploid mep2Δ/mep2Δ cells that contained either low or high copy plasmids expressing MEP2 (A); wild-type cells grown with either glucose or galactose as the carbon source (B), wild-type cells grown with or without galactose-regulated MEP2 were grown on synthetic medium with galactose as the carbon source and high levels of ammonium (D); and diploid mep2Δ/mep2Δ cells or diploid mep1,2,3Δ/mep1,2,3Δ cells expressing the Mep2T288A variant (E). The levels of Mep2-FLAG within wild-type cells grown on different carbon sources and low (50 μM)-ammonium sulfate (C) or Mep2T288A-FLAG within diploid mep2Δ/mep2Δ cells and diploid mep1,2,3Δ/mep1,2,3Δ cells grown in low (50 μM)-ammonium sulfate medium (F) were quantified by Western analysis. Levels of the plasma membrane H+-ATPase Pma1 served as a loading control.
These findings, together with studies on C. albicans Mep2, suggested that uncoupling MEP2 expression from nitrogen catabolite repression might induce pseudohyphal growth under noninducing conditions. To test this hypothesis, a diploid wild-type strain was transformed with a plasmid expressing MEP2 from a galactose-inducible promoter. Limited, but distinct, pseudohyphae were observed in cells grown in galactose nitrogen-replete conditions, whereas no pseudohyphae were observed in control cells (Figure 6D). This establishes that ammonium limitation per se is not required for the induction of the ammonium-responsive dimorphic switch in S. cerevisiae.
The influence of Mep2 expression levels suggested an explanation for an intriguing observation with the Mep2T288A mutant protein. Mep2T288A lacks a putative PKA phosphorylation site and does not induce pseudohyphal growth in mep2 mutant cells (Smith et al., 2003) (Figure 6E). However, when the Mep2T288A protein is expressed in cells lacking all three ammonium permeases pseudohyphal growth is induced at the wild-type level (Figure 6E). One potential explanation for this phenotype is that cells lacking all three ammonium permeases are more starved for nitrogen and therefore induce the expression of Mep2-T288A to a higher level. This increased expression could compensate for the reduced activity of the Mep2T288A protein. When levels of Mep2-FLAG and Mep2T288A-FLAG were analyzed in mep2 versus mep1, -2, and -3 mutant cells, both Mep2 proteins were more highly expressed in the mep1,2,3Δ strain (Figure 6F), supporting the interpretation that increased levels of the signaling compromised T288A mutant enable signaling function.
MEP2 Overexpression Induces Transcription of Genes Implicated in Filamentous Growth and Known Targets of Ste12
One hypothesis that is consistent with the role of Mep2 as a sensor of ammonium availability is that it interacts with a signal transduction pathway that activates downstream effectors that control gene expression. To test this hypothesis, the transcriptional profile of S288C haploid cells expressing the galactose-inducible MEP2 allele was assessed during growth in ammonium-rich medium. Initially we determined whether the polymorphisms in the ∑1278b genome would affect hybridizations on our yeast open reading frame (ORF) microarrays by comparing global gene expression between S288C and ∑1278b diploid wild-type strains. Only 24 and 43 genes exhibited a greater than twofold increase and decrease in expression, respectively, between the two genetic backgrounds. Among the differentially regulated genes, the only functional enrichment observed was for glycolysis genes (CDC19, TDH1–3) that were down-regulated greater than twofold in ∑1278b (Gene Ontology: 0006096; p = 2.7 × 10−6). These results indicate that the polymorphisms in the ∑1278b strains have minimal effects on hybridization of our yeast ORF microarrays.
The high expression of MEP2 under ammonium-rich conditions compared with cells with normal endogenous MEP2 expression levels resulted in induction of 219 genes that exhibited a twofold or greater increase in expression or more (Supplemental Table 1). These include genes involved in the cell cycle (e.g., CLB1, STB1, FAR3, SIC1, and WHI5), cell wall maintenance (e.g., MOB2, SKG1, and EXG1) and signal transduction (e.g., CDC25, SDC25, STE12, RGA1, and RTS1). The most striking feature of the MEP2-induced profile in haploid cells was the induction of 35 transposable element genes, consisting mainly of members of the Ty1 family of retrotransposons (Supplemental Table 1). These mobile genetic elements replicate through an RNA intermediate and consist of two direct long terminal repeats that flank the TYA and TYB open reading frames. The promoter of Ty1 elements contains binding sites for multiple transcription factors. Important for this study, the Ty1 promoter contains a filamentation response element within its promoter that responds to nitrogen limitation via the combinatorial activity of Ste12 and Tec1 (Conte et al., 1998; Morillon et al., 2002). Interestingly, in addition to the Ty1 genes, other genes where Ste12 is known to bind to their promoters were also induced in response to constitutive MEP2 expression. These genes include AGA1, SCW11, HXK1, GAP1, PRY3, and PRM6 (Zeitlinger et al., 2003; Borneman et al., 2006, 2007). This profile, when considered in conjunction with those data that support a signaling role for Mep2, suggests that the Ste12 transcription factor is a downstream effector of Mep2 function.
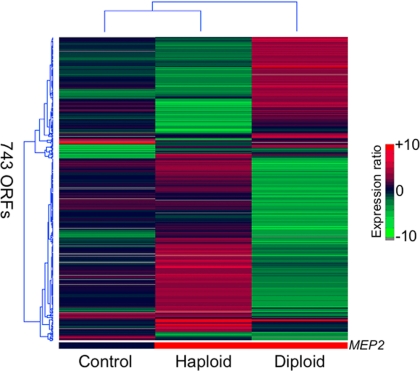
The identification of a Mep2-dependent transcriptional profile in haploid cells that included genes involved in pseudohyphal growth prompted similar experiments in pseudohyphal-competent Σ1278b diploid yeast cells. The transcriptional profile of wild-type diploid cells was compared with diploid cells expressing MEP2. Remarkably, the profile of genes exhibiting twofold or greater differential expression was the reciprocal of that identified in the haploid experiment. Therefore, in most cases, genes that were induced by the constitutive expression of MEP2 in haploid cells were repressed in diploid cells. Conversely, genes induced in diploid cells containing the GAL-MEP2 plasmid were repressed in haploid cells (Figure 7).
