Abstract
Inorganic polyphosphate (polyP) kinase was studied for its roles in physiological responses to nutritional deprivation in Escherichia coli. A mutant lacking polyP kinase exhibited an extended lag phase of growth, when shifted from a rich to a minimal medium (nutritional downshift). Supplementation of amino acids to the minimal medium abolished the extended growth lag of the mutant. Levels of the stringent response factor, guanosine 5′-diphosphate 3′-diphosphate, increased in response to the nutritional downshift, but, unlike in the wild type, the levels were sustained in the mutant. These results suggested that the mutant was impaired in the induction of amino acid biosynthetic enzymes. The expression of an amino acid biosynthetic gene, hisG, was examined by using a transcriptional lacZ fusion. Although the mutant did not express the fusion in response to the nutritional downshift, Northern blot analysis revealed a significant increase of hisG-lacZ mRNA. Amino acids generated by intracellular protein degradation are very important for the synthesis of enzymes at the onset of starvation. In the wild type, the rate of protein degradation increased in response to the nutritional downshift whereas it did not in the mutant. Supplementation of amino acids at low concentrations to the minimal medium enabled the mutant to express the hisG-lacZ fusion. Thus, the impaired regulation of protein degradation results in the adaptation defect, suggesting that polyP kinase is required to stimulate protein degradation.
Inorganic polyphosphate (polyP), a linear polymer of hundreds of orthophosphate residues linked by high-energy phosphoanhydride bonds, is ubiquitous in nature, having been found in microbes, fungi, plants, and animals examined (1–3). The principal enzyme responsible for the synthesis of polyP in Escherichia coli is polyP kinase (PPK), which uses the γ phosphate of ATP to extend the polymer (1, 4). Addition of serine hydroxamate to growing E. coli cells leads to amino acid starvation and stimulates polyP accumulation (5). Ault-Riché et al. showed that growing E. coli cells accumulated polyP during a nutritional downshift from a rich to a minimal medium and that reversal of these conditions resulted in a very rapid disappearance of the polyP (6). PolyP accumulation in response to nutrient deprivation has also been observed in many other microorganisms, including Pseudomonas aeruginosa, Klebsiella aerogenes, (A.K. and H.O., unpublished data), and Myxococcus xanthus (7). Important signals for nutritional stress responses in E. coli are guanosine 5′-triphosphate 3′-diphosphate (pppGpp) and guanosine 5′-diphosphate 3′-diphosphate (ppGpp), which accumulate in response to nutrient deficiencies as well as to undefined factors that operate during starvation and initiation of the stationary phase (8). E. coli mutants that fail to produce (p)ppGpp (e.g., relA and spoT) are deficient in the accumulation of polyP in response to amino acid starvation (5). It has been demonstrated in vitro that pppGpp, in particular, profoundly inhibits E. coli exopolyphosphatase activity, but not that of PPK, thus leading to accumulation of polyP (5). However, it is not known what roles PPK has in physiological responses to nutritional deprivation.
After amino acid starvation, protein degradation increases to provide amino acids needed for the synthesis of new enzymes such as amino acid biosynthetic enzymes (9). These metabolic responses are very important for cells to escape from amino acid starvation as well as to adapt to a new environmental condition (9). This paper will show that an E. coli ppk ppx mutant is severely impaired in recovery from the nutritional downshift. The PPK is important for cells to increase the rate of intracellular protein degradation and adapt rapidly to the nutritional deprivation.
Materials and Methods
Bacterial Strains and Plasmids.
E. coli strains used in this study were MG1655 (wild type) and CF5802 (Δppk-ppx∷km) (5). A plasmid pMWppk was constructed as follows: a 3-kb KpnI fragment containing the ppk gene of pBC10 (10) was inserted into a unique KpnI site of pMW119 (Nippon gene). The direction of transcription of ppk was opposite from that of pMW119 lac promoter. For construction of a plasmid pQFhis containing a hisG-lacZ transcriptional fusion, a 0.7-kb DNA fragment with the promoter region of the his operon (11) was amplified by PCR with primers his1 [5′-AGAGGATCCGTCTCTTCAGCGAGTTGTA-3′ (an added BamHI site is italicized)] and his2 [5′-CGGAAGCTTCTGCATAGCTATGCGTAACG-3′ (an added HindIII site is italicized)], which begin 627 bp upstream and 37 bp downstream of the hisG gene translational start codon, respectively. The amplified DNA fragment was digested with BamHI and HindIII and then was inserted into the BamHI and HindIII sites of pQF50 (a promoter-less lacZ plasmid) (12).
