Abstract
Background
Brucellosis, caused by members of the genus Brucella, remains one of the world's major zoonotic diseases. Six species have classically been recognised within the family Brucella largely based on a combination of classical microbiology and host specificity, although more recently additional isolations of novel Brucella have been reported from various marine mammals and voles. Classical identification to species level is based on a biotyping approach that is lengthy, requires extensive and hazardous culturing and can be difficult to interpret. Here we describe a simple and rapid approach to identification of Brucella isolates to the species level based on real-time PCR analysis of species-specific single nucleotide polymorphisms (SNPs) that were identified following a robust and extensive phylogenetic analysis of the genus.
Results
Seven pairs of short sequence Minor Groove Binding (MGB) probes were designed corresponding to SNPs shown to possess an allele specific for each of the six classical Brucella spp and the marine mammal Brucella. Assays were optimised to identical reaction parameters in order to give a multiple outcome assay that can differentiate all the classical species and Brucella isolated from marine mammals. The scope of the assay was confirmed by testing of over 300 isolates of Brucella, all of which typed as predicted when compared to other phenotypic and genotypic approaches. The assay is sensitive being capable of detecting and differentiating down to 15 genome equivalents. We further describe the design and testing of assays based on three additional SNPs located within the 16S rRNA gene that ensure positive discrimination of Brucella from close phylogenetic relatives on the same platform.
Conclusion
The multiple-outcome assay described represents a new tool for the rapid, simple and unambiguous characterisation of Brucella to the species level. Furthermore, being based on a robust phylogenetic framework, the assay provides a platform that can readily be extended in the future to incorporate newly identified Brucella groups, to further type at the subspecies level, or to include markers for additional useful characteristics.
Background
Brucellosis remains one of the world's major zoonotic disease problems and can lead to reproductive problems in a number of large livestock and other animals. In humans, brucellosis is associated with a broad spectrum of symptoms and can occasionally be fatal [1]. Whilst brucellosis is the global term for infection by organisms in the Gram-negative genus Brucella, there are actually a number of causative agents. Differentiation to species level within the genus is based on both different phenotypic characteristics and the host from which it was initially isolated [2]. These divisions are also supported by pulse field gel electrophoresis (PFGE)-based physical mapping, showing that each species has a specific physical organisation [3], and techniques such as multilocus sequencing [4] and variable number tandem repeat (VNTR) based typing [5,6]. Bovine infection is generally caused by B. abortus, B. melitensis infects predominantly sheep and goats, B. suis infects pigs, hares and reindeer, B. ovis infects sheep only, B. canis infects dogs, whilst B. neotomae has only been isolated from the desert wood rat. In addition to the six classically described species novel members of the genus have more recently been isolated from marine mammals, predominantly dolphins, porpoises and seals. These have very recently been classified as two additional species, B. ceti (cetaceans) and B. pinnipedialis (pinnipeds) [7]. In addition, a novel species of Brucella (named as Brucella microti), associated with voles, has recently been reported [8,9]. Some Brucella species are further sub-divided into groups known as biovars. Whilst these are of epidemiological interest in B. abortus and B. melitensis, the five biovars that are found in B. suis appear to relate to host preference. In humans, four of the classical species (not B. ovis or B. neotomae) have been shown to cause disease [2]. There is also evidence to suggest that humans can be infected by marine mammal Brucella further highlighting the importance of this group of organisms to human health [10-12].
Identification to species level is traditionally based on biotyping that examines antigenic, phenotypic and phage susceptibility profiles. While the approach has been useful for many years, it has a number of recognised drawbacks. Being culture based, biotyping requires specialist handling facilities for the initial isolation of a pure culture and then needs additional culture time for phenotype and phage susceptibility determination. In addition, some of the differentiating characteristics can be very subtle meaning that biotyping should only be carried out by highly experienced individuals. However, even with this caveat, interpretation of this method is acknowledged to be rather subjective. Furthermore there are frequent reports of atypical organisms that do not conform to the expected characteristics of particular species.
Molecular biology provides tools that can circumvent such problems associated with classical microbiology by using suitably validated genetic markers for objective and unambiguous identification of groups. As DNA is the target of this testing, culturing can be reduced or potentially bypassed, both removing the necessity for specialist handling facilities, and shortening time from sampling to identification. With the explosion of molecular sequence data and an expanding array of technological platforms the use of assays based on single nucleotide polymorphisms (SNPs) in microbial genotyping is becoming increasingly common. For example, point mutations in rpoB and katG have been used as markers for resistance to rifampicin and isoniazid in Mycobacterium tuberculosis [13] while SNPs have also been used to differentiate vaccine and field strains of canine parvovirus [14]. In terms of microbial diagnostics SNP-based genotyping has been applied to a number of organisms including members of the Mycobacterium tuberculosis complex [15], Bacillus anthracis [16] and Burkholderia species. [17].
We have recently undertaken extensive phylogenetic analysis of the Brucella group involving the sequencing of some 21 distinct gene fragments, from over 400 strains of Brucella, representing the known genetic diversity of the group [4, M.S. Koylass and A.M. Whatmore, unpublished data]. While this extensive analysis confirmed the long-standing view that the genus is highly homogeneous [18-20] it has nevertheless allowed us to confidently identify SNPs that we believe define each of the classically accepted Brucella species. Furthermore SNPs are particularly powerful markers in such a conserved group as they are highly unlikely to have occurred twice independently (i.e. in distinct evolutionary branches) and equally, having occurred, are unlikely to mutate again back to their ancestral state [21]. In addition, these studies revealed strong linkage disequilibrium in the distribution of alleles between Brucella species [4] implying that recombination is rare and giving increased confidence in the robustness and stability of species-specific SNPs selected as markers.
We have thus selected seven SNPs that we have worked into a multiple outcome assay to identify Brucella isolates at the species level. An assay based on these markers should deliver unambiguous results without the subjectivity associated with biotyping or problems associated with strains that are phenotypically aberrant. We recently described the development of a SNP-based assay identifying Brucella to the species level on a primer extension platform [22]. Here, we describe the development of an equivalent tool for the real-time PCR platform that has certain advantages in terms of speed, technical simplicity and potentially wider applicability being based on a cheaper, and more widely available, technological platform. We describe the use of a panel of short Taqman 5' labelled, 3' Minor Groove Binding (MGB), probes to interrogate species defining SNPs. Use of the MGB protein raises the melting temperature of probes meaning that a single base mismatch causes more destabilisation than would be the case with a longer probe [23]. This facilitates accurate SNP detection allowing us to use this approach in an assay to identify Brucella to the species level.
