Abstract
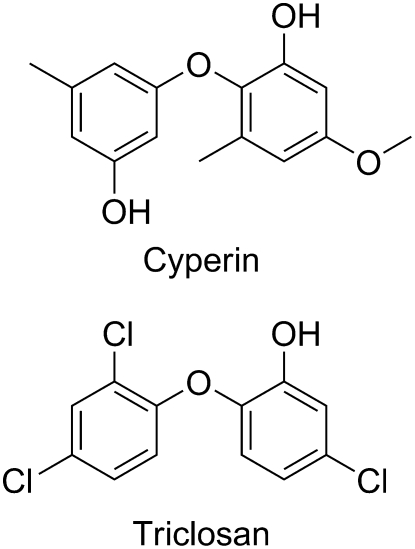
Cyperin is a natural diphenyl ether phytotoxin produced by several fungal plant pathogens. At high concentrations, this metabolite inhibits protoporphyrinogen oxidase, a key enzyme in porphyrin synthesis. However, unlike its herbicide structural analogs, the mode of action of cyperin is not light dependent, causing loss of membrane integrity in the dark. We report that this natural diphenyl ether inhibits Arabidopsis (Arabidopsis thaliana) enoyl (acyl carrier protein) reductase (ENR). This enzyme is also sensitive to triclosan, a synthetic antimicrobial diphenyl ether. Whereas cyperin was much less potent than triclosan on this target site, their ability to cause light-independent disruption of membrane integrity and inhibition of ENR is similar at their respective phytotoxic concentrations. The sequence of ENR is highly conserved within higher plants and a homology model of Arabidopsis ENR was derived from the crystal structure of the protein from Brassica napus. Cyperin mimicked the binding of triclosan in the binding pocket of ENR. Both molecules were stabilized by the π-π stacking interaction between one of their phenyl rings and the nicotinamide ring of the NAD+. Furthermore, the side chain of tyrosine is involved in hydrogen bonding with a phenolic hydroxy group of cyperin. Therefore, cyperin may contribute to the virulence of the pathogens by inhibiting ENR and destabilizing the membrane integrity of the cells surrounding the point of infection.
Interactions between pathogenic fungi and their host plants involve complex physical and chemical communications that lead to a series of biochemical actions and reactions by both the infecting microorganisms and the plants being infected (Vera-Estrella et al., 1994; Fray et al., 1999; Pedras et al., 2001). The virulence of an organism is sometimes enhanced by its ability to produce phytotoxins that kill cells in the tissue surrounding the point of infection (Baker et al., 1997). Fungi and other microorganisms that produce such biochemical compounds are being studied as potential biocontrol agents for weed management (Duke et al., 1996). These studies have led to the discovery that many pathogenic organisms, such as Preussia fleischhakii, Phoma sorghina, and Ascochyta cypericola, produce cyperin (Fig. 1; Weber and Gloer, 1988; Stierle et al., 1992; Venkatasubbaiah et al., 1992). These natural toxins are thought to play important roles in inhibiting the physiological processes in the plant cells surrounding the point of infection, enabling the spread of the disease (Feys and Parker, 2000; Staskawicz et al., 2001).
Figure 1.
Structures of the fungal phytotoxin cyperin and the synthetic antimicrobial diphenyl ether triclosan.
Cyperin is a natural diphenyl ether toxin with structural similarities with diphenyl ether herbicides (i.e. acifluorfen and oxyfluorfen), but it is not a strong inhibitor of the molecular target site of this chemical class, the enzyme protoporphyrinogen oxidase (Harrington et al., 1995; Dayan and Allen, 2000). Moreover, the mechanism of action of cyperin appears to be light independent (Harrington et al., 1995), whereas diphenyl ether herbicides require light to cause their phytotoxic effect (Dayan and Duke, 1997). These observations suggest that cyperin acts by another mechanism than that associated with protoporphyrinogen oxidase-inhibiting herbicides.
It was recently reported that enoyl (acyl carrier protein) reductases (ENRs) of the type II fatty acid synthase (plants and prokaryotes) were sensitive to the diphenyl ether triclosan (Fig. 1; McMurry et al., 1998; Roujeinikova et al., 1999). Inhibition of ENR interrupts the critical process of fatty acid elongation. It has also been reported that this compound can intercalate itself within the lipid bilayer and compromise the functional integrity of the cell membranes of microorganisms (Villalain et al., 2001; Paul et al., 2004). The objective of this study was to determine whether cyperin had the same molecular target site as triclosan. We report that cyperin also causes light-independent loss of membrane integrity and inhibits plant ENR. The binding orientation of this reversible inhibitor to the catalytic site of ENR is similar to that of the irreversible inhibitor triclosan. We conclude that ENR may be a good target site to develop new natural product-derived herbicides.
RESULTS
Phytotoxicity of Cyperin and Triclosan
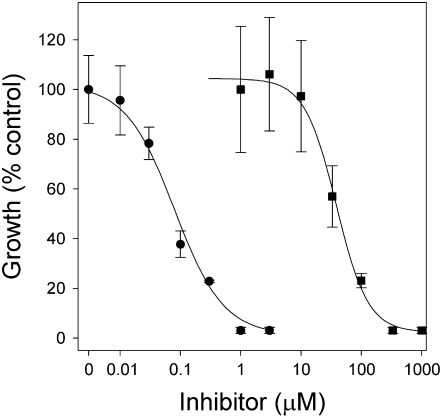
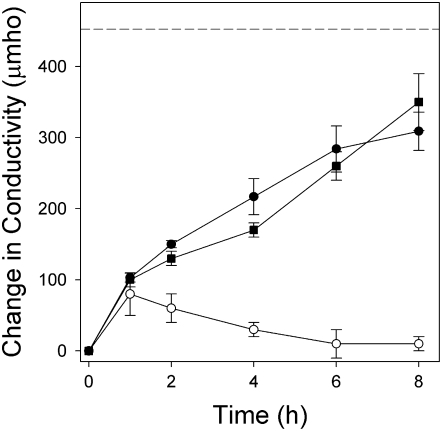
Cyperin was phytotoxic to Arabidopsis (Arabidopsis thaliana) and inhibited growth in a dose-dependent manner (Fig. 2), with an I50 of 38.4 μm (Table I). The effect was most evident on root length, but the leaves were also small and chlorotic at the higher concentrations. Triclosan was much more active on Arabidopsis than cyperin, with an I50 of 0.08 μm. Both cyperin and triclosan caused loss of membrane integrity of cucumber (Cucumis sativus) cotyledons in the absence of light, resulting in electrolyte leakage measured as increased conductivity of the medium (Fig. 3). The rate of electrolyte leakage was similar for the two compounds, when tested at 333 and 10 μm for cyperin and triclosan, respectively. After 8 h of incubation in the dark, the level of electrolyte leakage was equivalent to approximately 75% of the total leakage measured after boiling the leaf discs (Table I), suggesting a significant membranotropic effect of these compounds. Similar results were obtained when the experiment was carried out in the presence of light (300 μmol m−2 s−1 of light), indicating that the mode of action of these compounds was not modulated by photon flux or light-dependent physiological or biochemical processes (data not shown).
