Abstract
Many viruses regulate protein synthesis by −1 ribosomal frameshifting using an RNA pseudoknot. Frameshifting is vital for viral reproduction. Using the information gained from the recent high-resolution crystal structure of the beet western yellow virus pseudoknot, a systematic mutational analysis has been carried out in vitro and in vivo. We find that specific nucleotide tertiary interactions at the junction between the two stems of the pseudoknot are crucial. A triplex is found between stem 1 and loop 2, and triplex interactions are required for frameshifting function. For some mutations, loss of one hydrogen bond is sufficient to abolish frameshifting. Furthermore, mutations near the 5′ end of the pseudoknot can increase frameshifting by nearly 300%, possibly by modifying ribosomal contacts. It is likely that the selection of suitable mutations can thus allow viruses to adjust frameshifting efficiencies and thereby regulate protein synthesis in response to environmental change.
Keywords: RNA structure, hydrogen bond, RNA triplex, ribosome movement
Many viruses translate more than one polypeptide from polycistronic mRNA through −1 ribosomal frameshifting. It occurs in most retroviruses (1–3), coronaviruses (4), yeast (5) and plant viruses (6), and even in bacterial systems (7). In viral systems, the relative levels of the two proteins are translationally regulated. The upstream protein is translated in the conventional reading frame. At a frequency of between 1 and over 30%, a −1 frameshift occurs before reaching the termination codon of the first protein, resulting in a fusion protein linked to a second protein. Two cis-elements are involved in this unusual process. One is the slippery sequence, the heptanucleotide X XXY YYN (the bases X and Y can be identical), where the actual −1 frameshift takes place (1). The other component is a downstream RNA structural element, either a simple hairpin structure or, more frequently, a pseudoknot usually located 6 to 8 nt downstream of the slippery sequence.
In many cases, the RNA pseudoknot is essential for producing high-level frameshifting in viruses (2–4). This conformation consists of two double-stranded stem regions that form a quasicontinuous helix with two connecting loops (8), which cross the grooves of the stems [see supplemental Fig. 5 on the PNAS web site (www.pnas.org)]. In frameshifting pseudoknots, loop 1 crossing the major groove is short compared with loop 2, which crosses the minor groove. It is believed that ribosomal frameshifting occurs when the ribosome simultaneously engages the slippery sequence and the 5′ strand of stem 1 in the pseudoknot (9). The mRNA translocation machinery pauses at the pseudoknot barrier and increases the likelihood that the aminoacyl and peptidyl tRNA realigns in the 5′ direction on the mRNA (1). However, the basic mechanism and the pseudoknot participation in this process are not understood.
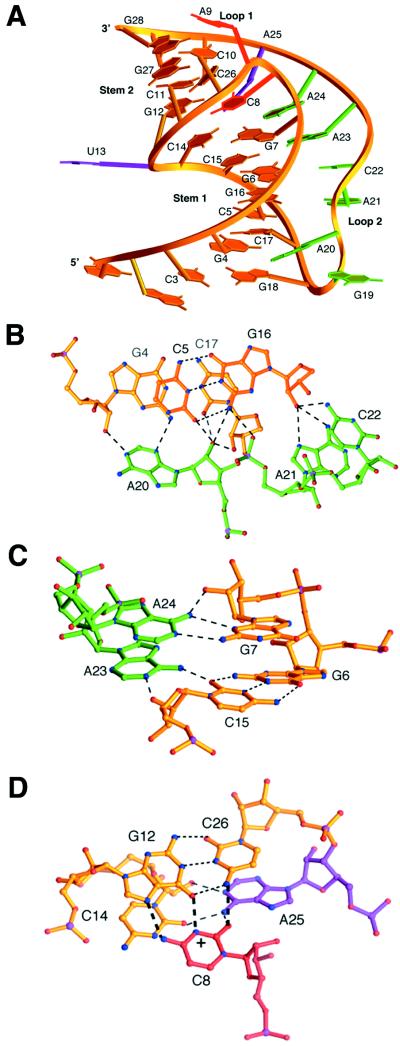
Certain mutations at the junction of the two stems or loop regions can strongly diminish frameshifting (10, 11). Because of the lack of precise structural information, the contribution of each region of the pseudoknot to frameshifting has been unclear. NMR structures of pseudoknots (12–14) show that the two stems have a bent conformation. Recently, the first crystallographic structure of an RNA pseudoknot involved in −1 frameshifting was solved at 1.6-Å resolution from beet western yellow virus (BWYV) (15). The crystal structure reveals a 48° rotation and a 25° bend at the junction of the two stems (Fig. 1), and a triplex is formed by loop 2 in the minor groove of stem 1 (Fig. 1 B and C). A quadruple base interaction between loop 1 and stem 2 is also near the junction (Fig. 1D). This structure reveals several unique features of a frameshifting pseudoknot and shows how the conformation is organized to stabilize its folding. The structure is stabilized by more tertiary hydrogen bond interactions than secondary interactions in Watson–Crick base pairs.