Results
Assay design
Previous multilocus sequence analysis (MLSA) studies, performed by us, identified a number of SNPs that are specific for known Brucella spp. These form the basis of the assay described here (see Table 1). Point changes specific for B. melitensis, B. ovis, B. canis and the marine mammal Brucella were described previously [22] based on multi-locus sequence analysis of 160 strains. The SNPs specific for B. abortus, B. suis, and B. neotomae used in this assay, were identified by extension of this work to examine a total of 21 gene fragments from over 400 strains (M. Koylass and A. M. Whatmore, unpublished data). In the case of B. suis it was not possible to identify a single SNP specific to the classical taxonomic group. This reflects the fact that B. suis biovar (bv) 5 is genetically distinct from the remaining B. suis biovars [4,6,24]. The SNP used in the assay described here therefore identifies only B. suis bvs 1–4. There are SNPs specific to B. suis bv 5 [4] that could be used in an assay to discriminate this biovar, although in practical terms it has only rarely been isolated from rodents and is of no diagnostic significance. The B. suis bvs 1–4 SNP in prpE is shared with B. canis reflecting the fact that B. canis is embedded within the B. suis phylogenetic cluster. However the presence of a SNP confined to B. canis in omp25 allows unambiguous separation of these two species [4]. In previous work, where we used primer extension to determine species, we had not incorporated a target to positively identify B. suis but rather used a process of elimination to identify members of this species. However, with the isolation of novel species such as B. microti, it became necessary to redress this problem. Brucellae isolated from marine mammals have recently been formally designated as two distinct species, B. pinnipedialis and B. ceti [7]. However, this designation is proving controversial as it is inconsistent with apparent genetic divisions within the marine mammal group [25,26]. Reflecting this, it has not been possible to identify SNPs that specifically identify these two species. Thus the SNP chosen for use in this assay, in trpE, is universal to all marine mammal isolates [4].
Table 1.
Gene targets, primers and allele-specific probes used in this study.
| Species | Gene target | Location in B. abortus 9–941 | Primers | Probes | ||
| B. abortus | fbaA | AE017224 | F | TGACATCATGCTCCGTCACATG | VIC | ATGCCGTGGCGGAA |
| 360225–361289 | R | CAGACCGGAATATGCGGATAGAT | FAM | ATGCCGTGACGGAA | ||
| B. melitensis | gap | AE017223 | F | GGCTCAGGTTCTCAACGATACTATC | VIC | CGTGGTCATAAAGC |
| 1684721–1685728 | R | TCGCCCGTATAGGAGTGGAT | FAM | CGTGGTCATGAAGC | ||
| B. ovis | aroA | AE017223 | F | CGACCACCGCATCGC | VIC | CCATGACAAGGAAAC |
| 29246–30598 | R | CCGGCTTTTCCGATGCAA | FAM | CATGACGAGGAAAC | ||
| B. suis bv1–4 | prpE | AE017223 | F | GCGACCGCATCCTCATCTATATG | VIC | CAAGCGTGGCAACC |
| 1687718–1689625 | R | CGCCGAATACGACGGAATGAAT | FAM | CAAGCATGGCAACC | ||
| Marine mammal Brucellae | trpE | AE017223 | F | CGAGGATTCCTTCGTCCATACG | VIC | CCAATTATTTCCACCAGACG |
| 1537355–1539550 | R | ACGCACGGTGGAAACCTT | FAM | CCAATTATTTCCGCCAGACG | ||
| B. canis | omp25 | AE017223 | F | GCTGGCGCCTTTGCT | VIC | AACTTCCAGAAGGACC |
| 710024–710625 | R | GGCCGTCCTTGGACTTCTTG | FAM | AACTTCCAGCAGGACC | ||
| B. neotomae | putative oxido-reductase | AE017223 | F | GGTTTTCCATGCGGTTTATTTGC | VIC | CATTGAGTGGCCCGAT |
| 1989869–1990870 | R | GGCATCATGCACAGTGATATCGA | FAM | ATTGAGCGGCCCGAT | ||
| 16SrRNA771/778 | 16SrRNA | - | F | CGCCGTAAACGATGAATGTTAGC | VIC | CGAAGTGTAAACACCCCGA |
| R | GCGGAATGTTTAATGCGTTAGCT | FAM | CGAAGAGTAAACTCCCCGA | |||
| 16SrRNA1055 | 16SrRNA | - | F | GGGTTAAGTCCCGCAACGA | VIC | ACCCTCGCCTTTAGTT |
| R | ACGTCATCCCCACCTTCCT | FAM | CCCTCGCCCTTAGTT | |||
Gene targets, primers and allele-specific probes used in this study. Brucella species-specific SNPs and non-specific SNPs (both underlined) are shown within the relevant probes. Gene target locations are given relative to the sequenced B. abortus 9-941 strain.
Individual assays for each SNP are based on the use of two alternative probes, one that preferentially binds to the species-specific polymorphism, while the other binds to the alternative allele state present in members of all other species. In each case the species-specific probe is labelled with VIC while the alternative state probe is labelled with FAM (see Table 1). By following a strategy of optimising each SNP discrimination assay to identical cycling conditions all seven species-defining assays can be analysed concurrently in a multiple outcome assay.
Optimisation
The single-tube assay, as provided by Applied Biosystems, is provided with a standard probe and primer concentration. However upon initial testing, certain assays displayed background with some probes not predicted to react in a particular sample, giving fluorescence values above those expected. To overcome this we obtained individual assay components and optimised primer and probe concentrations to maximise discrimination (data not shown). Details of final probe and primer concentrations selected based on this work are presented in Table 2.
Table 2.
Optimised primer and probe concentrations determined for the seven species defining MGB assays.
| MGB assay | Final Forward primer concentration (nM) | Final Reverse primer concentration (nM) | Final VIC labelled probe concentration (nM) | Final FAM labelled probe concentration (nM) |
| B. abortus | 500 | 700 | 300 | 100 |
| B. melitensis | 700 | 1100 | 200 | 100 |
| B. ovis | 300 | 300 | 250 | 50 |
| B. suis bv1–4 | 300 | 300 | 200 | 100 |
| B. canis | 700 | 1100 | 250 | 200 |
| Marine mammal Brucella | 500 | 1100 | 300 | 50 |
| B. neotomae | 500 | 700 | 200 | 100 |
Primer and probe concentrations were optimised to give the greatest amount of discrimination with the least background using the minimum amounts of the various components.