Figure 2.
Dose-response curves of triclosan (•) and cyperin (▪) against the root growth of Arabidopsis. Error bars = ±1 sd. Curves were analyzed with the drc add-on package for dose-response curves in R version 2.2.1 (R Development Core Team, 2005) using a four-parameter logistic function.
Table I.
Physiological and biochemical characterization of the effects of cyperin and triclosan on Arabidopsis and its ENR
| Compound | Cyperin | Triclosan |
|---|---|---|
| Inhibition of root growth I50 (μm)a | 38.4 ± 3.8 | 0.08 ± 0.007 |
| Electrolyte leakage % of totalb | 77.7 | 68.6 |
| Inhibition of ENR I50 (μm)a | 89.0 ± 15.1 | 0.046 ± 0.005 |
| Mechanism of inhibition | Reversible noncompetitive | Slow tight binding |
| Kia (μm) | 13.6 ± 2.2 | – |
I50 values were obtained from the regression curves of dose-response experiments analyzed with R version 2.2.1. Ki value was obtained using the enzyme kinetic module of SigmaPlot version 10. Each value is the mean of three independent measurements followed by sd.
Percentage of total is calculated as the electrolyte leakage caused by the treatment divided by the electrolyte leakage after boiling samples (complete loss of membrane integrity) multiplied by 100.
Figure 3.
Electrolyte leakage caused by 10 μm triclosan (•) and 333 μm cyperin (▪) on Arabidopsis leaf discs, as compared to the solvent control (○). Error bars = ±1 sd. The dotted line indicates maximal conductivity in the bathing medium following complete lysis of the membranes by boiling the samples.
Levels of C18:3 and C18:2 fatty acids were significantly lower in Arabidopsis exposed to sublethal concentrations of cyperin and triclosan (33 and 0.03 μm, respectively) than in control treatments (Table II). These phenotypic responses suggest that both of these phytotoxic diphenyl ethers share a similar mode of action.
Table II.
Fatty acid analysis of Arabidopsis as expressed as percentage of total fatty acid extracted
Each value is the mean of three independent measurements followed by sd. Numbers in columns followed by the same letter are not different at P < 0.05 according to Duncan multiple range analysis test.
| C16:0 | C16:1 | C16:3 | C18:0 | C18:1 | C18:2 | C18:3 | |
|---|---|---|---|---|---|---|---|
| Control | 25.3 ± 4.5 a | 3.7 ± 0.4 a | 7.0 ± 1.5 a | 1.4 ± 0.6 a | 1.5 ± 0.5 a | 15.6 ± 1.8 a | 29.2 ± 6.2 a |
| Triclosan | 31.3 ± 5.4 a | 4.8 ± 0.9 ab | 5.5 ± 0.6 ab | 1.6 ± 0.1 a | 1.7 ± 0.1 a | 13.6 ± 1.5 ab | 21.1 ± 3.3 b |
| Cyperin | 30.3 ± 1.7 a | 4.9 ± 0.2 b | 4.7 ± 0.1 b | 1.6 ± 0.1 a | 1.2 ± 0.2 a | 12.4 ± 0.3 b | 19.5 ± 0.7 b |
Biochemical Characterization of Arabidopsis ENR Inhibition
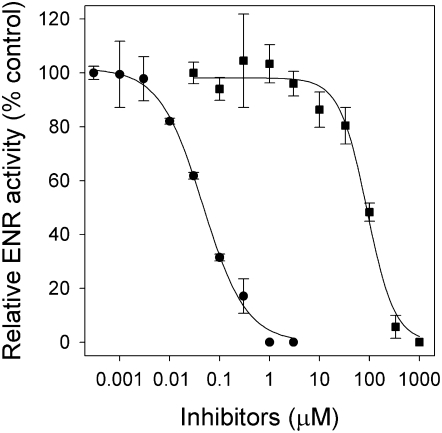
Arabidopsis ENR was overexpressed with a poly-His tag in Escherichia coli and purified using a nickel-activated column. The activity of ENR was measured spectrophotometrically by monitoring oxidation of NADH in the presence of crotonyl-CoA. Both cyperin and triclosan inhibited ENR catalytic activity (Fig. 4). However, as observed in vivo and with the electrolyte leakage experiment, triclosan was much more active than cyperin on this enzyme, with I50 values of 0.046 and 89.0 μm, respectively (Table I).
Figure 4.
Inhibitory activity of triclosan (•) and cyperin (▪) against purified Arabidopsis ENR. Error bars = ±1 sd. Curves were analyzed with the drc add-on package for dose-response curves in R version 2.2.1 (R Development Core Team, 2005) using a four-parameter logistic function.
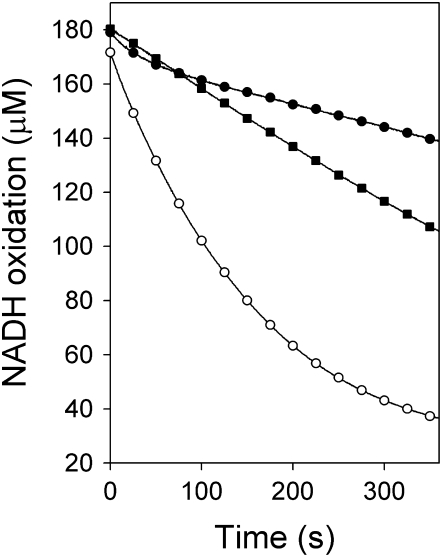
Examination of the reaction curves of the time-dependent assays revealed a clear difference between the inhibition of ENR activity by triclosan and cyperin (Fig. 5). The initial inhibition of ENR activity in the presence of 8.5 nm triclosan was similar to that observed in the presence of 20 μm cyperin, at 72.7% and 71.7% inhibition relative to control. However, inhibition of ENR activity by triclosan increased over the duration of the assay (88.5% inhibition at the end of the assay), which indicates that triclosan binds irreversibly to the enzyme. On the other hand, inhibition of ENR catalytic activity by cyperin remained constant during the entire assay (74.1% inhibition at the end of the assay), suggesting that this natural product is a reversible inhibitor of ENR.
Figure 5.
Time-dependent inhibition of Arabidopsis ENR by 8.5 nm triclosan (•) and 20 μm cyperin (▪) relative to uninhibited control (○). Control sample received equivalent volume of solvent. This experiment was repeated three times with similar results.