Figure 1.
Tertiary structure of BWYV RNA pseudoknot (A) Ribbon diagram of the overall fold of the pseudoknot. (B) Triplex hydrogen bonding interactions in the minor groove involving A20, A21, and C22. loop 2 is green, stem 1 is gold. The adenine base of A20 contacts G4 at the 2′-OH and N2 position. The 2′-OH of A20 forms a bifurcating hydrogen bonding network with two layers of base pairs. An additional bond (not shown) goes from the 2′-OH of C5 to N7 of A20 (15). A21 and C22 both bond to G16 at the 2′-OH position, and the amino group of G16 makes a phosphate contact to A21. In the crystal structure, a sodium ion (not shown) was found making base–base contacts between G16, A21, and C22. (C) Triplex interactions of A23 and A24 in the junction region. A23 and A24 use their Watson–Crick faces to interact with the minor groove side of the bases and 2′-OH groups of G7 and C15, which are in different strands of stem 1. (D) Junctional core interaction involving the four bases. The protonated C8 (indicated by +) simultaneously interacts with three other bases, G12, C26, and A25. C8 is on the same level as the G12⋅C26 base pair. Thick dashed lines represent the hydrogen bonding on the top layer, and gray dashed lines represent hydrogen bonds in the lower layer. Junctional base A25 tilts between C14 and C8. C26 propeller twists in its base pair to stack on A25.
The detailed knowledge arising from the crystallographic analysis has stimulated us to carry out a systematic series of mutations on a known structure that will target each tertiary element and provide information on structure and function. The results clearly show that the minor groove triplex formed at the 5′-half of the molecule is an important feature in frameshifting, especially near the junction of the two stems. Likewise, maintaining the specific quadruple base interactions and the conformation at the junction region is crucial. In addition, certain mutations at the joining of stem 1 to loop 2 dramatically enhance the efficiency of frameshifting.
Materials and Methods
Template Construct for Frameshifting Assay.
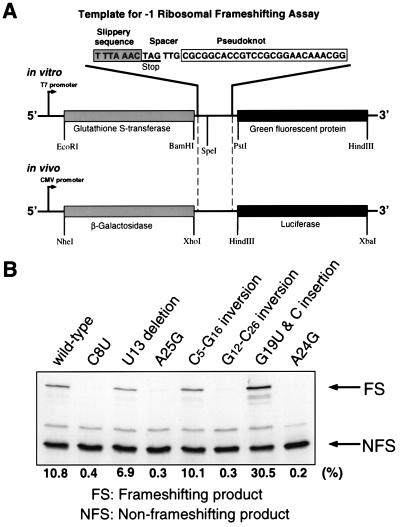
The glutathione S-transferase (GST) gene and the green fluorescent protein (GFP) downstream of the GST gene were inserted at EcoRI/BamHI and PstI/HindIII in the pGEM-3Z vector (Promega), respectively. The resulting vector construct containing the GST and GFP genes was named pGEM-GG. The slippery and pseudoknot sequences were inserted into pGEM-GG at the BamHI and PstI restriction sites by using annealed duplex DNA oligomers. This DNA insert introduced a SpeI restriction site between the slippery sequence and pseudoknot sequence. Separate mutations in the pseudoknot sequence or the slippery sequence were introduced by inserting duplex oligomers containing mutant sequences at SpeI/PstI for the pseudoknot and BamHI/SpeI for the slippery sequence (Fig. 2A). All wild-type and mutant constructs were confirmed by dideoxy DNA sequencing. To compare the mutational effects on a quantitative scale, the wild-type GGGAAAC slippery sequence, yielding 3.9% frameshifting in the in vitro system, was replaced by the more efficient UUUAAAC slippery sequence from infectious bronchitis virus (16).
Figure 2.
(A) Template construct for ribosomal frameshifting. A stop codon is found immediately after the slippery sequence. Because of the requirement for restriction sites, the spacer sequence differs from the wild type. The in vitro template is transcribed by using T7 RNA polymerase at the T7 promoter, and the resulting transcripts are translated by a rabbit reticulocyte lysate in the same reaction tube. For the in vivo ribosomal frameshifting assay, the −1 ribosomal frameshifting elements, including the slippery sequence and the pseudoknot, are inserted between the 5′ β-galactosidase gene and the luciferase gene. A CMV promoter is used in human embryonic kidney cells. The nonframeshifted product is β-galactosidase, whereas frameshifting yields a fusion protein with luciferase. (B) SDS/PAGE analysis of [35S]methionine-labeled translation products from ribosomal frameshifting assay of wild type and selected mutants in the reticulocyte lysate. Translation products are labeled with incoporation of [35S]methionine. The nonframeshifting product (NFS) is the GST protein, and the −1 frameshift product (FS) is a GST–GFP fusion protein.