Figure 7.
Transcriptional profiles of MEP2 overexpression in a S288C haploid (middle) and ∑1278b diploid (right) wild-type strain. Genes that are differentially regulated greater than twofold in at least one of the microarray experiments are shown in the clustergram. The control experiment (left) compares S288C and ∑1278b diploid wild-type strains to determine whether hybridizations differ between the two genetic backgrounds on the yeast ORF microarrays. Only 77 genes were differentially regulated greater than twofold between the two genetic backgrounds. The bottom bar indicates MEP2 expression in each microarray experiment.
The genes induced in diploid cells by MEP2 expression are involved in similar cellular processes as those identified in haploid cells, although the same individual genes are rarely induced in both experiments. Therefore, genes involved in cell wall maintenance, the cell cycle, and mitochondrial function were induced in response to MEP2 overexpression in diploid cells (Supplemental Table 2). In contrast to the haploid profile, however, the transposable element genes were not collectively induced. This observation may be due in part to lower transcription rates of Ty1 elements reported in diploid cells compared with haploid cells (Morillon et al., 2002). In addition, a large number of genes involved in glucose, methionine, and sulfur metabolism were uniquely induced in diploid cells. Several genes linked to dimorphic growth were induced, including BEM4 (bud emergence), CBK1 (protein kinase), and with just under a twofold induction, GIC2 (bud emergence), FLO11 (cell surface glycoprotein), and DIA1 (invasive growth). Interestingly, FLO11 (MUC1), which is essential for pseudohyphal growth, is induced to a higher level in diploid cells (1.8-fold increase) than in haploid cells (1.3-fold increase) in response to Mep2 overexpression. Similar to the Mep2 induced profile in haploid cells, many genes that are targets of Ste12 were induced in diploid cells. These genes include GIC2, CDC19, SCH9, FLO11, CWP2, and AGA1 (Zeitlinger et al., 2003; Borneman et al., 2006, 2007).
The Mep2 transcriptional profile also included genes that have a role in cell signaling in addition to the MAPK–Ste12 pathway. TSC11 encodes a member of the TORC2 complex. Consistent with the involvement of Tor signaling, the gene encoding the Tsc11 subunit of the TORC2, which is proposed to regulate actin cytoskeleton polarization and cell integrity, is induced by Mep2 expression in diploid cells (Barbet et al., 1996; Schmidt et al., 1996; Schmidt et al., 1997; Cutler et al., 2001; Wedaman et al., 2003). SCH9 encodes a protein kinase recently shown to be a major target of TORC1 for regulating ribosome biogenesis, translation initiation, and entry into G0 phase, was similarly induced (Urban et al., 2007). The product of RTS3 is predicted to form part of the protein phosphatase type 2A complex (PP2A) and physically interacts with catalytic and regulatory subunits of protein phosphatase 2A and the PP2A-related phosphatase Sit4–Sap185 complex (Ho et al., 2002; Samanta and Liang, 2003; Gavin et al., 2006). Interestingly, RTS3 is one of the few genes to be induced in both haploid and diploid cells overexpressing Mep2. In addition, the functional homologue of RTS3, RTS1, and the regulator of PP2A activity, RRD2, were induced by Mep2 expression in haploid and diploid cells, respectively. Rrd2 interacts with the Tap42-PP2Ac complex that is required for a Tor-dependent filamentous signaling pathway (Cutler et al., 2001; Zheng and Jiang, 2005). This suggests that Mep2 expression under nitrogen-sufficient conditions results in the differential regulation of PP2A activity, which might explain the number of identified genes that are involved in both the cell cycle and cell wall integrity and cAMP pathways (Cutler et al., 2001).
Constitutive Mep2 Expression Restores Pseudohyphal Growth in Mutants Lacking cAMP–PKA Signaling but Not the MAP Kinase-regulated Transcription Factor Ste12
The established role of the MAP kinase pathway in pseudohyphal growth suggested that Mep2-dependent induction of Ty1 elements resulted from activity of the Ste12/Tec1 transcription factors. The absence of induction of the equivalent elements in diploid cells could be the result of loss of Ste12/Tec1 activity in diploid cells or to a cell-type–specific regulatory mechanism. The pleiotropic transcriptional regulator Mcm1 is a MADS box protein involved in cell cycle progression, arginine metabolism, cell wall maintenance, cell type determination, pheromone response, and mating (Sengupta and Cochran, 1990; Lydall et al., 1991; Messenguy and Dubois, 1993). The transcriptional activation of a-specific genes results from dimerization of Ste12 and Mcm1 at target promoters (Sengupta and Cochran, 1990; Mueller and Nordheim, 1991). Conversely, Matα2–Mcm1 complexes repress the expression of a-specific genes in diploid cells (Keleher et al., 1989). In addition, Mcm1 regulates Ty1 expression and binds to regulatory sequences independent of Ste12, unlike the pheromone-induced genes that are regulated by a Mcm1–Ste12 heterodimer (Errede, 1993). The cell-type–specific response to Mep2 expression observed in the microarray studies and the identification of genes regulated by Ste12 and the cell cycle suggested that Ste12 and/or Mcm1 could be downstream effectors of Mep2.