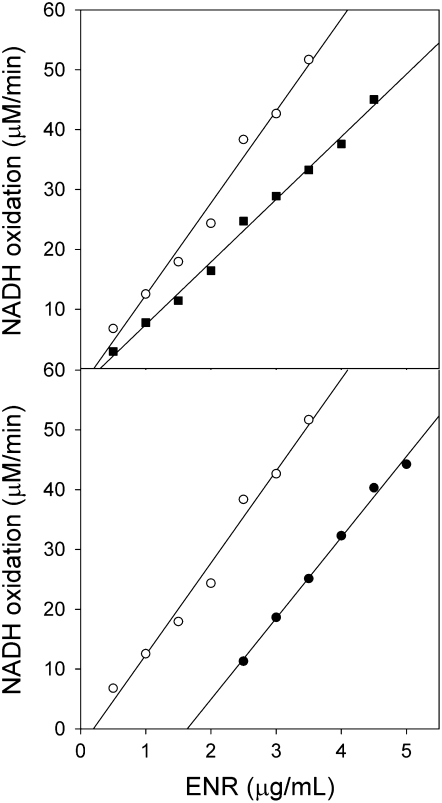
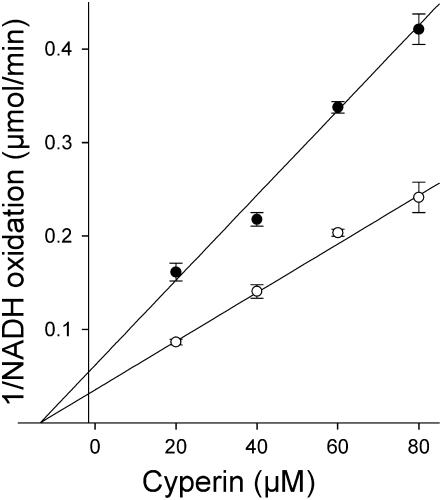
This difference in binding mechanism to ENR was confirmed by titrating ENR concentration with or without inhibitors (Fig. 6). The convergence of the regression lines of the assays performed with and without cyperin is indicative of reversible inhibition of ENR (Fig. 6A). The parallel lines observed in the case of the experiments performed with and without triclosan are typical of slow tight-binding inhibitors. Kinetic analysis determined that ENR had a Km of 288 ± 97 μm for crotonyl-CoA and that the binding of cyperin was noncompetitive with respect to crotonyl-CoA, with a Ki of 13.6 ± 2.2 μm (Fig. 7).
Figure 6.
Representative plot of the effect of ENR concentration upon the oxidation of NADH in the presence of 8.5 nm triclosan (•) and 20 μm cyperin (▪) relative to uninhibited control (○). Control sample received equivalent volume of solvent.
Figure 7.
Dixon plot showing noncompetitive inhibition of Arabidopsis ENR by cyperin in the presence of 100 (○) and 50 μm (•) crotonyl-CoA. Control sample received equivalent volume of solvent. Each point represents the average of three independent observations and error bars = ±1 sd. Kinetic parameters obtained are Km of 288 ± 97 μm for crotonyl-CoA and a Ki of 13.6 ± 2.2 μm for cyperin.
Modeling of Arabidopsis ENR and Binding of Cyperin to the Catalytic Site
The homology between the amino acid sequence of Arabidopsis ENR and nine other plant ENRs ranged from 63.5% to 94.9% amino acid identity. The sequence homology was significantly less with the ENR sequences of nonplant origin (four protozoal and 12 bacterial species), ranging from 32.0% to 68.2% amino acid identity. However, the catalytic domain of ENR is highly conserved across all species (Table III).
Table III.
Homology between amino acid residues involved in the binding pocket of ENR of Arabidopsis with respect to the sequence of this enzyme in other organismsa
Residue numbers are based on the sequence from Arabidopsis mature protein.
| Residue | Conservation | Other Residues at That Position |
|---|---|---|
| % | ||
| Ala-139 | 90 | Gly in Salmonella, E. coli |
| Asn-140 | 65 | Phe in seven bacteria |
| Gly-141 | 50 | Ala in P. falciparum and seven bacteria, and Ser in Chlamydophila and Chlamydia |
| Val-144 | 55 | Asp, Glu, or Ile in nine bacteria |
| Tyr-189 | 100 | – |
| Tyr-199 | 100 | – |
| Met-203 | 100 | – |
| Lys-207 | 100 | – |
| Pro-236 | 100 | – |
| Phe-247 | 65 | Gly or Asp in seven bacteria |
| Ile-248 | 70 | Phe in six bacteria |
Search for sequence homology using the BLASTp function in the NCBI protein database yielded seven higher plant sequences (two monocotyledonous and five dicotyledonous), one alga, and one cyanobacterial sequence, three protozoal parasite sequences, and eight bacterial sequences. GenBank accession numbers of the sequences and complete alignment are available as Supplemental Figure S1. Protein sequences were aligned by using the ClustalW program (Thompson et al., 1994) running under the software Lasergene version 6 (DNASTAR), with default parameters (gap penalty 10, gap length penalty 0.2).
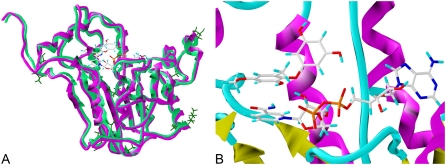
The homology model of Arabidopsis ENR derived from the crystal structure of Brassica napus was analyzed to assess its structural quality. The geometry and stereochemistry, solvent-accessible surface areas, side chain conformational probabilities, and backbone and side chain conformation were checked. No major discrepancies were found. A Ramachandran plot of the model indicated that, as expected, most Φ angles are negative and Ψ angles are positive. The few outliers with positive Φ were mostly Gly residues that are not subject to the strong conformational constraints due to a lack of side chain. Analysis of the Ramachandran plot showed 95% of the non-Gly residues in most favored regions. Alignment of the homology model to crystal structure of ENR B. napus has a root mean square distance of 0.872 Å, underlining the high degree of similarity between the two enzymes (Fig. 8A).
Figure 8.
A, Overlay of the homology model of Arabidopsis ENR (light-green) and the crystal structure of B. napus ENR (magenta). Amino acids in Arabidopsis that differ from those of B. napus are shown in dark green to identify their positions relative to the highly conserved catalytic site where cyperin and NAD+ are located (top center of protein model). B, Homology model of Arabidopsis ENR catalytic site showing the position of the cofactor NAD+ and cyperin. Cyperin was prepositioned in the catalytic site based on the coordinates of the bound pharmacophore in the crystal structure of B. napus. Cyperin was then docked to the catalytic pocket using FlexiDock as described in “Materials and Methods.” Notice the π-π stacking interaction between one of the rings of cyperin and NAD+.