Assay performance
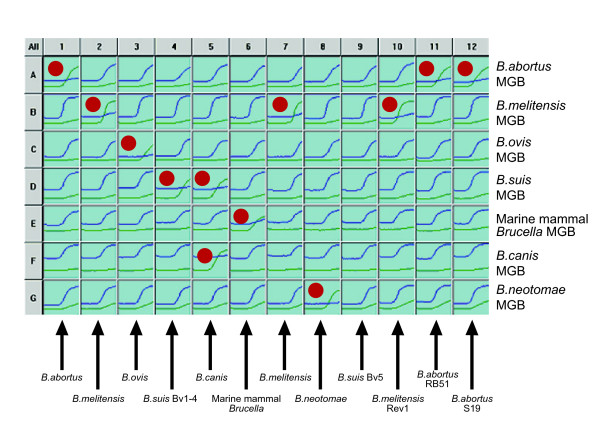
The optimised assay was run as shown in Figure 1 with each individual SNP discrimination assay running in rows (e.g. A1–A12 = B. abortus, B1–B12 = B. melitensis etc.). This enables typing of twelve isolates on a single 96-well plate. Reaction plots with the species-specific VIC labelled probes are in green, with the alternative FAM labelled probe in blue. In most cases each sample should generate a strong curve with the VIC labelled probe in only one well, and should react preferentially with the FAM-labelled probe in the remaining six wells. In this manner the sample in column one is identified as B. abortus, that in column 2 as B. melitensis and so on. Reflecting their phylogenetic position described above, B. canis isolates react with the VIC probes for both B. suis and B. canis (see column 5). The isolate in column 9 is B. suis bv 5 which as described above lacks any of the species defining SNPs and thus reacts only with all seven FAM-labelled alternative state probes. This is also the reaction outcome expected with any novel isolates that are not members of classically recognised species or the marine mammal Brucella groups.
Figure 1.
Seven Species defining MGB assay profile. Example of the multiple outcome assay used to identify twelve Brucella isolates. Each species-defining assay is run in rows (A-G) with samples run in columns. The green PCR profiles represent reactions with the VIC labelled probe, representing the species-specific probe in each probe pair. The blue PCR profiles represent reactions with the FAM labelled probes, representing the reaction with the alternate state (non species-specific) allele probe in each probe pair. The identity of each of the isolates 1–12 is indicated by a red dot where an isolate generates a positive PCR reaction with a VIC-labelled probe.
Assay validation
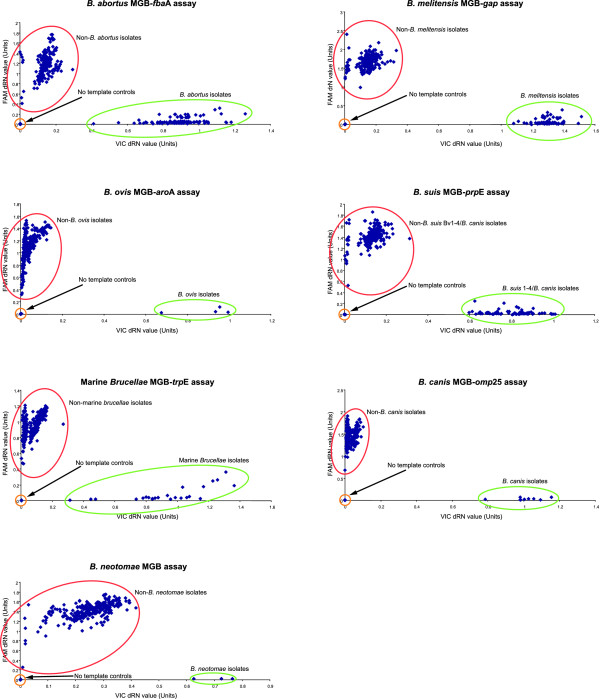
Assay validation was performed using 303 Brucella isolates encompassing all species and biovar reference strains and a collection of geographically and temporally distinct field isolates comprising members of all known species (Table 3). Alongside representatives of all species and biovars, the vaccine strains B. abortus S19 and RB51 as well as B. melitensis Rev1 were also included. In addition, strains that had both genotypes and phenotypes that did not correspond to any of the known classical Brucella species but were shown to be Brucella by 16S rRNA typing were also tested. When all seven individual assays were compared the predicted species identity of the 303 strains was consistent with identification based on other molecular and phenotypic methods. Based on this analysis allele discrimination plots were generated for each SNP assay (Figure 2). Each data-point represents the normalised end-point fluorescence reading for both VIC and FAM labelled probes plotted on the X-axis and Y-axis respectively. Each plot shows that there is unambiguous discrimination between genotypes with two distinct populations. One population clusters around the X-axis and consists of isolates that would be classified as a member of the species targeted by this one specific SNP. The second population clustering around the Y-axis represents members of the remaining species. No template controls cluster around the plot origin with negligible reaction with either probe. The spread of data-points in the scatter plot reflects the fact that DNA concentrations applied to the assay were not normalised.
Table 3.
Phenotype based identity of the 303 isolates used to validate the species defining MGB assay.
| Species/Biovar | Numbers present |
| B. abortus unknown biovar | 22 |
| B. abortus bv1 | 55 |
| B. abortus bv2 | 6 |
| B. abortus bv3 | 11 |
| B. abortus bv4 | 8 |
| B. abortus bv5 | 4 |
| B. abortus bv6 | 11 |
| B. abortus bv7 | 5 |
| B. abortus bv9 | 1 |
| B. melitensis unknown biovar | 23 |
| B. melitensis bv1 | 7 |
| B. melitensis bv2 | 2 |
| B. melitensis bv3 | 32 |
| B. ovis | 5 |
| B. suis unknown biovar | 30 |
| B. suis bv1 | 13 |
| B. suis bv2 | 8 |
| B. suis bv3 | 3 |
| B. suis bv4 | 5 |
| B. suis bv5 | 2 |
| Marine Mammal Brucellae | 30 |
| B. canis | 12 |
| B. neotomae | 3 |
| Novel phenotype/Brucella by 16S rRNA | 5 |
Figure 2.
Discrimination of the seven species defining assays. Allele discrimination plots generated by each species-defining MGB assay when applied to 303 Brucella isolates. Each assay was read after 40 cycles.
A number of isolates that represent likely new Brucella groups, based on 16S rRNA sequencing and other extensive analysis, were tested in the assay including recently described B. microti [8,9] and an atypical isolate from a human infection [27]. As with B. suis bv 5, seven non-specific curves were generated showing that this assay would identify isolates such as these as Brucella, but recognise that they did not belong to any of the currently described species. Within the 303 isolates tested were three reference strains (one of B. abortus, B. melitensis and B. suis) each cultured in vitro through up to 35 passages. Testing of these isolates showed that the target SNPs were stable even on extensive in vitro culture.