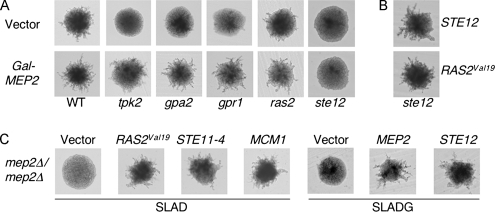
To examine this hypothesis, the ability of the GAL-MEP2 allele to induce pseudohyphal growth in mutants lacking components of the PKA pathway or Ste12 was analyzed. MEP2 overexpression restored pseudohyphal growth in mutants lacking Tpk2, Gpa2, Gpr1, or Ras2 (Figure 8A). However, MEP2 overexpression did not restore pseudohyphal growth in a strain lacking Ste12 (Figure 8A). As a control, complementation of the ste12Δ mutant with a plasmid expressing STE12 or a hyperactive RAS2 allele restored wild-type pseudohyphal growth in the ste12Δ strain (Figure 8B). Analogous experiments were conducted to determine whether activation of either the PKA or the MAP kinase pathway restores pseudohyphal growth in a mep2Δ mutant. Expression of alleles that constitutively activate the PKA pathway, the MAP kinase pathway, as well as overexpression of Ste12, all restored pseudohyphal growth of the mep2Δ mutant (Figure 8C). Together, these experiments are consistent with Mep2 and Ste12 acting in either the same pathway during pseudohyphal growth or in parallel interdependent pathways. Overexpression of the Ste12 interacting transcription factor Mcm1 also restored pseudohyphal growth in the mep2Δ strain, providing further functional evidence linking Mep2 and the MAP kinase pathway (Figure 8C).
Figure 8.
Epistasis analysis provides evidence that Mep2 and Ste12 are functionally related. After growth on galactose low-ammonium sulfate (50 μM) plates for 6 d at 30°C, pseudohyphal growth was assayed in diploid mutants lacking Tpk2, Gpa2, Gpr1, Ras2 and Ste12 that expressed MEP2 from a galactose-inducible promoter (A) and diploid ste12Δ/ste12Δ cells that contained either a galactose-inducible STE12 gene or the RAS2Val19 allele (B). (C) Cells lacking Mep2 and containing plasmids expressing MCM1, the RAS2Val19 allele, the STE11-4 allele, GAL-MEP2, or GAL-STE12.
DISCUSSION
Nitrogen availability regulates the initiation of diploid filamentous growth, haploid invasive growth, and mating in different model fungi. These developmental processes occur when ammonium is limiting, and they are dependent on members of the AmtB/Mep/Rh family of ammonium permeases. Here, we present evidence that supports the hypothesis that Mep2 functions as an ammonium sensor; importantly, we document that the transport activity of Mep2 is linked to both differentiation and the control of gene expression.
The localization and oligomerization state of Mep2 are consistent with other members of the AmtB/Mep2/Rh family of transporters. We have demonstrated that Mep2 monomers interact to form a trimeric complex. Our data doe not exclude the possibility that Mep2 could form heterotrimeric complexes with the nonpseudohyphal-inducing Mep1 and Mep3 ammonium permeases. When initially expressed, Mep2 is localized to the growing bud or the bud neck, a process that is dependent on the exocyst complex. Thereafter, Mep2 is localized throughout the plasma membrane.
A central question relating to Mep2 function is the extent to which ammonium binding, transport, or both are necessary to induce pseudohyphal growth. In previous studies, it was reported that Mep2-dependent pseudohyphal growth occurs only with limiting ammonium as a nitrogen source (Lorenz and Heitman, 1998a). This suggested that ammonium binding/transport via Mep2 is important for its regulatory function. Contrary to this, studies with C. albicans found that Mep2-dependent pseudohyphal growth occurs when this yeast is grown on a variety of nitrogen sources (Biswas and Morschhäuser, 2005). Similarly we find that S. cerevisiae also undergoes Mep2-dependent pseudohyphal growth in response to growth on glutamate and proline. In our previous S. cerevisiae studies, growth and filamentation assays were conducted on noble agar or agarose (Lorenz and Heitman, 1998a). Under these conditions, pseudohyphal growth is more limited, and even more so with glutamine or proline as the nitrogen source in which there was no clear obligate role for Mep2 in pseudohyphal growth. When growth assays are conducted on standard SLAD medium containing agar (including contaminating minor nitrogen sources), more robust pseudohyphal growth occurs and under these conditions Mep2 plays a clearly demonstrable role in promoting pseudohyphal differentiation, even with glutamine and proline as nitrogen source. During the course of this work, Marini and colleagues published similar findings implicating Mep2 in physiological roles during growth on nitrogen sources other than ammonium (Boeckstaens et al., 2007). They found that cells leak ammonium when grown on a variety of nitrogen sources, and they advanced the hypothesis that one role for Mep2 is to reimport ammonium that exits the cell during growth on alternative nitrogen sources. In the context of this model, pseudohyphal growth on nitrogen sources such as glutamine and proline still requires the ability of Mep2 to transport and sense ammonia, in this case involving ammonia that exits the cell from internal sources.
Mutational analysis of Mep2 supports the hypothesis that ammonium transport through Mep2 is required for the induction of pseudohyphal growth. A Mep2D186E variant that is correctly expressed and localized to the plasma membrane fails to transport ammonium and does not induce pseudohyphal growth (Marini et al., 2006). Our present study establishes that residues H194 and H348, which are conserved residues lining the pore of Mep2, are important in the initiation of pseudohyphal growth. The conservation of these residues throughout the AmtB/Mep/Rh family, and their position within the hydrophobic pore, suggest that their function is to bind ammonium as it translocates through the protein (Khademi et al., 2004). The importance of these residues for ammonium transport has been underscored by mutational analysis of the equivalent residues in AmtB from E. coli (Javelle et al., 2006). In S. cerevisiae, the Mep2H194A and Mep2H348A variants are stably expressed and localized to the cell membrane. The Mep2H194A protein has a significantly reduced ability to transport ammonium and does not induce pseudohyphal growth. The Mep2H348A protein transports ammonium but does not signal pseudohyphal growth. Somewhat unexpectedly, the ability of Mep2H348A to support growth on low ammonium contrasts to the lack of transport activity resulting from the equivalent mutation in AmtB from E. coli. Together, these data suggest ammonium translocation through Mep2 is required to initiate pseudohyphal growth and that mutation of residues D186 and H194 prevents this signaling process. The phenotype of the Mep2H348A protein suggests that the binding of ammonium ions to this residue could cause a conformational change in Mep2 that initiates pseudohyphal growth. Consistent with this, a Mep2 mutant that is hyperactive for both ammonium transport and pseudohyphal growth contains a mutation in the adjacent residue, G349 (Boeckstaens et al., 2007).