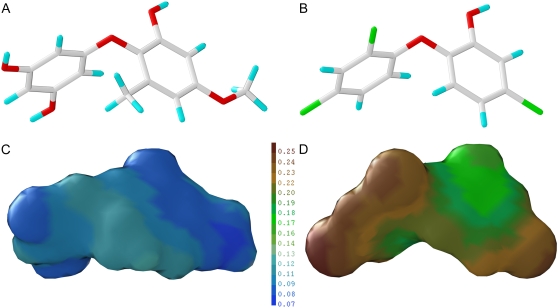
Binding of triclosan and cyperin modeled by FlexiDock provided an estimation of the position of the ligands in the catalytic site. Most of the amino acid residues found in the binding site are conserved across all species known (Table III). The amino acid residue Tyr-199 in the catalytic pocket is known to be involved in hydrogen bonding with the phenolic hydroxy group of triclosan. This residue also participated in the stabilization of cyperin. The tetrasubstituted hydroxyphenyl ring of cyperin is also stabilized via π-π stacking with the nicotinamide ring of the NAD+ (Fig. 8B) in a manner similar to that observed with triclosan. Modeling of cyperin and triclosan showed that these two structures share some structural similarities, such as their diphenyl ether backbone and molecular volumes (Fig. 9, A and B; Table IV). However, the presence of several hydroxy groups on cyperin renders it much more hydrophilic than triclosan (Fig. 9, C and D; Table IV).
Figure 9.
A and B, 3-D conformation of cyperin (A) and triclosan (B) showing the similarity in carbon skeleton between the two diphenyl ethers. C and D, Lipophilic maps of cyperin (C) and triclosan (D) showing the large difference in lipophilic properties between the two molecules. Lipophilic potentials ranged from 0.07 to 0.25 for more hydrophilic (blue) to more lipophilic (brown) values, respectively. The 3-D conformation of triclosan and cyperin are based on the bioactive conformation of triclosan within the crystal structure of B. napus followed by minimization as described in “Materials and Methods.”
Table IV.
Molecular properties of cyperin and triclosan
ConnollyVOLUME, Connolly solvent-accessible surface volume as calculated by Connolly (1983); cLogP, calculated log value of the octanol/water partition coefficient, as described in www.biobyte.com; LPMAX and LPMIN, maximum and minimum lipophilic potentials calculated by Ghose et al. (1988); EPMAX and EPMIN, maximum and minimum electrostatic potentials.
| Parameters | Cyperin | Triclosan |
|---|---|---|
| Molecular weight | 260.29 | 289.54 |
| ConnollyVOLUME, Å3 | 211.7 | 196.5 |
| Dipole, Debye | 2.12 | 2.11 |
| Ionization potential, eV | 8.95 | 9.21 |
| cLogP | 3.75 | 5.53 |
| LPMAX | −9.31 | −9.15 |
| LPMIN | −0.42 | −0.19 |
| EPMAX, eV | 32.32 | 35.93 |
| EPMIN, eV | −40.67 | −32.93 |
DISCUSSION
The ability of a pathogen to infect and invade a compatible host may be facilitated by the production of toxins that induce cell death in the proximity of the invading organism (Baker et al., 1997; Dangl and Jones, 2001). Because several microbial disease agents, such as P. fleischhakii, P. sorghina, and A. cypericola, produce cyperin (Weber and Gloer, 1988; Stierle et al., 1992; Venkatasubbaiah et al., 1992), it is possible that this natural diphenyl ether may also play a role in the virulence of these organisms.
Previous work showed that cyperin was an inhibitor of protoporphyrinogen oxidase (Harrington et al., 1995), the penultimate step in the production of porphyrin that is the target of certain diphenyl ether herbicides. However, the mechanism of action of protoporphyrinogen oxidase inhibitors is light dependent (Dayan and Duke, 1997), whereas cyperin caused loss of membrane integrity in darkness, indicating that its mode of action in plants was light independent (Fig. 3). Therefore, the primary mode of action of cyperin cannot be directly related to inhibition of protoporphyrinogen oxidase, although this molecular target site may still be inhibited at higher concentrations.
Triclosan is a relatively small antimicrobial diphenyl ether that is absorbed via diffusion into the bacterial cell wall and causes light-independent disruption of the cellular membranes (Guillén et al., 2004; Paul et al., 2004). The first evidence that this diphenyl ether is a strong inhibitor of fatty acid biosynthesis came when genetic analysis of an E. coli strain resistant to triclosan linked the resistance to the type II fatty acid synthase gene, which encodes for ENR (McMurry et al., 1998).
ENR is an NAD+-dependent enzyme involved in the reduction of a trans-2,3 enoyl moiety to a saturated acyl chain. Functional plant ENR is a homotetramer and each polypeptide chain forms a single domain consisting of a seven-stranded parallel β-sheet surrounded by seven α-helices (Rafferty et al., 1995). Such a Rossmann fold structural motif is often present in NAD+-binding proteins (Rao and Rossmann, 1973).
Subsequent extensive biochemical and structural studies have confirmed that triclosan is a specific inhibitor of ENR in plants and bacteria (Heath et al., 1998, 1999; Levy et al., 1999; Stewart et al., 1999; Ward et al., 1999), and several synthesis efforts are under way to discover new inhibitors of ENR (Heath et al., 1998; Stone et al., 2004; Rafi et al., 2006; Nicola et al., 2007). Phylogenetic analysis of the known ENR sequences indicates that the catalytic domain of ENR is highly conserved species (Table III). Additionally, an overlay of the crystal structures of ENR-triclosan complexes available (Helicobacter pylori, E. coli, Mycobacterium tuberculosis, B. napus, and Plasmodium falciparum) illustrates the highly conserved tertiary structure of these enzymes and nearly identical orientations of triclosan binding (Qiu et al., 1999; Roujeinikova et al., 1999; Kuo et al., 2003; Pidugu et al., 2004; Lee et al., 2007).
The structural similarity between cyperin and the antimicrobial diphenyl ether triclosan (Fig. 1), and the fact that both diphenyl ethers cause light-independent disruption of membrane integrity (Fig. 3) and similar changes to the fatty acid profile of Arabidopsis (Table II), suggest that these two molecules may share the same molecular target site. Cyperin inhibited purified Arabidopsis ENR. Its potency was much lower than that of triclosan (Fig. 4), but was commensurate with its in vivo activity (Fig. 2). Time-dependent enzyme assays indicated that cyperin inhibited ENR reversibly, whereas triclosan behaved like a slow tight-binding inhibitor (Fig. 5). This difference in binding mechanism was confirmed by enzyme titration assays (Fig. 6). Kinetic analysis determined that cyperin was a reversible inhibitor of ENR and its binding is noncompetitive with respect to crotonyl-CoA, with a Ki of 13.6 ± 2.2 μm (Table I). The lethality of inhibiting plant ENR with natural diphenyl ether cyperin is similar to that observed in the mod1 Arabidopsis mutant line, which is deficient in MOD1, a gene encoding an ENR (Mou et al., 2000; Serrano et al., 2007).