Chemicals.
[32P]Phosphate (9,000 Ci/mmol) and l-[14C(U)] leucine (310 mCi/mmol) were purchased from DuPont. Adenosine triphosphate (ATP) and adenosine diphosphate (ADP) were purchased from Boehringer Mannheim. Thin layer chromatography (TLC) plates coated with polyethylenimine-cellulose were purchased from Merck.
Nutritional Downshift.
E. coli MG1655 and CF5802 cells grown to mid-log phase (2 hr) in 2 × YT rich medium (1.6% peptone/1.0% yeast extract/0.5% NaCl) (13) with shaking at 37°C were collected by centrifugation. The pellet was washed once with a Mops minimal medium (14) containing 22.2 mM glucose, 40 mM potassium morpholinopropanesulfonate (Mops) (pH 7.2), 50 mM NaCl, 9.52 mM NH4Cl, 4 mM Tricine, 2 mM K2HPO4, 0.523 mM MgCl2, 0.276 mM K2SO4, 0.01 mM FeSO4, 0.0005 mM CaCl2, and trace metals (14) and was resuspended in the same medium. To prepare cells for β-galactosidase assays and RNA isolation, 50 mM glucose was added to the 2 × YT medium.
Measurement of polyP, ppGpp, and ATP.
PolyP was extracted and measured by the method of Ault-Riché et al. (6). An E. coli pellet was dissolved in 4 M guanidine isothiocyanate, and cells were lysed by heat (90°C), SDS, and sonication. After addition of ethanol, polyP was precipitated with Glassmilk (Bio 101) and was washed with NEW Wash (Bio 101). After DNase and RNase treatment, polyP was readsorbed to the Glassmilk and was extracted with water. PolyP was measured as an amount of ATP generated by the reaction with PPK and ADP. ATP was measured by the ATP Bioluminescence assay kit (Boehringer Mannheim). Concentrations of polyP were given in terms of phosphate residues. The stringent factor, ppGpp, was extracted and measured by the method described by Kuroda et al. (5). E. coli labeled with 32P-phosphate (10 μCi/ml) was disrupted in 2 M formic acid. After precipitation of the cell debris, 1 μl of the supernatant was spotted on a polyethylenimine-cellulose/TLC plate, which was developed with 1.5 M KH2PO4 (pH 3.5). The dried plate was exposed to a screen and was visualized in a BAS1000 system (Fuji). Extraction of ATP from cells was performed as follows. Cell culture samples (80 μl) were removed at the time indicated and immediately were mixed with 20 μl of 5 M formic acid. After incubation on ice for 2 hr, cell debris was removed by centrifugation. The supernatants were neutralized with 50 μl of 2 M NaOH and 20 μl of 1 M Tris⋅HCl (pH 8.0). ATP was measured as described above.
Northern Blot Analysis.
Total RNA was extracted from E. coli cells as described by Chuang et al. (15). Cell culture samples (10 ml) were collected and resuspended in 0.6 ml of ice-cold buffer (10 mM KCl/5 mM MgCl2/10 mM Tris, pH 7.4) and then immediately were added to 0.6 ml of hot lysis buffer (0.4 M NaCl/40 mM EDTA/1% β-mercaptoethanol/1% SDS/20 mM Tris, pH 7.4) containing 100 μl of phenol. The mixture was incubated in a boiling water bath for 40 seconds. Cell debris was removed by centrifugation (8,000 × g, 10 min, 20°C). The supernatant was extracted with phenol-chloroform five times, was precipitated by ethanol, and was dried. RNA was dissolved in water, and its concentration was calculated based on A260. Northern blot analysis was performed by the methods described by Sambrook et al. (13). Labeling a probe DNA fragment with fluorescein and detection was performed according to the instructions provided by the manufacturer (Amersham Pharmacia).