The requirement for Mep2 transport activity may explain the inability of mutants lacking the Npr1 kinase to undergo pseudohyphal growth (Lorenz and Heitman, 1998a). Our data establish that Mep2 is clearly localized to the plasma membrane and highly expressed in mutants lacking the Npr1 kinase. However, in the absence of Npr1, Mep2 does not transport ammonium (Boeckstaens et al., 2007). The Mep2G349C variant has increased transport activity and restores pseudohyphal growth in an npr1Δ strain (Boeckstaens et al., 2007). This is consistent with Npr1 being required to activate Mep2-dependent ammonium transport, but not expression or localization, which then enables a Mep2-dependent induction of pseudohyphal growth. Interestingly, Mep2 is also correctly localized in a mutant lacking the Ure2 transcriptional repressor. Further studies will be required to determine whether the inability of a ure2Δ mutant to initiate dimorphic growth is a result of a loss of Mep2 transport function or some other regulatory process in which constitutive induction of the nitrogen catabolite repression response blocks pseudohyphal growth.
We have established a correlation between the level of Mep2 expression and the degree of pseudohyphal growth in S. cerevisiae. Increased expression of Mep2 results in greater pseudohyphal growth and a modest increase in Mep2 expression can compensate for the pseudohyphal growth induction defect in cells expressing the Mep2T288A protein. In C. albicans, high expression levels of Mep2 are required for its ability to induce hyphal growth (Biswas and Morschhäuser, 2005). It is interesting to note that in each yeast that undergoes ammonium-dependent development, the relevant inducing permease is the most highly expressed of the family of ammonium permeases (Marini et al., 1997; Smith et al., 2003; Biswas and Morschhäuser, 2005; Rutherford et al., 2008). Mep2 is required for the induction of haploid invasive growth in response to low-ammonium growth, similar to other haploid yeasts. This takes place under conditions that are distinct from the invasive growth that occurs in response to carbon source availability and the presence of fusel alcohols (Lorenz et al., 2000b), which probably accounts for the novel finding of the involvement of Mep2 in this process. Similar to the diploid Mep2 pseudohyphal pathway, the Tpk2 catalytic subunit of PKA, the Npr1 kinase, and the Ste12 and Tec1 transcription factors are required for haploid invasive growth. The Tpk1/Tpk3 catalytic subunits of PKA repress ammonium-responsive invasive growth and Mep2 mutants altered in the conserved H194 and H348 residues do not undergo invasive growth. Cells that have undergone invasive growth in response to ammonium limitation can adopt an elongated morphology similar to that observed during diploid pseudohyphal growth. Together, these data support the view that the Mep2-dependent signaling processes that occur during haploid invasive growth are similar to those occurring during pseudohyphal growth. Our data contradict a previous study that reported that Mep2 is not required for haploid invasive growth when cells are grown on medium lacking or containing minimal (100 μM) ammonium (Van Nuland et al., 2006). The complementation of the mep2Δ strain with MEP2 expressed from a plasmid confirms that under these conditions, Mep2 is required for ammonium induced haploid invasive growth. Ammonium permease haploid invasive growth may be widely conserved within yeast, because this conservation has now been observed in haploid S. pombe, C. neoformans, and S. cerevisiae.
The expression of Mep2 under conditions that do not normally induce pseudohyphal growth generates a transcriptional profile that includes genes that are involved in cellular processes that are likely to be differentially regulated during pseudohyphal growth. Of particular interest is a group of genes controlled by Ste12 involved in the cell wall (AGA1, SED1, and CWP2), cell cycle (CLB1 and SCH9), MAP kinase pathway (GIC2), PKA pathway (SCH9), and pseudohyphal growth (STE12, FLO11, and HMS1). Also of interest is the induction of genes involved in the activity of Cdc42, a GTPase that establishes cell polarity and is also a component of the MAP kinase pathway (CDC25, SDC25, RGA1, and GIC2). Therefore, Mep2 expression results in the induction of a subset of Ste12-regulated genes and genes involved in the regulation of the MAP kinase pathway, indicative of Mep2-dependent differential activation of the pathway.
Consistent with a link between Ste12 and Mep2, activation of the MAP kinase pathway, through either overexpression of Ste12 or the hyperactive STE11-4 allele restored pseudohyphal growth in a diploid strain lacking Mep2, in contrast to a previous report (Lorenz and Heitman, 1998a). In addition, overexpression of Mep2 from the GAL promoter restored pseudohyphal growth in mutants lacking elements of the RAS–cAMP pathway but not in a mutant that lacks Ste12. This evidence strongly supports a link between Ste12 activity and the putative transceptor role of Mep2 during pseudohyphal growth. This, together with previous data that pseudohyphal growth in a S. cerevisiae strain lacking Mep2 can be restored by the exogenous addition of cAMP to cells (Lorenz and Heitman, 1998a), is similar to the epistatic relationship of C. albicans Mep2 and the PKA and MAP kinase pathways during filamentous growth. This mechanistic conservation in different fungi is consistent with the finding that the expression of certain Mep2 fungal homologues restores pseudohyphal growth in an S. cerevisiae strain lacking the endogenous Mep2, suggesting that significant aspects of the signaling role of these homologues is conserved in some fungal species (Montanini et al., 2002; Javelle et al., 2003; Smith et al., 2003).