Limits of detection of genotyping assay
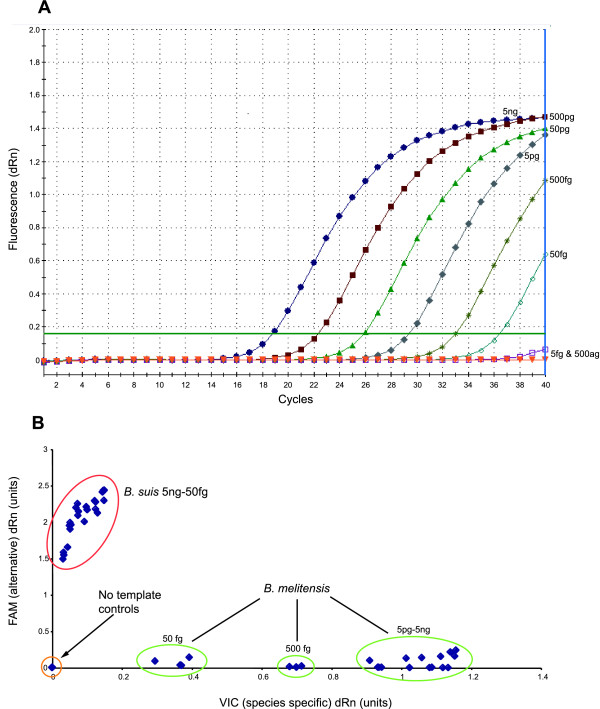
The analytical sensitivity of each MGB dual probe assay was firstly tested at least four times for each serial dilution from 5 ng (1.5 × 106 genome equivalents) to 0.5 fg (1.5 × 10-1 genome equivalents) of its target species/type. A representative example is shown in Figure 3a. Each individual assay could reliably detect DNA down to 50 fg, or approximately 15 cells from all the assays. Samples containing 5 fg could only be detected sporadically. Analytical sensitivity was in line with work done on Bacillus anthracis, where the lowest detection levels were 100 fg or 17 cells [16]. Allelic discrimination plots for each individual SNP assay comparing serial dilutions of target and non-target species illustrated that there was still clear discrimination even at 50 fg of DNA (for example see Figure 3b).
Figure 3.
Sensitivity and limits of discrimination using MGB assay. Sensitivity of the species defining assays. The data presented here relates to the B. melitensis gap assay with the remaining six assays giving equivalent results. A. Titrations of B. melitensis 16 M DNA from 5 ng to 500 ag. B. Allele discrimination plot showing performance of the B. melitensis assay in distinguishing B. melitensis from non-B. melitensis (B. suis) down to 50 fg of DNA.
Specificity
Initial optimisation work was carried out using the Taqman Universal mix which was effective for all seven assays although fluorescence levels were lower for the B. abortus probe pair. This led to lower sensitivity at 40 cycles for this probe pair, although allelic discrimination was unaffected. In order to check specificity of the assay five type strains of Ochrobactrum, the closest phylogenetic neighbour of Brucella [28], were tested in duplicate. They were shown not to give fluorescence readings greater than the background with all seven probe pairs, which is the very least we would see if these samples were Brucella. However, to improve upon the low fluorescence and sensitivity associated with the B. abortus probes, we switched to an improved reaction mix (TaqMan Genotyping mix) and then repeated the work. Upon repetition of the Ochrobactrum work with the new mix, it was noted that we were now seeing a seven probe profile for the five species that had not been seen previously. However, the kinetics of these reactions were clearly very different to those seen with comparable concentrations of Brucella DNA. Reactions were weak with very low endpoint fluorescence values (data not shown). When the targets of SNP assay were compared against the published O. anthropi ATCC49188 genome sequence, it was found that there were polymorphisms in most of the primers and probes used in this assay that might account for these differences in kinetics (data not shown). This assay was not designed as a diagnostic assay to distinguish Brucella from other bacteria, and there are other real time PCR assays available that can do this. For example, the insertion sequence IS711 is considered specific for organisms of the genus Brucella. Brucellae can be identified through amplification of this element as demonstrated by Ouahrani-Bettache et al. [29] or in the widely used Abortus-Melitensis-Ovis-Suis PCR (AMOS-PCR) assay [30]. In addition, there are genus specific real time PCR assays based around the conserved bcsp31 target that can fulfil this role [31,32]. However, in light of this observation, we sought to include additional markers that would differentiate Brucella from its nearest phylogenetic neighbours. To do this, we looked at the 16S rRNA sequence as the target most commonly used for identification of bacteria to the genus level [33,34]. Alignments of 16S rRNA sequences of Brucella spp. with Ochrobactrum spp. sequences deposited in GenBank, as well as equivalent sequences for other related α-proteobacteriawere constructed. It should be noted at this point that there are a number of Genbank entries annotated as Brucella but which clearly represent Ochrobactrum.
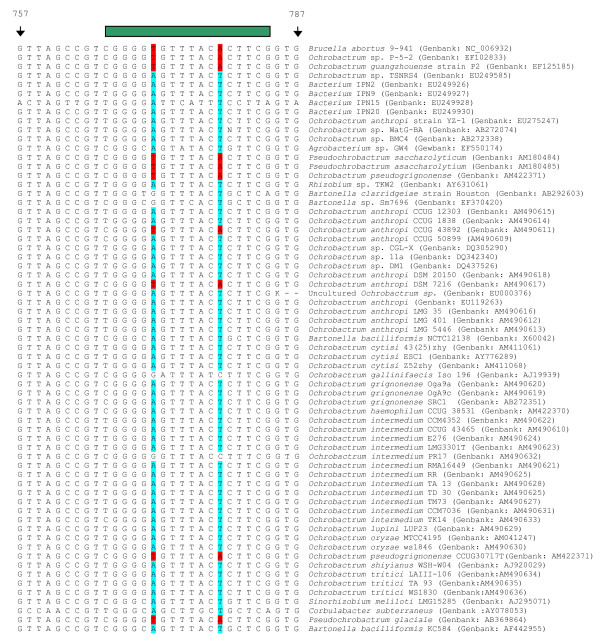
On this basis three SNPs were identified that when used in conjunction can distinguish Brucella from other α-proteobacteria based on the sequences deposited in Genbank currently (December, 2007) (Figures 4 and 5). The three SNPs correspond to positions 771 (16S rRNA771), 778 (16S rRNA778), and 1055 (16S rRNA1055) in the B. abortus 9–941 ribosomal RNA sequence. MGB probes were designed to discriminate alleles at these three sites, one probing 16S rRNA771 and 16S rRNA778, the other probing 16S rRNA1055 (see Table 1 and Figures 4 and 5). These were tested against all Brucella species and biovars as well as the five Ochrobactrum type strains previously tested and two additional Ochrobactrum anthropi strains (ATCC49188 and ATCC49237). Using this combination of three SNPs manages to distinguish Brucella isolates from non-Brucella isolates (Figures 6a and 6b).
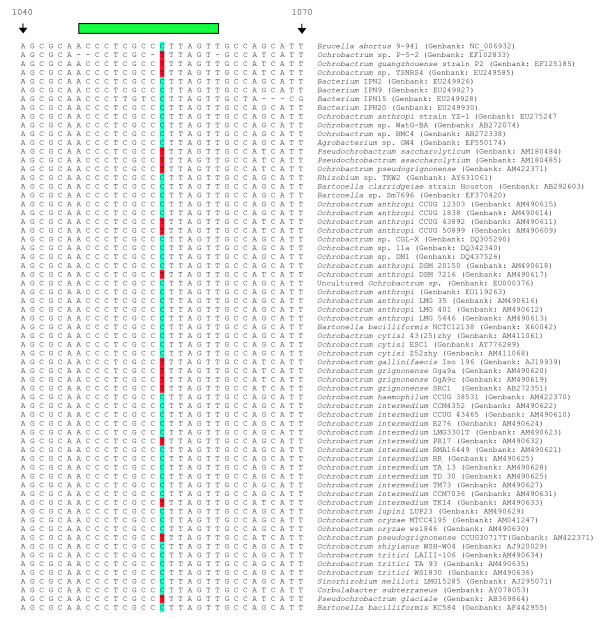
Figure 4.