Knowledge-based modeling enabled the construction of a three-dimensional (3-D) model of Arabidopsis ENR. It is established that the root mean square deviation of the Cαg atoms for protein sharing 40% amino acid identity is approximately 1 Å for 90% of backbone atoms (those having the most influence on the secondary and ternary conformation; Chothia and Lesk, 1989; Sali et al., 1995). Considering the high similarity of the amino acid sequences of B. napus and Arabidopsis ENR (94.9%) and the absence of significant conformational problems in the model, it is anticipated that the structure of the Arabidopsis ENR may be similar to a low-resolution crystal structure of this enzyme. Furthermore, residues in the catalytic sites are extremely conserved across all living organisms (Table III; identical between Arabidopsis and B. napus), and the regions where the model differs from the crystal structure are relatively distant from the catalytic site (Fig. 8A). The resolution of such models is generally sufficient to compare the binding of a substrate and an inhibitor.
The orientation of cyperin in the binding pocket of ENR obtained using the FlexiDock method is similar to that of triclosan in the B. napus catalytic site (Fig. 8B). Only minor shifts in the conformation of the amino acid side chains are necessary to accommodate the binding of the inhibitor. Cyperin appears to interact noncovalently with the Arabidopsis protein and the NAD+ cofactor in the substrate binding site. As with triclosan, cyperin appears to mimic the structure of the enoyl moiety of the substrate, and the oxygen atom of the ether bridge of cyperin is positioned where the carbonyl oxygen of the thioester of the substrate is normally localized (Pidugu et al., 2004).
The presence of one of the phenolic hydroxy groups on cyperin is in an identical position as in triclosan, which may provide some stabilization of the natural product via hydrogen bonding with Tyr-199 amino acid. Indeed, the presence of the hydroxy group of triclosan is essential for activity. It forms hydrogen bonds with the hydroxy group of Tyr-156 in E. coli that corresponds to Tyr-199 in higher plant ENR (Heath et al., 1999). Removing this hydroxy group decreases the affinity by more than 10,000-fold (Ward et al., 1999). As well, the tetrasubstituted hydroxyphenyl ring of cyperin is also stabilized via π-π stacking with the nicotinamide ring of the NAD+ with an interplanar distance of 3.48 Å (Fig. 8B). This interplanar distance was 3.4 Å in the case of triclosan (Heath et al., 1999).
Whereas cyperin and triclosan have some structural similarity and both caused light-independent loss of membrane integrity (Fig. 3), there was a large difference in their potency (Table I). The absence of chloro groups on cyperin may also contribute to the lower inhibition of ENR and the difference in their binding mechanism. The 5-chloro group of the phenolic moiety of triclosan is known to be very important for tight binding and removal of this group reduced the binding 450,000-fold (Sivaraman et al., 2003, 2004). The low enzyme inhibitory activity and reversible binding of cyperin to ENR is probably due to the absence of similar halogen functionality.
In addition, cyperin is much less lipophilic than triclosan (Fig. 9; Table IV). Consistent with the analysis of several ENR crystal structures (Lee et al., 2007), the Arabidopsis ligand-binding pocket adjacent to the NAD+ cofactor is composed of hydrophobic residues (Ala-214, Tyr-264, Ile-265, Ala-266, Ile-271, and Phe-246); therefore, the ability of this polar phytotoxin to interact with ENR may be reduced relative to the nonpolar triclosan, although still sufficient to induce electrolyte leakage in the absence of light at high concentrations. Furthermore, the lower clogP of cyperin may hinder its intercalation into the lipid bilayer (relative to triclosan), but may still be involved in the nonspecific perturbation of subcellular membrane structure as was reported with triclosan (Paul et al., 2004; Gomez Escalada et al., 2005).
CONCLUSION
The biochemical interactions between pathogenic microorganisms and their host plants are very intricate, often resulting from a dynamic and continually evolving process. The ability of certain pathogenic fungi to produce the natural diphenyl ether cyperin in the tissues of infected plants is likely to contribute to the virulence of these disease agents. Whereas the mechanism(s) by which the producing organisms are not affected by cyperin remains to be determined, this article suggests that ENR may be a valid target for the development of new natural product-based herbicides.
MATERIALS AND METHODS
Materials and Synthesis
Cyperin [2-(3-hydroxy-5-methylphenoxy)-5-methoxy-3-methyl phenol; CAS: 33716–82–4; Fig. 1] was synthesized (>95% purity) according to the method reported by Harrington et al. (1995). The structure was confirmed by comparing 1H-NMR and mass spectrometry data with literature values. Triclosan [5-chloro-2-(2,4-dichlorophenoxy)-phenol; CAS: 3380–34–5] and all other reagents were purchased from Sigma-Aldrich.
Phytotoxicity of Cyperin and Triclosan
Arabidopsis (Arabidopsis thaliana Columbia [Col-0] wild type; Lehle Seeds) seeds were surfaced sterilized in a solution of 2.5% (v/v) sodium hypochlorite with 0.05% (v/v) Tween 20 for 10 min and rinsed four times with sterile deionized water. Twenty seeds per dish were plated onto square culture dishes (L100 × W100 × H15 mm) containing one-half-strength Murashige and Skoog growth medium in agar (2.7 g L−1) with concentrations ranging from 3 to 1,000 μm for cyperin and from 0.01 to 3 μm for triclosan. Controls received equivalent amounts of acetone (0.1% [v/v]). Plates were placed vertically in a CU-32 L plant growth chamber (Percival Scientific) at 21°C with a 16-h day photoperiod (approximately 300 μmol m−2 s−1). Root lengths of 15 seedlings were measured for all treatments after 12 d of growth. The experiment was repeated three times.
Effect of Cyperin and Triclosan on Fatty Acid Profile
Arabidopsis seeds were surfaced sterilized as before and placed in six-well culture plates containing 5 mL of medium (Murashige and Skoog) per well containing either 0.03 μm triclosan or 33 μm cyperin. The control contained a similar amount of acetone (0.1% [v/v]). Plates were cultured for 15 d as described above. Approximately 60 mg of tissue from each well was harvested for extraction. Total fatty acids were extracted according to Sasser (1990) and samples were analyzed on a Hewlett-Packard 5890 series II gas chromatograph with a cross-linked 5% phenyl methyl siloxane column (25-m × 0.2-mm × 0.3-μm film thickness; Agilent Technologies).