The UAG termination codon of the GST gene is located immediately after the slippery sequence. If a −1 frameshift occurs at the slippery sequence, the termination codon of the GST gene is not read, and translation proceeds through the GFP gene, resulting in the production of a GST–GFP fusion protein.
In Vitro Frameshifting Assay.
All plasmids were isolated by alkaline lysis. Plasmid DNA was further purified by phenol/chloroform extraction and ethanol precipitation. The lyophilized DNA was dissolved in TE buffer (10 mM Tris⋅Cl, pH 8.0/1 mM EDTA). The TNT T7-coupled transcription/translation system (Promega) was used according to the manufacturer’s protocol. Template DNAs (400 ng) were used in 20-μl reactions containing 10 μl reticulocyte lysate and 0.8 μl of 10 μCi/μl [35S]-labeled methionine (NEN).
The GST–GFP fusion product yields a 58-kDa protein containing 14 methionine residues, whereas the nonframeshifting GST protein product is 30 kDa with 9 methionines. To separate the GST–GFP fusion protein from the nonframeshifting (GST) product, the samples were run on 12% SDS polyacrylamide gels. After electrophoresis, gels were dried, exposed to PhosphorImager screens, and signals were quantified (Molecular Dynamics). Frameshifting efficiencies were calculated with the formula (I[FS]/14)/[(I[FS]/14) + (I[NFS]/9)], where I[FS] is the signal intensity of the frameshifting product and I[NFS], the signal intensity of the nonframeshifting product. The product identity was confirmed by Western blot analysis with an anti-GST antibody (Sigma). All individual in vitro assays were accompanied by wild-type controls and repeated three times or more to determine the average frameshifting efficiencies. The frameshifting efficiency of all wild-type reactions was 10.8 ± 1.6%.
Mammalian Cell Culture Transfection and in Vivo Assay.
HEK 293 cells (American Type Culture Collection CRL1573) were cultured in Dulbecco’s modified essential medium containing penicillin (100 units/ml) and streptomycin (100 μg/ml) and supplemented with 10% fetal calf serum. Reporter plasmid (2 μg) was transfected onto a half-confluent 9.6 cm2 culture dish by cationic liposome-mediated transfection (17) using the lipid reagent Tfx-20 (Promega). The cells were assayed for transient expression of the reporter genes 48 h after transfection. For each construct, four independent transfection experiments with independent DNA preparations were performed.
To prepare crude extracts, cells were washed twice with PBS, scraped out with 190 μl of 1× reporter lysis buffer (Promega), lysed by one freeze/thaw cycle, vortexed, and spun at 25°C to pellet cell debris. Luciferase and β-galactosidase activities were measured from the same extract (18). Supernatant (20 μl) was placed in a luminometer (MGM Instruments, Hampden, CT), and the reaction was started by injection of 100 μl luciferase assay reagent (Promega). Light emission was recorded for 10 s. The luciferase background activity is 200–300 light units in 20 μl lysate of mock-transfected cells. To assay the β-galactosidase activity, 20 μl of supernatant was incubated with 2 μl of 60 mM Na2HPO4/40 mM NaH2PO4/10 mM KCl/1 mM MgCl2/50 mM β-mercaptoethanol/200 μl of 2 mg/ml o-nitrophenyl-β-d-galactopyranoside at 37°C for 40 min. The reactions were stopped by adding 500 μl of 1 M NaCO3, and the colorimetric change was measured in a spectrophotometer at 420 nm. To quantitate frameshifting efficiency in vivo, luciferase activity in the cellular extract was determined with β-galactosidase activity as an internal control for normalization. A construct containing the luciferase gene and the β-galactosidase gene in frame was used as 100% efficiency standard as previously described (18).
Results
Frameshifting was measured in two different systems: an in vitro rabbit reticulocyte lysate and in vivo by using human embryonic kidney cells. Most experiments were carried out initially in vitro. The in vivo experiments confirmed the pattern of mutational effects seen in vitro.
Adenosine-Rich Minor Groove Triplex Is an Important Feature.
It is likely that frameshifting occurs when the pseudoknot resists unraveling by the moving ribosome. The stem 1-loop 2 triplex stabilizes stem 1 by interacting with both strands of the stem. The pseudoknot stability and conformation involving certain conserved sequences are likely to be the features that are important for frameshifting efficiency. The conservation of adenosine residues in the leuteovirus loop 2 sequence AACAA (6) and the high adenosine content found in other frameshifting pseudoknots (2, 3) suggest that there might be a common sequence specificity in the triplex interactions.