Although the constitutive expression of Mep2 induces pseudohyphal growth under normally repressing conditions, overexpression of Ste12 under the same conditions does not (Borneman et al., 2007). This may be due to the increased turnover of Ste12 in nitrogen-sufficient conditions, a process regulated by the Srb10 cyclin-dependent kinase, which itself becomes depleted during nitrogen limitation (Nelson et al., 2003). This therefore argues against a simple model whereby Mep2 only activates the MAP kinase pathway. Mep2 could influence the turnover of Ste12 and/or regulate pathways that influence Ste12 activity. It is possible that mechanisms that regulate the differential output of the MAP kinase in haploid and diploid cells are responsible for the transcriptional profiles that we have identified. Interestingly, we have shown here and in previous work that Mcm1 and Tec1, two transcription factors that interact with Ste12 to modulate its activity, suppress the loss of pseudohyphal growth in cells lacking Mep2 (Lorenz and Heitman, 1998a). Additional candidates for signaling pathways that Mep2 might interact with were also identified in microarray experiments including PP2A and the Tor kinases.
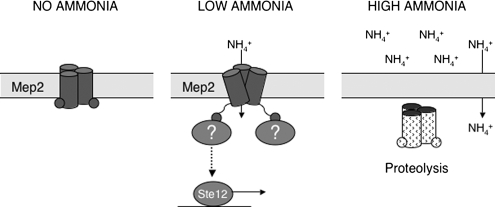
Together, our data suggest a novel model of Mep2 function (Figure 9). The requirement for ammonium transport by Mep2 suggests that the lack of ammonium in the environment is not the initiating signal for pseudohyphal growth as has been proposed. This is supported by the Mep2 dependent induction of pseudohyphal growth under ammonium-sufficient growth because presumably, under these conditions, Mep2 is transporting ammonium into the cell. This strongly suggests that the importance of Mep2 does not relate to its influence on the internal levels of nitrogen, because it will induce pseudohyphal growth irrespective of ammonium levels when expressed, further supporting the designation of Mep2 as a transceptor. We propose that when ammonium is absent from the external environment, Mep2 is expressed and localized on the plasma membrane (Figure 9). Under these conditions, Mep2 does not transport ammonium and the lowering of internal nitrogen levels prevents growth. When low levels of ammonium become available, the transport of ammonium through Mep2 triggers conformational changes that most likely involve the C-terminal domains, which signal to the cell that there is sufficient ammonium for growth. Support for the conformational fluidity of Mep2 comes from the finding that the C-terminal domain of each monomer of the Arabidopsis thaliana Mep2 homologue Amt1;1 interacts with the adjacent monomer to confer allosteric regulation (Loqué et al., 2007). The physical act of transport through Mep2 enables the permease to engage signaling components that result in Ste12 activation and the possible regulation of the Tor and PP2A pathways. Together, these result in the induction of pseudohyphal growth. If increased levels of ammonium become available, the transcription of the MEP2 gene is repressed via catabolite repression (Marini et al., 1997), and Mep2 is internalized, localized to the vacuole, and degraded (Zurita-Martinez et al., 2007). Under these conditions, ammonium is able to diffuse through the plasma membrane, and the loss of the pseudohyphal-inducing signal from Mep2 results in the repression of pseudohyphal growth.
Figure 9.
Model of Mep2 signaling during pseudohyphal growth. Mep2 forms a trimer within the membrane and acts as a sensor of ammonium availability. When no ammonium is present in the environment the signaling function of Mep2 is repressed. When limiting ammonium becomes available, its transport causes a conformational change in Mep2, which enables the permease to engage signaling partners that initiate pseudohyphal growth. High levels of ammonium result in the loss of Mep2 signaling because Mep2 is removed from the membrane and MEP2 transcription is repressed.
Ammonium permeases have been implicated in regulating haploid invasive growth, diploid pseudohyphal growth, and mating in different model fungal systems. Members of the Amt/Mep/Rh family are also required for certain aspects of the development of Dictyostelium discoideum (Kirsten et al., 2005; Singleton et al., 2006). Together, these systems suggest that ammonium permease-mediated control of development may be widespread throughout unicellular, and possibly also multicellular, eukaryotic organisms. The molecular mechanisms that link Mep2 to pseudohyphal differentiation may therefore be conserved and relevant for studies in other model developmental systems. The present study establishes for the first time, Mep2-dependent regulation of gene expression that likely involves, at least in part, the transcription factor Ste12. The challenge now is to identify the putative regulatory proteins that physically interact with Mep2 to govern transcriptional cascades that evoke novel cellular fates in response to external nutrient sources.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mike Perlin and Ralf-Peter Jansen for useful discussions and Mike Perlin for the gift of pYES2.1-MEP2. This research was funded by a National Institutes of Health/National Institute of Allergy and Infectious Diseases grant AI39115 and Department of Defense award W81xwh-04-01-0208 (to J.H.). Support was also provided in part by grant CA114107 from the National Cancer Institute (to M.E.C.) and by a grant from Genome Canada and the Natural Sciences and Engineering Research Council (to T.H.). G.C. was supported by a Charles H. Best postdoctoral fellowship, and J.C.R. is supported by a Research Councils (United Kingdom) Academic Fellowship.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-01-0033) on April 23, 2008.