Effect of Cyperin and Triclosan on Membrane Integrity and Photosynthetic Efficiency
Tissues were treated with 333 μm cyperin or 10 μm triclosan and their effect on membrane integrity was tested by monitoring electrolyte leakage as described by Kenyon et al. (1985). Fifty 4-mm cucumber (Cucumis sativus) cotyledon discs (approximately 0.1 g fresh weight) were placed in a 6-cm-diameter disposable petri dish containing 5 mL of 2% (w/v) Suc and 1 mm MES (pH 6.5) with or without the test compounds. Test compounds were dissolved in acetone. Control tissues were exposed to the same amount of acetone as treated tissues, but without the test compounds. The final concentration of acetone in the dishes was 1% (v/v). The cotyledon discs were incubated at 25°C in darkness. Cellular damage was determined over time by measuring electrolyte leakage into the bathing medium with a conductivity meter (model 1056; Amber Science) capable of assaying 1 mL of bathing medium (Kenyon et al., 1985). All measurements were done under dim green light (5 μmol m−2 s−1). Because of differences in background conductivity between different treatment solutions, results are expressed as a change in the conductivity. All treatments for electrolyte leakage measurements were in triplicate and the experiment was repeated.
Cloning and Purification of ENR
Total RNA was extracted from Arabidopsis leaves using TRIzol reagent and converted to first-strand cDNA using Superscript II RNase H− reverse transcriptase according to the manufacturer's instruction (Invitrogen). The open reading frame of fab1 was amplified from the cDNA by using PCR. The PCR reaction was performed using PfuTurbo DNA polymerase (Stratagene) according to the manufacturer's instructions with upstream primer 5′-ctagccATGGCTGAATCCAGTGAAAACAAGGC-3′ and downstream primer 5′-ccgctcgagATTCTTGCTGTTAAGGTCTTTGAAC-3′. The PCR product was digested with NcoI and XhoI that were introduced into the primers at the 5′ and 3′ ends of the primers, respectively, and cloned into similarly prepared pET28b. The clone was sequenced to confirm without errors. This plasmid was named pETFab and transformed BL21(DE3) cells for expression.
Cells were collected by centrifugation, resuspended in cold lysing buffer (50 mm Tris-HCl, pH 7.5, 1 m NaCl, 5 mm imidazole, 10% [v/v] glycerol, 1 μg mL−1 leupeptin), and lysed with a French Press (SIM-AMINCO). Benzonase (1 μL mL−1 of 25 units μL−1) and 1 mm phenylmethylsulfonyl fluoride were added immediately to the lysate. After 15-min incubation at room temperature, the lysate was centrifuged in a Sorvall SS34 rotor at 15,000g for 20 min. The supernatant was saved for purification of ENR.
A HisTrap HP column (GE Healthcare Bio-Sciences) was activated with 2 mL of 0.1 m NiSO4 and washed column with 10 mL of distilled water. The column was equilibrated with 10 mL of buffer A (20 mm Tris-HCl, pH 8.0, 500 mm NaCl, 5 mm imidazole). ENR was eluted from the column with elution buffer (20 mm Tris-HCl, pH 8.0, 500 mm NaCl, 250 mm imidazole). The ENR-containing fraction was desalted on a PD-10 column equilibrated with cold desalting buffer (10 mm sodium phosphate, pH 7.2, 10 mm dithiothreitol, 10% [v/v] glycerol). Protein concentration was determined using Bio-Rad protein reagent.
Assay of ENR and Inhibition Kinetics
The assay buffer consisted of 10 mm sodium phosphate, pH 7.2. The I50 values (concentration of inhibitor required for 50% inhibition of activity) of cyperin and triclosan on Arabidopsis ENR were determined by testing the inhibitors at concentrations ranging from 0.1 to 1,000 μm for cyperin and 0.001 to 3 μm for triclosan. All inhibitors were dissolved in acetone and control samples received equivalent amounts of acetone (1% [v/v] final concentration). ENR (24 nm or 1 ng mL−1) was incubated with the compounds in the presence of 100 μm NAD+ for 30 min at room temperature prior to assay. Crotonyl-CoA (100 μm) was added to the mixture and the reaction was initiated by the addition of 100 μm NADH. The time-dependent and enzyme titration experiments were performed with 240 nm ENR (10 ng mL−1) and 200 μm each of crotonyl-CoA, NADH, and NAD+. The kinetic study to determine the Km of crotonyl-CoA and the Ki of cyperin was done with 120 nm ENR (5 ng mL−1) and 100 μm of NADH and NAD+. The experiment was carried out in the presence of 2, 4, 6, and 8 μm cyperin with either 50 or 100 μm crotonyl-CoA using the enzyme assay conditions described by Sivaraman et al. (2004). The kinetic parameters were calculated from a Dixon plot. The reactions were started by addition of NADH. The oxidation of NADH was monitored for 60 s by measuring change in A340 in a Shimadzu model UV3101PC spectrophotometer with the cell thermostabilized at 25°C. Spectrophotometric measurements were converted to concentrations of NADH oxidized using ɛ = 6.3 mm−1 cm−1 (Ward et al., 1999).
Binding Mechanism of Cyperin and Triclosan to Arabidopsis ENR
The time dependence of ENR inhibition by cyperin and triclosan was determined by conduction enzyme assays over 6 min. The nature of the interaction between cyperin and triclosan and their binding site on ENR was further investigated by modifying the method of Ellis et al. (1995). This was done by incubating various concentrations of ENR for 5 min with either 20 μm cyperin or 8.5 nm triclosan at room temperature prior to conduction ENR assay as described above.
Modeling of Arabidopsis ENR and Binding of Cyperin and Triclosan
The National Center for Biotechnology Information (NCBI) Arabidopsis genomic database was searched using BLAST (http://www.ncbi.nlm.nih.gov/BLAST) with sequence of Brassica napus (GenBank accession no. P80030) as query. Twelve cDNA sequences were identified by this search. Analysis of these sequences with the software MegAlign (DNASTAR) showed that 11 were identical. One sequence (GenBank accession no. NM_126612) was used for this study.
The x-ray crystal coordinates of B. napus ENR/NAD+/triclosan complex at a resolution of 1.9 Å from Roujeinikova et al. (1999) were obtained from the Brookhaven Protein Database (1D7O.pdb). The significant sequence homology between B. napus ENR (Roujeinikova et al., 1999) and ENR of other organisms (as discussed later) enabled the construction of a homology model of the Arabidopsis ENR using the biopolymer functions of the computer modeling software Sybyl 7.2 (Tripos Associates) on a Silicon Graphics O2 250 MHz R10000 workstation. A total of 11 amino acid substitutions were introduced to the B. napus ENR to obtain a homology model corresponding to the Arabidopsis ENR sequence. The amino acid substitutions made (based on the numbering of the B. napus sequence) were Val-109 to Ala, Pro-160 to Ala, Ala-177 to Thr, Arg-197 to Lys, Gln-198 to Lys, Ser-244 to Arg, Phe-291 to Tyr, Gln-297 to Ser, Ala-331 to Gly, Val-230 to Ala, and Ser-370 to Ala. Pro Φ angles were fixed at 70°, side chain amides were checked to maximize potential H-bonding, side chains were checked for close Van der Waals contacts, and essential hydrogens were added. The model was checked for conformational problems using the module ProTable from Sybyl. Ramachandran plot, local geometry, and the location of buried polar residues/exposed nonpolar residues were examined. The model was then subjected to energy minimization following the gradient termination of the Powell method for 3,000 iterations using Kollman united force field with NB cutoff set at 9.0 and the dielectric constant set at 4.0.