Protein Degradation.
The rate of intracellular protein degradation was measured by the method as described by Sussman and Gilvarg (16). An E. coli culture was grown for 2 hr on the 2 × YT medium (2 ml) containing 14C-leucine (5 μCi/ml) and 50 mM glucose. The cells were collected by centrifugation (8,000 × g, 10 min, 20°C), were washed with 2 ml of the Mops medium, and were resuspended in 2 ml of either of the Mops medium or the 2 × YT medium. Both media contained 300 μg/ml nonradiolabeled leucine. The cell culture (180-μl aliquots) was removed at the time indicated, was immediately mixed with 50% trichloroacetic acid, and was incubated for 30 min at 20°C. After centrifugation, 180 μl of the supernatant was added to 20 μl of 2 mg/ml BSA. After subsequent centrifugation, 180 μl of the supernatant was removed, and the radioactivity was measured in the scintillation counter (Packard, TRI-CARB 2300TR).
Other Methods.
PPK activity was measured by the method as described by Ahn and Kornberg (4). The β-galactosidase activity was measured as described by Miller (17). DNA manipulations were done as described by Sambrook et al. (13).
Results
A ppk ppx Mutant Exhibited an Extended Growth Lag on a Nutritional Downshift.
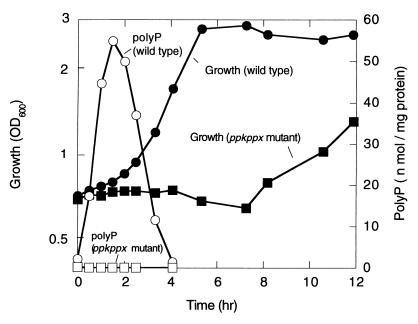
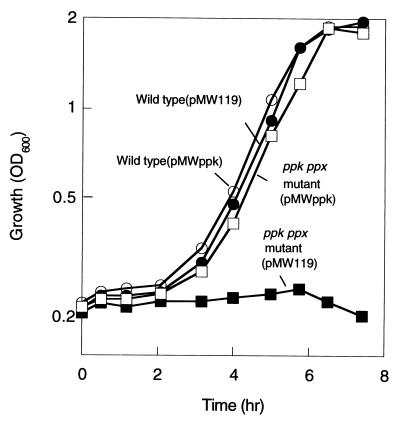
E. coli CF5802, which lacks both the ppk and ppx genes, grew on the Mops minimal medium, as did the wild-type strain MG1655, when it was precultured in the same minimal medium (data not shown). However, the ppk ppx mutant showed an extended lag upon recovery to logarithmic growth when exponentially growing cells on the 2 × YT medium were transferred to the Mops medium (nutritional downshift) (Fig. 1). The wild type accumulated polyP transiently at the onset of growth on the Mops medium whereas the ppk ppx mutant did not (<0.2 nmol/mg protein). Introduction of a low copy plasmid, pMWppk, harboring only the ppk gene abolished this extended lag phase (Fig. 2). The mutant harboring pMWppk restored the PPK activity (4,200 units/mg protein, two-fold higher than the wild type) as well as polyP accumulation (150 nmol/mg protein), but not exopolyphosphatase activity (200 units/mg protein).
Figure 1.
Cell growth and polyP accumulation during the nutritional downshift. E. coli MG1655 (wild type) and CF5802 (ppk ppx mutant) cells were subjected to the nutritional downshift from the 2 × YT to the Mops medium. PolyP accumulated in response to the downshift in the wild type (open circle) but not in the mutant (open square). Growth was measured as the optical density at 600 nm (OD600) in the wild type (filled circle) and the ppk ppx mutant (filled square).
Figure 2.
Introduction of the polyP kinase gene into the mutant abolished the growth lag during the nutritional downshift. Growth after the nutritional downshift was measured as the OD600 in the wild type harboring a vector pMW119 (filled circle) and pMWppk (open circle) and in the mutants harboring pMW119 (filled square) and pMWppk (open square).