REFERENCES
- Andrade S. L., Einsle O. The Amt/Mep/Rh family of ammonium transport proteins. Mol. Membr. Biol. 2007;24:357–365. doi: 10.1080/09687680701388423. [DOI] [PubMed] [Google Scholar]
- Andrade S. L., Dickmanns A., Ficner R., Einsle O. Crystal structure of the archaeal ammonium transporter Amt-1 from Archaeoglobus fulgidus. Proc. Natl. Acad. Sci. USA. 2005;102:14994–14999. doi: 10.1073/pnas.0506254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnat M., Simons K. Cell surface polarization during yeast mating. Proc. Natl. Acad. Sci. USA. 2002;99:14183–14188. doi: 10.1073/pnas.172517799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbet N. C., Schneider U., Helliwell S. B., Stansfield I., Tuite M. F., Hall M. N. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas K., Morschhäuser J. The Mep2 ammonium permease controls nitrogen starvation-induced filamentous growth in Candida albicans. Mol. Microbiol. 2005;56:649–669. doi: 10.1111/j.1365-2958.2005.04576.x. [DOI] [PubMed] [Google Scholar]
- Blinder D., Coschigano P. W., Magasanik B. Interaction of the GATA factor Gln3p with the nitrogen regulator Ure2p in Saccharomyces cerevisiae. J. Bacteriol. 1996;178:4734–4736. doi: 10.1128/jb.178.15.4734-4736.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckstaens M., André B., Marini A. M. The yeast ammonium transport protein Mep2 and its positive regulator, the Npr1 kinase, play an important role in normal and pseudohyphal growth on various nitrogen media through retrieval of excreted ammonium. Mol. Microbiol. 2007;64:534–546. doi: 10.1111/j.1365-2958.2007.05681.x. [DOI] [PubMed] [Google Scholar]
- Borneman A. R., Gianoulis T. A., Zhang Z. D., Yu H., Rozowsky J., Seringhaus M. R., Wang L. Y., Gerstein M., Snyder M. Divergence of transcription factor binding sites across related yeast species. Science. 2007;317:815–819. doi: 10.1126/science.1140748. [DOI] [PubMed] [Google Scholar]
- Borneman A. R., Leigh-Bell J. A., Yu H., Bertone P., Gerstein M., Snyder M. Target hub proteins serve as master regulators of development in yeast. Genes Dev. 2006;20:435–448. doi: 10.1101/gad.1389306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas M. E., Laroche T., Gasser S. M. The composition and morphology of yeast nuclear scaffolds. J. Cell Sci. 1990;96:439–450. doi: 10.1242/jcs.96.3.439. [DOI] [PubMed] [Google Scholar]
- Conroy M. J., Jamieson S. J., Blakey D., Kaufmann T., Engel A., Fotiadis D., Merrick M., Bullough P. A. Electron and atomic force microscopy of the trimeric ammonium transporter AmtB. EMBO Rep. 2004;5:1153–1158. doi: 10.1038/sj.embor.7400296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte D, Jr., Barber E., Banerjee M., Garfinkel D. J., Curcio M. J. Posttranslational regulation of Ty1 retrotransposition by mitogen-activated protein kinase Fus3. Mol. Cell. Biol. 1998;18:2502–2513. doi: 10.1128/mcb.18.5.2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler N. S., Pan X., Heitman J., Cardenas M. E. The TOR signal transduction cascade controls cellular differentiation in response to nutrients. Mol. Biol. Cell. 2001;12:4103–4113. doi: 10.1091/mbc.12.12.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Craene J. O., Soetens O., André B. The Npr1 kinase controls biosynthetic and endocytic sorting of the yeast Gap1 permease. J. Biol. Chem. 2001;276:43939–43948. doi: 10.1074/jbc.M102944200. [DOI] [PubMed] [Google Scholar]
- Errede B. MCM1 binds to a transcriptional control element in Ty1. Mol. Cell. Biol. 1993;13:57–62. doi: 10.1128/mcb.13.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S., Herskowitz I. Regulation by the yeast mating-type locus of STE12, a gene required for cell-type-specific expression. Mol. Cell. Biol. 1987;7:3818–3821. doi: 10.1128/mcb.7.10.3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan J. M., Moreau V., André B., Volland C., Haguenauer-Tsapis R. Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J. Biol. Chem. 1996;271:10946–10952. doi: 10.1074/jbc.271.18.10946. [DOI] [PubMed] [Google Scholar]
- Gavin A. C., et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Grigull J., Mnaimneh S., Pootoolal J., Robinson M. D., Hughes T. R. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol. Cell. Biol. 2004;24:5534–5547. doi: 10.1128/MCB.24.12.5534-5547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima T., Heitman J. The Galpha protein Gpa2 controls yeast differentiation by interacting with kelch repeat proteins that mimic Gbeta subunits. Mol. Cell. 2002;10:163–173. doi: 10.1016/s1097-2765(02)00569-5. [DOI] [PubMed] [Google Scholar]
- Harashima T., Anderson S., Yates J. R., 3rd., Heitman J. The kelch proteins Gpb1 and Gpb2 inhibit Ras activity via association with the yeast RasGAP neurofibromin homologs Ira1 and Ira2. Mol. Cell. 2006;22:819–830. doi: 10.1016/j.molcel.2006.05.011. [DOI] [PubMed] [Google Scholar]
- Ho Y., et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Holsbeeks I., Lagatie O., Van Nuland A., Van de Velde S., Thevelein J. M. The eukaryotic plasma membrane as a nutrient-sensing device. Trends Biochem. Sci. 2004;29:556–564. doi: 10.1016/j.tibs.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Ishikita H., Knapp E. W. Protonation states of ammonia/ammonium in the hydrophobic pore of ammonia transporter protein AmtB. J. Am. Chem. Soc. 2007;129:1210–1215. doi: 10.1021/ja066208n. [DOI] [PubMed] [Google Scholar]
- Jackson C. L., Hartwell L. H. Courtship in S. cerevisiae: both cell types choose mating partners by responding to the strongest pheromone signal. Cell. 1990;63:1039–1051. doi: 10.1016/0092-8674(90)90507-b. [DOI] [PubMed] [Google Scholar]
- Javelle A., Morel M., Rodríguez-Pastrana B. R., Botton B., André B., Marini A. M., Brun A., Chalot M. Molecular characterization, function and regulation of ammonium transporters (Amt) and ammonium-metabolizing enzymes (GS, NADP-GDH) in the ectomycorrhizal fungus Hebeloma cylindrosporum. Mol. Microbiol. 2003;47:411–430. doi: 10.1046/j.1365-2958.2003.03303.x. [DOI] [PubMed] [Google Scholar]