Alignments of various 16SrRNA sequences around the location of the 16S rRNA771/778 probes. Alignments of fragments of 16S rRNA centred around SNPs at bases 771 and 778 relative to B. abortus 9–941, used in combination with the 16S rRNA1055 SNP to separate Brucella from closely related bacteria. The alignment contains 65 sequences taken directly from Genbank and includes many examples of Ochrobactrum, the closest phylogentic neighbour of Brucella as well as other, less-closely related, members of the α-proteobacteria. In this figure, the targetted Brucella specific SNPs are highlighted in red and the non-Brucella alternatives in blue. The green bar above both figures represents the location of probe hybridisation. The alignment includes only one Brucella sequence as there is 100% identity in the 16SrRNA sequences between all Brucella [34]. Using this assay on its own will discriminate most but not all of the non- Brucella shown from Brucella organisms.
Figure 5.
Alignments of various 16SrRNA sequences around the location of the 16S rRNA1055 probes. Alignments of fragments of 16S rRNA centred around SNP at base 1055 relative to B. abortus 9–941 showing location of the SNP, used in combination with the 16S rRNA771/778 SNPs to separate Brucella from closely related bacteria. The alignment contains 65 sequences taken directly from Genbank and includes many examples of Ochrobactrum, the closest phylogentic neighbour of Brucella as well as other, less-closely related, members of the α-proteobacteria. In this figure, the targetted Brucella specific SNP is highlighted in blue and the non-Brucella alternatives in red. The green bar above both figures represents the location of probe hybridisation. The alignment includes only one Brucella sequence as there is 100% identity in the 16SrRNA sequences between all Brucella [34]. Whilst this assay on its own is not as discriminatory as 16S rRNA771/778 assay, it crucially distinguishes non-Brucella not detected by the afore mentioned assay.
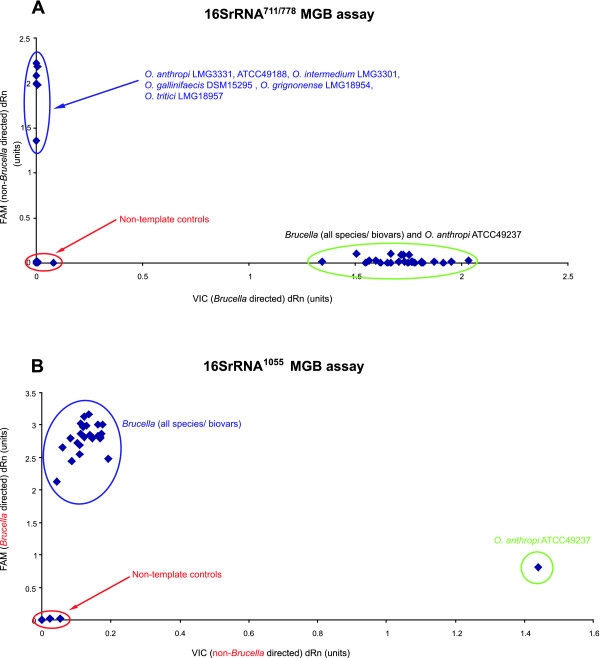
Figure 6.
Discrimination of the Brucella genus defining assays. Application of the two 16S rRNA based probe pairs in distinguishing isolates from the genus Brucellae from other α-proteobacteria. a. Results generated by the 16SrRNA771/778 probe pair when used against 5 species of Ochrobactrum (including three strains of O. anthropi) and representatives of all the known Brucella species and biovars. This reaction separates Brucella isolates from all other except O. anthropi ATCC49237. b. Results generated with the application of 16SrRNA1055 probe pair illustrating the use of this assay to further separate O. anthropi ATCC49237 from Brucella isolates.
Discussion
The aim of this work was to use the robust phylogenetic framework provided by existing MLSA studies to develop a rapid, unambiguous assay for the real-time PCR platform capable of identifying Brucella isolates to species level. The approach is based on a series of discrimination assays interrogating SNPs that we have shown to be specific to a particular Brucella species. As each individual SNP assay will always give one of two results, species determination is straightforward and unambiguous. Amplification profiles, representing binding of either the VIC- or FAM-labelled probe, are generated in all seven individual discrimination assays. One of the seven individual assays should give a positive reaction with the VIC-labelled probe (or two in the case of B. canis), with a reaction with the FAM-labelled probe apparent in all the remaining assays. An assay which did not generate a positive PCR result from each of the seven individual probe pairs would be considered to have failed. This approach avoids any danger of 'false-negatives', where lack of a PCR product could be indicative of PCR inhibition rather than absence of target. Furthermore, although any signals generated by non-Brucella organisms in this study were weak, we sought to absolutely ensure the specificity of the assay by identifying three SNPs in 16S rRNA that, when used in conjunction, differentiate Brucella from closely related bacteria. Allele discrimination assays based on these three markers were shown to clearly differentiate Brucella from closely related α-proteobacteria examined.
As a SNP based speciation assay the approach described here has a number of potential advantages, comparable to those of our primer extension based SNP typing assay [22], when compared to classical biotyping approaches. Both methods overcome the subjectivity associated with biotyping with approaches that are relatively technically straightforward, unambiguous, and have substantial advantages in terms of simplicity and speed. In addition, both techniques reduce the potential exposure to live Brucella. This is desirable given the ease of acquisition of Brucella in the absence of stringent bio-containment facilities [35,36].
Both assays also have substantial advantages over currently used molecular assays. They can be carried out on crude bacterial lysates bypassing the need for lengthy DNA extraction procedures as required by some molecular typing approaches [20,37] and, as PCR based methods, both techniques should theoretically be applicable directly to field material obviating the need for any culture. In contrast to some extensively used existing assays such as AMOS-PCR [30] these assays are all encompassing identifying all relevant biovars of all species. Of the classically recognised Brucella species and biovars only B. suis bv 5 will not be positively identified as a particular Brucella species by the real-time PCR assay, reflecting its unique phylogenetic position within the group apparently distinct from other B. suis. However, like other potentially novel Brucella groups, such isolates are positively identified as Brucella displaying seven clear non species-specific FAM signals. Both techniques lend themselves to future expansion to include markers for such new groups. For example, after this manuscript was submitted a novel species, B. microti, was described. We have already described a SNP specific to this species based on MLSA [9] and probes interrogating this SNP could readily be incorporated into this assay. This capacity for rapid inclusion of newly identified species is a potential advantage over some other molecular assays such as the multiplex PCR assay proposed by Garcia-Yoldi et al [38]. Both the primer extension assay and the real-time PCR based assay described here have the advantage of potentially increased sensitivity associated with PCR based techniques. We have shown that the real-time PCR assay described here can detect and differentiate as little as 15 genome equivalents suggesting that it could possibly identify the organism in relevant field material such as foetal tissue or vaginal swabs.