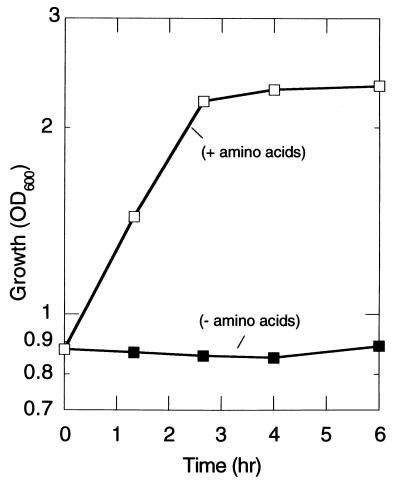
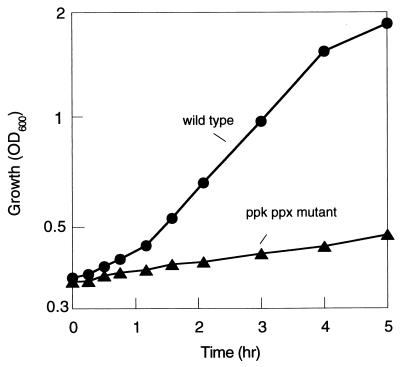
In the nutritional downshift experiment, supplementation of all amino acids to the Mops medium abolished the extended lag phase of the mutant (Fig. 3). Addition of individual amino acids to the medium has no effect on the lag, but supplementation of histidine, isoleucine, leucine, valine, cysteine, methionine, asparagine, and threonine together partially abolished it. The mutant as well as the wild type grew on the Mops medium supplemented with amino acids. Shifting from this medium to that without amino acids led to a significant reduction in the growth rate in the mutant (Fig. 4). Under this condition, transient polyP accumulation (20 nmol/mg protein) was observed only in the wild type (data not shown).
Figure 3.
Supplementation of amino acids to the Mops medium abolished the growth lag. Growth after the nutritional downshift was measured as the OD600 in the ppk ppx mutant in the absence (filled square) and presence (open square) of amino acids (50 mg/ml), respectively.
Figure 4.
Effect of removal of amino acids from the medium on the growth of the wild type and the ppk ppx mutant. E. coli MG1655 and CF5802 cells grown to mid-log phase in the Mops medium with amino acids (50 mg/liter) were collected and resuspended in the Mops medium without amino acids. Growth was measured as OD600 in the wild type (filled circle) and the ppk ppx mutant (filled triangle).
Levels of ppGpp in the Wild Type and the ppk ppx Mutant.
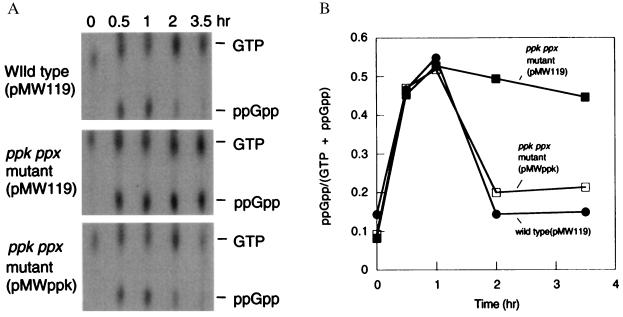
Both the mutant and the wild type accumulated ppGpp in response to the nutritional downshift (Fig. 5). The levels of ppGpp decreased after 1 hr in the wild type whereas those in the mutant decreased only slightly over 3.5 hr (Fig. 5). The mutant harboring the pMWppk accumulated ppGpp transiently as did the wild type (Fig. 5). Levels of ppGpp increase in proportion to the ratios of uncharged to charged amino acyl-tRNAs (8). These results suggested that the mutant was starved for amino acids during the nutritional downshift.
Figure 5.
Accumulation of ppGpp during the nutritional downshift. (A) ppGpp was detected on the polyethylenimine-cellulose/TLC plate in the wild type harboring pMW119 and the ppk ppx mutant harboring pMW119 or pMWppk. (B) Relative amounts of ppGpp were indicated as the intensity of ppGpp divided by that of ppGpp plus GTP.