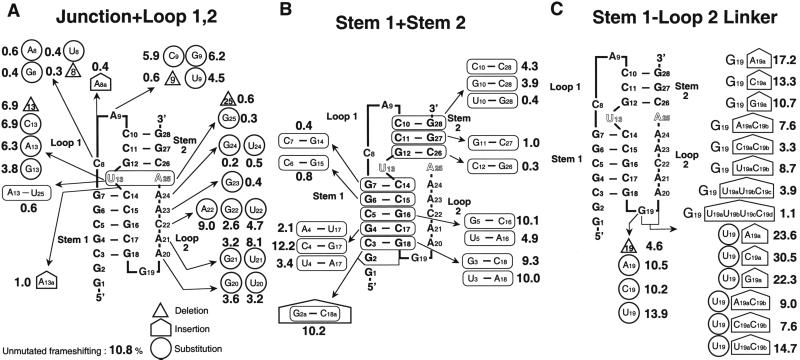
A series of mutations were made of the bases in loop 2, and the efficiency of ribosomal frameshifting was measured [Fig. 3A and supplemental Table 1 (see www.pnas.org)]. Adenines of loop 2 were systematically changed to guanines or uridines (Fig. 3A). Mutations show that there is a preference for adenosine residues at each position, as shown by the decreased frameshifting. Compared with pyrimidines, adenines can form stacking platforms and interact with the shallow minor groove, yet still have hydrogen donors and acceptors available for other contacts. Adenosines also have the advantage over the stackable guanosines, in which steric problems arise from the additional O6 group (19).
Figure 3.
Mutations in the BWYV pseudoknot and their effects on −1 ribosomal frameshifting activity. The unmutated frameshifting efficiency in this experiment is 10.8%. The numbers near the mutations correspond to the frameshifting efficiency. (A) Mutations in the junction and loops, 1 and 2. (B) Mutations and inversions in stem 1 and stem 2 base pairs. (C) Mutations at the stem 1–loop 2 linker.
Frameshifting was not abolished by the mutating A20. Substitution of adenosine residues with uridines may result in the loss of optimal stacking or disruption of triplex interactions. The replacement with G residues will result in the disruption of specific triple-base interactions because of altered hydrogen bonding associated with the additional N2 amino group as in the A20 position (Fig. 1B). In contrast, 2′-OH-mediated interactions will not be affected. Other triplex interactions with different geometries could form with the substitution of U or G but would require rearrangement in the backbone or altered stacking conformation. To further probe the loop 2–stem 1 interaction, A⋅U base pairs were substituted for G⋅C base pairs in stem 1 (Fig. 3B). A4⋅U17 and U5⋅A16 mutations result in the loss of the hydrogen bond to A20 involving N2 of G, and they show reduced frameshiftings. In contrast, the U3⋅A18 mutation has no effect. This supports the importance of triplex interactions in frameshifting, because the C3⋅G18 base pair is not involved in tertiary interactions. It is possible that mutations in loop 2 may also interfere with possible contacts to the ribosome or other protein factors involving the free donor/acceptor groups of the bases. This is suggested from mutations in A21 (to G21) and C22 (to U22), in which no loss of tertiary interactions is anticipated, but rather a change in the donor/acceptor hydrogen bonding pattern. Nonetheless, these substitutions show a negative effect on frameshifting (Fig. 3A) with loss of two-thirds of the activity. Fig. 3B also shows the effect of introducing other mutations in the lower part of stem 1. Inverting the three lower 5′ C⋅G base pairs does not affect frameshifting significantly, and there is even a suggestion that inverting the G4⋅C17 base pair increases frameshifting slightly. Inverting G4⋅C17 is likely to maintain the interactions with A20. Furthermore, adding another base pair to the bottom of stem 1 (G2a⋅C18a in Fig. 3B) does not affect frameshifting. This is in accord with the results of Napthine et al. (20), who showed that lengthening stem 1 of a frameshifting pseudoknot left it with full activity.
In contrast, great sensitivity to mutation is observed near the junction between the two stems. A23 and A24 form hydrogen bonds with adjacent base pairs of the minor groove but are associated with different strands of the duplex; A23 is bound to C15, whereas A24 binds to G7 (Fig. 1C). Substituting either A23 to G or A24 to G or U abolishes frameshifting to background levels (Fig. 3A). U23 was not used because it created a stop codon. G23 replacing A23 can form a hydrogen bond with the 2′OH of C15 by using the N1-H as a hydrogen bond donor. However, loss of the hydrogen bond interacting with O2 of C15 appears to be sufficient to abolish frameshifting activity. The sensitivity of these interactions is seen in further experiments in which the stem base pairs G6⋅C15 and G7⋅C14 are inverted (Fig. 3B), because those mutations also abolish frameshifting. The effect of these inversions might be partly associated with changes in stacking energy perturbing adjacent base-stacking interaction (21), but it is probably because of an asymmetry in the minor groove. There is a slight change in the directionality of the guanine N2-H hydrogen donor after inversion. However, the distribution of orbitals that are used for hydrogen bonding is not symmetric. This has been emphasized in the work of Kielkopf et al. (22), who have shown that it is possible to differentiate between inverted base pairs with minor groove binding molecules. Our results are in accord with those of Liphardt et al. (23), who showed in infectious bronchitis virus that in the position analogous to A24, good frameshifting required an adenine, and inverting the base pairs analogous to G7⋅C14 and G6⋅C15 also adversely affected frameshifting. Additional experiments with both substitutions and inversions are listed in supplemental Table 1. They show that the frameshifting efficiency is usually the product of the efficiencies of the individual component changes, and triplex interactions near the stem junction are crucial to frameshifting activity.