There are, however, substantial advantages in the use of a real-time PCR platform, compared to the primer extension format that led us to investigate and report this alternative approach. The primer extension approach requires a DNA sequencer, while the approach described in this manuscript requires only a real-time PCR machine. Indeed, it could feasibly be undertaken in a simpler format with a conventional thermocycler and a basic fluorescence plate reader. Thus, while the primer extension assay might be appropriate in larger reference laboratories, the technique described in this manuscript is likely to prove far more applicable in routine diagnostics. This is particularly so as real-time PCR machines become more widely available in areas of the world where brucellosis remains a major public and animal health issue. In contrast, the uptake of sequencing based technologies, requiring a substantially greater capital input, is likely to be much slower in these areas. Furthermore, the real-time PCR approach is also technically simpler, faster and cheaper than the primer extension based approach. As amplification and detection is carried out concurrently the real-time assay described here can characterise isolates in around two hours. This compares to the day required for the primer extension based approach that also involves a much less straightforward experimental procedure. Although both primer extension and real-time assays could be further expanded the number of markers that can be included in a single primer extension assay is limited by the need to space markers (ideally by at least 5 bp) between the 13 bp and 88 bp size standards. There is no such limitation with the real-time PCR described here where additional markers can be interrogated simply by the addition of an additional probe pair in another well of a 96 well plate. This will make the latter assay our approach of choice for incorporation of additional markers to type at the sub-species level. Finally, the primer extension assay we described did not include a specific marker for B. suis, with this species being recognised by process of elimination (i.e. lack of any of the markers specific for the other classical Brucella species). While this was appropriate at the time, the recent description of a novel species of Brucella (B. microti) indicates the added value of including a specific marker for each species. The inclusion of a marker for B. suis in the real-time assay ensures that all species are identified 'positively' by the recognition of a SNP shown, by the extensive population genetic analysis that supports this work, to be specific for the B. suis/canis group.
Conclusion
We have described a simple and unambiguous multiple outcome SNP assay based on a robust phylogenetic framework that can characterise Brucella isolates to the species level. The approach is based on a sensitive and rapid real-time PCR platform that will be widely applicable in diagnostic laboratories and is readily expandable as knowledge of the genus increases. Further work will focus on the expansion of the assay to include markers to discriminate live vaccine strains and to type at the sub-species level.
Methods
Bacterial isolates
All Brucella examined in this study came from the Veterinary Laboratories Agency strain archive. In total, 303 crude extractions from Brucella isolates were used to validate the complete assay. These consisted of either boiled cells or crude methanol extractions prepared as described previously [5]. The purified genomic DNA extracts used for sensitivity determination were isolated from B. abortus 544, B. melitensis 16 M, B. suis 1330, B. ovis F10/B7/02, B. canis 79/92, B. neotomae 65/196, and VLA04/72 (isolated in a porpoise from South West England). The non-Brucella used in this work were Ochrobactrum anthropi (LMG3301, ATCC49188 and ATCC49237), O. intermedium (LMG 3331), O. gallinifaecis (DSM15295), O. grignonense (LMG18954), and O. tritici (LMG18957).
SNP identification
Identification of SNPs was based on an extension of previously discussed MLSA work [4] to examine twenty-one distinct gene fragments from over 400 isolates of Brucella. In addition to the SNPs isolated in four genes gap, omp25, trpE, and aroA identified in this previous study [4], SNPs located in three further genes, fbaA, prpE, and a putative oxidoreductase encoding gene (see Table 1) were used in the setting up of this assay. The locations of all SNPs in relation to the sequenced strain, B. abortus 9–941 is shown in Table 1. In the case of the assay to distinguish Brucella spp. from near neighbour organisms 16S rRNA sequence data from a number of different members of the α-proteobacteria group were downloaded from Genbank and aligned with a number of known Brucella species using the ClustalW algorithm on the Lasergene 7 Megalign programme (DNAstar, Madison, Wisconsin, USA). Many of these sequences were recently described by Scholz et al., [39] who demonstrated separation of Brucella from Ochrobactrum based on them. Polymorphisms were selected based on the ability to separate Brucella sequences as a complete entity from within the remaining α-proteobacteria. In total, three polymorphisms were found that were able to distinguish Brucella isolates from their genetic "near neighbours".
Design of multiple-outcome assay to type Brucella species
The design of this multiple-outcome assay involved the development of seven individual allelic discrimination assays to determine bases present at the seven species defining SNP sites interrogated. Using the MLSA data available, sequences of around 170 bp containing the species defining SNP were edited using the Applied Biosystems File Builder 3.1 software. Probes and primers were designed and provided in a single tube, with the species-specific probe labelled with VIC, and the alternative state probe (reacting with all other species) with FAM. The sequences of both primers and both probes are shown in Table 1. All assays were first checked for species specificity on a small panel of isolates of 27 methanol isolates. All real time PCR reactions were initially run in a final volume of 12.5 μl, comprising of 6.25 μl Taqman universal master mix (Applied Biosystems, Warrington, UK), 900 nM final concentration of each primer, 200 nM final concentration of each probe, and an arbitrary volume of 0.5 μl sample (DNA not quantified). All PCR reactions were run on the Stratagene MX3000P platform (Stratagene, La Jolla, USA) as follows: 1 cycle at 50°C for 2 mins, followed by 1 cycle at 95°C for 10 mins, followed by 40 cycles of 92°C for 15 secs, and 60°C for 1 min. The concentrations of primers and probes were then optimised to maximise discrimination with the least amount of reagents (see Table 2). For all real time reactions involving the validation on 303 isolates and the sensitivity work, the Taqman universal mix was replaced by Taqman Genotyping mix (Applied Biosystems, Warrington, UK) but otherwise, the reactions were the same. For the work involving assay sensitivity genomic DNA, extracted by a phenol/chloroform method [20], was quantified by spectrophotometer (Smartspec Plus, Bio-Rad, Hemel Hempstead, UK). This DNA was then diluted to the required concentration in DNA/RNA free sterile water (Ambion, Huntingdon, UK) prior to replicate testing. For the Brucella genus specific MGB dual probe assays, probes and primers were designed around the three SNPs of interest in the 16S rRNA sequence (Table 1). Real time PCR reactions for this part of the study were run as for the validation and sensitivity work for the seven SNP species defining assays.
Authors' contributions
KKG oversaw all practical work, analyzed the data and drafted the manuscript. MSK contributed to sequencing work that led to the identification of species-specific SNPs. CJS carried out some of the assay development. AMW conceived the study, and participated in its design and coordination, and helped to draft the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
Work described in this manuscript was funded by the UK Department for the Environment, Food and Rural Affairs (Defra). We would like to thank Dr Scott Clark from the University of Waterloo, Canada for providing genomic DNA from O. anthropi ATCC49188 and ATCC49237. This work was presented in part at the 107th General meeting of the American Society of Microbiology (ASM) held on May 21–25, 2007 in Toronto, Canada.