The ppk ppx Mutant Fails To Express an Amino Acid Biosynthetic Gene.
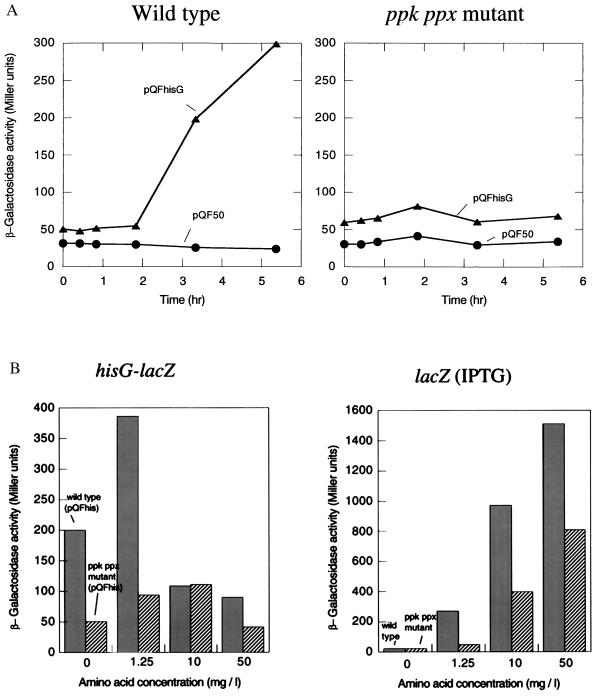
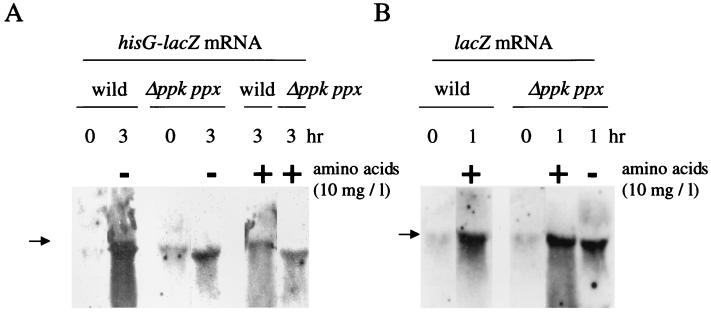
To know whether the ppk ppx mutant affected the expression of an amino acid biosynthetic gene, we constructed the pQFhis plasmid, which contains the hisG-lacZ transcriptional fusion. Induction of β-galactosidase expression by the nutritional downshift was observed in the wild type but not in the mutant within 5 hr (Fig. 6A). Yet, Northern blot analysis revealed that the mutant transcribed the hisG-lacZ gene at ≈20% that of the wild type after 3 hr of the nutritional downshift (without amino acids) (Fig. 7A). No significant signals were detected in the strains harboring pQF50. Addition of amino acids to the Mops medium at a low concentration (1.25 mg/liter) resulted in induction of the hisG-lacZ expression in the mutant (Fig. 6B Left). β-galactosidase activities in strains harboring pQF50 did not increase with the supplementation of amino acids. The hisG-lacZ expression decreased with a higher concentration of amino acids. This may be attributable to inactivation of the hisG promoter (hisP1) and transcriptional termination at the attenuator control site in the presence of histidine (11). Indeed addition of amino acids reduced the transcription of the hisG-lacZ gene in the wild type (Fig. 7A). On the other hand, isopropyl-1-thio-β-d-galactoside induced a chromosomal lacZ gene expression in proportion to the concentration of amino acids during the nutritional downshift (Fig. 6B Right). Northern blot analysis showed that the lacZ mRNA was present in the mutant irrespective of the presence of amino acids (Fig. 7B). The mutant showed β-galactosidase activities of 120 Miller units after 1 hr of the downshift in the presence of amino acids and <2 units in the absence of amino acids. These results suggested that availability of amino acids is important for enzyme production after the nutritional downshift.
Figure 6.