- Javelle A., Severi E., Thornton J., Merrick M. Ammonium sensing in Escherichia coli. Role of the ammonium transporter AmtB and AmtB-GlnK complex formation. J. Biol. Chem. 2004;279:8530–8538. doi: 10.1074/jbc.M312399200. [DOI] [PubMed] [Google Scholar]
- Javelle A., Lupo D., Zheng L., Li X. D., Winkler F. K., Merrick M. An unusual twin-his arrangement in the pore of ammonia channels is essential for substrate conductance. J. Biol. Chem. 2006;281:39492–39498. doi: 10.1074/jbc.M608325200. [DOI] [PubMed] [Google Scholar]
- Karpova T. S., Reck-Peterson S. L., Elkind N. B., Mooseker M. S., Novick P. J., Cooper J. A. Role of actin and Myo2p in polarized secretion and growth of Saccharomyces cerevisiae. Mol. Biol. Cell. 2000;11:1727–1737. doi: 10.1091/mbc.11.5.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleher C. A., Passmore S., Johnson A. D. Yeast repressor alpha 2 binds to its operator cooperatively with yeast protein Mcm1. Mol. Cell. Biol. 1989;9:5228–5230. doi: 10.1128/mcb.9.11.5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khademi S., O'Connell J., 3rd, Remis J., Robles-Colmenares Y., Miercke L. J., Stroud R. M. Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 Å. Science. 2004;305:1587–1594. doi: 10.1126/science.1101952. [DOI] [PubMed] [Google Scholar]
- Kirsten J. H., Xiong Y., Dunbar A. J., Rai M., Singleton C. K. Ammonium transporter C of Dictyostelium discoideum is required for correct prestalk gene expression and for regulating the choice between slug migration and culmination. Dev. Biol. 2005;287:146–156. doi: 10.1016/j.ydbio.2005.08.043. [DOI] [PubMed] [Google Scholar]
- Liu H., Styles C. A., Fink G. R. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- Long R. M., Gu W., Lorimer E., Singer R. H., Chartrand P. She2p is a novel RNA-binding protein that recruits the Myo4p-She3p complex to ASH1 mRNA. EMBO J. 2000;19:6592–6601. doi: 10.1093/emboj/19.23.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M. C., Heitman J. Yeast pseudohyphal growth is regulated by GPA2, a G protein alpha homolog. EMBO J. 1997;16:7008–7018. doi: 10.1093/emboj/16.23.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M. C., Heitman J. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 1998a;17:1236–1247. doi: 10.1093/emboj/17.5.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M. C., Heitman J. Regulators of pseudohyphal differentiation in Saccharomyces cerevisiae identified through multicopy suppressor analysis in ammonium permease mutant strains. Genetics. 1998b;150:1443–1457. doi: 10.1093/genetics/150.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M. C., Pan X., Harashima T., Cardenas M. E., Xue Y., Hirsch J. P., Heitman J. The G protein-coupled receptor Gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics. 2000a;154:609–622. doi: 10.1093/genetics/154.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M. C., Cutler N. S., Heitman J. Characterization of alcohol-induced filamentous growth in Saccharomyces cerevisiae. Mol. Biol. Cell. 2000b;11:183–199. doi: 10.1091/mbc.11.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loqué D., Lalonde S., Looger L. L., von Wirén N., Frommer W. B. A cytosolic trans-activation domain essential for ammonium uptake. Nature. 2007;446:195–198. doi: 10.1038/nature05579. [DOI] [PubMed] [Google Scholar]
- Lydall D., Ammerer G., Nasmyth K. A new role for MCM1 in yeast: cell cycle regulation of SW15 transcription. Genes Dev. 1991;5:2405–2419. doi: 10.1101/gad.5.12b.2405. [DOI] [PubMed] [Google Scholar]
- Ma H., Kunes S., Schatz P. J., Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58:201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- Marini A. M., Boeckstaens M., Benjelloun F., Chérif-Zahar B., André B. Structural involvement in substrate recognition of an essential aspartate residue conserved in Mep/Amt and Rh-type ammonium transporters. Curr. Genet. 2006;49:364–374. doi: 10.1007/s00294-006-0062-5. [DOI] [PubMed] [Google Scholar]
- Marini A. M., Soussi-Boudekou S., Vissers S., André B. A family of ammonium transporters in Saccharomyces cerevisiae. Mol. Biol. Cell. 1997;17:4282–4293. doi: 10.1128/mcb.17.8.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini A. M., André B. In vivo N-glycosylation of the Mep2 high-affinity ammonium transporter of Saccharomyces cerevisiae reveals an extracytosolic N-terminus. Mol. Microbiol. 2000;38:552–564. doi: 10.1046/j.1365-2958.2000.02151.x. [DOI] [PubMed] [Google Scholar]
- Messenguy F., Dubois E. Genetic evidence for a role for MCM1 in the regulation of arginine metabolism in Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:2586–2592. doi: 10.1128/mcb.13.4.2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuzawa H. Ammonium transporter genes in the fission yeast Schizosaccharomyces pombe: role in ammonium uptake and a morphological transition. Genes Cells. 2006;11:1183–1195. doi: 10.1111/j.1365-2443.2006.01014.x. [DOI] [PubMed] [Google Scholar]
- Montanini B., Moretto N., Soragni E., Percudani R., Ottonello S. A high-affinity ammonium transporter from the mycorrhizal ascomycete Tuber borchii. Fungal Genet. Biol. 2002;36:22–34. doi: 10.1016/S1087-1845(02)00001-4. [DOI] [PubMed] [Google Scholar]
- Morillon A., Bénard L., Springer M., Lesage P. Differential effects of chromatin and Gcn4 on the 50-fold range of expression among individual yeast Ty1 retrotransposons. Mol. Cell. Biol. 2002;22:2078–2088. doi: 10.1128/MCB.22.7.2078-2088.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller C. G., Nordheim A. A protein domain conserved between yeast MCM1 and human SRF directs ternary complex formation. EMBO J. 1991;10:4219–4229. doi: 10.1002/j.1460-2075.1991.tb05000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C., Goto S., Lund K., Hung W., Sadowski I. Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature. 2003;421:187–190. doi: 10.1038/nature01243. [DOI] [PubMed] [Google Scholar]