Stem Junctional Interactions Are Essential in Frameshifting.
The other striking feature in the BWYV pseudoknot is the organization at the junction region between C8 of loop 1 hydrogen bonding in a quadruple interaction to the G12⋅C26 base pair of stem 2, and A25 (Fig. 1D). The loop 1 base C8 is deeply inserted into the major groove of stem 2 and stacks on G7 (Fig. 1A) (15). This interaction holds together nucleotides from loop 1, stem 2, and the nearby stem 1–loop 2 minor groove triplex. The region is likely to influence the equilibrium between folded and unfolded forms of the pseudoknot. The observed quadruple base interaction is sequence specific, and every base is essential for function. As shown in Fig. 3 B and C, deletion or substitution of C8 or A25 or inversion of the G12⋅C26 base pair completely destroys frameshifting. All of these mutations, except C8 to U8, are expected to disrupt the observed interactions in the structure because of steric clashes (C8 to A8 or G8) or unfavorable hydrogen donor/acceptor contacts. The severe decrease in frameshifting at first appears unexpected for the mutation C8 to U8, which results in the loss of only one hydrogen bond to the N7 group of G12. Because C8 is deeply buried in the major groove of stem 2, it is unlikely to be accessible for higher-order contact (Fig. 1A). However, considering the accumulative negative charges from the surrounding phosphate groups, the positive charge on the protonated cytosine residue may play an important electrostatic role and thereby stabilize the overall pseudoknot conformation.
The junctional A25 platform is also a key element, because not only is it involved in the minor groove triplex interactions binding to C14 (15), but it also continues the stacking of the triplex third strand into stem 2 (Fig. 1A). A25 is stacked between C26 of stem 2 and A24, the last base of loop 2. It is tilted between the C8 and C14 layers with multiple interaction (15). Mutating or deleting A25 largely eliminates frameshifting (Fig. 3A). These interactions may have a central organizing influence on the pseudoknot conformation. There is less impairment of frameshifting as we move away from the junction. Inverting base pair C10⋅G28 leaves 40% of frameshifting activity, whereas inversions in stem 2 closer to the junction are more deleterious (Fig. 3B).
Comparing the BWYV junctional sequences with other frameshifting pseudoknots: in Mouse mammary tumor virus, there is a U⋅G pair at the U13⋅A25 position (2); in Simian retrovirus (3), the junctional pair is inverted to A⋅U. However, the identity of the loop 1 base analogous to C8 is reported to be not crucial for frameshifting in those systems (4, 10, 11). Therefore, the junctional interactions are likely to be quite different in those systems. Tinoco and coworkers have proposed that a bent conformation induced by an intercalated adenosine between the two stems might be important for frameshifting in mouse mammary tumor virus (14). In BWYV, mutations were carried out in an attempt to mimic the higher efficiency frameshifting pseudoknots. An adenosine (A13a) insertion between the two stems next to U13 or inversion of the U13⋅A25 pair results in a severe decrease in frameshifting efficiencies (Fig. 3A). This suggests that the junction region is an integral part of the BWYV molecule related to the overall geometry of the stems.
Exposed Regions Not Involved in Tertiary Contact Can Affect Frameshifting.
Extruded U13.
In the crystal structure, U13 is not base paired to A25 as predicted. Several biochemical and NMR studies suggest that these junctional base pairs may also not be formed for the frameshifting function in other systems (24, 25). The extrusion of U13 from the stem region widens the major groove and allows insertion of the C8 base into stem 2 (Fig. 1A). In comparison to the previous mutations at the junction, deletion or substitutions of U13 still maintains about 60% of wild-type frameshifting (Fig. 3A). This is consistent with the fact that U13 is neither involved in secondary interactions with A25 nor engaged in tertiary interactions. Deletion of U13 is likely to introduce some tension in the backbone of the corresponding strand and result in subtle changes in the junctional interactions. Surprisingly, compared with the deletion, the substitution of U13 to A or C equally reduces the efficiency (Fig. 3B). This suggests that U13 may be in contact with the ribosomal machinery, or it could promote the transition from an unstructured hairpin to the pseudoknot conformation by transiently base pairing with A25.
A9 capping region.