Induction of β-galactosidase during the nutritional downshift. (A) β-galactosidase activities from the hisG-lacZ fusion were measured in the wild type (Left) and the mutant (Right) harboring pQFhis (the hisG-lacZ fusion), respectively. Strains harboring pQF50 (a promoterless lacZ vector) were used as a control (circle). (B) Effects of amino acid supplementation on β-galactosidase production. The wild type (filled bar) and the mutant (striped bar) were downshifted to the Mops medium supplemented with all 20 amino acids at concentrations of 0, 1.25, 10, and 50 mg/liter, respectively. The left panel shows β-galactosidase activities in strains harboring pQFhis. The right panel indicates β-galactosidase activities from the chromosomal lacZ gene in the wild type and the mutant. The chromosomal lacZ gene was induced by adding isopropyl-1-thio-β-d-galactoside (1 mM) to the Mops medium. β-galactosidase activities were measured 3 hr after the nutritional downshift.
Figure 7.
Northern blot analysis of the hisG-lacZ and the lacZ mRNA. (A) RNA was isolated from the wild type and the mutant harboring pQFhis after 0 and 3 hr of the nutritional downshift. Addition of amino acids (10 mg/ml) to the Mops medium was indicated above. RNA (10 μg) was used for hybridization. Arrows indicate the position of 23S rRNA. The fluorescein-labeled lacZ probe was used. (B) RNA was isolated from the wild type and the mutant after 0 and 1 hr of the nutritional downshift. Isopropyl-1-thio-β-d-galactoside (1 mM) was added to the Mops medium to induce the chromosomal lacZ gene.
PolyP Kinase Is Important for the Activation of Protein Degradation.
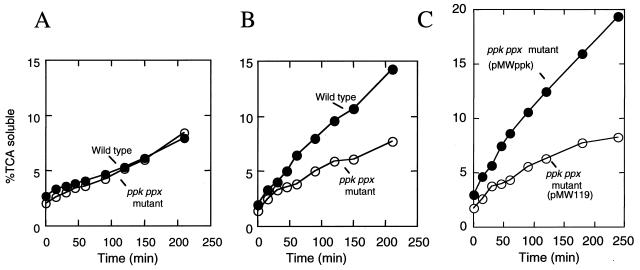
Protein degradation is stimulated during amino acid starvation to provide amino acids for the synthesis of proteins required for adaptation under starvation or nutritional downshift (16). The wild type demonstrated increased rates of protein degradation in response to the nutritional downshift in contrast to the mutant (Fig. 8 A and B). The mutant harboring pMWppk restored the ability to increase the rates of protein degradation whereas the mutant harboring pMW119 failed (Fig. 8C). To know whether the intracellular ATP concentration in the mutant altered during the nutritional downshift, the ATP levels were measured by a bioluminescence method. The intracellular ATP concentrations remained stable in both the wild type and the mutant during the downshift (data not shown).
Figure 8.
Degradation of intracellular protein during the nutritional downshift. [14C]Leucine was incorporated during exponential growth. The wild type (filled circle) and the ppk ppx mutant (open circle) were resuspended in the 2 × YT rich medium (A) and the Mops minimal medium (B). The ppk ppx mutant harboring pMWppk (filled circle) and pMW119 (open circle) were resuspended in the Mops minimal medium (C). Protein degradation was measured as described in Materials and Methods. Trichloroacetic acid soluble counts at a given time are then expressed as a percentage of the total initially incorporated counts.
Discussion
E. coli accumulates polyP in response to a nutritional downshift from a rich to a minimum medium (6, 18). We have shown that the E. coli mutant lacking PPK and exopolyphosphatase failed to accumulate polyP and exhibited an extended lag phase of growth recovery during a nutritional downshift. Introduction of the ppk gene on a low-copy plasmid into the mutant abolished the lag, suggesting that PPK is important for this growth recovery. This phenotype of the mutant can be attributed to an impaired adaptation to amino acid starvation. Support for this hypothesis was provided by the following observations: (i) supplementation of amino acids to the Mops medium abolished the extended lag phase; (ii) the mutant was significantly reduced in growth rate when only amino acids were removed from the medium; (iii) levels of ppGpp remained high in the mutant during the nutrient downshift. Once the mutant adapted to the Mops medium, it grew as well as did the wild type. Therefore, PPK is important for the rapid recovery from amino acid starvation.