- Novick P., Field C., Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Pan X., Heitman J. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:4874–4887. doi: 10.1128/mcb.19.7.4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Heitman J. Sok2 regulates yeast pseudohyphal differentiation via a transcription factor cascade that regulates cell-cell adhesion. Mol. Cell. Biol. 2000;20:8364–8372. doi: 10.1128/mcb.20.22.8364-8372.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Harashima T., Heitman J. Signal transduction cascades regulating pseudohyphal differentiation of Saccharomyces cerevisiae. Curr. Opin. Microbiol. 2000;3:567–572. doi: 10.1016/s1369-5274(00)00142-9. [DOI] [PubMed] [Google Scholar]
- Pruyne D. W., Schott D. H., Bretscher A. Tropomyosin-containing actin cables direct the Myo2p-dependent polarized delivery of secretory vesicles in budding yeast. J. Cell Biol. 1998;143:1931–1945. doi: 10.1083/jcb.143.7.1931. [DOI] [PubMed] [Google Scholar]
- Roberts R. L., Fink G. R. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 1994;15:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- Rutherford J. C., Lin X., Nielsen K., Heitman J. Amt2 permease is required to induce ammonium responsive invasive growth and mating in Cryptococcus neoformans. Eukaryot. Cell. 2008;7:237–246. doi: 10.1128/EC.00079-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta M. P., Liang S. Predicting protein functions from redundancies in large-scale protein interaction networks. Proc. Natl. Acad. Sci. USA. 2003;100:12579–12583. doi: 10.1073/pnas.2132527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schiestl R. H., Gietz R. D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Schmidt A., Kunz J., Hall M. N. TOR2 is required for organization of the actin cytoskeleton in yeast. Proc. Natl. Acad. Sci. USA. 1996;93:13780–13785. doi: 10.1073/pnas.93.24.13780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A., Bickle M., Beck T., Hall M. N. The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell. 1997;88:531–542. doi: 10.1016/s0092-8674(00)81893-0. [DOI] [PubMed] [Google Scholar]
- Sengupta P., Cochran B. H. The PRE and PQ box are functionally distinct yeast pheromone response elements. Mol. Cell. Biol. 1990;10:6809–6812. doi: 10.1128/mcb.10.12.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton C. K., Kirsten J. H., Dinsmore C. J. Function of ammonium transporter A in the initiation of culmination of development in Dictyostelium discoideum. Eukaryot. Cell. 2006;5:991–996. doi: 10.1128/EC.00058-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. G., Garcia-Pedrajas M. D., Gold S. E., Perlin M. H. Isolation and characterization from pathogenic fungi of genes encoding ammonium permeases and their roles in dimorphism. Mol. Microbiol. 2003;50:259–275. doi: 10.1046/j.1365-2958.2003.03680.x. [DOI] [PubMed] [Google Scholar]
- Stevenson B. J., Rhodes N., Errede B., Sprague G. F., Jr. Constitutive mutants of the protein kinase STE11 activate the yeast pheromone response pathway in the absence of the G protein. Genes Dev. 1992;6:1293–1304. doi: 10.1101/gad.6.7.1293. [DOI] [PubMed] [Google Scholar]
- Takizawa P. A., Sil A., Swedlow J. R., Herskowitz I., Vale R. D. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature. 1997;389:90–93. doi: 10.1038/38015. [DOI] [PubMed] [Google Scholar]
- TerBush D. R., Maurice T., Roth D., Novick P. The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- TerBush D. R., Novick P. Sec6, Sec8, and Sec15 are components of a multisubunit complex which localizes to small bud tips in Saccharomyces cerevisiae. J. Cell Biol. 1995;130:299–312. doi: 10.1083/jcb.130.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban J., et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell. 2007;26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Van Nuland A., Vandormael P., Donaton M., Alenquer M., Lourenço A., Quintino E., Versele M., Thevelein J. M. Ammonium permease based sensing mechanism for rapid ammonium activation of the protein kinase A pathway in yeast. Mol. Microbiol. 2006;59:1485–1505. doi: 10.1111/j.1365-2958.2005.05043.x. [DOI] [PubMed] [Google Scholar]
- Ward M. P., Gimeno C. J., Fink G. R., Garrett S. SOK2 may regulate cyclic AMP-dependent protein kinase-stimulated growth and pseudohyphal development by repressing transcription. Mol. Cell. Biol. 1995;15:6854–6863. doi: 10.1128/mcb.15.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedaman K. P., Reinke A., Anderson S., Yates J., III, McCaffery J. M., Powers T. Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:1204–1220. doi: 10.1091/mbc.E02-09-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J., Simon I., Harbison C. T., Hannett N. M., Volkert T. L., Fink G. R., Young R. A. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell. 2003;113:395–404. doi: 10.1016/s0092-8674(03)00301-5. [DOI] [PubMed] [Google Scholar]
- Zheng L., Kostrewa D., Bernèche S., Winkler F. K., Li X. D. The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli. Proc. Natl. Acad. Sci. USA. 2004;101:17090–17095. doi: 10.1073/pnas.0406475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Jiang Y. The yeast phosphotyrosyl phosphatase activator is part of the Tap42-phosphatase complexes. Mol. Biol. Cell. 2005;16:2119–2127. doi: 10.1091/mbc.E04-09-0797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurita-Martinez S. A., Puria R., Pan X., Boeke J. D., Cardenas M. E. Efficient Tor signaling requires a functional class C Vps protein complex in Saccharomyces cerevisiae. Genetics. 2007;176:2139–2150. doi: 10.1534/genetics.107.072835. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.