A9 of loop 1 partially stacks on the cytosine 10 base in the so-called “C turn” but does not engage in tertiary interactions. The “C turn” is stabilized by a hydrogen bond from N4 of C10 to the phosphate oxygen of A9 in the minor groove of stem 2, as well as through an intricate network of water molecules (15). Mutation of adenine 9 decreased frameshifting by 40–50%, depending on the substituted base (Fig. 3A). A related observation was reported in the mutation of the helix cap at the 3′-end of the T4 gene 32 pseudoknot that affected its stability (26). The decrease in frameshifting observed in the A9 to C9 or U9 mutation might be explained by the loss in optimal purine stacking. Nonetheless, the G9 mutation does not alter stacking, and the efficiency decrease is unexpected. This suggests the possibility that pseudoknot stability is not the only factor contributing to frameshifting efficiency. To study this further, mutations were performed on the adjacent C10⋅G28 pair [supplemental Table 1 (see www.pnas.org)]. However, the combined mutations of A9 (C9/G9/U9) and the G10⋅C28 inversion failed to rescue full frameshifting, implying that an adenosine in the 9 position is more suitable.
It is not surprising that deletion of A9 abolishes frameshifting (Fig. 3A), because that will make it difficult for C8 to cross the groove and is likely to cause disruption in the junctional interactions. On the other hand, insertion of a nucleotide (A8a in Fig. 4A) may also destabilize by introducing more distance between C8 and C10, leading to loss of overwinding and a less constrained stem 2.
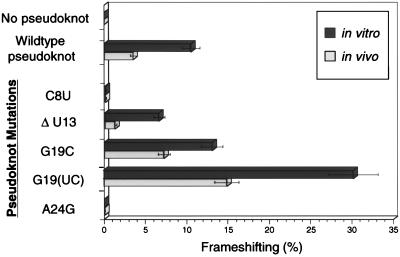
Figure 4.
Comparison of in vivo and in vitro ribosomal frameshifting efficiencies of selected BWYV pseudoknot mutants. The G19(UC) mutation has a UC insertion instead of G19.
At the 3′-end base pair, mutation to U10⋅G28 abolishes frameshifting. If a U10⋅G28 base pair forms, the guanine or uridine must rotate relative to C10⋅G28, to form two hydrogen bonds. This may alter helical stacking and lead to destabilization. Furthermore, the “C turn” will not be stabilized because of the loss of the amino group in a cytosine. The G10⋅C28 mutation also results in the loss of this interaction with the C turn, but there is less loss of frameshifting compared with the U10⋅G28 mutation. Disruption of a stem 2 base pair (C10⋅C28 mutation) does not entirely abolish frameshifting (Fig. 3B). Although it is possible that C10⋅C28 can form protonated base pairs, the general observation is consistent with previous studies, in which the stability of stem 2 was reported to be less crucial in frameshifting than that of stem 1 (11).
Stem 1–loop 2 junction
In the BWYV conformation, most of the loop 2 residues form a stacking ladder, progressing upward to interact with each base pair layer of stem 1 (Fig. 1A). Each loop 2 nucleotide contributes to the stability of the structure and seems indispensable. Adenosine residues are involved in sequence-specific interactions, and other residues such as G19 and C22 may serve as space linkers. A sharp turn occurs at residue G19 at the stem 1–loop 2 junction, and G19 is bulged out in the crystal structure (Fig. 1A). The only stabilizing feature is that its ends are fixed, G18 is base paired, and A20 is involved in a network of seven hydrogen bonds (Fig. 2B).
As expected, deletion of G19 decreases frameshifting by 50%, possibly through disrupting the A20 interactions. However, the insertion of a base (A19a, G19a, or C19a) after G19 increased the frameshifting efficiency, but adding two to four pyrimidines gradually decreased the efficiency (Fig. 3C). This suggests that, whereas one base insertion may relieve some tension at the sharp turn or provide an additional loop base to interact with another surface, additional insertions may introduce flanking nucleotides that are energetically unfavorable. Similar observations have been observed in the simian retrovirus −1, in which deletion of nonconserved nucleotides in a longer loop 2 notably increased frameshifting (11).
Base substitutions were also carried out on G19, and the 30% increase in frameshifting for mutation to U19 was not anticipated, whereas A/C mutations gave wild-type efficiency (Fig. 3C). Strikingly, the G19 to U19C19a substitution and insertion lead to an unprecedented almost 3-fold (30.5%) increase in frameshifting. This phenomenon was observed both in the reticulocyte translation system and in a wheat germ extract system (data not shown). Comparing the effects of mutations G19C19a (13.3%), U19C19a (30.5%), and U19A19a (23.6%) shows that a single U at position 19 can produce a dramatic effect in frameshifting. U19C19a may stabilize the pseudoknot structure, but an increase in frameshifting of this magnitude suggests that the 5′ end region of the molecule may be in a position to make contact with the moving ribosome. Because the ribosome approaches the pseudoknot along the 5′ end, the bulge created by the U19C19a residues may be the first ribosomal contact. The mutation U19C19a may introduce nucleotides projecting out from the pseudoknot, and the loop 2 triplex interactions are likely to remain in place. The size and composition of the protruding nucleotides also seems to be important. C19a is more effective than either A19a or G19a.