Upon starvation for amino acids, E. coli induces expression of amino acid biosynthetic enzymes (8, 11). This physiological response is necessary for the cells to adapt to this condition. The eight structural histidine biosynthesis genes are transcribed into a single, polycistronic mRNA that extends from the primary promoter (hisP1) to the rho-independent terminator (11). The hisG gene is the first structural gene of the operon and follows the his attenuation control leader region. Because the ppk ppx mutant did not induce β-galactosidase expression from the hisG-lacZ fusion gene, one might assume that the mutant failed to transcribe the histidine biosynthesis genes. The mutant also failed to induce β-galactosidase from the ilvG-lacZ and trpE-lacZ transcriptional fusion under this condition (A.K. and H.O., unpublished results). However, Northern blot analysis revealed that the mutant synthesized hisG-lacZ mRNA to a significant level, ≈20% that of the wild type. This result indicated that polyP accumulation might have affected the initiation of the hisG transcription but was apparently insufficient to explain why the mutant showed no expression of β-galactosidase from the hisG-lacZ transcriptional fusion. We found that supplementation of amino acids restored the hisG-lacZ as well as the chromosomal lacZ expression. Starvation for amino acids stimulates intracellular protein degradation (16). Multiple peptidase-deficient mutants showed an extended lag phase compared with that of their parents in a downshift experiment (19). Therefore, one of the functions of protein degradation is to provide amino acids for the synthesis of proteins required for adaptation under downshifted conditions (9, 19). A plausible explanation for the ppk defect in the hisG-lacZ expression is that the mutant failed to increase the rate of protein degradation and thereby supply amino acids for enzyme production. Consequently, the mutant experienced the extended growth lag in the downshift experiment.
An E. coli strain carrying a relA mutation could not increase the rate of protein degradation in response to amino acid starvation (16). Thus, ppGpp plays a direct or indirect role in regulating protein degradation (9, 16). Here, we have shown that the ppk ppx mutant could not stimulate protein degradation despite the presence of ppGpp. The stringent factors, (p)ppGpp, are important for the accumulation of polyP (5). E. coli mutants that fail to produce (p)ppGpp are deficient in the accumulation of polyP (5). The failure of the relA mutant to increase the rate of protein degradation could thus be ascribed to the defect in polyP accumulation. Whether polyP affects the rate of protein degradation directly or indirectly remains to be determined. Furthermore, little is known about the degradation pathway that is activated in response to nutritional deprivation (9). Levels of polyP reached 20 mM in phosphate residues, which could be used to synthesize ATP as well as other nucleoside triphosphates by PPK activity (20, 21). At least 40 enzymes catalyze the hydrolysis of peptide bonds, and some of them are ATP-dependent (9). Levels of intracellular ATP can affect the rate of protein degradation (22). However, we observed that polyP accumulation did not affect the intracellular concentration of ATP significantly in this condition.
An increase in expression of the his operon in cells subjected to histidine starvation is markedly less in relA mutants than that in the wild type, suggesting that ppGpp is a positive effector of the his operon (11). In the ppk ppx mutant, the his transcription was reduced compared with the wild type, although it did accumulate ppGpp. PolyP is also involved in expression of the rpoS gene (23, 24). Although a functional interaction of RNA polymerase with polyP (25) and a multiple complex of the RNA degradosome with PPK (26) have been reported, details of the reactions and regulatory roles of polyP in RNA transcription and degradation also remained obscure.
Acknowledgments
We thank Dr. A. Kornberg for providing an E. coli ppk gene and its deletion strain and for critical reading of this manuscript. We also thank Drs. N. N. Rao, C. D. Fraley, and M. H. Rashid for helpful advise on manuscript preparation. This work was supported in part by a grant from Research Institute of Innovative Technology for the Earth (RITE) and a grant for scientific research from the Ministry of Education, Science and Culture of Japan.
