Abstract
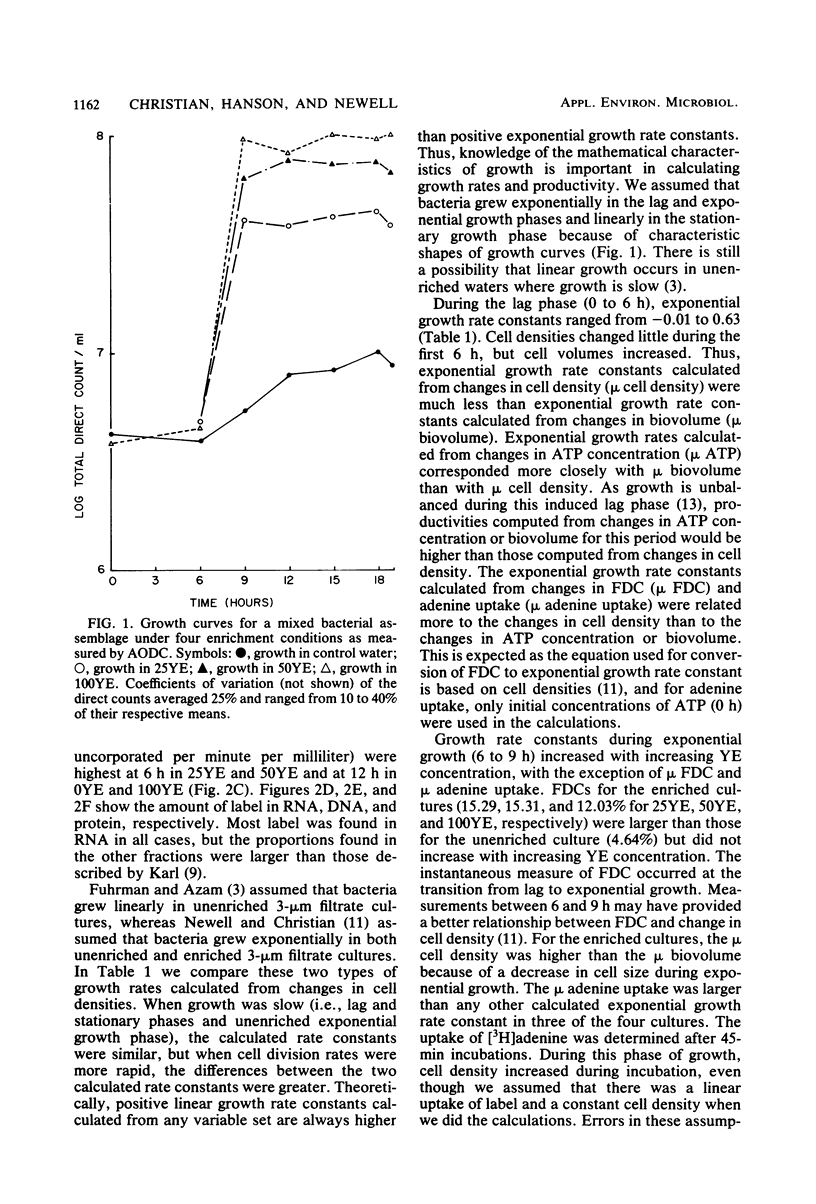
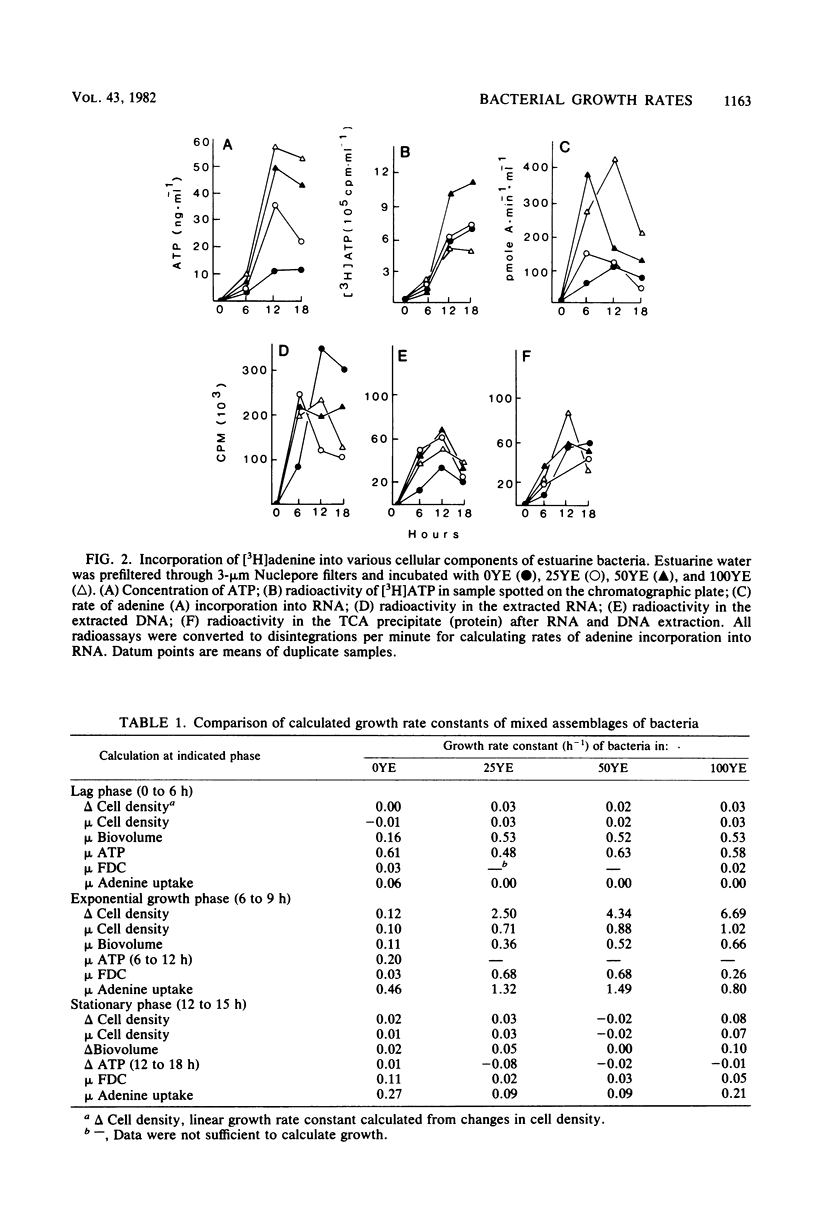
Eight methods of assessing growth rate constants of bacteria were compared in batch cultures of 3-micrometers-filtered estuarine water from the Skidaway River in Ga. Mixed assemblages of bacteria were grown under four nutrient regimes of added yeast extract ranging from 0 to 100 mg/liter. Linear and exponential growth rate constants were computed from changes in cell densities, biovolumes, and ATP concentrations. Exponential growth rate constants were obtained from the frequency of dividing cells and RNA synthesis as measured by [3H]adenine uptake. Rate constants obtained during lag, exponential, and stationary growth phases depended largely on the method used. Constants calculated from changes in cell densities, frequency of dividing cells, and adenine uptake correlated most closely with each other, whereas constants calculated from changes in ATP concentrations and biovolumes correlated best with each other. Estimates of in situ bacterial productivity and growth vary depending on the method used and the assumptions made regarding the growth state of bacteria.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brock T. D. Microbial growth rates in nature. Bacteriol Rev. 1971 Mar;35(1):39–58. doi: 10.1128/br.35.1.39-58.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman A. G., Fall L., Atkinson D. E. Adenylate energy charge in Escherichia coli during growth and starvation. J Bacteriol. 1971 Dec;108(3):1072–1086. doi: 10.1128/jb.108.3.1072-1086.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman J. A., Azam F. Bacterioplankton secondary production estimates for coastal waters of british columbia, antarctica, and california. Appl Environ Microbiol. 