The 3-D structure of triclosan was obtained from its x-ray crystal coordinates found in the 1D7O.pdb file. The structure of cyperin was derived from the coordinate of triclosan. The binding of either cyperin or triclosan was adjusted within the binding site using the FlexiDock module of Sybyl 7.2. The ligands were prepositioned in the binding pocket by using the coordinates of triclosan in the crystal structure of B. napus ENR. The binding pocket was defined by selecting all the residues within a 4 Å boundary around the ligand. Briefly, partial charges were assigned to the atoms. All donor and receptor atoms and rotatable bonds were selected in the binding pocket and the ligands to optimize the interaction. The distances between atoms involved in hydrogen bonding in the ligand-receptor interaction were measured in the resulting models.
Statistical Analysis
Data from dose-response experiments were analyzed with the add-on package for dose-response curves (drc; Ritz and Streibig, 2005) for R version 2.2.1 (R Development Core Team, 2005) using a four-parameter logistic function. Means and sd were obtained using the raw data, and I50 values were one of the parameters in the regression curves. The regression curves were imported into SigmaPlot version 10 (Systat Software). The kinetic analysis was performed with the enzyme kinetic module of SigmaPlot version 10. Statistical analysis of the fatty acid profiles was performed using SAS version 9.1 (SAS, 2004).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers P80030 and NM_126612.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. ENR sequence alignment of 20 organisms.
Supplementary Material
Acknowledgments
We are grateful for the excellent technical support provided by the staff of the Computer Modeling Laboratory of the National Center for Natural Products Research of the University of Mississippi. We also extend our thanks to Mrs. Susan Watson, Mrs. J'Lynn Howell, and Mrs. Marilyn Ruscoe for their technical assistance in the laboratory.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Franck E. Dayan (franck.dayan@ars.usda.gov).
The online version of this article contains Web-only data.
References
- Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar SP (1997) Signaling in plant-microbe interactions. Science 276 726–733 [DOI] [PubMed] [Google Scholar]
- Chothia C, Lesk AM (1989) The relation between the divergence of sequence and structure in proteins. EMBO J 5 823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly ML (1983) Analytical molecular surface calculations. J Appl Cryst 16 548–558 [Google Scholar]
- Dangl JL, Jones JDG (2001) Plant pathogens and integrated defense responses to infection. Nature 411 826–833 [DOI] [PubMed] [Google Scholar]
- Dayan FE, Allen SN (2000) Predicting the activity of the natural phytotoxic diphenyl ether cyperine using comparative molecular field analysis. Pest Manag Sci 56 717–722 [Google Scholar]
- Dayan FE, Duke SO (1997) Phytotoxicity of protoporphyrinogen oxidase inhibitors: phenomenology, mode of action and mechanisms of resistance. In RM Roe, JD Burton, RJ Kuhr, eds, Herbicide Activity: Toxicology, Biochemistry and Molecular Biology. IOS Press, Washington, DC, pp 11–35
- Duke SO, Abbas HK, Duke MV, Lee HJ, Vaughn KC, Amagasa T, Tanaka T (1996) Microbial toxins as potential herbicides. J Environ Sci Health B 31 427–434 [Google Scholar]
- Ellis MK, Whitfield AC, Gowans LA, Auton TR, Provan WM, Lock EA, Smith LL (1995) Inhibition of 4-hydroxyphenylpyruvate dioxygenase by 2-(2-nitro-4-trifluoromethylbenzoyl)-cyclohexane-1,3-dione and 2-(2-chloro-4-methanesulfonylbenzoyl)-cyclohexane-1,3-dione. Toxicol Appl Pharmacol 133 12–19 [DOI] [PubMed] [Google Scholar]
- Feys BJ, Parker JE (2000) Interplay of signaling pathways in plant disease resistance. Trends Genet 16 449–455 [DOI] [PubMed] [Google Scholar]
- Fray RG, Throup JP, Daykin M, Wallace A, Williams P, Stewart GSAB, Grierson D (1999) Plants genetically modified to produce N-acylhomoserine lactones communicate with bacteria. Nat Biotechnol 17 1017–1020 [DOI] [PubMed] [Google Scholar]
- Ghose AK, Pritchett A, Crippen GM (1988) Atomic physicochemical parameters for three dimensional structure directed quantitative structure-activity relationships III: modeling hydrophobic interactions. J Comput Chem 9 80–90 [DOI] [PubMed] [Google Scholar]
- Gomez Escalada M, Russell AD, Maillard J-Y, Ochs D (2005) Triclosan–bacteria interactions: single or multiple target sites? Lett Appl Microbiol 41 476–481 [DOI] [PubMed] [Google Scholar]
- Guillén J, Bernabeu A, Shapiro S, Villalaín J (2004) Location and orientation of triclosan in phospholipid model membranes. Eur Biophys J 33 448–453 [DOI] [PubMed] [Google Scholar]
- Harrington PM, Singh BK, Szamosi IT, Birk JH (1995) Synthesis and herbicidal activity of cyperin. J Agric Food Chem 43 804–808 [Google Scholar]
- Heath RJ, Rubin JR, Holland DR, Zhang E, Snow ME, Rock CO (1999) Mechanism of triclosan inhibition of bacterial fatty acid synthesis. J Biol Chem 274 11110–11114 [DOI] [PubMed] [Google Scholar]
- Heath RJ, Shapiro MA, Olson E, Rock CO (1998) Broad spectrum antimicrobial biocides target the FabI component of fatty acid synthesis. J Biol Chem 273 30316–30320 [DOI] [PubMed] [Google Scholar]
- Kenyon WH, Duke SO, Vaughn KC (1985) Sequences of effects of acifluorfen on physiological and ultrastructural parameters in cucumber cotyledon discs. Pestic Biochem Physiol 24 240–250 [Google Scholar]
- Kuo MR, Morbidoni HR, Alland D, Sneddon SF, Gourlie BB, Staveski MM, Leonard M, Gregory JS, Janjigian AD, Yee C, et al (2003) Targeting tuberculosis and malaria through inhibition of enoyl reductase: compound activity and structural data. J Biol Chem 278 20851–20859 [DOI] [PubMed] [Google Scholar]
- Lee HH, Moon J, Suh SW (2007) Crystal structure of the Helicobacter pylori enoyl-acyl carrier protein reductase in complex with hydroxydiphenyl ether compounds, triclosan and diclosan. Proteins 69 691–694 [DOI] [PubMed] [Google Scholar]