Abbreviations
- polyP
inorganic polyphosphate
- PPK
polyphosphate kinase
- ppGpp
guanosine 5′-diphosphate 3′-diphosphate
- pppGpp
guanosine 5′-triphosphate 3′-diphosphate
References
- 1.Kornberg A. J Bacteriol. 1995;177:491–496. doi: 10.1128/jb.177.3.491-496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kulaev I S. The Biochemistry of Inorganic Polyphosphates. New York: Wiley; 1979. [DOI] [PubMed] [Google Scholar]
- 3.Wood H G, Clark J E. Annu Rev Biochem. 1988;57:235–260. doi: 10.1146/annurev.bi.57.070188.001315. [DOI] [PubMed] [Google Scholar]
- 4.Ahn K, Kornberg A. J Biol Chem. 1990;265:11734–11739. [PubMed] [Google Scholar]
- 5.Kuroda A, Murphy H, Cashel M, Kornberg A. J Biol Chem. 1997;272:21240–21243. doi: 10.1074/jbc.272.34.21240. [DOI] [PubMed] [Google Scholar]
- 6.Ault-Riché D, Fraley C D, Tzeng C-M, Kornberg A. J Bacteriol. 1998;180:1841–1847. doi: 10.1128/jb.180.7.1841-1847.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voelz H, Voelz U, Ortigoza R O. Arch Mikrobiol. 1966;53:371–388. doi: 10.1007/BF00409874. [DOI] [PubMed] [Google Scholar]
- 8.Cashel M, Gentry D R, Hernandez V J, Vinella D. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1458–1496. [Google Scholar]
- 9.Miller C G. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 938–954. [Google Scholar]
- 10.Akiyama M, Crooke E, Kornberg A. J Biol Chem. 1992;267:22556–22561. [PubMed] [Google Scholar]
- 11.Winkler M E. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 485–505. [Google Scholar]
- 12.Farinha M A, Kropinski A M. J Bacteriol. 1990;172:3496–3499. doi: 10.1128/jb.172.6.3496-3499.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 14.Neidhardt F C, Block P L, Smith D F. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chuang S-E, Daniels D L, Blattner F R. J Bacteriol. 1993;175:2026–2036. doi: 10.1128/jb.175.7.2026-2036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sussman A J, Gilvarg C. J Biol Chem. 1969;244:6304–6308. [PubMed] [Google Scholar]
- 17.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 18.Rao N N, Liu S, Kornberg A. J Bacteriol. 1998;180:2186–2193. doi: 10.1128/jb.180.8.2186-2193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yen C, Green L, Miller C G. J Mol Biol. 1980;143:21–33. doi: 10.1016/0022-2836(80)90122-9. [DOI] [PubMed] [Google Scholar]
- 20.Kornberg S R. Biochim Biophys Acta. 1957;26:294–300. doi: 10.1016/0006-3002(57)90008-2. [DOI] [PubMed] [Google Scholar]
- 21.Kuroda A, Kornberg A. Proc Natl Acad Sci USA. 1997;94:439–442. doi: 10.1073/pnas.94.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.St. John A C, Goldberg A L. J Biol Chem. 1978;253:2705–2711. [PubMed] [Google Scholar]
- 23.Rao N N, Kornberg A. J Bacteriol. 1996;178:1394–1400. doi: 10.1128/jb.178.5.1394-1400.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shiba T, Tsutsumi K, Yano H, Ihara Y, Kameda A, Tanaka K, Takahashi H, Masanobu M, Rao N N, Kornberg A. Proc Natl Acad Sci USA. 1997;94:11210–11215. doi: 10.1073/pnas.94.21.11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kusano S, Ishihama A. Genes Cells. 1997;2:433–441. doi: 10.1046/j.1365-2443.1997.13203301320330.x. [DOI] [PubMed] [Google Scholar]
- 26.Blum E, Py B, Carpousis A J, Higgins C F. Mol Microbiol. 1997;26:387–398. doi: 10.1046/j.1365-2958.1997.5901947.x. [DOI] [PubMed] [Google Scholar]