Contributor Information
Krishna K Gopaul, Email: k.gopaul@vla.defra.gsi.gov.uk.
Mark S Koylass, Email: m.koylass@vla.defra.gov.gsi.gov.uk.
Catherine J Smith, Email: c.j.smith@vla.defra.gov.gsi.gov.uk.
Adrian M Whatmore, Email: a.whatmore@vla.defra.gov.gsi.gov.uk.
References
- Godfroid J, Cloeckaert A, Liautard JP, Kohler S, Fretin D, Walravens K, Garin-Bastuji B, Letesson JJ. From the discovery of the Malta fever's agent to the discovery of a marine mammal reservoir, brucellosis has continuously been a re-emerging zoonosis. Vet Res. 2005;36:313–326. doi: 10.1051/vetres:2005003. [DOI] [PubMed] [Google Scholar]
- Moreno E, Cloeckaert A, Moriyón I. Brucella evolution and taxonomy. Vet Microbiol. 2002;90:209–227. doi: 10.1016/S0378-1135(02)00210-9. [DOI] [PubMed] [Google Scholar]
- Michaux-Charachon S, Bourg G, Jumas-Bilak E, Guigue-Talet P, Allardet-Servent A, O'Callaghan D, Ramuz M. Genome structure and phylogeny in the genus Brucella. J Bacteriol. 1997;179:3244–3249. doi: 10.1128/jb.179.10.3244-3249.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whatmore AM, Perrett LL, MacMillan AP. Characterisation of the genetic diversity of Brucella by multilocus sequencing. BMC Microbiol. 2007;7:34. doi: 10.1186/1471-2180-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whatmore AM, Shankster SJ, Perrett LL, Murphy TJ, Brew SD, Thirlwall RE, Cutler SJ, MacMillan AP. Identification and characterization of variable-number tandem-repeat markers for typing of Brucella spp. J Clin Microbiol. 2006;44:1982–1993. doi: 10.1128/JCM.02039-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Flèche P, Jacques I, Grayon M, Al Dahouk S, Bouchon P, Denoeud F, Nöckler K, Neubauer H, Guilloteau LA, Vergnaud G. Evaluation and selection of tandem repeat loci for a Brucella MLVA typing assay. BMC Microbiol. 2006;6:9. doi: 10.1186/1471-2180-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster G, Osterman BS, Godfroid J, Jacques I, Cloeckaert A. Brucella ceti sp. nov. and Brucella pinnipedialis sp. nov. for Brucella strains with cetaceans and seals as their preferred hosts. Int J Syst Evol Microbiol. 2007;57:2688–2693. doi: 10.1099/ijs.0.65269-0. [DOI] [PubMed] [Google Scholar]
- Hubálek Z, Scholz HC, Sedláèek I, Melzer F, Sanogo YO, Nesvadbová J. Brucellosis of the common vole (Microtus arvalis) Vector Borne Zoonotic Dis. 2007;7:679–687. doi: 10.1089/vbz.2007.0143. [DOI] [PubMed] [Google Scholar]
- Scholz HC, Hubalek Z, Sedlácek I, Vergnaud G, Tomaso H, Al Dahouk S, Melzer F, Kämpfer P, Neubauer H, Cloeckaert A, Maquart M, Zygmunt MS, Whatmore AM, Falsen E, Bahn P, Göllner C, Pfeffer M, Huber B, Busse HJ, Nöckler K. Brucella microti sp. nov., isolated from the common vole Microtus arvalis. Int J Syst Evol Microbiol. 2008;58:375–382. doi: 10.1099/ijs.0.65356-0. [DOI] [PubMed] [Google Scholar]
- McDonald WL, Jamaludin R, Mackereth G, Hansen M, Humphrey S, Short P, Taylor T, Swingler J, Dawson CE, Whatmore AM, Stubberfield E, Perrett LL, Simmons G. Characterization of a Brucella sp. strain as a marine-mammal type despite isolation from a patient with spinal osteomyelitis in New Zealand. J Clin Microbiol. 2006;44:4363–4370. doi: 10.1128/JCM.00680-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn AH, Probert WS, Glaser CA, Gupta N, Bollen AW, Wong JD, Grace EM, McDonald WC. Human neurobrucellosis with intracerebral granuloma caused by a marine mammal Brucella spp. Emerg Infect Dis. 2003;9:485–488. doi: 10.3201/eid0904.020576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whatmore AM, Dawson CE, Groussaud P, Koylass M, King AC, Shankster S, Sohn AH, Probert WS, McDonald WL. A marine mammal Brucella genotype associated with zoonotic infection. Emerg Infect Dis. 2008;14:517–518. doi: 10.3201/eid1403.070829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yesilkaya H, Meacci F, Niemann S, Hillemann D, Rüsch-Gerdes S, LONG DRUG Study Group. Barer MR, Andrew PW, Oggioni MR. Evaluation of molecular-Beacon, TaqMan, and fluorescence resonance energy transfer probes for detection of antibiotic resistance-conferring single nucleotide polymorphisms in mixed Mycobacterium tuberculosis DNA extracts. J Clin Microbiol. 2006;44:3826–3829. doi: 10.1128/JCM.00225-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N, Elia G, Martella V, Campolo M, Desario C, Camero M, Cirone F, Lorusso E, Lucente MS, Narcisi D, Scalia P, Buonavoglia C. Characterisation of the canine parvovirus type 2 variants using minor groove binder probe technology. J Virol Methods. 2006;133:92–99. doi: 10.1016/j.jviromet.2005.10.026. [DOI] [PubMed] [Google Scholar]
- Huard RC, Fabre M, de Haas P, Lazzarini LC, van Soolingen D, Cousins D, Ho JL. Novel genetic polymorphisms that further delineate the phylogeny of the Mycobacterium tuberculosis complex. J Bacteriol. 2006;188:4271–4287. doi: 10.1128/JB.01783-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ert MN, Easterday WR, Simonson TS, U'Ren JM, Pearson T, Kenefic LJ, Busch JD, Huynh LY, Dukerich M, Trim CB, Beaudry J, Welty-Bernard A, Read T, Fraser CM, Ravel J, Keim P. Strain-specific single-nucleotide polymorphism assays for the Bacillus anthracis Ames strain. J Clin Microbiol. 2007;45:47–53. doi: 10.1128/JCM.01233-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U'Ren JM, Van Ert MN, Schupp JM, Easterday WR, Simonson TS, Okinaka RT, Pearson T, Keim P. Use of a real-time PCR TaqMan assay for rapid identification and differentiation of Burkholderia pseudomallei and Burkholderia mallei. J Clin Microbiol. 2005;43:5771–5774. doi: 10.1128/JCM.43.11.5771-5774.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verger JM, Grimont F, Grimont PAD, Grayon M. Brucella, a monospecific genus as shown by deoxyribonucleic acid hybridization. Int J Syst Bacteriol. 1985;35:292–295. [Google Scholar]