- Levy CW, Roujeinikova A, Sedelnikova S, Baker PJ, Stuitje AR (1999) Molecular basis of triclosan activity. Nature 398 383–384 [DOI] [PubMed] [Google Scholar]
- McMurry LM, McDermott PF, Levy SB (1998) Triclosan targets lipid synthesis. Nature 394 531–532 [DOI] [PubMed] [Google Scholar]
- Mou Z, He Y, Dai Y, Liu X, Li J (2000) Deficiency in fatty acid synthase leads to premature cell death and dramatic alterations in plant morphology. Plant Cell 12 405–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicola G, Smith CA, Lucumi E, Kuo MR, Karagyozov L, Fidock DA, Sacchettini JC, Abagyan R (2007) Discovery of novel inhibitors targeting enoyl–acyl carrier protein reductase in Plasmodium falciparum by structure-based virtual screening. Biochem Biophys Res Commun 258 686–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul KS, Bacchi CJ, Englund PT (2004) Multiple triclosan targets in Trypanosoma brucei. Eukaryot Cell 3 855–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedras MSC, Zaharia IL, Gai Y, Zhou Y, Ward DE (2001) In planta sequential hydroxylation and glycosylation of a fungal phytotoxin: avoiding cell death and overcoming the fungal invader. Proc Natl Acad Sci USA 98 747–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidugu LS, Kapoor M, Surolia N, Surolia A, Suguna K (2004) Structural basis for the variation in triclosan affinity to enoyl reductases. J Mol Biol 343 147–155 [DOI] [PubMed] [Google Scholar]
- Qiu X, Janson CA, Court RI, Smyth MG, Payne DJ, Abdel-Meguid SS (1999) Molecular basis for triclosan activity involves a flipping loop in the active site. Protein Sci 8 2529–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2005) R: a language and environment for statistical computing. In Ed 2.2.1. R Foundation for Statistical Computing, Vienna
- Rafferty B, Simon JW, Baldock C, Artymiuk PJ, Baker PJ, Stuije AR, Slabas AR, Rice DW (1995) Common themes in redox chemistry emerge from the X-ray structure of oilseed rape (Brassica napus) enoyl acyl carrier protein reductase. Structure 3 927–938 [DOI] [PubMed] [Google Scholar]
- Rafi SB, Cui G, Song K, Cheng X, Tonge PJ, Simmerling C (2006) Insight through molecular mechanics Poisson-Boltzmann surface area calculations into the binding affinity of triclosan and three analogues for FabI, the E. coli enoyl reductase. J Med Chem 49 4574–4580 [DOI] [PubMed] [Google Scholar]
- Rao ST, Rossmann MG (1973) Comparison of super-secondary structures in proteins. J Mol Biol 76 241–256 [DOI] [PubMed] [Google Scholar]
- Ritz C, Streibig JC (2005) Bioassay analysis using R. J Stat Softw 12 1–22 [Google Scholar]
- Roujeinikova A, Levy CW, Rowsell S, Sedelnikova S, Baker PJ, Minshull CA, Mistry A, Colls JG, Camble R, Stuitje AR, et al (1999) Crystallographic analysis of triclosan bound to enoyl reductase. J Mol Biol 294 527–535 [DOI] [PubMed] [Google Scholar]
- Sali A, Potterton L, Yuan F, Van Vlijmen H, Karplus M (1995) Evaluation of comparative protein modeling by MODELLER. Proteins 23 318–326 [DOI] [PubMed] [Google Scholar]
- SAS (2004) Statistical Analysis Systems. Release 9.1. Statistical Analysis System Institute, Cary, NC
- Sasser M (1990) Identification of bacteria through fatty acid analysis. In Z Klement, K Rudolph, D Sands, eds, Methods in Phytobacteriology. Akademiai Kiado, Budapest, pp 199–204
- Serrano M, Robatzek S, Torres M, Kombrink E, Somssich IE, Robinson M, Schulze-Lefert P (2007) Chemical interference of pathogen-associated molecular pattern-triggered immune responses in Arabidopsis reveals a potential role for fatty-acid synthase type II complex-derived lipid signals. J Biol Chem 282 6803–6811 [DOI] [PubMed] [Google Scholar]
- Sivaraman S, Sullivan TJ, Johnson F, Novichenok P, Cui G, Simmerling C, Tonge PJ (2004) Inhibition of the bacterial enoyl reductase FabI by triclosan: a structure-reactivity analysis of FabI inhibition by triclosan analogues. J Med Chem 47 509–518 [DOI] [PubMed] [Google Scholar]
- Sivaraman S, Zwahlen J, Bell AF, Hedstrom L, Tonge PJ (2003) Structure-activity studies of the inhibition of FabI, the enoyl reductase from Escherichia coli, by triclosan: kinetic analysis of mutant FabIs. Biochemistry 42 4406–4413 [DOI] [PubMed] [Google Scholar]
- Staskawicz BJ, Mudgett MB, Dangl JL, Galan JE (2001) Common and contrasting themes of plant and animal diseases. Science 292 2285–2289 [DOI] [PubMed] [Google Scholar]
- Stewart MJ, Parikh S, Xiao G, Tonge PJ, Kisker C (1999) Structural basis and mechanism of enoyl reductase inhibition by triclosan. J Mol Biol 290 859–865 [DOI] [PubMed] [Google Scholar]
- Stierle A, Upadhyay R, Strobel G (1992) Cyperine, a phytotoxin produced by Ascochyta cypericola, a fungal pathogen of Cyperus rotundus. Phytochemistry 30 2191–2192 [Google Scholar]
- Stone GW, Zhang Q, Castillo R, Doppalapudi VR, Bueno AR, Lee JY, Li Q, Sergeeva M, Khambatta G, Georgopapadakou NH (2004) Mechanism of action of NB2001 and NB2030, novel antibacterial agents activated by β-lactamases. Antimicrob Agents Chemother 48 477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 11 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatasubbaiah P, Van Dyke CG, Chilton WS (1992) Phytotoxic metabolites of Phoma sorghina, a new foliar pathogen of pokeweed. Mycologia 84 715–723 [Google Scholar]
- Vera-Estrella R, Barkla BJ, Higgins VJ, Blumwald E (1994) Plant defense response to fungal pathogens. Plant Physiol 104 209–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalain J, Mateo R, Aranda FJ, Shapiro S, Micol V (2001) Membranotropic effects of the antibacterial agent triclosan. Arch Biochem Biophys 390 128–136 [DOI] [PubMed] [Google Scholar]
- Ward WHJ, Holdgate GA, Rowsell S, McLean EG, Pauptit RA, Clayton E, Nichols WW, Colls JG, Minshull CA, Jude DA, et al (1999) Kinetic and structural characteristics of the inhibition of enoyl (acyl carrier protein) reductase by triclosan. Biochemistry 38 12514–12525 [DOI] [PubMed] [Google Scholar]
- Weber HA, Gloer JB (1988) Interference competition among natural fungal competitors: an antifungal metabolite from the coprophilous fungus Preussia Fleischhakii. J Nat Prod 51 879–883 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.