1980 Jun;39(6):1085–1095. doi: 10.1128/aem.39.6.1085-1095.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagström A., Larsson U., Hörstedt P., Normark S. Frequency of dividing cells, a new approach to the determination of bacterial growth rates in aquatic environments. Appl Environ Microbiol. 1979 May;37(5):805–812. doi: 10.1128/aem.37.5.805-812.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmstetter C., Cooper S., Pierucci O., Revelas E. On the bacterial life sequence. Cold Spring Harb Symp Quant Biol. 1968;33:809–822. doi: 10.1101/sqb.1968.033.01.093. [DOI] [PubMed] [Google Scholar]
- Hobbie J. E., Daley R. J., Jasper S. Use of nuclepore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977 May;33(5):1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl D. M. Cellular nucleotide measurements and applications in microbial ecology. Microbiol Rev. 1980 Dec;44(4):739–796. doi: 10.1128/mr.44.4.739-796.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl D. M. Measurement of microbial activity and growth in the ocean by rates of stable ribonucleic Acid synthesis. Appl Environ Microbiol. 1979 Nov;38(5):850–860. doi: 10.1128/aem.38.5.850-860.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell S. Y., Christian R. R. Frequency of dividing cells as an estimator of bacterial productivity. Appl Environ Microbiol. 1981 Jul;42(1):23–31. doi: 10.1128/aem.42.1.23-31.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldringh C. L. Morphological analysis of nuclear separation and cell division during the life cycle of Escherichia coli. J Bacteriol. 1976 Jan;125(1):248–257. doi: 10.1128/jb.125.1.248-257.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]