In Vivo Frameshifting.
Some of the results of the in vitro experiments using a rabbit reticulocyte lysate were unexpected, especially in the enhancement of frameshifting. It was felt important to confirm the pattern of results in vivo. For these experiments, human embryonic kidney cells were transfected with the construct shown in Fig. 2A, in which the nonframeshifted product was β-galactosidase, and frameshifting produced β-galactosidase-luciferase fusion protein, both of which can be readily assayed (18). The results of these assays are illustrated in Fig. 4, comparing the in vitro and in vivo frameshifting. It is well known that in vivo frameshifting results are generally less than those measured in vitro, probably because of the presence of other factors that influence ribosomal mRNA interactions (18, 27). However, what is important are the relative patterns in the two systems. Mutations that abolish frameshifting in vitro act the same way in vivo (C8U, A24G), reinforcing our conclusions about the importance of the junction region. The wild-type in vivo frameshifting is approximately 40% of the in vitro results. The U13 mutation is somewhat more deleterious in vivo, but it still retains some frameshifting activity. The enhancement of frameshifting for the mutation G19C or substituting (U19C19a) for G19 is seen in vivo. Thus the broad pattern of in vitro results is mirrored by the in vivo experiments.
Discussion
Although there have been many studies of the role of pseudoknots in ribosomal frameshifting, we do not yet understand the process in detail. The pseudoknot is a unique tertiary motif that has a complicated topology. In our studies, mutations that are likely to disrupt or alter the observed tertiary interactions in the BWYV crystal structure generally show a decrease or complete loss in frameshifting efficiencies. It has been suggested that there may be a ribosome-associated RNA helicase that opens up mRNA secondary structure; however, there is no evidence for such factors. As the ribosome moves along the mRNA, it first encounters the 5′ end of the pseudoknot and may encounter the minor groove triplex, which may impede the process of unraveling by cross-strand interactions with stem 1. The adenosine-specific triplex motifs may be a general feature in other frameshifting pseudoknots as well. We have carried out A to G or U mutations on the loop 2 adenines of the simian retrovirus-1 and mouse mammary tumor virus pseudoknots, and there is a significant decrease in frameshifting (data not shown). This is consistent with other pseudoknot studies in which deletion of the adenine-rich sequence in simian retrovirus-1 (11) and murine leukemia virus (28) significantly decreased frameshifting or readthrough, respectively.
Although stability is one element that plays a role in this process, the resistance against deformation by the translocation machinery from a topological point of view should also be considered. In studies of Somogyi et al. (29) a hairpin loop of equal or higher stability was found to induce ribosomal pausing, but little frameshifting was promoted. Compared with the hairpin loop structure, the pseudoknot has two regions where there are changes in 5′-3′ directionality. This, along with the intricate loop–stem interactions, probably makes the pseudoknot topologically more difficult to unravel in a linear fashion. The sensitivity of the junction region may be related to the fact that it resists deformations because of the changes in strand directions that are associated with the rotation and bend of the molecule at that point.
On the other hand, a particular conformation or certain specific residues may have dynamic interactions with the ribosome or with auxiliary protein factors needed to trigger the −1 slippage. There are several locations for such higher-order contacts, and mutations of these regions may change the frameshifting efficiencies, not by altering the stability of the pseudoknot structure, but by modifying contacts with ribosomes or ribosome associated factors. In particular, the stem 1–loop 2 junction region is a good candidate area for investigating such interactions. Thermodynamic studies of these mutants may differentiate those that destabilize the structure from those in which interactions with ribosomes and or other factors influence changes in frameshifting efficiency.
In our study, we show how mutations in the pseudoknot sequence can dramatically increase or decrease frameshifting efficiencies. This raises the possibility that throughout evolution, viral systems could have adjusted the translation levels of proteins through the changes or addition of nucleotides in the pseudoknot sequence leading to optimized reproduction. As we learn more of the fundamental features of this process, we may in principle learn how to modify frameshifting. Because higher eukaryotes apparently do not use −1 ribosomal frameshifting, this system is potentially an attractive drug target that will allow us to develop means of partially or wholly inactivating these viruses.
Supplementary Material
Acknowledgments
We have profited from discussions with Drs. K. Lowenhaupt, M. Egli, and B. Brown. This research was supported by grants from the National Institutes of Health, the National Science Foundation, and the National Foundation for Cancer Research. S.M. was supported by a long-term fellowship of the European Molecular Biology Organization, and A.O. was supported by the Massachusetts Institute of Technology Undergraduate Research Opportunities Program.