- Gándara B, Merino AL, Rogel MA, Martínez-Romero E. Limited genetic diversity of Brucella spp. J Clin Microbiol. 2001;39:235–240. doi: 10.1128/JCM.39.1.235-240.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whatmore AM, Murphy TJ, Shankster S, Young E, Cutler SJ, Macmillan AP. Use of amplified fragment length polymorphism to identify and type Brucella isolates of medical and veterinary interest. J Clin Microbiol. 2005;43:761–769. doi: 10.1128/JCM.43.2.761-769.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keim P, Van Ert MN, Pearson T, Vogler AJ, Huynh LY, Wagner DM. Anthrax molecular epidemiology and forensics: using the appropriate marker for different evolutionary scales. Infect Genet Evol. 2004;4:205–213. doi: 10.1016/j.meegid.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Scott JC, Koylass MS, Stubberfield MR, Whatmore AM. Multiplex assay based on single-nucleotide polymorphisms for rapid identification of Brucella isolates at the species level. Appl Environ Microbiol. 2007;73:7331–7337. doi: 10.1128/AEM.00976-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutyavin IV, Lukhtanov EA, Gamper HB, Meyer RB. Oligonucleotides with conjugated dihydropyrroloindole tripeptides: base composition and backbone effects on hybridization. Nucleic Acids Res. 1997;25:3718–3723. doi: 10.1093/nar/25.18.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavigne JP, Vergunst AC, Bourg G, O'Callaghan D. The IncP island in the genome of Brucella suis 1330 was acquired by site-specific integration. Infect Immun. 2005;73:7779–7783. doi: 10.1128/IAI.73.11.7779-7783.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groussaud P, Shankster SJ, Koylass MS, Whatmore AM. Molecular typing divides marine mammal strains of Brucella into at least three groups with distinct host preferences. J Med Microbiol. 2007;56:1512–1518. doi: 10.1099/jmm.0.47330-0. [DOI] [PubMed] [Google Scholar]
- Bourg G, O'Callaghan D, Boschiroli ML. The genomic structure of Brucella strains isolated from marine mammals gives clues to evolutionary history within the genus. Vet Microbiol. 2007;125:375–80. doi: 10.1016/j.vetmic.2007.06.002. [DOI] [PubMed] [Google Scholar]
- De BK, Stauffer L, Koylass MS, Sharp SE, Gee JE, Helsel LO, Steigerwalt AG, Vega R, Clark TA, Daneshvar MI, Wilkins PP, Whatmore AM. Characterization of a novel Brucella strain (BO1) associated with a prosthetic breast implant infection. J Clin Microbiol. 2008;46:43–49. doi: 10.1128/JCM.01494-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco J, Romero C, López-Goñi I, Leiva J, Díaz R, Moriyón I. Evaluation of the relatedness of Brucella spp. and Ochrobactrum anthropi and description of Ochrobactrum intermedium sp. nov., a new species with a closer relationship to Brucella spp. Int J Syst Bacteriol. 1998;48:759–768. doi: 10.1099/00207713-48-3-759. [DOI] [PubMed] [Google Scholar]
- Ouahrani-Bettache S, Soubrier MP, Liautard JP. IS 6501-anchored PCR for the detection and identification of Brucella species and strains. J Appl Bacteriol. 1996;81:154–160. doi: 10.1111/j.1365-2672.1996.tb04493.x. [DOI] [PubMed] [Google Scholar]
- Bricker BJ, Halling SM. Differentiation of Brucella abortus bv. 1, 2, and 4, Brucella melitensis, Brucella ovis, and Brucella suis bv. 1 by PCR. J Clin Microbiol. 1994;32:2660–2666. doi: 10.1128/jcm.32.11.2660-2666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probert WS, Schrader KN, Khuong NY, Bystrom SL, Graves MH. Real-time multiplex PCR assay for detection of Brucella spp., B. abortus, and B. melitensis. J Clin Microbiol. 2004;42:1290–1293. doi: 10.1128/JCM.42.3.1290-1293.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Dahouk S, Nöckler K, C Scholz H, Pfeffer M, Neubauer H, Tomaso H. Evaluation of genus-specific and species-specific real-time PCR assays for the identification of Brucella spp. Clin Chem Lab Med. 2007;45:1464–1470. doi: 10.1515/CCLM.2007.305. [DOI] [PubMed] [Google Scholar]
- Gevers D, Dawyndt P, Vandamme P, Willems A, Vancanneyt M, Swings J, De Vos P. Stepping stones towards a new prokaryotic taxonomy. Philos Trans R Soc Lond B Biol Sci. 2006;361:1911–1916. doi: 10.1098/rstb.2006.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee JE, De BK, Levett PN, Whitney AM, Novak RT, Popovic T. Use of 16S rRNA gene sequencing for rapid confirmatory identification of Brucella isolates. J Clin Microbiol. 2004;42:3649–3654. doi: 10.1128/JCM.42.8.3649-3654.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouza E, Sánchez-Carrillo C, Hernangómez S, González MJ. Laboratory-acquired brucellosis: a Spanish national survey. J Hosp Infect. 2005;61:80–83. doi: 10.1016/j.jhin.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Baron EJ, Miller JM. Bacterial and fungal infections among diagnostic laboratory workers: evaluating the risks. Diagn Microbiol Infect Dis. 2008;60:241–246. doi: 10.1016/j.diagmicrobio.2007.09.016. [DOI] [PubMed] [Google Scholar]
- Ouahrani S, Michaux S, Sri Widada J, Bourg G, Tournebize R, Ramuz M, Liautard JP. Identification and sequence analysis of IS6501, an insertion sequence in Brucella spp.: relationship between genomic structure and the number of IS6501 copies. J Gen Microbiol. 1993;139:3265–3273. doi: 10.1099/00221287-139-12-3265. [DOI] [PubMed] [Google Scholar]
- García-Yoldi D, Marín CM, de Miguel MJ, Muñoz PM, Vizmanos JL, López-Goñi I. Multiplex PCR assay for the identification and differentiation of all Brucella species and the vaccine strains Brucella abortus S19 and RB51 and Brucella melitensis Rev1. Clin Chem. 2006;52:779–781. doi: 10.1373/clinchem.2005.062596. [DOI] [PubMed] [Google Scholar]
- Scholz HC, Al Dahouk S, Tomaso H, Neubauer H, Witte A, Schloter M, Kämpfer P, Falsen E, Pfeffer M, Engel M. Genetic diversity and phylogenetic relationships of bacteria belonging to the Ochrobactrum-Brucella group by rec A and 16S rRNA gene-based comparative sequence analysis. Syst Appl Microbio. 2008;31:1–16. doi: 10.1016/j.syapm.2007.10.004. [DOI] [PubMed] [Google Scholar]