Abbreviations
- BWYV
beet western yellow virus
- GST
glutathione S-transferase
- GFP
green fluorescent protein
Footnotes
See commentary on page 14177.
References
- 1.Jacks T, Madhani H D, Masiarz F R, Varmus H E. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chamorro M, Parkin N, Varmus H E. Proc Natl Acad Sci USA. 1992;89:713–717. doi: 10.1073/pnas.89.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.ten Dam E, Brierley I, Inglis S, Pleij C. Nucleic Acids Res. 1994;22:2304–2310. doi: 10.1093/nar/22.12.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brierley I, Rolley N J, Jenner A J, Inglis S C. J Mol Biol. 1991;220:889–902. doi: 10.1016/0022-2836(91)90361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tzeng T H, Tu C L, Bruenn J A. J Virol. 1992;66:999–1006. doi: 10.1128/jvi.66.2.999-1006.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller W A, Dinesh-Kumar S P, Paul C P. Crit Rev Plant Sci. 1995;14:179–211. [Google Scholar]
- 7.Gesteland R F, Atkins J F. Annu Rev Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- 8.Pleij C W, Rietveld K, Bosch L. Nucleic Acids Res. 1985;13:1717–1731. doi: 10.1093/nar/13.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tu C, Tzeng T H, Bruenn J A. Proc Natl Acad Sci USA. 1992;89:8636–8640. doi: 10.1073/pnas.89.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L, Cai L, Zhang X, Rich A. Biochemistry. 1994;33:13540–13546. doi: 10.1021/bi00250a005. [DOI] [PubMed] [Google Scholar]
- 11.ten Dam E B, Verlaan P W, Pleij C W. RNA. 1995;1:146–154. [PMC free article] [PubMed] [Google Scholar]
- 12.Shen L X, Tinoco I., Jr J Mol Biol. 1995;247:963–978. doi: 10.1006/jmbi.1995.0193. [DOI] [PubMed] [Google Scholar]
- 13.Du Z, Giedroc D P, Hoffman D W. Biochemistry. 1996;35:4187–4198. doi: 10.1021/bi9527350. [DOI] [PubMed] [Google Scholar]
- 14.Kang H, Tinoco I., Jr Nucleic Acids Res. 1997;25:1943–1949. doi: 10.1093/nar/25.10.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su L, Chen L, Egli M, Berger J M, Rich A. Nat Struct Biol. 1999;6:285–292. doi: 10.1038/6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia A, van Duin J, Pleij C W. Nucleic Acids Res. 1993;21:401–406. doi: 10.1093/nar/21.3.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felgner P L, Ringold G M. Nature (London) 1989;337:387–388. doi: 10.1038/337387a0. [DOI] [PubMed] [Google Scholar]
- 18.Reil H, Kollmus H, Weidle U H, Hauser H. J Virol. 1993;67:5579–5584. doi: 10.1128/jvi.67.9.5579-5584.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pley H W, Flaherty K M, McKay D B. Nature (London) 1994;372:111–113. doi: 10.1038/372111a0. [DOI] [PubMed] [Google Scholar]
- 20.Napthine S, Liphardt J, Bloys A, Routledge S, Brierley I. J Mol Biol. 1999;288:305–320. doi: 10.1006/jmbi.1999.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saenger W. Principles of Nucleic Acid Structure. New York: Springer; 1984. pp. 242–252. [Google Scholar]
- 22.Kielkopf C L, White S, Szewczyk J W, Turner J M, Baird E E, Dervan P B, Rees D C. Science. 1998;282:111–115. doi: 10.1126/science.282.5386.111. [DOI] [PubMed] [Google Scholar]
- 23.Liphardt J, Napthine S, Kontos H, Brierley I. J Mol Biol. 1999;288:321–335. doi: 10.1006/jmbi.1999.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sung D, Kang H. Nucleic Acids Res. 1998;26:1369–1372. doi: 10.1093/nar/26.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X, Chamorro M, Lee S I, Shen L X, Hines J V, Tinoco I, Jr, Varmus H E. EMBO J. 1995;14:842–852. doi: 10.1002/j.1460-2075.1995.tb07062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Theimer C A, Wang Y, Hoffman D W, Krisch H M, Giedroc D P. J Mol Biol. 1998;279:545–564. doi: 10.1006/jmbi.1998.1812. [DOI] [PubMed] [Google Scholar]
- 27.Parkin N T, Chamorro M, Varmus H E. J Virol. 1992;66:5147–5151. doi: 10.1128/jvi.66.8.5147-5151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wills N M, Gesteland R F, Atkins J F. EMBO J. 1994;13:4137–4144. doi: 10.1002/j.1460-2075.1994.tb06731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Somogyi P, Jenner A J, Brierley I, Inglis S C. Mol Cell Biol. 1993;13:6931–6940. doi: 10.1128/mcb.13.11.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.